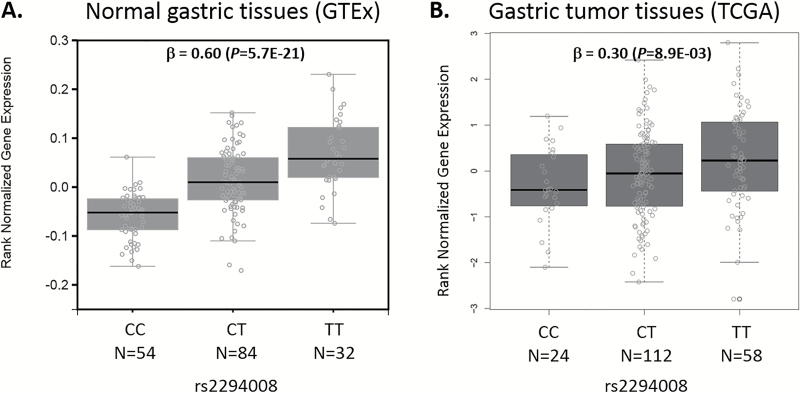
The association of germline variants with prognosis in GC patients suggests a heritable component in cancer survival. The ability of rs2294008 as a cis-expression quantitative trait loci for PSCA mRNA in both tumor and normal tissues may act in the underlying mechanism.
Abstract
Eleven high-evidence single-nucleotide polymorphisms (SNPs) at nine loci for gastric cancer (GC) risk were reported, but their associations with survival remain unknown. In this study, we examined associations between SNP and GC survival by anatomic location and histology among 1147 incident cases from the Shanxi Upper Gastrointestinal Genetics Project. We further examined whether SNPs were expression quantitative trait loci in normal and tumor gastric tissues, and whether tumor versus normal somatic mRNA differences in 126 cases were associated with survival. No SNPs were associated with GC survival overall. However, subtype-specific associations were observed for gastric cardia adenocarcinomas at MUC1/TRIM46/1q22 rs2070803 [HRAA versus GA+GG = 2.16; 95% confidence interval (CI) = 1.24–3.78; P = 0.0068] and LTA/TNF/6p21.33 rs1799724 (HRTT+CT versus CC = 1.30; 95% CI = 1.07–1.57; P = 0.0077), and for diffuse-type GC at PSCA/8q24.3 rs2294008 (HRTT versus CT+CC = 1.99; 95% CI = 1.33–2.97; P = 7.8E-04). Rs2294008T was a cis-expression quantitative trait loci for PSCA, upregulating mRNA in normal gastric (β = 0.60; P = 5.7E-21) and GC (β = 0.30; P = 0.0089) tissues. Cases in the highest quartile (the smallest downregulation of tumor PSCA) had shortest survival than cases with the most downregulated PSCA (median survival of 0.47 years in the highest quartile versus 3.73 years in the lowest quartile; hazard ratio = 9.70; 95% CI = 2.46–38.4; P = 0.0012). Less striking effects for mRNA levels were observed for MTX1 at 1q22 in gastric cardia adenocarcinoma and for JRK at 8q24.3 in diffuse GC. Our results suggest three high-evidence GC risk loci have prognostic importance in GC subtypes. Future studies in well-characterized independent populations are warranted to validate our findings and further investigate the clinical utility of these variants in predicting GC prognosis.
Introduction
A population-based study showed familial concordance in cancer survival among breast, lung, colorectal and prostate cancers, and suggested that these genetic determinants might be also involved in cancer survival (1). However, germline associations with cancer survival remain largely unknown. Although strong evidence for genetic susceptibility has been found in relation to cancer risk, there is little evidence linking germline variants to survival. Reports relating germline variants to survival or recurrence are limited to breast (2–4), non-small (5,6) or small cell lung cancer (7), glioblastoma (8), pancreatic cancer (9), colorectal cancer (CRC) (10), esophageal squamous-cell carcinoma (11) and thyroid cancer (12). To our knowledge, no promising germline loci have been reported to have prognostic significance for gastric cancer (GC).
As one approach to the evaluation of genetic susceptibility to GC survival, we hypothesized that some cancer predisposing variants might also be associated with prognosis. This hypothesis has been examined previously in several cancers including breast, prostate, and CRC (13–17). A review found substantial overlap between germline cancer predisposition genes and somatically mutated genes (18), suggesting that germline variants may contribute not only to cancer risk but also to cancer progression and survival. Germline variants can act as normal tissue expression quantitative trait loci (eQTLs) influencing the expression of a specific gene(s) in normal tissue. They may also impact cancer progression and survival by acting as eQTLs in premalignant and tumor tissues (tumor eQTLs) (19). Clinical implications attributable to somatic gene expression changes in tumors as independent prognostic factors have been reported for many cancers, including GC (20).
In a recent comprehensive meta-analysis of GC risk susceptibility variants, Mocellin et al. (21) nominated 11 single-nucleotide polymorphisms (SNPs) at 9 loci conferring a high-level of evidence for association with GC risk. Five of the nine loci (MUC1 at 1q22, PKLR at 1q22, CASP8 at 2q33.1, TGFBR2 at 3p22, and GSTP1 at 11q13.2) came from candidate gene approaches, whereas the remaining four loci (PRKAA1 at 5p13, TNF at 6p21.3, PSCA at 8q24.2 and PLCE1 at 10q23) were identified through genome-wide association studies (GWAS) (MTX1 at 1q22 was also confirmed through GWAS). Using in silico and eQTL analysis of the Genotype-Tissue Expression (GTEx) data (Version 4; Release date 4 April 2014), we previously found that the majority of these SNPs located to regulatory regions, and that more than half were normal tissue eQTLs for nearby genes (22). However, the prognostic significance of these SNPs remains unknown. In this study, we evaluated each of these SNPs for their associations with GC survival in a high-risk region of China, accounting for anatomic and histologic subtypes. Furthermore, we evaluated the influence of selected SNPs on mRNA expression of nearby genes (cis-eQTL) in both normal and tumor gastric tissues. We also linked differentially expressed mRNAs in tumor versus normal gastric tissues to GC survival.
Materials and methods
Study population
Detailed methods for the Shanxi Upper Gastrointestinal Genetics Project have been published elsewhere (23). The Shanxi Project was conducted between 1994 and 2007 and had several components, including a case–control study and a case-only study. Study participants for the current study were histologically confirmed incident GC cases who underwent surgical resection at the Shanxi Cancer Hospital and Institute in Taiyuan, Shanxi Province in China. A total of 1161 cases with GWAS genotyping data and follow-up for survival were available for evaluation. After exclusion of 14 cases without age, 1147 cases were included in the final analysis. Clinical characteristics including tumor location, invasiveness, grade, regional lymph node, distant metastasis and Lauren classification (diffuse-type or intestinal-type) were collected from medical abstract forms and subsequently confirmed by a pathologist at the National Cancer Institute (NCI). Cancer treatment after surgery (radiation, chemotherapy, herbs and others) was also collected during survival follow-up. The Shanxi Project was approved by the Institutional Review Boards (IRB) of the Shanxi Cancer Hospital and Institute and the NCI Special Studies IRB. All participants provided written informed consent prior to joining the study. The NCI Special Studies IRB also approved the overall GWAS.
SNP selection
Candidate SNPs were selected based on a previous meta-analysis that nominated 11 SNPs as high-evidence genetic susceptibility markers for GC (21). Details of these 11 SNPs are presented in Supplementary Table 1. Genotyping results were obtained from existing GWAS data (four SNPs were genotyped and seven were imputed), which is available through the database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/gap/) Authorized Access (dbGaP Study Accession: phs000361.v1.p1). For CASP8 rs3834129, we used rs6747918 (r2 = 0.95 with rs3834129 in the 1000 Genomes East Asian population) as a surrogate as no genotyped or imputed data were available. All 11 SNPs had a minor allele frequency of >0.05 and passed Hardy–Weinberg equilibrium threshold (P > 1E-06) among study participants. Average genotyping call rates for the four SNPs were 100% and imputation quality (r2) for the seven imputed SNPs was 0.98.
Cis-eQTL analyses in tumor and normal gastric tissues
For eQTL analyses in normal gastric tissue, we compiled summary statistics of linear regression results from the updated version of GTEx (Version 6; Release date 5 October 2015: dbGaP Accession phs000424.v6.p1; http://www.gtexportal.org/home/). For detection of cis-eQTLs, we collated all genes within ±1 Mb of the index SNP. Linear regressions were performed among 170 normal gastric tissues with the genotype of each SNP and log and quantile normalized mRNA expression levels measured by RNA-Seq of candidate genes. Covariates included three genotyping principle components, 15 peer factors, and sex. To identify tumor eQTLs, we used data from The Cancer Genome Atlas (TCGA). Genotype data (Affymetrix Genome-Wide Human SNP Array 6.0 platform, level 2; broad.mit.edu_STAD.Genome_Wide_SNP_6.sdrf.txt), RNA-Seq data (Illumina HiSeq 2000, level 3; unc.edu_STAD.IlluminaHiSeq_RNASeqV2.1.0.0.sdrf.txt), and phenotypes were downloaded from TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). We excluded two samples whose sex information was missing, resulting in 196 GC samples for inclusion in tumor eQTL analyses. Transcript reads/counts for each gene were quantile normalized and regressed against the imputed dosage of the minor allele for each SNP (MTX1 for rs2070803 and rs2075570; PSCA and JRK for rs2294008 and rs2976392). The regression model was adjusted for age, sex, and copy number variation (CNV).
Differential gene expression in tumor versus normal gastric tissues
Differential expression or fold-change (fc) difference of mRNA between tumor and histologically confirmed adjacent normal gastric tissues was examined for association with survival in GC cases from the Shanxi project. Fold-change was defined as the ratio of logarithmic normalized mRNA value in tumor compared with normal. A total of 126 cases with paired tumor and normal mRNA expression and survival follow-up data were included in this analysis. Selection of subjects for the tissue study was based solely on the availability of appropriate tissues for RNA testing [i.e. consecutive testing of cases with available frozen tissue, tumor samples that were predominantly (>50%) tumor, histologically confirmed normal paired tissues, and RNA quality/quantity adequate for testing]. All tissues were snap-frozen in liquid nitrogen and stored at −130°C until required for RNA extraction. RNA extraction and mRNA microarray analyses were previously described (24,25). Gene expression data are publicly available from Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE29272. Candidate genes included were either annotated as genes proximal to survival-associated SNPs (five SNPs in three loci) in the current study (MUC1/TRIM46/MTX1 at 1q22, LTA/TNF at 6p21.33, and PSCA/JRK at 8q24.3) or as genes known to be influenced by the SNP (eQTL) in normal gastric tissues (GBAP1/THBS3/RP11-263K19.4/GBA at 1q22, MICA1/BAG6 at 6p21.33 and PSCA/JRK/LYNX1 at 8q24.3) based on the GTEx Project. Among these candidates, gene expression data were available for nine genes that were included in the final analysis: MUC1, TRIM46, MTX1, TNF, LTA, PSCA, JRK, MICA, and THBS3.
Statistical analysis
The primary study outcome was overall survival (OS). OS time was defined as the time from surgery until the date of death from any cause or to the date of the last follow-up (31 May 2013), whichever came first. Estimation of OS was performed using the Kaplan–Meier method and differences between survival curves by SNP genotypes or quartile levels of mRNA expression as tumor/normal fc were assessed using the log-rank test. Cox’s proportional hazard regression models were used to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) for SNPs or mRNA expression levels adjusted for potential confounding factors. The proportional hazard assumption of the Cox model was examined by graphic evaluation of Schoenfeld residual plots. Covariates included in the final model were age at diagnosis (10-year interval), sex, primary tumor location [gastric cardia adenocarcinoma (GCA) or gastric noncardia adenocarcinoma (GNCA)], Lauren classification (intestinal or diffuse), histologic grade (well/moderately differentiated or poorly differentiated/undifferentiated), tumor invasiveness (T1–T2 or T3–T4), and distant metastasis (M0 or M1). Other covariates considered, but not included, in the final model were lymph node invasion and treatment group because these variables did not alter HRs significantly after adjusting for other covariates (statistical significance for inclusion of the final model was set at P < 0.05). Subgroup analyses were performed by anatomical subtype (GCA and GNCA) and Lauren classification.
For SNP analysis, different genetic models (additive, dominant and recessive) for each SNP were tested and the model with the smallest P-value was selected as the best-fitting model. The Bonferroni-corrected threshold for SNP analyses was P = 0.0015 (0.05/33 tests based on 11 SNPs and three genetic models [33 tests] for each subgroup). Fold-changes in mRNA expression between tumor versus normal tissue in each case were ranked and HRs were estimated for each quartile with the lowest quartile (q1) as the reference. Statistical analyses were conducted using Stata version 13.0 (Statacorp, College Station, TX), SAS version 9.3 (SAS Institute, Cary, NC) and R (R Foundation for Statistical Computing, Vienna, Austria; http://www.R-project.org/).
Results
Association of SNPs with GC survival
Table 1 shows baseline characteristics at the time of diagnosis and estimated HRs for OS by each covariate. Among 1147 cases, 828 were deceased. Median follow-up duration was 1.72 years (5.12 years for living cases and 1.17 for deceased cases). GC cases included here were predominantly male (82%), GCA (62%) and intestinal-type cases (55%). Most cases were diagnosed at an advanced stage or with poor prognostic characteristics: 67% of tumors were poorly differentiated or undifferentiated, 93% invaded (sub)serosal connective tissue or adjacent structures (T3–T4), 79% had positive lymph nodes and 72% had distant metastasis. In multivariate models, older age, GCA tumor location, diffuse-type histology, higher grade, invasiveness and distant metastasis all remained independently associated (P < 0.05) with poor survival (Table 1).
Table 1.
Characteristics of GC cases of survival study in the Shanxi Upper Gastrointestinal Genetics Project
| Overall (n = 1147) | Alive (n = 319) | Deceased (n = 828) | |||||
|---|---|---|---|---|---|---|---|
| n (%) | n (%) | n (%) | HR (95% CI) | P a | HR (95% CI) | P b | |
| Age, mean (SD) | 58.1 (9.2) | 56.5 (9.37) | 58.7 (9.06) | 1.17 (1.08–1.26) | <0.0001 | 1.15 (1.06–1.24) | 5.0E-4 |
| Sex | |||||||
| Male | 939 (81.9) | 267 (83.7) | 672 (81.2) | 1 (Reference) | 1 (Reference) | ||
| Female | 208 (18.1) | 52 (16.3) | 156 (18.8) | 1.06 (0.89–1.26) | 0.53 | 1.11 (0.93–1.32) | 0.26 |
| Follow-up years, Median | 1.72 | 5.12 | 1.17 | ||||
| Primary tumor location | |||||||
| Non-cardia | 435 (37.9) | 144 (45.1) | 291 (35.1) | 1 (Reference) | 1 (Reference) | ||
| Cardia | 712 (62.1) | 175 (54.9) | 537 (64.9) | 1.30 (1.12–1.52) | 0.001 | 1.20 (1.03–1.40) | 0.02 |
| Lauren classification | |||||||
| Intestinal | 495 (55.3) | 145 (59.4) | 350 (53.8) | 1 (Reference) | 1 (Reference) | ||
| Diffuse | 400 (44.7) | 99 (40.6) | 301 (46.2) | 1.21 (1.03–1.43) | 0.02 | 1.20 (1.02–1.42) | 0.03 |
| Missing | 252 | 75 | 177 | ||||
| Grade | |||||||
| Well or moderately differentiated | 371 (33.0) | 133 (42.8) | 238 (29.2) | 1 (Reference) | 1 (Reference) | ||
| Poorly or undifferentiated | 755 (67.1) | 178 (57.2) | 577 (70.8) | 1.29 (1.10–1.52) | 0.002 | 1.34 (1.14–1.57) | 4.0E-4 |
| Missing | 21 | 8 | 13 | ||||
| T stage | |||||||
| T1–T2 | 83 (7.3) | 52 (16.4) | 31 (3.8) | 1 (Reference) | 1 (Reference) | ||
| T3–T4 | 1056 (92.7) | 265 (83.6) | 791 (96.2) | 1.94 (1.34–2.82) | 0.001 | 2.11 (1.46–3.06) | <0.0001 |
| Missing | 8 | 2 | 6 | ||||
| Regional lymph nodes (N) | |||||||
| N0 | 213 (20.8) | 109 (41.6) | 104 (13.6) | 1 (Reference) | – | ||
| N1 | 247 (24.1) | 71 (27.1) | 176 (23.0) | 0.41 (0.12–1.39) | 0.15 | – | |
| N2 | 319 (31.1) | 61 (23.3) | 258 (33.8) | 0.58 (0.17–1.99) | 0.39 | – | |
| N3 | 247 (24.1) | 21 (8.0) | 226 (29.6) | 0.98 (0.29–3.32) | 0.98 | – | |
| Missing | 121 | 57 | 64 | ||||
| Distant metastasis (M) | |||||||
| M0 | 322 (28.5) | 162 (51.8) | 160 (19.6) | 1 (Reference) | 1 (Reference) | ||
| M1 | 809 (71.5) | 151 (48.2) | 658 (80.4) | 3.89 (1.16–13.0) | 0.03 | 2.39 (2.00–2.86) | <0.0001 |
| Missing | 16 | 6 | 10 | ||||
| Cancer treatment group after surgery | |||||||
| Surgery only | 588 (51.3) | 166 (52.0) | 422 (51.0) | 1 (Reference) | |||
| Chemotherapy | 453 (39.5) | 131 (41.1) | 322 (38.9) | 0.92 (0.68–1.25) | 0.59 | – | |
| Radiotherapy | 17 (1.5) | 2 (0.6) | 15 (1.8) | 0.90 (0.66–1.22) | 0.49 | – | |
| Chemotherapy and radiotherapy | 57 (5.0) | 10 (3.1) | 47 (5.7) | 0.78 (0.47–1.31) | 0.35 | – | |
| Herbs or others | 31 (2.7) | 10 (3.1) | 21 (2.5) | 1.71 (0.95–3.06) | 0.07 | – | |
| Missing | 1 | 1 | |||||
Follow-up years: years from date of surgery to date of death or end-of-follow-up.
aAdjusted for age (10-year frequency), sex, primary tumor location, lauren classification, grade, T stage, regional lymph nodes, and distant metastasis.
bAdjusted for age (10-year frequency), sex, primary tumor location, lauren classification, grade, T stage, and distant metastasis.
SD, standard deviation.
We first evaluated the association between each SNP and survival. Among the 11 SNPs at 9 loci examined, no SNP was associated with overall GC survival (Table 2). PSCA rs2294008T was associated with overall poor survival in a recessive model (HRTT versus CT+CC = 1.32; 95% CI = 1.03–1.69; P = 0.03) but the association did not reach the Bonferroni threshold after considering multiple comparisons. Subgroup analyses revealed three loci (1q22, 6p21.33 and 8q24.3) that were associated with survival in specific subtypes of GC. Supplementary Figure 1 shows Kaplan–Meier curves for OS stratified by SNPs at each locus based on its best-fitting model: rs2070803 intergenic to MUC1/TRIM46 at 1q22 among GCA cases (PLog-rank = 0.0081), rs1799724 intergenic to LTA/TNF at 6p21.33 among GCA cases (PLog-rank = 0.0055) and rs2294008 in PSCA at 8q24.3 among diffuse-type cases (PLog-rank = 0.0080). The multivariable models adjusted for potential confounders showed that the subtype-specific association remained for PSCA/8q24.3 after multiple comparison adjustment, but was just above the Bonferroni threshold for MUC1/TRIM46/1q22 and LTA/TNF/6p21.33 (Pthreshold = 0.0015, Table 2). Two of the 11 SNPs examined here were at the PSCA/8q24.3 locus where both highly correlated (r2 = 1.0), and both were associated with worse survival in diffuse-type GC in recessive models: rs2294008T HRTT versus CT+CC = 1.99; 95% CI = 1.33–2.97; P=7.8E-04, and rs2976392A HRAA versus GA+GG = 1.93; 95% CI = 1.32–2.81; P=6.7E-04. Two other highly correlated SNPs in our population (rs2070803 and rs2075570, r2 = 0.89) at 1q22 were also associated with GCA survival in recessive models: MUC1 rs2070803 HRAA versus GA+GG = 2.16; 95% CI = 1.24–3.78; P=0.0068 and MTX1 rs2075570 HRCC versus TC+TT = 2.48; 95% CI = 1.27–4.84; P=0.0079. Finally, LTA/TNF rs1799724T at 6p21.33 was associated with GCA survival in a dominant model (HRTT+CT versus CC = 1.30; 95% CI = 1.07–1.57; P=0.0077). Results for each SNP by tumor subtype and genetic model are shown in Supplementary Table 2.
Table 2.
Association of GC risk SNPs with GC survival by tumor subtype
| Chr | Nearby genes | SNP | Major|Minor allele | MAF | Best-fitting model | Overall (n = 1161) | Cardia (n = 712) | Noncardia (n = 435) | Intestinal (n = 495) | Diffuse (n = 400) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P a | HR (95% CI) | P b | HR (95% CI) | P b | HR (95% CI) | P c | HR (95% CI) | P c | ||||||
| 1q22 | MUC1/TRIM46/ | rs2070803d | G|A | 0.17 | Recessive | 1.37 (0.83–2.29) | 0.22 | 2.16 (1.24–3.78) | 0.0068 | 0.59 (0.17–2.04) | 0.40 | 1.27 (0.64–2.53) | 0.49 | 1.48 (0.34–6.39) | 0.60 |
| MTX1 | rs2075570 | T|C | 0.15 | Recessive | 1.68 (0.93–3.03) | 0.09 | 2.48 (1.27–4.84) | 0.0079 | 0.95 (0.26–3.44) | 0.93 | 1.47 (0.67–3.23) | 0.34 | 2.05 (0.28–14.99) | 0.48 | |
| 1q22 | PKLR | rs3762272 | T|C | 0.23 | Dominant | 0.95 (0.82–1.09) | 0.45 | 0.93 (0.78–1.11) | 0.44 | 0.93 (0.73–1.19) | 0.56 | 0.92 (0.74–1.15) | 0.48 | 0.95 (0.75–1.20) | 0.64 |
| 2q33.1 | CASP8 | rs6747918 | G|A | 0.23 | Recessive | 0.91 (0.68–1.21) | 0.51 | 0.87 (0.61–1.22) | 0.41 | 1.09 (0.63–1.89) | 0.75 | 0.91 (0.59–1.39) | 0.65 | 0.72 (0.43–1.22) | 0.22 |
| 3p24.1 | TGFBR2 | rs3087465 | G|A | 0.20 | Additive | 0.95 (0.84–1.07) | 0.40 | 0.97 (0.83–1.12) | 0.65 | 0.92 (0.74–1.14) | 0.44 | 0.95 (0.78–1.15) | 0.57 | 0.93 (0.76–1.13) | 0.46 |
| 5p13.1 | PRKAA1 | rs13361707 | C|T | 0.48 | Dominant | 1.02 (0.88–1.19) | 0.76 | 1.01 (0.84–1.22) | 0.92 | 1.07 (0.83–1.38) | 0.59 | 1.06 (0.84–1.35) | 0.60 | 0.99 (0.77–1.28) | 0.96 |
| 6p21.33 | LTA/TNF | rs1799724d | C|T | 0.14 | Dominant | 1.15 (0.99–1.34) | 0.08 | 1.30 (1.07–1.57) | 0.0077 | 0.94 (0.72–1.22) | 0.64 | 1.31 (1.04–1.66) | 0.02 | 1.02 (0.80–1.32) | 0.85 |
| 8q24.3 | PSCA | rs2294008 | C|T | 0.30 | Recessive | 1.32 (1.03–1.69) | 0.03 | 1.13 (0.81–1.57) | 0.47 | 1.50 (1.00–2.24) | 0.05 | 1.33 (0.89–2.00) | 0.17 | 1.99 (1.33–2.97) | 7.8E-4 |
| rs2976392 | G|A | 0.30 | Recessive | 1.25 (0.98–1.58) | 0.07 | 1.11 (0.81–1.51) | 0.51 | 1.35 (0.92–1.98) | 0.12 | 1.11 (0.75–1.65) | 0.59 | 1.93 (1.32–2.81) | 6.7E-4 | ||
| 10q23.33 | PLCE1 | rs2274223 | A|G | 0.18 | Recessive | 1.06 (0.82–1.37) | 0.64 | 1.07 (0.78–1.45) | 0.68 | 1.09 (0.69–1.72) | 0.72 | 1.28 (0.87–1.88) | 0.20 | 0.94 (0.61–1.45) | 0.77 |
| 11q13.2 | GSTP1 | rs1695 | A|G | 0.21 | Recessive | 1.23 (0.93–1.63) | 0.15 | 1.12 (0.79–1.59) | 0.52 | 1.50 (0.92–2.43) | 0.10 | 1.24 (0.78–1.99) | 0.36 | 1.63 (1.03–2.58) | 0.04 |
aAdjusted for age (10-year frequency), sex, primary tumor location, lauren classification, grade, T stage, regional lymph nodes, and distant metastasis.
Chr, chromosome; MAF, minor allele frequency.
bAdjusted for age (10-year frequency), sex, lauren classification, grade, T stage, regional lymph nodes, and distant metastasis.
cAdjusted for age (10-year frequency), sex, primary tumor location, grade, T stage, regional lymph nodes, and distant metastasis.
dIntergenic SNP.
Survival-associated SNPs can act as tumor cis-eQTLs
Using the newly released GTEx v6 data generated from 170 samples of normal gastric mucosa, we evaluated the five potential survival-associated SNPs (rs2070803, rs2075570, rs1799724, rs2294008, and rs2976392) as possible cis-eQTLs (i.e. SNPs that can influence the expression of a gene within a 1 MB distance of the index SNP position) in normal gastric tissues. Supplementary Table 3 shows the normal eQTL results for each of the five SNPs and their potential influences on the expression of all genes in cis. We observed that the following SNPs were correlated with mRNA expression of genes in cis: rs2070803 and GBAP1, rs2075570 and GBAP1/THBS3/RP11-263K19.4/GBA, rs1799724 and MICA/BAG6, and rs2294008 and rs2976392 and PSCA/JRK/LYNX1 (Supplementary Table 3). We also used TCGA data to evaluate these same SNPs as potential tumor eQTLs and found that rs2294008 and rs2976392 were positive eQTLs for PSCA mRNA in gastric tumors after adjusting for CNVs (Supplementary Table 4). Figure 1 shows that rs2294008T is a positive cis-eQTL for PSCA, increasing mRNA expression in both normal (β = 0.60; P = 5.7E-21) and tumor (β = 0.30; P =8.9E-03) gastric tissues.
Figure 1.
Normal and tumor eQTL analysis between rs2294008 genotypes and PSCA mRNA expression. (A) Normal gastric tissues (n = 170) from cancer-free donors; β and P-values were obtained from GTEx Analysis Release V6 (dbGaP Accession phs000424.v6.p1) using linear regression between rs2294008-T allele and log and quantile normalized PSCA mRNA expression levels (RPKM) measured by RNA-Seq Gencode ID, ENSG00000167653.4; Transcript ID, ENST00000301258 and ENST00000513264. Covariates included three genotyping principle components, 15 peer factors, and sex. (B) Gastric tumor tissues (n = 194); Rs2284008 and RNA-Seq data were downloaded from TCGA data portal. Transcript reads/counts of PSCA were quantile normalized and regressed against rs2284008-T allele. Covariates included age, sex, and CNV.
Association of somatic mRNA levels with GC survival
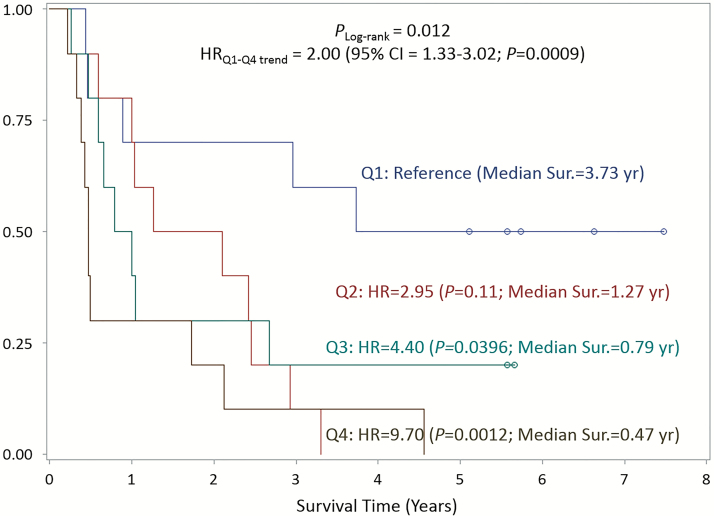
On the basis of our SNP association and eQTL results, we examined somatic mRNA levels of putative target genes [MUC1/TRIM46/MTX1/THBS3 (1q22), LTA/TNF/MICA (6p21.33), and PSCA/JRK (8q24.3)] for their association with survival (Supplementary Table 5). Table 3 shows the median fc in mRNA expression between tumor and normal tissues in each quartile by subtype and their association with survival. A higher fc in MTX1 mRNA was associated with worse survival among GCA cases: cases in the highest quartile had shorter survival than cases in the lowest quartile (HR = 3.51; 95% CI = 1.34–9.18; P = 0.010), and this association also showed a strong trend, with HRq1-q4 estimated at 1.45 (95% CI = 1.09–1.93; P = 0.011). In addition, higher PSCA mRNA in tumor compared with normal was associated with markedly decreased survival rates in GNCA and diffuse-type cancer cases (Figure 2). The HR for the highest versus lowest quartile was 4.04 for GNCA cases (95% CI = 1.39–22.74; P = 0.010) and 9.70 for diffuse-type cases (95% CI = 2.46–38.35; P = 0.0012) (Table 3; Figure 2). Monotonic trends were evident for both of the associations as well: the HR for GNCA was 1.60 (95% CI = 1.15–2.23; Ptrend = 0.0052) and the HR for diffuse was 2.00 (95% CI = 1.33–3.02; Ptrend = 9.0E-04). A higher fc in JRK mRNA was also associated with poorer survival among diffuse-type cancer cases with HR of the fourth quartile estimated at 3.19 (95% CI = 1.05–9.72; P = 0.041).
Table 3.
Association of mRNA tumor/normal (T/N) fc levels with GC survival overall and by subtype
| Gene Probe | Overall (n = 126) | Cardia (n = 55) | Noncardia (n = 71) | Intestinal (n = 64) | Diffuse (n = 40) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TN fc, median | HR (95% CI) | P a | TN fc, median | HR (95% CI) | P b | TN fc, median | HR (95% CI) | P b | TN fc, median | HR (95% CI) | P c | TN fc, median | HR (95% CI) | P c | |
| MTX1 210386_s_at | |||||||||||||||
| quartile1 | 0.911 | 1 (Reference) | 0.963 | 1 (Reference) | 0.872 | 1 (Reference) | 0.940 | 1 (Reference) | 0.863 | 1 (reference) | |||||
| quartile2 | 1.190 | 1.06 (0.58–1.96) | 0.85 | 1.228 | 2.34 (0.82–6.71) | 0.11 | 1.104 | 1.51 (0.63–3.61) | 0.35 | 1.252 | 1.36 (0.51–3.62) | 0.54 | 1.119 | 0.85 (0.30–2.43) | 0.76 |
| quartile3 | 1.527 | 1.05 (0.58–1.91) | 0.87 | 1.610 | 2.50 (0.94–6.66) | 0.07 | 1.384 | 0.79 (0.31–2.03) | 0.63 | 1.593 | 1.80 (0.74–4.34) | 0.19 | 1.421 | 0.37 (0.10–1.38) | 0.14 |
| quartile4 | 2.416 | 0.92 (0.50–1.70) | 0.80 | 2.501 | 3.51 (1.34–9.18) | 0.010 | 2.157 | 0.91 (0.35–2.35) | 0.84 | 2.622 | 1.28 (0.52–3.13) | 0.59 | 2.153 | 1.24 (0.38–4.04) | 0.72 |
| HRtrend | 0.98 (0.81–1.18) | 0.80 | 1.45 (1.09–1.93) | 0.011 | 0.93 (0.68–1.27) | 0.64 | 1.10 (0.84–1.44) | 0.47 | 1.01 (0.67–1.53) | 0.96 | |||||
| PSCA 205319_at | |||||||||||||||
| quartile1 | 0.004 | 1 (Reference) | 0.007 | 1 (Reference) | 0.003 | 1 (Reference) | 0.005 | 1 (Reference) | 0.003 | 1.00 (Reference) | |||||
| quartile2 | 0.015 | 1.13 (0.59–2.16) | 0.71 | 0.018 | 0.63 (0.25–1.61) | 0.34 | 0.014 | 1.99 (0.76–5.21) | 0.16 | 0.017 | 0.45 (0.17–1.16) | 0.10 | 0.013 | 2.95 (0.77–11.3) | 0.11 |
| quartile3 | 0.047 | 1.58 (0.83–3.01) | 0.16 | 0.068 | 0.79 (0.33–1.89) | 0.60 | 0.036 | 3.67 (1.27–10.6) | 0.016 | 0.054 | 1.26 (0.49–3.27) | 0.63 | 0.027 | 4.40 (1.07–18.0) | 0.040 |
| quartile4 | 0.226 | 1.87 (0.94–3.74) | 0.074 | 0.261 | 0.91 (0.35–2.33) | 0.84 | 0.203 | 4.04 (1.39–11.7) | 0.010 | 0.189 | 0.81 (0.30–2.17) | 0.68 | 0.155 | 9.70 (2.46–38.4) | 0.0012 |
| HRtrend | 1.26 (1.01–1.56) | 0.037 | 1.00 (0.73–1.35) | 0.98 | 1.60 (1.15–2.23) | 0.0052 | 1.02 (0.73–1.42) | 0.91 | 2.00 (1.33–3.0) | 9.0E-4 | |||||
| JRK 216309_x_at | |||||||||||||||
| quartile1 | 0.787 | 1 (Reference) | 0.817 | 1 (Reference) | 0.771 | 1 (Reference) | 0.814 | 1 (Reference) | 0.884 | 1.00 (Reference) | |||||
| quartile2 | 0.994 | 0.69 (0.38–1.29) | 0.25 | 1.002 | 0.93 (0.36–2.38) | 0.88 | 0.969 | 0.59 (0.25–1.41) | 0.23 | 1.012 | 0.90 (0.36–2.22) | 0.82 | 1.024 | 0.50 (0.14–1.77) | 0.28 |
| quartile3 | 1.146 | 1.09 (0.59–2.00) | 0.79 | 1.166 | 0.53 (0.19–1.49) | 0.23 | 1.136 | 1.22 (0.56–2.65) | 0.62 | 1.189 | 0.52 (0.19–1.40) | 0.19 | 1.180 | 3.07 (1.01–9.35) | 0.048 |
| quartile4 | 1.349 | 1.12 (0.62–2.02) | 0.71 | 1.404 | 0.95 (0.39–2.35) | 0.92 | 1.344 | 0.74 (0.32–1.74) | 0.49 | 1.404 | 0.84 (0.37–1.92) | 0.68 | 1.346 | 3.19 (1.05–9.72) | 0.041 |
| HRtrend | 1.09 (0.89–1.32) | 0.41 | 0.96 (0.71–1.31) | 0.79 | 1.17 (0.89–1.55) | 0.26 | 0.92 (0.69–1.21) | 0.55 | 1.64 (1.13–2.37) | 0.0096 | |||||
TC fc, tumor versus normal fc.
aAdjusted for age (10-year frequency), sex, primary tumor location, lauren classification, grade, T stage, regional lymph nodes and distant metastasis.
bAdjusted for age (10-year frequency), sex, lauren classification, grade, T stage, regional lymph nodes and distant metastasis.
cAdjusted for age (10-year frequency), sex, primary tumor location, grade, T stage, regional lymph nodes and distant metastasis.
Figure 2.
Kaplan–Meier curves and HR for OS by quartiles of PSCA tumor/normal mRNA fc among diffuse-type GC patients (n = 40).
Discussion
We evaluated 11 high-evidence SNPs identified as risk susceptibility loci in GC for their potential impact on survival, and found that three loci were associated with survival. Variants at MUC1/MTX1/1q22 and LTA/TNF/6p21.33 were associated with prognosis among GCA cases, and variants at PSCA/8q24.3 were associated with survival among diffuse-type cases. Somatic mRNA expression levels for genes (MTX1, PSCA and JRK) at two of these loci (1q22 and 8q24.3) were also associated with survival. The most striking survival difference was in diffuse-type GC where median survival was strongly dependent on PSCA fc in mRNA expression (0.47 years for cases in the highest quartile versus 3.73 years for cases in the lowest quartile). Importantly, we found that PSCA rs2294008T can act as an eQTL not only in normal stomach but also in gastric tumor tissues, which may be one mechanism underlying the observed associations between rs2294008T and somatic changes in PSCA with survival.
Somewhat unexpectedly, we found that rs2070803A and rs2075570C (at 1q22), which were associated with reduced GC risk, were associated with poor survival among GCA cases. Opposing effects for cancer susceptibility and prognosis are not uncommon and similar findings have been reported for breast (15), colorectal (17) and thyroid cancers (12). For breast cancer, the opposing effect of rs3817198 was explained by two independent mechanisms associated with the variant. Although the association of rs3817198C with mammographic density may explain elevated cancer risk (26), correlation with increased expression of the tumor suppressor cyclin-dependent kinase inhibitor 1C (CDKN1C) was postulated to explain the association with favorable clinical outcome (15). Similarly, in CRC, the C allele of rs446148 in SMAD7 involved in TGF-β and Wnt signaling was associated with increased risk of CRC, but better survival (17) and this paradoxical role was suggested to involve the differential role of TGF-β in tumor initiation as tumor suppressor and in progression as a tumor promoter (17,27). It is plausible to hypothesize that potential target genes relevant to 1q22 associations may have dual effects in cancer initiation (predisposing cancer risk) and progression (relevant to clinical outcome).
Rs2070803 is located 3′ of MUC1 and rs2075570 is an MTX1 intronic SNP, and both SNPs are highly correlated (r2 = 0.89). Similar to TGF-β in normal epithelium, MUC1 is known to be protective against potentially tumorigenic environmental insults, but after significant epithelial damage to membrane protein, becomes a promoter of cell growth and survival (28). However, our eQTL analysis does not support MUC1 as a potential target gene for rs2070803. Instead, we found that both SNPs at 1q22 and somatic mRNA expression of the nearby gene, MTX1, were associated with prognosis. It is intriguing that both associations were only seen among GCA cases. However, it is unlikely that MTX1 is the functionally relevant gene mediating 1q22 association. In a previous evaluation which used an earlier version (v4) of the GTEx data, we found that rs2070803A was associated with decreased MTX1 mRNA expression in normal gastric tissues, while MUC1 expression was not decreased (22). In the current study, which used GTEx v6 data, rs2070803A was not a normal eQTL for MTX1. Instead, rs2070803 or rs2075570 did influence the expression of other nearby genes, including GBAP1, THBS3, RP11-263K19.4, and GBA (Supplementary Table 3). Also, neither rs2070803A nor rs2075570C were tumor eQTLs for MTX1 (Supplementary Table 4). Therefore, functional relevance of this locus may be linked to other target genes which need further investigation. Consistent with previous somatic GC expression data from a Chinese population (29), MTX1 mRNA was overexpressed in GC compared with normal tissues (fc = 1.25; P = 1.0E-15; Supplementary Table 5), and cases with higher somatic MTX1 mRNA expression had poorer prognosis. Although MTX1 is known to encode metaxin-1, which is a mitochondrial protein involved in tumor necrosis factor-induced cell death (30), its function in relation to GC is not known. Also, it is not clear how MTX1 expression is regulated. Given the lack of significant eQTLs or SNPs associated with MTX1 in normal gastric tissues (based on GTEx V6 data), this suggests that the observed differential expression of MTX1 in tumor compared with normal is not influenced by germline variants, but likely involves other somatic mechanisms, such as mutation or structural variants (31). Thus, the association of rs2070803/rs2075570 and the potential role of tumor MTX1 levels (independent of a potential germline genetic influence) with GCA survival remain to be elucidated.
We found that the GC risk allele of rs1799724T at 6p21.33 was also associated with poorer survival among GCA cases. Rs1799724 is located 5′ of TNF and 3′ of LTA, encoding tumor necrosis factor-alpha and lymphotoxin-alpha, respectively. Both are proinflammatory cytokines implicated in cancer and autoimmune disease (32). We observed downregulation of LTA (fc=0.85; P=3.0E-09) but not TNF (fc=0.98; P=2.7E-01) in GC compared with normal tissues. However, there was no evidence that rs1799724 was an eQTL for either LTA or TNF. Instead, we found that rs1799724T was associated with decreased and increased expression of more distant genes, MICA (159 kb downstream) and BAG6 (64 kb upstream), respectively, in normal gastric tissues (Supplementary Table 3). Indeed, rs1799724 locates to open chromatin and enhancer-like (15_EnhP) region in gastric smooth muscle tissue (22). MICA encodes a major histocompatibility complex class I chain-related protein A, which activates cytolytic activities against tumor cells. High MICA expression determined by immunohistochemistry has been inversely correlated with tumor stage and histological grade (33) and associated with longer survival among GC patients treated with adjuvant chemotherapy and immunotherapy, but not chemotherapy alone (33). In the current study, differential expression of MICA was not associated with GC survival in this predominantly treatment-naive sample. Thus, investigating rs1799724 in the context of different treatments remains an interesting question for future investigation.
Since the first GWAS reported PSCA variants as GC risk loci (34), rs2976392/rs2294008 has been replicated in other studies. However, risk associations by ethnicity, tumor location and histology remain inconsistent (21,35), and studies in relation to survival are more contradictory. Two studies conducted among Chinese populations reported opposite subtype-specific results for rs2294008. Wang et al. (36) showed that diffuse-type cases with CT or TT genotypes survived better than the cases with CC. In contrast, Zeng et al. (37) observed poorer survival for intestinal-type cases (subtype analysis was not shown, but >75% were intestinal GC) with the TT genotype. Inconsistent results across these studies may be due to differences in minor allele frequency (range, 23–49%) and proportions of subtypes (diffuse-type cases, 25–57%). We observed worse survival among diffuse-type cases with TT genotype compared to cases with CT or CC genotypes. In support of this result, a recent study conducted in a Spanish population also showed worse survival among diffuse-type cases carrying rs2294008T in a dominant model (38). However, although the T-allele of rs2294008 was associated with worse survival in three of four studies (including our own), associations and subtype specificity require further elucidation in future studies.
Our results for OS also showed that cases in the highest quartile of differential PSCA mRNA fc had worst prognosis: cases that characterized the smallest downregulation of tumor PSCA mRNA compared with normal tissues were more likely to die than cases with the greatest downregulation of tumor PSCA. This was particularly true for GCs with diffuse-type histology where cases in the highest quartile of expression were nearly 10-fold more likely to die than cases in the lowest quartile. The fact that rs2294008T can act as a positive eQTL for PSCA mRNA in GC tumors may help explain worse survival, especially among rs2294008T carriers. In other words, the association between the T-allele and worse survival may be mediated by the T-allele effect on sustained PSCA mRNA levels in gastric tumors. These results parallel findings from bladder cancer where the GWAS risk SNP rs2294008T was a strong predictor of both PSCA mRNA and protein expression levels (39). Interestingly, both the germline tumor eQTL and somatic mRNA associations appear to be specific for survival among diffuse-type GC cases.
Cumulative evidence indicates that PSCA has an important function in both tumor development and suppression in different cancers (40). However, although reduced PSCA mRNA expression and protein levels in GC in vitro and in vivo have been reported in several studies (34,41), including our own (22), the consequences of PSCA dysregulation and its role in GC carcinogenesis and/or survival are not known. Given the silencing of PSCA in primary gastric tumor tissues (34,41), coupled with the cell-proliferation inhibitory activity of PSCA in vitro, PSCA has been considered to have a tumor-suppressing effect in GC development (34). However, our findings suggest that the risk predisposing and worse prognosis T-allele of rs2294008 is associated with increased mRNA expression, suggesting a potential oncogenic role for PSCA in GC rather than tumor suppressing. A possible explanation for this is that the functional role of PSCA is dependent on its protein localization and stability (42).
Data from in vitro studies in two different cancers (42,43) suggest that rs2294008T and C variants can directly influence PSCA protein function (independent of mRNA alterations) by changing stability and cellular localization. In brief, rs2294008T produces a long growth-promoting PSCA (with N-terminal signal peptide) that is membrane bound, whereas rs2294008C produces a short cytosolic PSCA without the signal peptide. Moreover, the short PSCA is more susceptible to proteasomal degradation than long PSCA (42). Notably, it has been shown that the expression of PSCA rs2294008T and rs2294008C cDNAs in vitro induced inhibition of cell growth, with no detectable difference in activity (34). However, it is interesting that proteins expressed from the rs2294008T and rs2294008C cDNAs in GC cells localized to the cytoplasm, and that the in vitro-translated proteins from the cDNAs were the same size. In fact, these data (34) are contrary to expectation based on what we know about the larger glycosylated membrane-bound PSCA (long PSCA) encoded by rs2294008T (42). Thus, the protein expressed from rs2294008T cDNA (34) may not have had the signal peptide for membrane localization and/or may not have been glycosylated, and these data might reflect characteristics of the short cytosolic PSCA encoded by rs2294008C cDNA. Indeed, growth-promoting or oncogenic effects of PSCA have been suggested in other types of cancers, including prostate, pancreatic, and bladder (40,44) In addition, monoclonal antibodies against PSCA show substantial antioncogenic effects in vitro indicating a role for cell surface PSCA in cell growth (45) and anti-PSCA immunotherapy is currently in clinical trials for prostate and pancreatic cancers (46,47). Whether the associations of rs2294008T and PSCA mRNA with GC survival observed in this study are also linked to membrane localization and increased proliferation in GC in vivo remains to be investigated. Importantly, if confirmed, rs2294008T may have the potential to predict both quantitative (expression levels) and qualitative (cellular localization) changes to PSCA protein in GC from cases with poor prognosis that might benefit from targeted immunotherapy or precision medicine.
Previously, we demonstrated that rs2294008T was a positive cis-eQTL for PSCA in normal gastric tissue (22). However, in vitro studies in GC cell lines reported that rs2294008T results in transcriptional repression of PSCA and inhibition of tumor suppressor functions (34,48). We proposed that rs2294008T may have opposing effects on PSCA transcription in normal compared with neoplastic gastric epithelium that is determined by the presence and binding affinity of the repressor Yin Yang 1 in early-stage GC, but not normal epithelia (22). Yin Yang 1 is implicated in the early stage of GC carcinogenesis and in progression independent of PSCA. However, in further contrast, we report here for the first time, that rs2294008T also has a positive influence on PSCA expression in GC in vivo after correcting for CNVs. We also observed a significant positive correlation (without adjusting for CNVs) between rs2294008T and PSCA mRNA in tumor tissues from Shanxi study (n = 89; correlation coefficient r2 = 0.27; P = 0.0097). However, there was no significant association of T-allele with PSCA mRNA expression in a small number of diffuse-type GC cases (n = 28; β per number of T-allele = 0.016; P=0.71); this result may reflect limited power. Alternatively, we note that our PSCA mRNA and worse survival results may simply reflect other somatic alterations in these cases not related to PSCA.
Limited data suggest that JRK can facilitate Wnt/Wingless signaling in colon cancer cells (49); however, its function in carcinogenesis is unknown. We previously reported that rs2976392A and rs2294008T were correlated with higher JRK mRNA expression in normal stomach tissue (22). However, we found no evidence for these variants as tumor eQTLs. Thus, the association of JRK mRNA with survival reported here likely reflects acquired somatic changes/events in JRK transcription during GC progression.
Our study has several strengths and limitations. The major strength of our study is sample size. To our knowledge, this study is the largest genetic association study evaluating GC survival. However, despite the large sample size, power was limited for subgroup analyses and further studies are needed to replicate our findings. Study strengths include high genotyping quality, completeness of tumor/patient characteristics, and long-term follow-up. As a result, we were able to adjust for established prognostic factors related to tumor characteristics. Our study population consists of ethnically and geographically homogeneous participants who were enrolled at a single institute, treated with standardized management, and followed-up for mortality in a uniform and centralized manner. Therefore, observed associations should be largely free from potential biases. However, because our study population consisted primarily of advanced GC cases, we could not address heterogeneity of the associations by tumor burden (i.e. tumor invasiveness, grade, local/distant disease, etc.). Finally, our results may not be generalizable to other populations. Future studies are warranted to validate our findings. Selecting a well-characterized patient groups with high mortality will be a useful strategy for replicating our findings. Replication studies will provide important data on the potential clinical utility of these germline variants for risk prediction and personalized treatment options.
In summary, we evaluated high-evidence GC risk variants for associations with prognosis and found three risk loci that were also associated with survival. Tissue mRNA studies supported the association of specific genes with survival, including PSCA. Finally, a potential heritable component to PSCA mRNA levels in normal and GC tissues was also suggested highlighting PSCA as a potential target for antibody immunotherapy in a subset of GCs.
Funding
This research was funded by Intramural Research Program of the National Institutes of Health, National Cancer Institute, Division of Cancer Epidemiology and Genetics.
Conflict of Interest Statement: None declared.
Supplementary Material
Abbreviations
- CI
confidence interval
- eQTL
expression quantitative trait loci
- fc
fold-change
- GC
gastric cancer
- GCA
gastric cardia adenocarcinoma
- GNCA
gastric noncardia adenocarcinoma
- GTEx
Genotype-Tissue Expression
- HR
hazard ratio
- OS
overall survival
- SNP
single-nucleotide polymorphism
References
- 1. Lindström L.S., et al. (2007) Familial concordance in cancer survival: a Swedish population-based study. Lancet. Oncol., 8, 1001–1006. [DOI] [PubMed] [Google Scholar]
- 2. Shu X.O., et al. (2012) Novel genetic markers of breast cancer survival identified by a genome-wide association study. Cancer Res., 72, 1182–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rafiq S., et al. (2014) A genome wide meta-analysis study for identification of common variation associated with breast cancer prognosis. PLoS One, 9, e101488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao L., et al. (2015) STAT3 and STAT5b polymorphism contributes to breast cancer risk and clinical outcomes. Int. J. Clin. Exp. Pathol., 8, 2033–2038. [PMC free article] [PubMed] [Google Scholar]
- 5. Tang S., et al. (2015) Genome-wide association study of survival in early-stage non-small cell lung cancer. Ann. Surg. Oncol., 22, 630–635. [DOI] [PubMed] [Google Scholar]
- 6. Yoon K.A., et al. (2014) Genetic variations associated with postoperative recurrence in stage I non-small cell lung cancer. Clin. Cancer Res., 20, 3272–3279. [DOI] [PubMed] [Google Scholar]
- 7. Wu C., et al. (2010) Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res., 70, 9721–9729. [DOI] [PubMed] [Google Scholar]
- 8. Xiao Y., et al. (2012) SSBP2 variants are associated with survival in glioblastoma patients. Clin. Cancer Res., 18, 3154–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu C., et al. (2014) Genome-wide association study of survival in patients with pancreatic adenocarcinoma. Gut, 63, 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Phipps A.I., et al. (2016) Common genetic variation and survival after colorectal cancer diagnosis: a genome-wide analysis. Carcinogenesis, 37, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu C., et al. (2013) Genome-wide association study identifies common variants in SLC39A6 associated with length of survival in esophageal squamous-cell carcinoma. Nat. Genet., 45, 632–638. [DOI] [PubMed] [Google Scholar]
- 12. Świerniak M., et al. (2016) Association between GWAS-Derived rs966423 Genetic Variant and Overall Mortality in Patients with Differentiated Thyroid Cancer. Clin. Cancer Res., 22, 1111–1119. [DOI] [PubMed] [Google Scholar]
- 13. Fasching P.A., et al. ; kConFab Investigators. (2012) The role of genetic breast cancer susceptibility variants as prognostic factors. Hum. Mol. Genet., 21, 3926–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kote-Jarai Z., et al. ; UK Genetic Prostate Cancer Study Collaborators, and ProtecT Study Group. (2015) Prevalence of the HOXB13 G84E germline mutation in British men and correlation with prostate cancer risk, tumour characteristics and clinical outcomes. Ann. Oncol., 26, 756–761. [DOI] [PubMed] [Google Scholar]
- 15. Barrdahl M., et al. (2015) Association of breast cancer risk loci with breast cancer survival. Int. J. Cancer, 137, 2837–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris E.J., et al. (2015) A retrospective observational study of the relationship between single nucleotide polymorphisms associated with the risk of developing colorectal cancer and survival. PLoS One, 10, e0117816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dai J., et al. (2012) GWAS-identified colorectal cancer susceptibility loci associated with clinical outcomes. Carcinogenesis, 33, 1327–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rahman N. (2014) Realizing the promise of cancer predisposition genes. Nature, 505, 302–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ongen H., et al. (2014) Putative cis-regulatory drivers in colorectal cancer. Nature, 512, 87–90. [DOI] [PubMed] [Google Scholar]
- 20. Becker K.F., et al. (2000) The use of molecular biology in diagnosis and prognosis of gastric cancer. Surg. Oncol., 9, 5–11. [DOI] [PubMed] [Google Scholar]
- 21. Mocellin S., et al. (2015) Genetic variation and gastric cancer risk: a field synopsis and meta-analysis. Gut, 64, 1209–1219. [DOI] [PubMed] [Google Scholar]
- 22. Sung H., et al. (2016) Functional annotation of high-quality SNP biomarkers of gastric cancer susceptibility: the Yin Yang of PSCA rs2294008. Gut, 65, 361–364. [DOI] [PubMed] [Google Scholar]
- 23. Abnet C.C., et al. (2010) A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat. Genet., 42, 764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Su H., et al. (2011) Global gene expression profiling and validation in esophageal squamous cell carcinoma and its association with clinical phenotypes. Clin. Cancer Res., 17, 2955–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang G., et al. (2013) Comparison of global gene expression of gastric cardia and noncardia cancers from a high-risk population in china. PLoS One, 8, e63826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vachon C.M., et al. (2012) Common breast cancer susceptibility variants in LSP1 and RAD51L1 are associated with mammographic density measures that predict breast cancer risk. Cancer Epidemiol. Biomarkers Prev., 21, 1156–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu Y., et al. (2007) TGF-beta signaling alterations and susceptibility to colorectal cancer. Hum. Mol. Genet., 16 Spec No 1, R14–R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kufe D.W. (2009) Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer, 9, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X.Q., et al. (2006) [Gene expression profiling of diffuse-type gastric cancer by cDNA microarray]. Zhonghua Zhong Liu Za Zhi, 28, 116–119. [PubMed] [Google Scholar]
- 30. Wang X., et al. (2001) Metaxin is required for tumor necrosis factor-induced cell death. EMBO Rep., 2, 628–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Collins M., et al. (1996) SP1-binding elements, within the common metaxin-thrombospondin 3 intergenic region, participate in the regulation of the metaxin gene. Nucleic Acids Res., 24, 3661–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balkwill F. (2009) Tumour necrosis factor and cancer. Nat. Rev. Cancer, 9, 361–371. [DOI] [PubMed] [Google Scholar]
- 33. Chen Y., et al. (2015) MHC I-related chain a expression in gastric carcinoma and the efficacy of immunotherapy with cytokine-induced killer cells. Am. J. Cancer Res., 5, 3221–3230. [PMC free article] [PubMed] [Google Scholar]
- 34. Sakamoto H., et al. (2008) Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet., 40, 730–740. [DOI] [PubMed] [Google Scholar]
- 35. Zhang T., et al. (2012) Effect of PSCA gene polymorphisms on gastric cancer risk and survival prediction: A meta-analysis. Exp. Ther. Med., 4, 158–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang M., et al. (2011) Genetic variant in PSCA predicts survival of diffuse-type gastric cancer in a Chinese population. Int. J. Cancer, 129, 1207–1213. [DOI] [PubMed] [Google Scholar]
- 37. Zeng Z., et al. (2011) Polymorphisms in prostate stem cell antigen gene rs2294008 increase gastric cancer risk in Chinese. Mol. Carcinog., 50, 353–358. [DOI] [PubMed] [Google Scholar]
- 38. García-González M.A., et al. (2015) Association of PSCA rs2294008 gene variants with poor prognosis and increased susceptibility to gastric cancer and decreased risk of duodenal ulcer disease. Int. J. Cancer, 137, 1362–1373. [DOI] [PubMed] [Google Scholar]
- 39. Kohaar I., et al. (2013) Genetic variant as a selection marker for anti-prostate stem cell antigen immunotherapy of bladder cancer. J. Natl. Cancer Inst., 105, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Saeki N., et al. (2010) Prostate stem cell antigen: a Jekyll and Hyde molecule? Clin. Cancer Res., 16, 3533–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bahrenberg G., et al. (2000) Reduced expression of PSCA, a member of the LY-6 family of cell surface antigens, in bladder, esophagus, and stomach tumors. Biochem. Biophys. Res. Commun., 275, 783–788. [DOI] [PubMed] [Google Scholar]
- 42. Tanikawa C., et al. (2012) A genome-wide association study identifies two susceptibility loci for duodenal ulcer in the Japanese population. Nat. Genet., 44, 430–4, S1. [DOI] [PubMed] [Google Scholar]
- 43. Mumy A., et al. (2011) Prostate stem cell antigen (PSCA) and risk of bladder cancer: linking genotypes to functional mechanisms. Genome Biol., 12, 11–12. [Google Scholar]
- 44. Marra E., et al. (2010) Growth delay of human bladder cancer cells by Prostate Stem Cell Antigen downregulation is associated with activation of immune signaling pathways. BMC Cancer, 10, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saffran D.C., et al. (2001) Anti-PSCA mAbs inhibit tumor growth and metastasis formation and prolong the survival of mice bearing human prostate cancer xenografts. Proc. Natl. Acad. Sci. U. S. A., 98, 2658–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris M.J., et al. (2012) A phase I/IIA study of AGS-PSCA for castration-resistant prostate cancer. Ann. Oncol., 23, 2714–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wolpin B.M., et al. (2011) Global, multicenter, open-label, randomized phase II trial comparing gemcitabine (G) with. G plus AGS-1C4D4 (A) in patients (pts) with metastatic pancreatic cancer (mPC). J Clin Oncol., 29, (suppl 15), 4031. [Google Scholar]
- 48. Saeki N., et al. (2010) Prostate stem cell antigen: a Jekyll and Hyde molecule? Clin. Cancer Res., 16, 3533–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Benchabane H., et al. (2011) Jerky/Earthbound facilitates cell-specific Wnt/Wingless signalling by modulating β-catenin-TCF activity. EMBO J., 30, 1444–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.