Summary
We show that sulforaphane, a diet-derived cancer prevention agent, reduces PRMPT5/MEP50 function to reduce cancer cell survival.
Abstract
Protein arginine methyltransferase 5 (PRMT5) cooperates with methylosome protein 50 (MEP50) to arginine methylate histone H3 and H4 to silence gene expression, and increased PRMT5 activity is associated with enhanced cancer cell survival. We have studied the role of PRMT5 and MEP50 in epidermal squamous cell carcinoma. We show that knockdown of PRMT5 or MEP50 results in reduced H4R3me2s formation, and reduced cell proliferation, invasion, migration and tumor formation. We further show that treatment with sulforaphane (SFN), a cancer preventive agent derived from cruciferous vegetables, reduces PRMT5 and MEP50 level and H4R3me2s formation, and this is associated with reduced cell proliferation, invasion and migration. The SFN-dependent reduction in PRMT5 and MEP50 level requires proteasome activity. Moreover, SFN-mediated responses are partially reversed by forced PRMT5 or MEP50 expression. SFN treatment of tumors results in reduced MEP50 level and H4R3me2s formation, confirming that that SFN impacts this complex in vivo. These studies suggest that the PRMT5/MEP50 is required for tumor growth and that reduced expression of this complex is a part of the mechanism of SFN suppression of tumor formation.
Introduction
PRMT5 is a protein arginine methyltransferase which catalyzes formation of symmetrically dimethylated arginine on a number of targets to modulate intracellular events (1,2). PRMT5 modulates EGFR-mediated ERK activation (3) and is required for p53 expression and induction of p53 targets (4). PRMT5 also binds to death receptor 4 (5), and is a component of the androgen receptor cofactor complex where it positively modulates androgen receptor-driven transcription (6,7). PRMT5 also controls RNA processing, signal transduction and transcription (8–15), and participates in the assembly of repressor complexes on various eukaryotic promoters (16). Histones H3 and H4 are important PRMT5 targets and PRMT5-dependent histone methylation is associated with increased cancer cell survival (11,17).
PRMT5 functions in conjunction with methylosome protein 50 (MEP50), a WD repeat (tryptophan-aspartic acid)-containing protein cofactor, that is required for PRMT5 activity (18,19). Elevated MEP50 and PRMT5 expression is associated with cancer development (20–24). We have shown that PRMT5 inhibits MAPK-dependent differentiation in normal human epidermal keratinocytes (25), a phenotype consistent with a role in enhancing cell survival. However, the impact of PRMT5 on epidermal squamous cell carcinoma has not been adequately explored.
Sulforaphane (1-isothiocyanato-4-(methylsulfinyl) butane, SFN) is a natural isothiocyanate derived from cruciferous vegetables and is an important cancer prevention agent (26). SFN is particularly appealing as a cancer prevention and treatment agent, as it is highly bioavailable in blood and tissues and has no known side effects (27–31). SFN suppresses skin cancer development and is regarded as an important potential skin cancer prevention/treatment agent (32–37). SFN is known to suppress activity of several epigenetic regulators leading to reduced cell survival, but its impact on PRMT5/MEP50 has not been explored.
In the present study, we examine the role of PRMT5 and MEP50 in epidermal squamous cell carcinoma. We show that PRMT5 or MEP50 knockdown reduces cell proliferation, invasion and migration, and tumor formation. SFN treatment reduces PRMT5 and MEP50 level via a mechanism that involves proteasome degradation and this reduction is required for suppression of cancer cell proliferation and invasion. Moreover, treatment of tumor xenografts with SFN reduces MEP50 level and H4R3me2s formation, showing that SFN impacts this complex in vivo. These studies suggest that the PRMT5/MEP50 is required for optimal squamous cell carcinoma formation and that a reduction in level/function of these proteins occurs in response to SFN treatment. Thus, PRMT5/MEP50 is an important epigenetic cell survival regulatory complex that may be a SFN cancer prevention target.
Materials and methods
Chemicals and reagents
R, S-Sulforaphane (SFN, #S8044) was purchased from LKT Laboratories (St. Paul, MN) and dimethyl sulfoxide was purchased from Sigma. SFN was dissolved in DMSO at a stock concentration of 100 mM and stored at −20°C. Matrigel (354234) and BD Biocoat cell inserts (353097) were purchased from BD Biosciences. Rabbit monoclonal antibody specific for p44/MEP50 (2823) was purchased from Cell Signaling Technology (Danvers, MA) and mouse polyclonal antibody specific for PRMT5 (sc-376937) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-H4R3me2s (ab5823), anti-H3 (ab12209) and anti-H4 (ab10158) were purchased from Abcam (Cambridge, MA). Anti-H3R8me2s was obtained from Thermo Scientific (PA5-27039, Rockfored, IL). Peroxidase-conjugated anti-mouse IgG (NXA931) and peroxidase conjugated anti-rabbit IgG (NA934V) were obtained from GE Healthcare. Mouse monoclonal β-actin antibody (A5441) and lactacystin were purchased from Sigma. Control-siRNA (D-001206-13-05), MEP50-siRNA (M-006895-01-0005) and PRMT5-siRNA (M-015817-02-005) were obtained from Dharmacon.
Cell culture, plasmids and viruses
Human squamous cell carcinoma cell line SCC-13, epidermoid carcinoma cell line A431 and HaCaT cells were purchased from American Type Culture Collection (ATCC, Rockville, MD). The cells were maintained in high glucose DMEM (Gibco, 11960-044) supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 μg/ml streptomycin and 5% fetal bovine serum (Sigma, St.Louis, MO). All cell lines were authenticated.
PRMT5 and MEP50 knockdown cell lines
SCC-13 cells (1 × 105) were allowed to attach overnight in 24-well cluster plates and then infected with PRMT5- or MEP50-shRNA encoding lentivirus in serum-free growth media for 24 h at 37°C. The PRMT5 shRNA (TRCN0000303447) and MEP50 shRNA (TRCN00000727810) lentiviral transduction particles were obtained from Sigma–Aldrich. The serum-free growth media contained 8 μg/ml polybrene. The medium was then replaced with 5% fetal calf serum supplemented with growth media and near-confluent cells were harvested, plated at low density in 100 mm dishes and selected for 2 weeks in the presence of 1 μg/ml puromycin. These cells were then infected a second time with the same virus and reselected. The resulting cells are a nonclonal population of cells called SCC13-PRMT5-shRNA2 and SCC13-MEP50-shRNA2. A control cell line (SCC13-Control-shRNA) was derived by double infection with control-shRNA (scrambled) lentivirus (Sigma–Aldrich, SHC001V).
Electroporation and cell proliferation assay
The AMAXA electroporator and VPD-1002 nucleofection kit were used for keratinocyte electroporation. Cells were harvested with trypsin and replated 1 day prior to electroporation. The cells were then re-harvested with trypsin, and 1 million cells were electroporated by suspension in 100 μl of keratinocyte nucleofection solution containing 3 μg of control-, MEP50-, or PRMT5-siRNA. The mixture was gently mixed, transferred to the electroporation cuvette and electroporated using the T-018 setting. Warm DMEM (500 μl) was added, followed by transfer to a 55 cm2 dishes containing 10 ml of DMEM. The cells were maintained for various time points before the extracts were prepared for mRNA or protein analysis. This method achieves electroporation efficiencies of >90% (38). In some cases, cells were double-electroporated. This involved an initial electroporation with 3 μg of appropriate siRNA, recovery in culture for 48 h, a repeat electroporation with 3 μg of siRNA, and 24 h of recovery in culture. In some experiments, cells were electroporated with 1–3 μg of empty vector or expression plasmids encoding MEP50 or PRMT5.
Immunoblot analysis
Subconfluent cultures of SCC-13, A431 and HaCaT cells were treated with 0 or 20 μM SFN and after 24 h washed with PBS followed by lysis in 20 mM Tris–HCl (pH 7.5) containing 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate, 1 mM sodium vanadate, 1 μg/ml leupeptin (Cell Signaling, 9803) and 1 mM phenylmethylsulphonylfluoride. Protein concentration was determined by Bradford assay. Equal amounts of protein were electrophoresed on a 10% denaturing polyacrylamide gel and transferred to nitrocellulose. The membranes were blocked with 5% skimmed milk in Tris-buffered saline (pH 7.5) containing 0.1% Tween 20 for 1 h. Subsequently, the blots were incubated overnight with primary antibody (p44/MEP50 1:1000, PRMT5 1:1000, β-actin 1:1000) followed by an appropriate horseradish peroxidase-conjugated secondary antibody for 2 h. Antibody binding was detected using chemiluminescence (Amersham Biosciences).
Quantitative RT-PCR
Cells were treated with vehicle or SFN as described above. After 24 h, total RNA was isolated using Illustra RNAspin Mini Kit (GE Healthcare) and 1 μg RNA was used for cDNA synthesis. Gene expression was measured by real time PCR using Light Cycler 480 SYBR Green I Master Mix (04-707 516 001) from Roche Diagnostics. The signals were normalized using cyclophillin A control primers. The gene specific primers used for detection of mRNA levels were as follows: p44/MEP50 (forward, 5′-TTG CTC AGC AGG TGG TAC TGA GTT; reverse, 5′-AAT CTG TGA TGC TGG CTT GGG ACA), PRMT5 (forward, 5′-TGA GGC CCA GTT TGA GAT GCC TTA; reverse, AGT AGC CGG CAA AGC CAT GTA GTA) and cyclophilin A (forward, 5′-CAT CTG CAC TGC CAA GAC TGA; reverse, 5′-TTC ATG CCT TCT TTC ACT TTG C).
Immunostaining
SCC-13 cells, on coverslips, were fixed for 20 min in phosphate-buffered saline containing 4% paraformaldehyde. The cells were then permeabilized with −20°C methanol and incubated with the appropriate primary and secondary antibody for 1 h each. The cells were incubated with Hoechst 33258 (1:2000) for 5 min, washed, and mounted on glass slides using Fluoromount (Sigma, catalog no. F4680). An Olympus OX81 spinning disc confocal microscope was used to visualize fluorescence. The skin malignant tissue array BC21014 was obtained from Biomax. MEP50 antibody (catalog no. ab5772) staining was detected using an appropriate biotinylated secondary antibody obtained as part of the mouse IgG Vectastain ABC kit (catalog no. PK-6102, Vector Laboratories, Burlingame, CA).
Cell invasion assay
Matrigel was diluted in 0.01 M Tris–HCl/0.7% NaCl, filter sterilized and 0.1 ml was added to the BD BioCoat cell inserts and permitted to solidify for 2 h. For assay, the matrigel-coated wells were seeded with 25000 cells in growth media supplemented with 1% FCS. Growth media, containing 10% FCS, was added to the lower chamber as an attractant, and the cells were incubated for 0–24 h at 37°C. The following day, cells that had migrated to the bottom of the membrane were fixed with 4% paraformaldehyde for 10 min, and then stained in 1 μg/ml DAPI for 10 min. The underside of the membrane was photographed with an inverted fluorescent microscope to count the number of nuclei per ×10 field.
Cell migration assay
SCC-13 cells (2 million) were plated on 10 cm dishes in growth media and allowed to attach overnight. Once confluent, a 10 μl pipette was used to create a wound and the released cells were removed. Fresh growth media was added and the cells were treated with an appropriate concentration of SFN and images were taken at 0–20 h (×10 magnification) to determine the extent of wound closure.
Tumor xenograft assays
Monolayer-derived cells were harvested with trypsin to prepare a single cell suspension. These cells were resuspended in phosphate buffered saline containing 30% Matrigel and 100 μl containing 0.4 million cells was injected subcutaneously at the two sites in each front flank of each of five female 6-week old NOD scid IL2 receptor gamma chain knockout mice (NSG mice) per group using a 26.5 gauge needle. Tumor growth was monitored by measuring diameter and calculating the tumor volume using the formula, volume = 4/3π × (diameter/2)3. Tumors were photographed and samples were harvested for immunostaining and immunoblot. Animal studies were conducted with approval of the Institutional Animal Care and Use Committee. Statistics were calculated using the t-test.
Results
MEP50 expression in squamous cell carcinoma
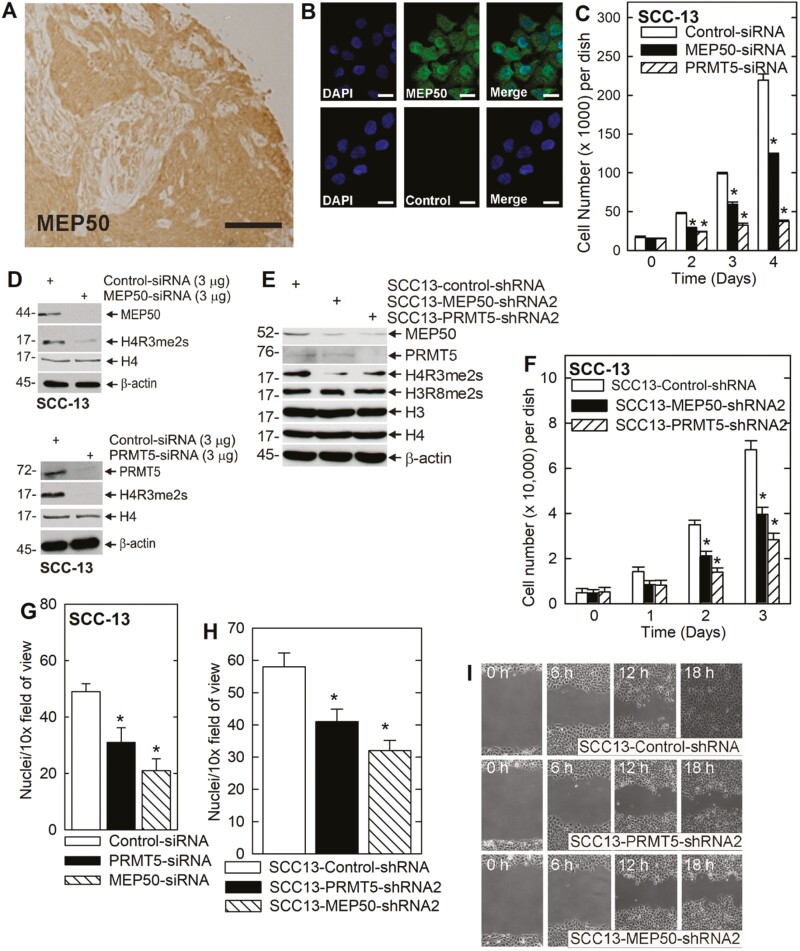
Recent studies show that MEP50 is expressed in cancer cells and is present in both the cytoplasm and nucleus (39,40). To localize MEP50 in squamous cell carcinoma tumor sections, we stained paraffin embedded squamous cell carcinoma tumor specimens and stained with anti-MEP50. Figure 1A identifies MEP50 as present in both the cytoplasm and nucleus in sections from a scalp-derived epidermal squamous cell carcinoma. Moreover, elevated MEP50 expression was observed in a large panel of squamous cell carcinoma sections derived from the Biomax Skin Tumor Array (not shown). The epithelial cells of the tumor display intense staining as compared to surrounding connective tissue. We next assessed the distribution in cultured cells. Figure 1B identifies MEP50 in both nuclear and cytoplasmic locations in SCC-13 epidermal squamous cell carcinoma cells, a distribution that is consistent with that observed in lung cancer cells (39,40).
Figure 1.
MEP50 and PRMT5 are required for SCC-13 proliferation and migration. (A) Skin cancer tissue array was stained with anti-MEP50 and binding was visualized using a peroxidase-conjugated secondary antibody. The section shown is from a scalp squamous cell carcinoma. Bar = 10 μm. Similar staining patterns were observed in 50 distinct cancer samples (not shown). (B) SCC-13 cells were seeded on coverslips and localization of endogenous MEP50 was visualized. The cells were fixed and co-stained with DAPI (blue) and anti-MEP50 (green). Similar results were observed in each of three experiments. The staining indicates distribution in the nucleus and cytoplasm. (C) SCC-13 cells were double electroporated with control-, MEP50- or PRMT5-siRNA, and then 15000 cells were plated per well. After an overnight attachment, cell number was determined (day zero) and at the indicated times thereafter. The values are mean ± SEM (n = 3). The asterisks indicate a significant difference (P < 0.005). (D) Immunoblot confirms a reduction in MEP50 and PRMT5 and reduced H4R3me2s level in cultures treated with the indicated siRNA. (E) SCC-13 cell lines stably expressing control-, MEP50- and PRMT5-shRNA were tested for detection of MEP50, PRMT5, H4R3me2s, H3R8me2s and the total histone. Similar results were obtained in each of the three experiments. (F) The stable cell lines were plated at a low density of 15000 cells/well. After overnight attachment, cell number was determined (day 0) and at the indicated times thereafter. The values are mean ± SEM (n = 3). The asterisks indicate a significant difference (P < 0.005). (G) SCC-13 cells were double electroporated with the indicated siRNA and 25000 cells were seeded on a matrigel layer in the upper well of a Transwell chamber and cell migration to the lower chamber was monitored over a 24 h period. Values are mean ± SEM (n = 3, P < 0.001). (H) The indicated cell lines were seeded on a matrigel layer in the upper well of a Transwell chamber (20000 cells per well) and cell migration to the lower chamber was monitored at 24 h. Values are mean ± SEM, n = 3 (P < 0.001). (I) The cell lines were grown to confluence, uniformly wounded, and migration to close the wound was monitored over 0–18 h.
MEP50 regulation of SCC-13 cell proliferation, invasion and migration
To assess the functional role of MEP50, tumor cells were double-electroporated with control-, MEP50- or PRMT5-siRNA to reduce levels of these targets. Figure 1C shows that MEP50 or PRMT5 knockdown reduces cell number. H4R3me2s is a biological marker of MEP50/PRMT5 action (2,41). As anticipated, knockdown of MEP50 or PRMT5 reduces H4R3me2s formation (Figure 1D). To test the effect of long-term PRMT5 and MEP50 silencing, we created stable knockdown cells using MEP50- or PRMT5-shRNA encoding lentiviruses. Figure 1E confirms the reduction in MEP50 and PRMT5 in the respective cell lines and an associated reduction in H4R3me2s formation. Interestingly, formation of H3R8me2s, another histone mark associated with PRMT5 activity, is not altered. We next examined the impact of reduced MEP50 or PRMT5 on biological endpoints. Figure 1F confirms that MEP50 and PRMT5 knockdown cell lines proliferate at a slower rate compared to the control-shRNA cells.
Enhanced tissue invasion/metastasis and migration are hallmarks of cancer cells (42). We therefore examined the impact of MEP50 and PRMT5 knockdown on invasion and migration. Figure 1G and H shows that transient or stable MEP50 or PRMT5 knockdown reduces matrigel invasion. To monitor the impact of PRMT5 and MEP50 on migration, uniform wounds were created in confluent cell monolayers and ability of the cells to migrate to close the wound was monitored. Figure 1I shows that loss of PRMT5 or MEP50 reduces wound closure. These studies suggest that MEP50 and PRMT5 are required for optimal cancer cell proliferation, invasion and migration.
PRMT5 and MEP50 impact on tumor formation
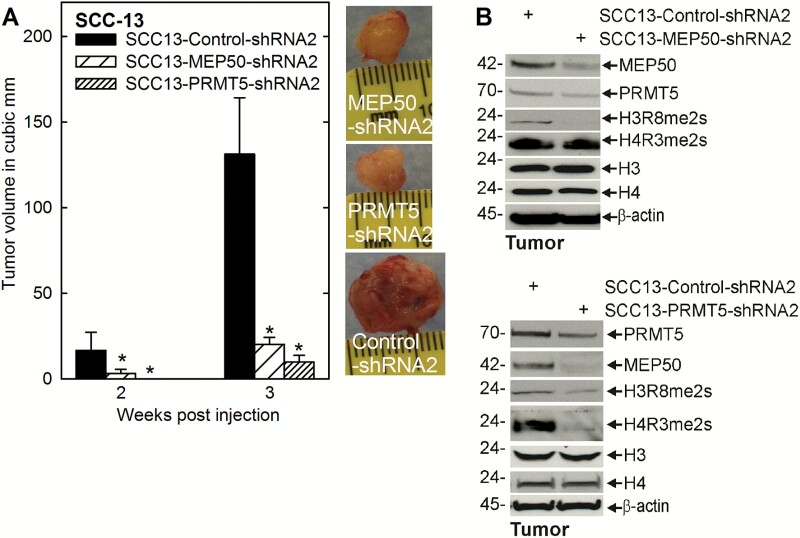
We next assessed whether PRMT5 and MEP50 are required for tumor formation. Control or PRMT5 or MEP50 knockdown cell lines were injected into each front flank in NSG mice and tumor formation was monitored over 3 weeks. PRMT5 or MEP50 knockdown produced a remarkable 80–90% reduction in tumor volume (Figure 2A). The tumor images reveal a marked reduction in vascularization as evidenced by the reduced redness. Immunoblot reveals that MEP50 knockdown cells, derived from tumors, show the expected reduction in MEP50 and a substantial reduction in H3R8me2s formation (Figure 2B). In contrast, the PRMT5 knockdown cells show a partial reduction in PRMT5 level accompanied by substantial reduction in MEP50 level, and H3R8me2s and H4R3me2s formation.
Figure 2.
PRMT5 and MEP50 impact tumor formation. (A) 0.4 million cells from each of the control-, MEP50- and PRMT5-shRNA cell lines were injected subcutaneously in the two front flanks in NSG mice. Tumor growth was monitored by measuring the diameter over 3 weeks. The values are mean ± SEM (n = 3). The asterisks indicate a significant difference (P < 0.005). Representative tumors from each group were photographed at 3 weeks. (B) The tumors were harvested at 3 weeks for immunoblot detection of the indicated markers.
PRMT5 and MEP50 are novel SFN targets
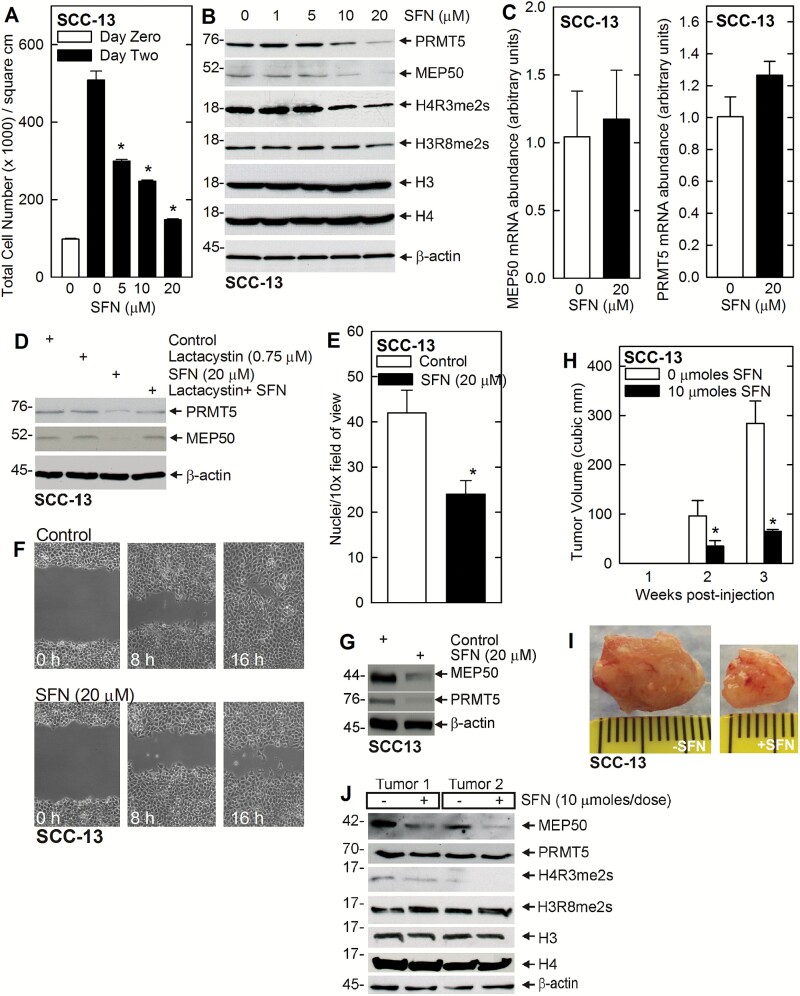
SFN is an important diet-derived cancer prevention agent (26,43–45). We have previously shown that SFN reduces skin cancer cell survival (32); however, it is not known whether reduced MEP50 and PRMT5 activity are associated with or required for SFN reduction of tumor formation. Figure 3A and B shows that SFN suppression of SCC-13 cell number is associated with reduced PRMT5 and MEP50 level, and reduced H4R3me2s formation. In contrast, SFN treatment does not markedly reduce H3R8me2s formation (Figure 3B). To gain insight regarding the mechanism of MEP50 and PRMT5 reduction, we studied the impact of SFN treatment on MEP50 and PRMT5 mRNA. SFN treatment did not result in any substantive change in MEP50 or PRMT5 mRNA levels, suggesting an absence of regulation (Figure 3C). Thus, SFN regulates PRMT5 and MEP50 at the protein level. To assess this further, we treated cells with SFN in the presence of 0 or 0.75 μM lactacystin, an inhibitor of proteasome activity. Lactacystin stabilizes PRMT5 and MEP50, suggesting that SFN promotes proteasome-dependent degradation of these proteins (Figure 3D). We next examined the impact of SFN treatment on biological endpoints, including cell invasion and migration. Figure 3G shows that SFN treatment reduces PRMT5 and MEP50 level and that this is associated with reduced tumor cell invasion and migration (Figure 3E and F).
Figure 3.
PRMT5 and MEP50 are SFN targets. (A) SFN reduces proliferation in SCC-13 cells. SCC-13 cells were treated with increasing doses of SFN for 24 h. The values are the mean ± SEM, n = 3. The asterisks indicate a significant difference (P < 0.005). (B) SFN suppresses MEP50 and PRMT5 protein levels and activity. SCC-13 cells were treated with 0–20 μM SFN for 24 h. Lysates were then collected for immunoblot. (C) SFN does not regulate MEP50 or PRMT5 mRNA level. SCC-13 (0.5 million) were treated with 20 μM SFN for 48 h and RNA was isolated for qRT-PCR detection of MEP50 and PRMT5 mRNA. The values are mean ± SEM, n = 3. (D) SFN reduces MEP50 and PRMT5 level via a proteasome-dependent pathway. SCC-13 cells were treated with 0 or 20 μM SFN with or without 0.75 μM lactacystin for 24 h, and lysates were prepared for detection of PRMT5 and MEP50. Similar results were obtained from three independent experiments. (E) SCC-13 cells were harvested and plated (25000 cells) on a matrigel-coated membrane, in a Millicell chamber, and 0 or 20 μM SFN was added. After 24 h, the membrane was collected and cell migration to the lower membrane surface was monitored using an inverted fluorescent microscope (mean ± SEM, n = 3, P < 0.005). (F) Confluent cultures of SCC-13 cells were uniformly wounded using pipette tip and then treated with 0 or 20 μM SFN and wound closure was monitored from 0 to 16 h. (G) SFN suppresses MEP50 and PRMT5 protein level. SCC-13 cells were treated with 0 or 20 μM SFN for 24 h. Lysates were collected for immunoblot detection of MEP50 and PRMT5. (H, I) SFN treatment reduces tumor volume growth. SCC-13 cells were injected into each front flank in NSG mice and treatment was initiated with 0 or 10 μmoles SFN per treatment administered by oral gavage on three alternate days per week. The values are mean ± SEM, n = 3. The asterisks indicate significant differences, P < 0.005. The tumors from the control and the SFN treated group were photographed at 3 weeks. (J) SFN regulates tumor levels of MEP50 and PRMT5. Extracts were prepared from control- and SFN-treated tumors at 3 weeks for immunoblot detection of the indicated proteins. Two representative tumors from each group are shown.
SFN reduces MEP50 and H4R3me2s level in vivo
If loss of MEP50 or PRMT5 function is required for SFN suppression of tumor formation, than SFN treatment should reduce the level of these proteins in tumors. To test this, SCC-13 cells were injected into each front flank in NSG mice and treatment was initiated with 0 or 10 μmole SFN per treatment administered by oral gavage on three alternate days per week. Tumor volume was monitored at 1, 2 and 3 weeks. Figure 3H and I shows that SFN treatment reduces tumor growth. To assess the impact of SFN treatment on MEP50 and PRMT5 levels, we prepared tumor extracts for immunoblot. Figure 3J shows that SFN treatment does not impact PRMT5 or H3R8me2s level, but that it does substantially reduce MEP50 level and H4R3me2s formation.
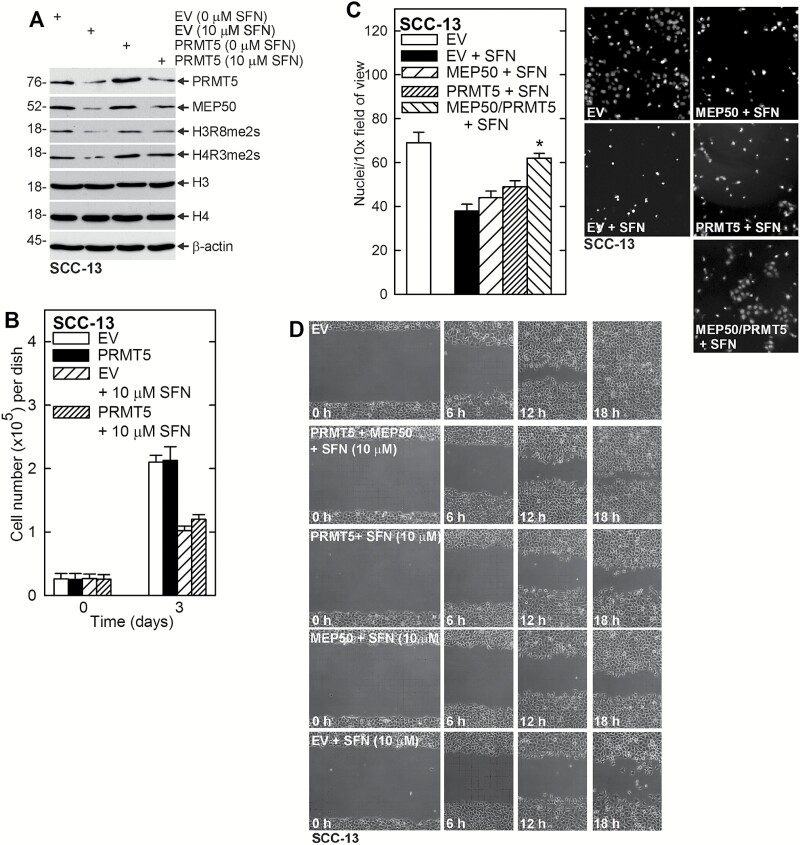
PRMT5 and MEP50 can partially reverse the SFN effect
If SFN reduction of tumor cell function requires loss of PRMT5/MEP50, maintaining expression of these proteins in SFN-challenged cells should partially reverse SFN biological activity. To test this, we forced maintenance of PRMT5 and then treated with SFN. Figure 4A shows that it is difficult to elevate PRMT5 level using an expression vector in SFN-treated cells, probably because SFN produces a strong proteasome-related stimulus to decrease the level of these proteins. However, PRMT5 level is slightly elevated and this is associated with increased MEP50 level and increased H3R8me3s and H4R3me2s formation.
Figure 4.
PRMT5 and MEP50 expression partially reverses SFN action. (A) SCC-13 cells were electroporated with empty vector (EV) or PRMT5 expression plasmid, plated in 100 mm dishes in growth medium and treated for 24 h with 0 or 10 μM SFN. Lysates were prepared for detection of the indicated proteins. (B) SCC-13 cells were electroporated with empty (EV) or PRMT5 expression vector and plated in 35 mm dishes in growth medium. The cells were then treated with 0 or 10 μM SFN and cell number was counted at 0 and 3 d. (C) SCC-13 cells were electroporated with empty (EV), MEP50 or PRMT5 expression vector and 25000 cells were plated on matrigel in Millicell chambers and treated with 0 or 10 μM SFN. Migrated cells were counted at 24 h. (D) SCC-13 cells (2 million) were electroporated with empty, MEP50 and/or PRMT5 expression vector and then plated as confluent monolayers followed by addition of 0 or 10 μM SFN, and wound closure was monitored over 0–18 h.
As shown in Figure 4B, forced expression of PRMT5 does not reverse SFN suppression of cell proliferation. However, forced expression did partially reverse the SFN suppression of invasion and migration. Figure 4C and D shows that forced MEP50 and PRMT5 expression partially attenuates SFN suppression of cell invasion and migration, and that combined MEP50/PRMT5 expression is more effective. These findings suggest that PRMT5/MEP50 reduction is required for SFN to suppress invasion and migration.
SFN impact on MEP50 and PRMT5 in other skin-derived cell lines
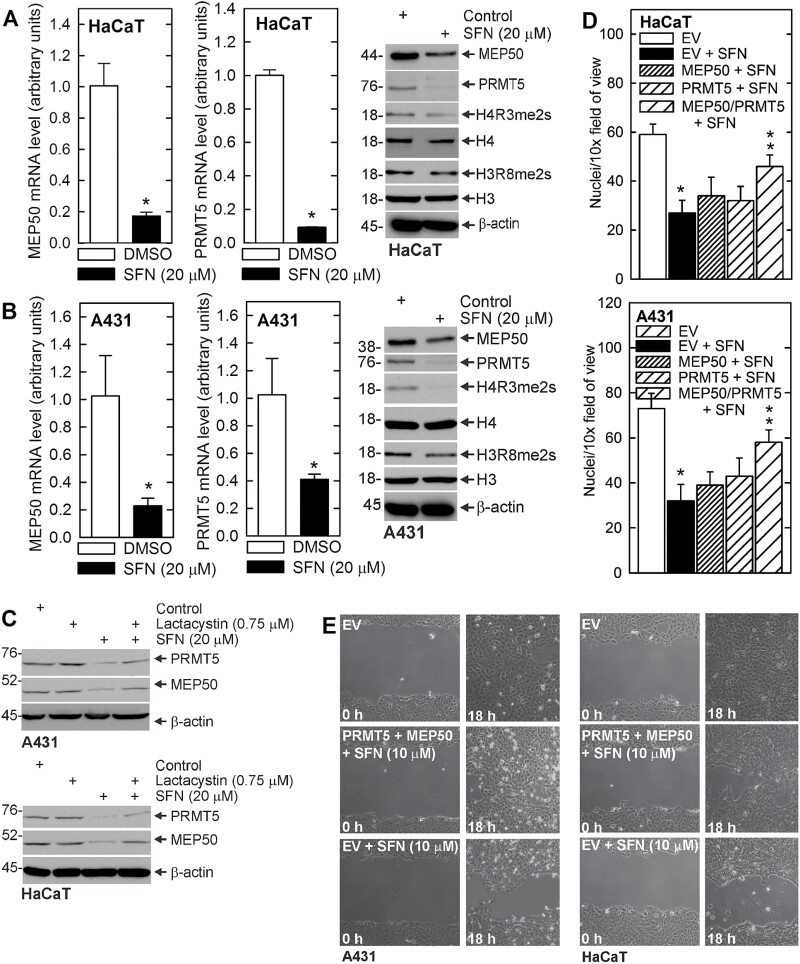
In order to assess the impact of SFN dependent regulation of MEP50 and PRMT5 in other skin-derived immortalized and transformed cells, we treated A431 and HaCaT cell lines with SFN. It is interesting that SFN does not regulate PRMT5 and MEP50 mRNA level in SCC-13 cells (Figure 3C), but it does suppress mRNA level in HaCaT and A431 cells (Figure 5A and B). This is associated with reduced MEP50, PRMT5 and H4R3me2s levels in HaCaT cells, and reduced MEP50, PRMT5, H4R3me2s and H3R8me2s levels in A431 cells. We also show that inhibition of proteasome activity partially reverses the SFN-dependent reduction in MEP50 and PRMT5 (Figure 5C). As shown in Figure 5D, forced expression of MEP50 or PRMT5 partially reverses SFN suppression of A431 and HaCaT cell invasion, and combined expression of MEP50/PRMT5 is the most effective. Figure 5E shows that MEP50/PRMT5 combined expression also reverses SFN suppression of cell migration. These findings suggest that loss of PRMT5 and MEP50 is required for SFN to suppress invasion and migration.
Figure 5.
SFN impacts MEP50 and PRMT5 level and activity in other skin-derived cells lines. (A, B) HaCaT and A431 cells were treated with 0 or 20 μM SFN for 48 h and protein and RNA samples were prepared for detection of the indicated mRNA and proteins. The values are mean ± SEM, n = 3, P < 0.005. (C) SFN suppression of MEP50 and PRMT5 requires proteasome activity. A431 and HaCaT were treated with lactacystin (0.75 μM) and/or SFN (20 μM) and after 24 h, extracts were prepared for detection of the indicated proteins. (D) A431 or HaCaT cells were electroporated with empty (EV), MEP50 or PRMT5 expression vector, and 25000 cells were plated on matrigel in Millicell chambers and treated with 0 or 10 μM SFN. Migrated cells were counted at 24 h. The values are mean ± SEM. The single asterisk indicates a significant reduction in EV + SFN treated versus EV cultures. The double asterisks indicate a significant increase as compared to the EV + SFN group (n = 3, P < 0.005). (E) A431 and HaCaT cells (2 million) were electroporated with empty (EV) or MEP50 + PRMT5 vectors, and then plated as confluent monolayers followed by addition of 0 or 10 μM SFN and wound closure was monitored over 0–18 h.
Discussion
PRMT5/MEP50 and cancer
Protein arginine methyltransferases transfer methyl groups from S-adenosylmethionine to the guanidine nitrogen of arginine in proteins (1). PRMT1, 2, 3, 4, 6 and 8 are Type I, while PRMT5 is a type II enzyme. Type I enzymes catalyze asymmetric dimethylarginine formation, and type II enzymes catalyze formation of symmetric dimethylarginine (46). Many of these enzymes display a limited tissue-specific distribution, but PRMT5 is expressed in many cell types (2). PRMT5 catalyzes symmetric dimethylation of arginine 3 on histone H2A and H4 (H2AR3me2s, H4R3me2s), and arginine 8 on histone H3 (H3R8me2s) (8,11,47), and these events are associated with silencing of gene expression (48).
PRMT5 and MEP50 have been established as having an important role in cancer cell survival, suggesting that they represent important cancer prevention targets. MEP50, a WD40 repeat-containing protein, directly binds and activates PRMT5 methyltransferase activity by enhancing affinity of the complex for substrate (18,19). The PRMT5/MEP50 complex is implicated in cancers including ovarian, lung, lymphoid, lymphoma, glioblastoma, melanoma, colon, gastric, bladder cancer and germ cell tumors (15,21,49–57). For example, decreased patient survival is associated with elevated PRMT5 in ovarian cancer (51), elevated PRMT5 and MEP50 expression is associated with reduced survival in nonsmall cell lung cancer (58), and MEP50 and PRMT5 enhance lung cancer tumorigenesis (59). Mouse studies also support a role in cancer, as overexpression of PRMT5 in immune-compromised mice causes tumor formation (55), and PRMT5 knockdown reduces cell proliferation in breast and lung cancer (4,11,54) and melanoma cells (52).
PRMT5/MEP50 and histone arginine dimethylation
In the present study, we examine the role of PRMT5 and MEP50 in epidermal squamous cell carcinoma. PRMT5 catalyzes formation of H4R3me2s and H3R8me2s chromatin marks that are associated with silencing of gene expression (41). For example, PRMT5 catalyzed H4R3me2s and H3R8me2s formation results in silencing of the RB1 gene (49). However, the literature indicates that it is not clear whether H3R4me2s and H4R8me2s formation always go hand-in-hand or if the marks can be independently applied. For example PRMT5 catalyzes formation of H3R8me2s at the ST7 and NM23 gene promoters leading to repression; however, changes in H4R3me2s formation at these loci could not be detected (11). We monitored both marks and found that H4R3me2s and H3R8me2s formation depended upon environment. For example PRMT5 or MEP50 knockdown in cultured SCC-13 cells results in reduced H4R3me2s formation, but no change in H3R8me2s level. When SCC-13 cells are placed in mice for tumor formation, PRMT5 knockdown resulted in reduced H4R3me2s and H3R8me2s levels. In contrast, MEP50 knockdown resulted in no change in H4R3me2s formation but a reduction in H3R8me2s level. Finally, SFN treatment of SCC-13 cell tumors resulted in a reduction in MEP50 and H4R3me2s, but no change in PRMT5 or H3R8me2s levels. These findings suggest that H4R3me2s formation is more frequently impacted, but that the level of either mark can be modulated in a context-dependent manner.
SFN control of PRMT5 and MEP50
SFN is an important candidate diet-derived cancer prevention agent (44). It is a particularly important agent for this purpose, as it does not produce any known side effects, is readily ingested, and displays high bioavailability in mice and humans (27,29,60). Moreover, it has been shown to reduce skin cancer formation in several types of models (35,44,61). However, the mechanism of action is not well understood, and there is an ongoing effort to identify key molecular targets (26,32,61–63). In the present study, we show that SFN treatment reduces PRMT5 and MEP50 level, and that this is associated with a consistent reduction in H4R3me2s formation. This reduction was observed in three independent cell lines, SCC-13, HaCaT and A431. However, it is interesting that the mechanism varied. In SCC-13 cells, SFN treatment reduced PRMT5 and MEP50 level is a proteasome-dependent mechanism. In contrast, in HaCaT and A431 cells, SFN treatment reduced the level of PRMT5- and MEP50-encoding mRNA, suggesting a transcriptional regulatory mechanism, although a proteasome-dependent mechanism is also involved. We also confirm that forced vector-mediated PRMT5 or MEP50 expression partially reverses the impact of SFN, which suggests that loss of these proteins is required for optimal SFN suppression of cell survival. This is consistent with recent studies in other cell types, showing that these proteins influence multiple cellular processes (64).
PRMT5/MEP50, SFN and tumor formation
The results of in vitro experiments are always more compelling when confirmed by in vivo studies. We were particularly interested in the role of MEP50 in tumor formation, because, as noted below, our studies revealed a reduction in MEP50 in SFN-treated tumors. To study the role of MEP50 in vivo, we monitored the impact of MEP50 knockdown on tumor formation. These studies reveal a substantial reduction in tumor size for tumors derived from MEP50 knockdown cells, suggesting that MEP50 is required for maximal tumor growth. Moreover, this was associated with a reduction in PRMT5 level and H3R8me2s, but no change in H4R3me2s formation. In contrast, PRMT5 knockdown resulted in a marked loss of MEP50 and a substantial reduction in H4R3me2s and H3R8me2s formation.
Another important finding is that SFN treatment reduces tumor size and that this is associated with reduced MEP50 level and reduced H4R3me2 formation. It is interesting that SFN treatment does not reduce PRMT5 level. However, since MEP50 and PRMT5 form a heterotetramer of four MEP50 and four PRMT5 subunits, and MEP50 is required for PRMT5 activity, the loss of MEP50 will clearly lead to reduced activity of the complex. This is evidenced by the reduction in H4R3me2s formation. SFN treatment also reduces PRMT5 and MEP50 levels in HaCaT and A431 cells. As in SCC-13 cells, MEP50 and PRMT5 loss is required for optimal SFN suppression of invasion and migration. This confirms that MEP50 and PRMT5 are SFN targets in multiple cell lines.
In conclusion, these studies suggest that the PRMT5/MEP50 complex is required for optimal growth and survival of epidermal squamous cell carcinoma cells and for optimal tumor formation. Additionally, the PRMT5/MEP50 complex is targeted as part of the mechanism responsible for SFN suppression of tumor formation. We further propose that PRMT5 inhibitors (65) may be useful for the treatment of epidermal squamous cell carcinoma.
Funding
This work was supported by grants from the Maryland Stem Cell Research Foundation (R.L.E.) and the National Institutes of Health (CA131074 and CA184027 to R.L.E.) and a pilot grant from the Greenebaum Cancer Center (P30 CA134274).
Conflict of Interest Statement: None declared.
Abbreviations
- MEP50
methylosome protein 50
- PRMT5
protein arginine methyltransferase 5
- SFN
sulforaphane.
References
- 1. Bedford, M.T., et al. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell, 33, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stopa, N., et al. (2015) The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell. Mol. Life Sci., 72, 2041–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsu, J.M., et al. (2011) Crosstalk between Arg 1175 methylation and Tyr 1173 phosphorylation negatively modulates EGFR-mediated ERK activation. Nat. Cell Biol., 13, 174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scoumanne, A., et al. (2009) PRMT5 is required for cell-cycle progression and p53 tumor suppressor function. Nucleic Acids Res., 37, 4965–4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanaka, H., et al. (2009) PRMT5, a novel TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis via nuclear factor-kappaB activation. Mol. Cancer Res., 7, 557–569. [DOI] [PubMed] [Google Scholar]
- 6. Hosohata, K., et al. (2003) Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription. Mol. Cell. Biol., 23, 7019–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Paik, W.K., et al. (2007) Historical review: the field of protein methylation. Trends Biochem. Sci., 32, 146–152. [DOI] [PubMed] [Google Scholar]
- 8. Pollack, B.P., et al. (1999) The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J. Biol. Chem., 274, 31531–31542. [DOI] [PubMed] [Google Scholar]
- 9. Friesen, W.J., et al. (2001) The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol., 21, 8289–8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabbrizio, E., et al. (2002) Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep., 3, 641–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pal, S., et al. (2004) Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol., 24, 9630–9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pal, S., et al. (2003) mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol., 23, 7475–7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ancelin, K., et al. (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol., 8, 623–630. [DOI] [PubMed] [Google Scholar]
- 14. Tee, W.W., et al. (2010) Prmt5 is essential for early mouse development and acts in the cytoplasm to maintain ES cell pluripotency. Genes Dev., 24, 2772–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pal, S., et al. (2007) Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J., 26, 3558–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cesaro, E., et al. (2009) The Kruppel-like zinc finger protein ZNF224 recruits the arginine methyltransferase PRMT5 on the transcriptional repressor complex of the aldolase A gene. J. Biol. Chem., 284, 32321–32330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dacwag, C.S., et al. (2007) The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol. Cell. Biol., 27, 384–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Antonysamy, S., et al. (2012) Crystal structure of the human PRMT5:MEP50 complex. Proc. Natl. Acad. Sci. U. S. A., 109, 17960–17965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ho, M.C., et al. (2013) Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity. PLoS One, 8, e57008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu, Z., et al. (2013) The p44/wdr77-dependent cellular proliferation process during lung development is reactivated in lung cancer. Oncogene, 32, 1888–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gu, Z., et al. (2012) Protein arginine methyltransferase 5 is essential for growth of lung cancer cells. Biochem. J., 446, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng, Y., et al. (2008) Distinct nuclear and cytoplasmic functions of androgen receptor cofactor p44 and association with androgen-independent prostate cancer. Proc. Natl. Acad. Sci. U. S. A., 105, 5236–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ligr, M., et al. (2011) Expression and function of androgen receptor coactivator p44/Mep50/WDR77 in ovarian cancer. PLoS One, 6, e26250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng, Y., et al. (2010) Androgen receptor coactivator p44/Mep50 in breast cancer growth and invasion. J. Cell. Mol. Med., 14, 2780–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanade, S.R., et al. (2012) Protein arginine methyltransferase 5 (PRMT5) signaling suppresses protein kinase Cδ- and p38δ-dependent signaling and keratinocyte differentiation. J. Biol. Chem., 287, 7313–7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clarke, J.D., et al. (2008) Multi-targeted prevention of cancer by sulforaphane. Cancer Lett., 269, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Egner, P.A., et al. (2014) Rapid and sustainable detoxication of airborne pollutants by broccoli sprout beverage: results of a randomized clinical trial in China. Cancer Prev. Res. (Phila)., 7, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ho, E., et al. (2009) Dietary sulforaphane, a histone deacetylase inhibitor for cancer prevention. J. Nutr., 139, 2393–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh, S.V., et al. (2009) Sulforaphane inhibits prostate carcinogenesis and pulmonary metastasis in TRAMP mice in association with increased cytotoxicity of natural killer cells. Cancer Res., 69, 2117–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li, Y., et al. (2013) Kinetics of sulforaphane in mice after consumption of sulforaphane-enriched broccoli sprout preparation. Mol. Nutr. Food Res., 57, 2128–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bricker, G.V., et al. (2014) Isothiocyanate metabolism, distribution, and interconversion in mice following consumption of thermally processed broccoli sprouts or purified sulforaphane. Mol. Nutr. Food Res., 58, 1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Balasubramanian, S., et al. (2011) Sulforaphane suppresses polycomb group protein level via a proteasome-dependent mechanism in skin cancer cells. Mol. Pharmacol., 80, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abel, E.L., et al. (2013) Sulforaphane induces phase II detoxication enzymes in mouse skin and prevents mutagenesis induced by a mustard gas analog. Toxicol. Appl. Pharmacol., 266, 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chew, Y.C., et al. (2012) Sulforaphane induction of p21(Cip1) cyclin-dependent kinase inhibitor expression requires p53 and Sp1 transcription factors and is p53-dependent. J. Biol. Chem., 287, 16168–16178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gills, J.J., et al. (2006) Sulforaphane prevents mouse skin tumorigenesis during the stage of promotion. Cancer Lett., 236, 72–79. [DOI] [PubMed] [Google Scholar]
- 36. Shibata, A., et al. (2010) Sulforaphane suppresses ultraviolet B-induced inflammation in HaCaT keratinocytes and HR-1 hairless mice. J. Nutr. Biochem., 21, 702–709. [DOI] [PubMed] [Google Scholar]
- 37. Zhu, M., et al. (2004) Phase II enzyme inducer, sulforaphane, inhibits UVB-induced AP-1 activation in human keratinocytes by a novel mechanism. Mol. Carcinog., 41, 179–186. [DOI] [PubMed] [Google Scholar]
- 38. Adhikary, G., et al. (2010) PKC-delta and -eta, MEKK-1, MEK-6, MEK-3, and p38-delta are essential mediators of the response of normal human epidermal keratinocytes to differentiating agents. J. Invest. Dermatol., 130, 2017–2030. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 39. Gu, Z., et al. (2012) Protein arginine methyltransferase 5 functions in opposite ways in the cytoplasm and nucleus of prostate cancer cells. PLoS One, 7, e44033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shilo, K., et al. (2013) Cellular localization of protein arginine methyltransferase-5 correlates with grade of lung tumors. Diagn. Pathol., 8, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karkhanis, V., et al. (2011) Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci., 36, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Radisky, E.S., et al. (2007) Stromal induction of breast cancer: inflammation and invasion. Rev. Endocr. Metab. Disord., 8, 279–287. [DOI] [PubMed] [Google Scholar]
- 43. Antosiewicz, J., et al. (2008) Role of reactive oxygen intermediates in cellular responses to dietary cancer chemopreventive agents. Planta Med., 74, 1570–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cheung, K.L., et al. (2010) Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J., 12, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singh, S.V., et al. (2004) Sulforaphane-induced G2/M phase cell cycle arrest involves checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C. J. Biol. Chem., 279, 25813–25822. [DOI] [PubMed] [Google Scholar]
- 46. Yang, Y., et al. (2013) Protein arginine methyltransferases and cancer. Nat. Rev. Cancer, 13, 37–50. [DOI] [PubMed] [Google Scholar]
- 47. Branscombe, T.L., et al. (2001) PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J. Biol. Chem., 276, 32971–32976. [DOI] [PubMed] [Google Scholar]
- 48. Jahan, S., et al. (2015) Protein arginine methyltransferases (PRMTs): role in chromatin organization. Adv. Biol. Regul., 57, 173–184. [DOI] [PubMed] [Google Scholar]
- 49. Wang, L., et al. (2008) Protein arginine methyltransferase 5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells. Mol. Cell. Biol., 28, 6262–6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eckert, D., et al. (2008) Expression of BLIMP1/PRMT5 and concurrent histone H2A/H4 arginine 3 dimethylation in fetal germ cells, CIS/IGCNU and germ cell tumors. BMC Dev. Biol., 8, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bao, X., et al. (2013) Overexpression of PRMT5 promotes tumor cell growth and is associated with poor disease prognosis in epithelial ovarian cancer. J. Histochem. Cytochem., 61, 206–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nicholas, C., et al. (2013) PRMT5 is upregulated in malignant and metastatic melanoma and regulates expression of MITF and p27(Kip1.). PLoS One, 8, e74710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim, J.M., et al. (2005) Identification of gastric cancer-related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clin. Cancer Res., 11(2 Pt 1), 473–482. [PubMed] [Google Scholar]
- 54. Wei, T.Y., et al. (2012) Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci., 103, 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Uzdensky, A., et al. (2014) Expression of proteins involved in epigenetic regulation in human cutaneous melanoma and peritumoral skin. Tumour Biol., 35, 8225–8233. [DOI] [PubMed] [Google Scholar]
- 56. Han, X., et al. (2014) Expression of PRMT5 correlates with malignant grade in gliomas and plays a pivotal role in tumor growth in vitro. J. Neurooncol., 118, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yan, F., et al. (2014) Genetic validation of the protein arginine methyltransferase PRMT5 as a candidate therapeutic target in glioblastoma. Cancer Res., 74, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Győrffy, B., et al. (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One, 8, e82241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wei, T.Y., et al. (2014) Methylosome protein 50 promotes androgen- and estrogen-independent tumorigenesis. Cell. Signal., 26, 2940–2950. [DOI] [PubMed] [Google Scholar]
- 60. Kensler, T.W., et al. (2012) Modulation of the metabolism of airborne pollutants by glucoraphanin-rich and sulforaphane-rich broccoli sprout beverages in Qidong, China. Carcinogenesis, 33, 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu, C., et al. (2006) Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res., 66, 8293–8296. [DOI] [PubMed] [Google Scholar]
- 62. Saw, C.L., et al. (2011) Impact of Nrf2 on UVB-induced skin inflammation/photoprotection and photoprotective effect of sulforaphane. Mol. Carcinog., 50, 479–486. [DOI] [PubMed] [Google Scholar]
- 63. Juge, N., et al. (2007) Molecular basis for chemoprevention by sulforaphane: a comprehensive review. Cell. Mol. Life Sci., 64, 1105–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ibrahim, R., et al. (2014) Expression of PRMT5 in lung adenocarcinoma and its significance in epithelial-mesenchymal transition. Hum. Pathol., 45, 1397–1405. [DOI] [PubMed] [Google Scholar]
- 65. Chan-Penebre, E., et al. (2015) A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat. Chem. Biol., 11, 432–437. [DOI] [PubMed] [Google Scholar]