Abstract
Systems biology models are rapidly increasing in complexity, size and numbers. When building large models, researchers rely on software tools for the retrieval, comparison, combination and merging of models, as well as for version control. These tools need to be able to quantify the differences and similarities between computational models. However, depending on the specific application, the notion of ‘similarity’ may greatly vary. A general notion of model similarity, applicable to various types of models, is still missing. Here we survey existing methods for the comparison of models, introduce quantitative measures for model similarity, and discuss potential applications of combined similarity measures. To frame model comparison as a general problem, we describe a theoretical approach to defining and computing similarities based on a combination of different model aspects. The six aspects that we define as potentially relevant for similarity are underlying encoding, references to biological entities, quantitative behaviour, qualitative behaviour, mathematical equations and parameters and network structure. We argue that future similarity measures will benefit from combining these model aspects in flexible, problem-specific ways to mimic users’ intuition about model similarity, and to support complex model searches in databases.
Keywords: systems biology, network model, model version control, information retrieval, SBML
Introduction
‘Over the past few decades, mathematical models of molecular and gene networks have become an important part of the research toolkit for the biosciences’ [1]. Mathematical models are formal representations of natural systems that can help answer questions about the complex system they represent [2]. According to Robert Rosen, a model establishes a modelling relation between a formal and a natural system: the formal system encodes the natural system, and inferences made in the formal system can be interpreted (decoded) as statements about the natural system [3]. Systems biology models serve as abstractions of biological systems. Biochemical models, for example, associate model components, such as mathematical expressions, objects or variables, with biochemical entities such as molecule species or chemical reactions. Depending on the scientific questions addressed and on available data, a biological system may be described by models of different scopes and levels of granularity, reflecting different views of the system.
In this article, we focus on systems biology models. Systems biology models can be based on a number of mathematical formalisms [1]. Here, again, the choice of a particular approach largely depends on the type of question asked and on the data available [2]. Metabolic and signalling pathways are usually modelled by ordinary differential equation systems, and the resulting models are known as kinetic models. Larger metabolic systems are typically described by constraint-based network models that capture stationary metabolic fluxes, but disregard enzyme kinetics. Gene expression dynamics can be modelled by kinetic models, stochastic processes or discrete dynamic processes such as Boolean networks [4]. Spatial cell models may even involve partial differential equations. In addition, the rise of synthetic biology and the simulation of entire organisms require hybrid modelling.
Because models are rapidly increasing in numbers and in complexity, many of them are now formally encoded in standard formats to be processed by and exchanged among different software tools. The Systems Biology Markup Language (SBML) [5] and CellML [6] are two XML-based de facto standards that encode the entities and interactions in biological models. Both enable software interoperability across the diverse landscape of modelling, visualization and simulation software [7]. To further standardize the representation of models and enable interoperability between them, the biological meaning of model components can be specified by semantic annotations that relate components (e.g. a variable representing glucose concentration) to common identifiers defined in ontologies or public databases (e.g. CHEBI:17234, which is the identifier for glucose in the ChEBI database [8]). Together, standard formats and semantic annotations foster the reuse of data in the biosciences [9]. In addition, the reuse of models across research groups and scientific use cases demand sophisticated model management strategies for storage, search, retrieval, version control and provenance [10].
All these tasks require, at least implicitly, mathematical notions of similarity between models. If similar models can be recognized by software, then they can also be found in databases, differences between models can be assessed and genealogies of model versions can be reconstructed. For any domain of knowledge, different notions of similarity can be used depending on the aspects between which similarity is determined. For example, to decide whether two persons are alike, we need to choose what features of the person to focus on, and we may assign priorities to selected features. Then, we may compare two persons by how tall they are, by the shapes of their faces, or by their behaviour—with different results. Certain groups of people may be easily distinguished by size (e.g. children versus adults), while the same criterion will fail to predict other distinctions (left-handed versus right-handed people). Also in computer science, data objects can be compared with regard to different aspects depending on the purpose of the comparison. For example, when comparing image files, typical features are the colours used, the objects shown or file size and type. The choice of the features and similarity measure depends on the intended application.
Model comparison has been implemented in a few software tools as part of their similarity searches. For example, public model repositories such as the CellML model repository [11] can use a retrieval system [12] that supports users in finding models relevant to their research. For such a model retrieval system, similarity measures are applied and adapted to yield a measure of model similarity based on various features of models [13]. Another example is semanticSBML [14], an online software tool that provides functionality for clustering, merging and comparison of SBML models based on semantic annotations.
We argue that model comparison, as a computational task, should be abstracted from specific model types, applications and software. Here we set out to study what is common to different similarity measures for systems biology models, and attempt to place them into a common mathematical frame. The practical aim of such an approach is to build a common software library that implements various similarity measures and can be integrated into systems biology software to provide flexibility in model comparison tasks. In this article we overview and categorize existing similarity measures between models and their components. These measures may rely on aspects such as the biological entities described, network structure, model assumptions, mathematical statements and parameter values, or the dynamic behaviour displayed in simulations. We discuss the different aspects and show how they can be incorporated into computable similarity measures. Furthermore, we show applications for these measures using model search, clustering and merging as examples, and we discuss open problems that should be addressed in future research.
Formal notions of similarity
Similarity measures
The general notion of similarity can be conceptualized by mathematical functions called similarity measures. Intuitively, a similarity measure, for some type of objects, assigns to each pair of objects a similarity value. Larger values signify greater similarity. If properties of the objects are represented in some property space, similarity resembles an inverse distance: objects with small distances are considered similar, objects with large distances dissimilar. Similarity measures are often normalized to yield values between 0 and 1. A similarity σ = 1 then implies that two objects are identical (or indistinguishable) with regard to the properties considered, while entirely different objects will have a similarity σ = 0. Formally, a normalized similarity measure for a set X is defined as a function which assigns x1, x2 ∈ X a value σ ∈ [0, 1]. This σ is called the ‘similarity value’. Most similarity measures are symmetrical and satisfy the triangle inequality.
Model similarity based on single aspects
Similarity measures for models refer to specific model aspects. For example, the similarity between two network models can be determined by aligning the networks and assessing their common overlap [15]. The result is a similarity value with respect to network structure. Alternatively, the similarity between two models can be calculated by comparing simulated time series [16], by comparing semantic annotations [12] or by identifying occurring patterns, etc. In the following, we define similarity with respect to certain model properties. We first define similarity with regard to a single aspect. Later in the article, we also discuss similarity with regard to a combination of aspects.
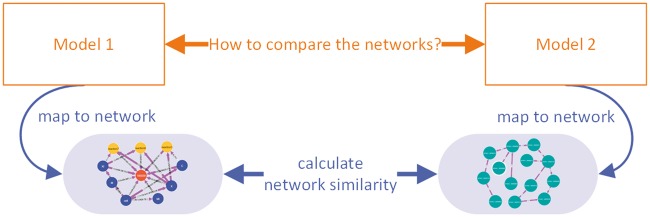
To formally express similarity with regard to a single aspect, we introduce the projection of a model M onto an aspect α, for example, the projection of a dynamical model onto the underlying network topology. All other model features, which do not belong to this aspect, are ignored. The similarity measure between the two projected models then determines the similarity of these models with respect to that aspect (see Figure 1). More formally, for a model aspect α the α-similarity between two models M1, M2, simα(M1, M2) is defined as σα(Πα(M1), Πα(M2)), where Πα is the projection of a model onto aspect α and σα is a similarity measure for aspect α. This scheme applies both to models as mathematical objects and to encoded models, i.e. text files representing these models, and allows for statements such as ‘model 1 and model 2 are similar with respect to aspect α’. This definition may leave some similarity measures undefined because we consider the projection function Πα a partial function. If a model lacks the aspect for which the similarity measure is defined, the similarity to other models remains undefined as well.
Figure 1.
Comparison of two systems biology models based on a model aspect. To compare two models, the models are mapped to a particular aspect (in this case, their network structure). Then, an appropriate similarity measure (e.g. for graph similarity) is applied [15].
With the same scheme, models can also be compared with other types of data, e.g. to sets of experimentally measured substance concentrations. To see whether a model and a data set refer to similar sets of substances, we may choose the sets of substances as the aspect we are focusing on. Mathematically, if a projection Πα(M) yields the aspect α of a model M and a related projection Π’α projects data sets D to the same aspect α, then an α-similarity function σα can be used to find data sets that resemble a model M with respect to α, by computing σα(Πα(M), Π’α(D)).
Combined similarity measures
Similarities arising from different model aspects can be combined to define more complex measures. For example, two models may only be considered similar if they describe similar biological entities and if they describe them by similar mathematical formulae. The combination of different similarity measures occurs frequently in model search, e.g. to enable users to find ‘models that describe aspects of the cell cycle and use similar parameter space’.
To perform such complex tasks, we need to aggregate multiple similarity scores into a single score. We can compose or decompose complex similarity measures by projecting models on individual aspects and then combining the resulting similarities between these aspects. Formally, we can express this as follows (without loss of generality limited to normalized similarity measures): simα°β (M1, M2) = σα°β(Πα°β(M1), Πα°β(M2)) = σα(Πα(M1), Πα(M2)) ° σβ (Πβ(M1), Πβ(M2)) where ° is a function °: [0, 1] × [0, 1] → [0, 1].
For larger numbers of properties to be combined, similarity measures can be conveniently defined by feature vectors. A feature vector represents different model properties by a list of numbers arranged in a vector. For example, the elements of the vector may be zeros or ones, denoting the occurrence of certain annotations in the model, or integer numbers denoting the frequencies of certain network motifs in the network. Once models have been translated into feature vectors, a variety of methods from multivariate data analysis can be applied, including supervised and unsupervised classification [17]. Simple similarity measures for feature vectors can be defined based on Euclidean distances, the Jaccard index or on normalized scalar products (i.e. the cosine of the angle between feature vectors). To weigh different features and to account for their known relationships, special metrics can be used, e.g. quadratic forms instead of simple scalar products. This can be useful to account for known logical connections between different features.
Model comparison based on specific aspects
Relevant aspects of models can be extracted (or calculated) from the encoded models. Depending on the goal of a comparison and on a model’s representation, the computable similarity measure is chosen. When models are presented as network graphs, it will be natural to compare them by network structure using a graph similarity measure. For example, the Systems Biology Graphical Notation [18] for graphical display supports comparisons of network structural and visual similarity. When only model equations are given, network similarity would be more difficult to recognize even if all necessary information is implicitly given. For example, SBML and CellML for model structure and formulae support comparisons of structural and mathematical similarity. The Systems Biology Ontology (SBO) [19] and other ontologies enable semantic similarity comparisons. According to our framework, the projection function Πα can be more or less complex to define (and evaluate) depending on the model representation. Consequently, certain similarity measures will be more natural given a certain model representation.
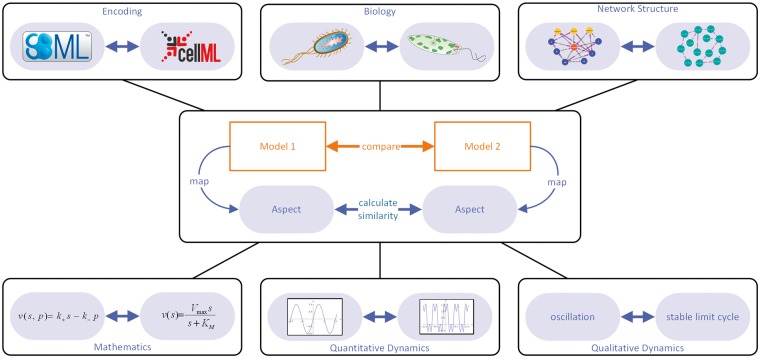
We propose to classify similarity measures based on five types of model aspects: (i) model encoding; (ii) biological meaning; (iii) network structure; (iv) mathematical statements and numerical values; and (v) quantitative and qualitative behaviour (compare Figure 2 and Table 1). Additional ‘meta-properties’ and provenance information further improve the comparison (e.g. information about file format or year of development).
Figure 2.
Similarity measures for models can be derived from similarity measures for model aspects. Automatic model comparison relies on quantitative similarity measures. A practical way of defining such measures is to project both models to some relevant aspect, e.g. network structure, for which a similarity measure has been defined. In an abstract scheme for model comparison, two models are mapped onto a specific focal aspect.
Table 1.
Model aspects and related similarity measures
| Model aspect | Existing measures | Tool support (selection) | Applications |
|---|---|---|---|
| Encoding | Similarity on the XML-encoding (SBML or CellML), Levenshtein distance | BiVeS [21], Unix Diff | Model version control |
| Biological meaning | Similarity regarding biological meaning of model components | SemanticSBML, Semantic Measures Library and Toolkit Model set comparison, MORRE [45] | Search, retrieval, comparison of model intentions and assumptions |
| Network structure | Graph similarity, stoichiometric similarity | Cytoscape [62], graph libraries | Model merging, extraction and comparison of submodels |
| Mathematical statements, quantitative and qualitative behaviour | Difference in statements and numbers; correlation between states, time-series, steady-states | Semantic Measures Library and Toolkit (for SBO annotations) | Compare dynamic behaviour; classification of models (e.g. oscillatory versus non-oscillatory glycolysis models) |
The table lists examples of similarity measures for the proposed aspects of a model, examples of software tools supporting these measures and practical application cases.
Model encoding
Models can be directly compared based on their encoding, i.e. a specific file format. If models are encoded as computer programs, e.g. in MATLAB, a comparison on the syntactic level can be performed using tools for difference detection in software code (e.g. diff). As computer programs, in general, do not share a predefined structure, such a comparison does rarely yield satisfactory results.
However, standardized formats such as SBML and CellML have well-defined XML-syntax that restrict the number of possible operations. On the one hand, this constitutes a limitation in what information can be encoded. On the other hand, the limited number of supported structures facilitates the implementation of domain-specific algorithms. These algorithms use the rich information about the encoded biological mechanisms, the components of models and their interactions and the biological meaning of (parts of) the model. Similarly, simulations of models, and the context of the simulation, can be encoded in standard formats such as the Simulation Experiment Description Markup Language (SED-ML [20]). Thus, the comparison of models can convey information about their biological meaning and about their behaviour.
Several algorithms use this reduced approach of determining the similarity of models (or model versions) by comparing their syntactic structure, e.g. [21–23]. Some software tools compare representations of a model, taking the specific syntactic structure and the corresponding biological meaning into account, while others compare the representations directly and subsequently interpret the results with respect to the biology. For example, XML patches [22] are generated to compare models solely at the XML level, while the BiVeS tool described in [21] also considers the structure of the actual model representation format.
Biological meaning
To compare models with respect to the biological system described, the meaning of components must be considered. The biological entities in a model (substances, reactions, cell compartments, etc.) must be explicitly associated with a biological description. To this end, biological entities are usually semantically annotated with terms from biological and biomedical ontologies [19]. For example, BioModels provides 1516 literature-based, SBML-encoded models. Among them are 613 curated (manually validated and annotated) models. In addition, BioModels offers 903 non-curated models (see http://www.ebi.ac.uk/biomodels-main/news as of 1 July 2016). A curated model has on average 30 species, 41 reactions, 40 parameters, 16 compartments, 10 rules and 3 events. Each entity in a curated model is on average linked to 1.86 ontology terms providing semantics and biological background. Model annotations may also point to entries in biological databases from which ontology based annotations can then be derived, e.g. UniProt [24].
Most models in open repositories are annotated with terms from ontologies such as the Gene Ontology (GO) [25], ChEBI [8] or the SBO [19]. Different ontologies cover different domains of knowledge and allow for model comparison related to these domains: model annotations referring to GO terms allow for defining similarity related to the biological function or cellular location of biological entities; annotations based on ChEBI allow for similarity based on chemical compounds; annotations based on the SBO allow for similarity based on biochemical rate laws or on the roles of chemical compounds in reaction mechanisms, etc. Similarity measures that compare models by their semantic annotations can be defined on the basis of existing similarity measures for ontology elements [26]. These measures help to identify similarities in model matching and retrieval tasks [12, 27–29]. For example, in regulatory pathways describing protein interactions, semantic similarity is traditionally assessed as a function of the shared annotation of proteins with GO terms [30].
Network structure
Biological network structures, and especially their statistical properties, have received large attention in systems biology research [31]. Network structures can be extracted from encoded models, and network alignments between models allow for detecting specific structural differences and similarities. Gay et al., for example, proposed graph matching techniques to assess similarities between models [32]. They reduced the models to directed, bipartite reaction graphs and searched for epimorphisms between the graph structures. Subsequently, they evaluated their method on SBML models from BioModels. Graphs have also been used to measure the similarity of models based on the motifs they contain [33]. Motifs are small partial subgraphs that are statistically overrepresented in a network. Motifs are often interpreted as small functional units, and Alon [34] showed that networks realizing similar tasks (e.g. signalling pathways) contain similar motifs. Therefore, comparing the motif distributions in biological networks could provide information about typical biological functions of these networks.
Mathematical statements and model behaviour
Models can be compared by their quantitative or qualitative dynamic behaviour. This behaviour can either be determined from their mathematical structure or from simulation runs. Similarity of behaviour has been associated with similar internal mechanisms and with dependencies on common extrinsic factors [16]. While we are not aware of any system that formally compares models with respect to dynamic behaviour, there are attempts to compare, for a given model, the simulation results obtained from different simulators [35]. The Functional Curation framework, for example, compares the dynamic effects of particular simulated perturbations [36, 37]. A comparison of qualitative model behaviour (e.g. oscillation, steady state) can be useful to identify similarities between biological mechanisms [16].
Annotations of reactions with terms from SBO may help to determine behavioural similarities in the future. SBO is, for example, already used to annotate mathematic rate laws and therefore a good candidate for a qualitative comparison of mathematical expressions. If mathematical expressions are similar, then the models may also show similar dynamic behaviour.
Comparison of two models
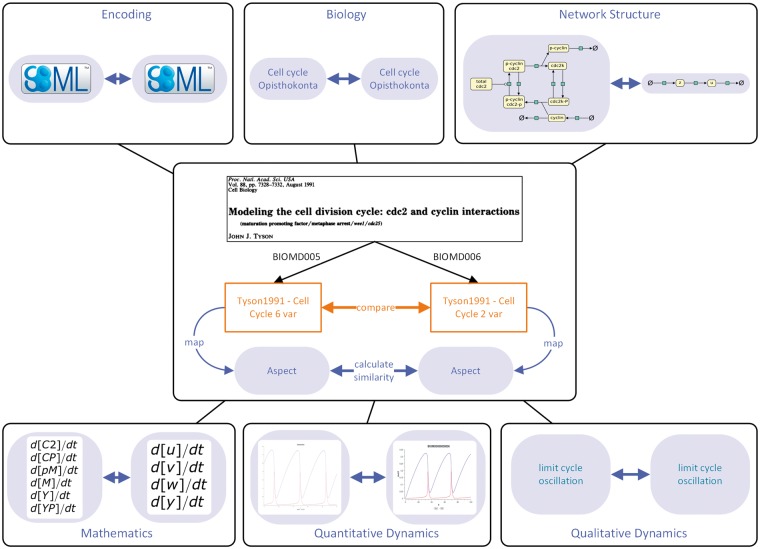
To illustrate the comparison of models by different aspects, we analyse two dynamical models that stem from the same publication [38] and study a cyclin and a cyclin-dependent kinase (see Figure 3). While the first model describes the phosphorylation of the two compounds by explicit reactions, the second model has been simplified and captures only the general dynamics. The model files, obtained from BioModels (models BIOMD0000000005 and BIOMD0000000006), are encoded in SBML and refer to the organism and biological pathway described. Owing to their different levels of resolution, the models differ in their network structures and mathematical statements. As BIOMD0000000006 is a simplification of BIOMD0000000005 in terms of mathematics, we start our example with the mathematics aspect.
Figure 3.
Comparison of two models describing the cell division cycle. Two SBML encoded models, obtained from BioModels, are compared by different aspects. Both models stem from the same publication describing the cell division cycle [38]. Each box represents an aspect and shows an excerpt of BIOMD0000000005 on the left and BIOMD0000000006 on the right side.
Mathematics
Mathematically, each of the models is represented by a system of differential equations. The first model encodes to six molecular species (including phosphorylated and unphosphorylated forms of the same compounds) and describes their dynamics by rate equations in which elementary reaction steps are clearly visible. The reaction rates depend on molecular species concentrations, two constant concentrations, and nine rate constants. In the second model, the equations have been radically simplified: by summing and normalizing the variables, we obtain four new variables whose differential equations do not have the form of simple rate equations anymore. In a next simplification step, by an approximation based on time scale separation, the number of variables is further reduced to two, and we are left with four parameters only. Could the similarities between our three equation systems be recognized automatically by inspecting the mathematical formulae? Between the six-variable and the four-variable model, the main problem is the change of variables: even though the second equation system is directly derived from the first one, the correspondence is hard to see if the definition of the new variables (in terms of the old ones) is not known. Especially, the new variable names do not give any clue about their relation to the old variables. Between the four-variable and the two-variable model, by contrast, two variables are abandoned, but the other two variables remain unchanged (same names and same meaning). The remaining two equations are formally similar to equations from the previous model variant, and only some details have changed (e.g. a term being omitted). This formal similarity could be recognized by comparing the formulae in a string or tree representation.
Quantitative dynamics
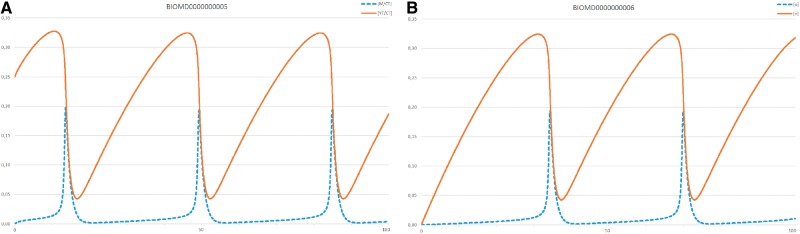
To compare the simulation results of the two models, one has to introduce two variables in BIOMD0000000005, [M]/[CT] and ([Y] + [pM] + [M])/[CT], which correspond to u and v in BIOMD0000000006, respectively. The two time courses are similar except for a phase shift of about 15 s and a different beginning part (compare Figure 4A and B). This shift is owing to different initial conditions. If one uses equal initial conditions the time courses will become identical.
Figure 4.
Time courses for BIOMD0000000005 (A) and BIOMD0000000006 (B). Both time courses (with a duration of 100 s) were generated with COPASI [39]. Model parameters and initial conditions remain as encoded in the SBML-files obtained from BioModels.
Qualitative dynamics
Both time courses can be characterized as stable limit cycles, i.e. as undamped oscillating behaviour. TEDDY, the TErminology for the Descriptions of DYnamics [19], provides the term TEDDY_0000114 to annotate these qualitative dynamics. See [40] for a more elaborate qualitative characterization of the dynamics of both models. A comparison of the qualitative dynamics of both models would reveal that the models are equal in this respect.
Encoding
As described in the aspect Mathematics, the second model contains fewer variables and equations. This is also reflected in the encoding. Both models are encoded in SBML Level 2, Version 4. The BiVeS algorithm for difference detection in computational models can be used to identify changes between the two SBML encodings. The BiVeS output is shown in Table 2. It provides information about insertion, deletion, updates and moves to convert one model into the other. A closer look at the table reveals that one model can be considered a simplification of the other. The number and types of identified changes, e.g. characterized with terms from COmputational MOdels DIffer (COMODI), an ontology describing possible changes on computational biology models [41], could be used as a similarity measure.
Table 2.
Comparison of encoded species
| Species | Changes |
|---|---|
| CT (total_cdc2) | Deleted |
| EmptySet | Attribute hasOnlySubstanceUnits was inserted: true |
| Attribute initialAmount has changed: 0→ 1 | |
| Attribute sboTerm was inserted: SBO:0000291 | |
| YP (p-cyclin) | Deleted |
| Y (cyclin) → v | Attribute boundaryCondition was inserted: true |
| Attribute hasOnlySubstanceUnits was inserted: true | |
| Attribute sboTerm was inserted: SBO:0000297 | |
| Attribute name was deleted: cyclin | |
| Attribute id has changed: Y → v | |
| YT (total_cyclin) | Deleted |
| CP (cdc2k-P) | Deleted |
| M (p-cyclin_cdc2) → z | Attribute boundaryCondition was inserted: true |
| Attribute hasOnlySubstanceUnits was inserted: true | |
| Attribute sboTerm was inserted: SBO:0000297 | |
| Attribute name was deleted: p-cyclin_cdc2 | |
| Attribute id has changed: M → z | |
| pM (p-cyclin_cdc2-p) | Deleted |
| C2 (cdc2k) → u | Attribute boundaryCondition was inserted: true |
| Attribute hasOnlySubstanceUnits was inserted: true | |
| Attribute sboTerm was inserted: SBO:0000297 | |
| Attribute name was deleted: cdc2k | |
| Attribute id has changed: C2 → u |
This table lists the changes for the SBML-specific species elements. BIOMD0000000005 encodes nine species, whereas BIOMD0000000006 only encodes six species. For each species in BIOMD0000000005 we list deletions, insertions or modification.
Biology
Based on two encoded models only, how could we know that the models describe the same biological pathway? Both models carry semantic annotations describing the cyclins and kinases involved. Using the original model BIOMD0000000005 as a query, we can screen BioModels for similar models by using the semanticSBML tool. We obtain a ranked list of result models, where the ranks depend on similarity scores computed by assessing the percentage of shared biological annotations. If we run this query on the manually curated models of BioModels Database, the query model itself appears on top of the list, whereas model BIOMD0000000006 appears only at rank 28. The models ranking above describe cell cycle and MAP-kinase pathways, containing many similar annotations. As BIOMD0000000006 is a simplification with few annotations on a more abstract level, there is only a moderate similarity in terms of biological annotations. In summary, the biological similarity of our two models can be detected, but there are other models that contain the same annotations and obtain similar, or even higher, similarity scores (in particular, Goldbeter’s minimal models of the mitotic oscillator [42]).
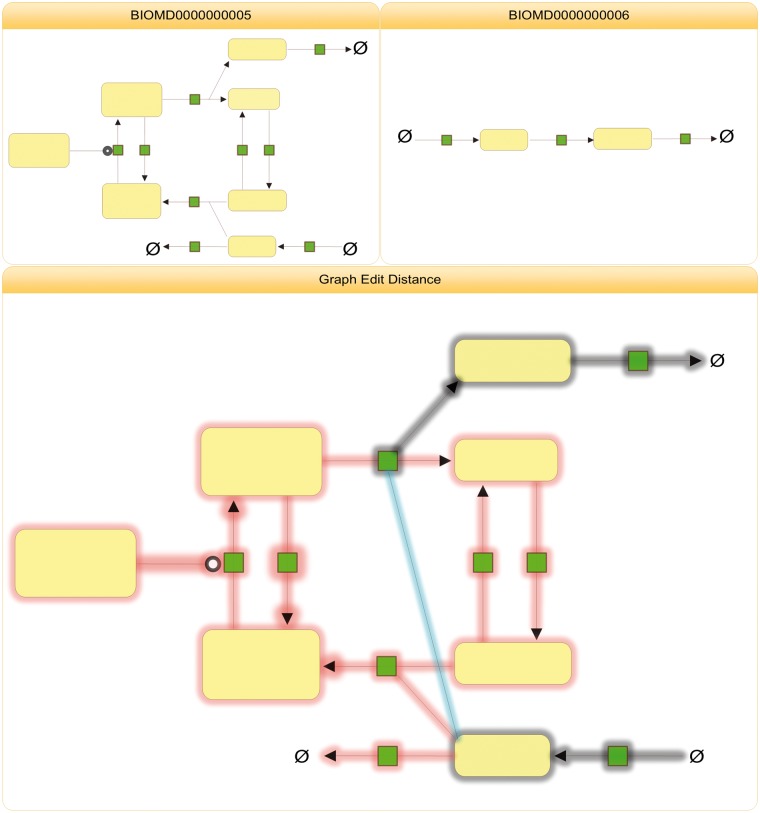
Network
A simple dissimilarity measure for networks is the graph edit distance [43]. The basic idea is to align two graphs with respect to the lowest edit costs, meaning to insert and delete as few nodes and edges as possible to transform one graph into the other:
where P(g1, g2) is the set of edit paths transforming g1 into g2 and c(e) > 0 is the cost of each graph edit operation e. In our example we disregard label changes. Figure 5 shows the necessary edits to transform the network of BIOMD0000000005 into the network of BIOMD0000000006.
Figure 5.
Graph edit distance. To transform the network of BIOMD0000000005 into the network of BIOMD0000000006, 28 edit operations had to be performed; 16 edges and 11 nodes are deleted, 1 edge is inserted. Deletions are shown in red, insertions in blue and unchanged elements in grey. The node ∅, representing the empty set, is displayed multiple times for clarity.
Practical applications of similarity measures
To profit from the wealth of published models, modellers need software that helps them explore, access, compare, simulate and combine models with minimal effort. We distinguish two possible lines of action: On the one hand, researchers may either analyse small, defined sets of models, and classify or align them; as an example, Figure 3 shows how a dynamical cell cycle model could be compared with a simplified variant of the model from the same publication based on model files available at BioModels. On the other hand, researchers may search for models in databases such as BioModels, possibly starting from some query model of interest. Methods for model comparison are equally important in both cases. We now describe basic use cases of model comparison as well as existing tools and methods devoted to these tasks.
Model search and clustering
Number and sizes of available models are increasing beyond what even the most well-read scholar can review or analyse, as exemplified in the growth of BioModels [44]. Probably the most common use of similarity measures is for model search, in which researchers query repositories to obtain models related to a given keyword, to a query model or to a data set.
In a keyword search, a user enters a set of terms to retrieve models that match these terms. In the simplest form, a model can be represented by a ‘bag of terms’, i.e. a list of relevant keywords or annotations, to which the user’s query terms can be matched. A similarity between query terms and model can then be defined using measures from information retrieval. For example, a similarity score can be calculated from the frequency with which a term occurs, or from semantic similarity measures between terms [45]. Instead of such ‘bag of term’ representations, models may also be represented in a structured form to incorporate network information, semantic annotations and other associated meta-data. Keyword search is state of the art in open model repositories. BioModels, for example, incorporates the aspects ‘model encoding’ and ‘biological meaning’ (see Table 1). A query by ‘model encoding’ matches exactly a set of terms associated with a model. A query by ‘biological meaning’ enables more sophisticated semantic searches. Search engines can be coupled with ranking algorithms to ensure that the most relevant search results appear in the top of the result list. The Physiome Model Repository, for example, uses a Lucene-based ranking algorithm that incorporates model encoding, biological meaning and other meta-data [12].
If the starting point of a search is a model, then the aim of a search is to find similar models. SemanticSBML allows users to provide their own model as an input query. The search engine then finds SBML models that resemble the input model with respect to semantic annotations [27]. The search operates on the openly available models from BioModels.
Similarity measures can also be used to calculate the similarity between models and query terms. Based on functions for model ranking [12, 27], similarity measures can also help to cluster models into similar sets. For example, a cluster may organize models into thematic sets such as ‘models describing metabolism’, ‘the cell cycle’ or ‘models showing calcium oscillations’. Thematic model sets may be characterized through semantic annotations [46] or through recurring structural patterns [47]. BioModels, for example, has implemented a web-based model browser that clusters models based on GO terms. In the future, models may also be clustered based on biological motifs in their networks [48]. Thematic sets can be searched and compared more easily, e.g. when constructing comprehensive models.
Network alignments
Network alignments provide a way to detect structural overlaps between pathways or networks. They are a basic tool in model merging and can be used to define similarity scores. For example, an alignment of kinetic pathway models with the metabolic network of yeast showed that large parts of the network are not yet covered, while central metabolism is heavily overrepresented in kinetic models [27]. Another study showed how Boolean models can be coupled if the models adhere to certain modelling standards [49]. The resulting integrated model of cell–cell interaction between hepatocytes and Kupffer cells provides deeper insights into how different cell types respond to apoptosis or proliferation, and how IL-6 (Interleukin 6) and TNF (tumor necrosis factor) influence the interactions between cells of different types. Further recent examples of successful model integration include the prediction of phenotypes from genotypes using a whole-cell model [50] and the global reconstruction of human metabolism [51].
These studies demonstrate how relative overlaps between networks can be quantified if the models contain a sufficient number of descriptive annotations to align the network components. However, if only few of the network components are annotated precisely, network alignment becomes a more difficult task: Only a few nodes can be matched based on the description of biological objects represented by network components, and the rest of the networks remain incomparable. One approach to address this problem is through semantic propagation [52], a method that distributes semantic information in the network structure, either to infer missing annotations or to fully align the networks. The algorithm effectively gathers, in each model component, semantic information about the component's neighbours, its second neighbours, and so on. It then compares the models’ components based on this neighbour information. In this context, neighbour elements are defined in an abstract sense. For example, reactions can be compared by annotations of their reactants, and cell compartments can be compared by the compounds they contain. Tests with blinded annotations have shown that model alignment and comparison can be strongly improved by using such propagated semantic information.
Model version control
New insights about a biological system may call for an adjustment of network structure, mathematical formulae or parameters of a model, resulting in new model versions. Another frequent reason for updates is error corrections. The comparison of model versions can help to keep track of the model’s evolution in time, and it identifies points at which a model underwent major changes [53]. BiVeS is a software library that aligns the XML encodings of two model versions, identifies and interprets changes and measures the changes’ impact. BiVeS also considers characteristics of the model encoding format and differentiates between SBML and CellML. Single diffs are automatically annotated to terms of COMODI.
Discussion
Many efforts were made in the past years to improve the reusability of systems biology models and the reproducibility of associated results [54–56]. Model repositories collect curated models ready to be reused, provide semantic annotations and offer instructions on how to simulate the models appropriately. With standardized, annotated models being available, automatic model comparison has become a feasible task. The search for models is one important application of comparison.
Key challenges in model search include defining appropriate relevance scores for search results, defining measures for the quality of retrieved models and building frameworks for the access of a model’s history. Such frameworks will enable researchers to follow a model’s evolution. For example, modellers can learn how the knowledge about the cell cycle evolved. Similarity measures are also valuable for merging existing pathway models into larger cell models. The map of Human Metabolism developed in the ReconX project, for instance, relies strongly on previously published models [51]. When two models are merged, parts of their networks may have to be exchanged or replaced by suitable alternatives, requiring tools that can compare both, models and model parts, and find the most similar matches. Because these tools need to solve similar problems and share similar difficulties, we propose to study model similarity as a general task.
As a key challenge, we identified the appropriate choice of a similarity measure. This leads to the following questions: How can we define computational measures that reflect a user’s expectations about similarity? And how can these measures, with various criteria, be applied in complex tasks such as search or merging? To create a general framework for model similarity and model comparison in future research, a number of issues need to be addressed.
Implement similarity measures for all model aspects. When comparing models, current software focuses on two of the aspects defined in this article, namely biological meaning and model encoding. Other model aspects are not yet commonly used. However, their implementation in existing algorithms is feasible and will improve model comparison. Dynamic behaviour, obtained from model simulations, could reveal similarities between biological processes during execution. These similarities will not show up when pathway structure alone is considered. One can directly compare simulated time series (as showcased by the Cardiac Physiology Web Lab [37]) or build a system that compares their semantic annotations with quantitative or qualitative behaviour observed in simulations. A valuable resource of terms for dynamic behaviour is the TEDDY ontology. Likewise, improved similarity measures could be obtained by more extensively exploring graph matching and graph similarity algorithms [57, 58], as suggested in [59]. Comparison of network structure can facilitate the matching of dynamical models to experimentally determined interaction networks. Equally, mathematical expressions could be directly compared to yield deeper insights into similarities of the models’ behaviour. Furthermore, information that is attached to the model may become relevant, such as the purpose of an investigation or the modellers’ intentions. However, this information must first be formalized—a new and interesting challenge.
Combine similarity measures. Today’s software tools typically compare models by a single model aspect. As different similarity measures have proven useful for specific applications, we expect that the combination of aspects would enable an even more powerful comparison of models, as in the following example: A scientist searches for a MAP kinase cascade model that contains regulatory feedback loops and shows dynamic oscillations. In a first step, a search tool could search for models of the specific biological system (using semantic comparison). Then, it could filter the intermediate results for specific network topologies. Next, a second filter could be applied to select models with oscillatory behaviour. Given sufficient additional information in the model repository, the remaining models could be compared based on their dynamic behaviour (e.g. by evaluating associated simulation descriptions in SED-ML format, and by comparing TEDDY terms therein). Eventually, an overall similarity measure joining the different aspects, could be defined. Such a procedure requires suitable functions for combining the individual similarities in a single formula, and it should be possible for users to specify weights, i.e. a relative importance, for each of the aspects.
Support multiscale models. Future systems biology models may be much more complex than today’s biochemical network models, ranging from spatial and stochastic cell simulations and modular whole-cell models to multiscale models of tissues, human physiology or populations of organisms. With different biological scopes and mathematical forms, new model aspects will become relevant (e.g. the way in which modules in a whole-cell model communicate with each other), and new similarity measures will be required. Some other aspects (e.g. the lists of molecular species described, or the occurrence of dynamic oscillations), known from our current network models, will still hold. A particular challenge will be posed by models that are internally and hierarchically structured, e.g. body models composed of organ models, which are composed of cell models, and so on. In these cases, similarities may first be defined on the level of individual modules, and then be propagated up to the level of entire models. We expect that software support for model comparison across multiple levels will become a requirement for flexible, semi-automatic model construction
Improve software support. To allow for extended similarity measures and to integrate them easily into software applications, new tools need to be developed. Interoperability through standard formats and common software libraries must be ensured. Such tools could incorporate a large set of existing similarity measures and provide functions for projecting models onto their relevant aspects. In addition, the scientific community has developed shared resources for models and associated metadata, widely accessible through graph databases [45] or publicly accessible Semantic Web resources [60]. Tools to process models and determine similarity should be able to access and interoperate with these shared resources.
Provide tools to align models and data by similarity. We discussed how similarity measures can link models to other models or to user queries. It is equally interesting to investigate how the process of linking models and experimental data can be improved. We envision that models and other data can be compared by projecting them onto a common aspect. For example, models and a patient’s cancer genomics data set can be projected to proteins appearing in both, model and data set. Subsequently, semantic annotations of biological processes can be compared between the model entities and the data items. Afterwards, similarity measures for biological meaning can help identify whether observations in the patient match a particular state predicted by a model.
Develop intuitive user interfaces. When offering purpose-driven similarity measures, it is important to communicate the details of the scores to the users. Consequently, there is a need to develop clear and intuitive user interfaces that show both, the results of a similarity score and the details of calculation. For example, a system for model retrieval should return a ranked list of models, with a detailed description of filtering processes and relationships between models and query. Some software tools already visualize element alignments between models as network graphs (e.g. BudHat [53], SemanticSBML, STON [61]), or present ranking scores for retrieved models (e.g. MASYMOS [45], SemanticSBML). However, an explanation of the steps leading to the similarity scores will increase the trust of the users clarify which of the search results are most relevant.
Conclusions
Similarity between models is assessed by a variety of software applications. We provided a systematic classification and a review of model similarity measures. We further introduced and discussed aspects that help with determining the similarity between models: the model encoding, when comparing versions of a model; the mathematical description of a model, when investigating the systems’ dynamic behaviour; the biological elements appearing in a model, when searching for models of a specific biological system or phenomenon; the network structure, when investigating the reuse of models as submodels in large networks; the parameter values in a model, during functional curation; and simulation outcomes, when comparing behaviour and sensitivity of a model. We envision a general framework for model similarity, based on a systematic treatment of models and their aspects. Such a framework will enhance the automated processing of models and may have numerous applications in computational systems biology.
Key Points
Computational models in biology today can be large and complex; they evolve over time.
Reuse of models demands sophisticated tools for comparison, retrieval, combination and version control.
Similarity between models can be determined with respect to encoded biology, network structure, mathematical equations or dynamic behaviour.
Software tools perform model comparison for various purposes, but generic aspects of model similarity have not been discussed.
Such aspects, and generic concepts for model comparison, will become crucial for dealing with increasingly complex models in the future.
Acknowledgements
The authors thank Wolfgang Müller, Matthias Lange and Martin Scharm for valuable discussions on similarity notions of systems biology models.
Funding
This article was drafted during a meeting that was organized by D.W. and funded through the BMBF e:Bio program (grant no. FKZ0316194). R.H. is funded by the German Federal Ministry of Education and Research (BMBF; grant number FKZ 031 A540A [de.NBI]). The Junior Research Group SEMS, BMBF e:Bio program (grant no. FKZ0316194 to D.W.). T.K. is funded by the German Federal Ministry of Education and Research (BMBF) via the Greifswald Approach to Individualized Medicine (GANI_MED; grant 03IS2061A) and by the Unternehmen Region as part of the ZIK-FunGene (grant 03Z1CN22). German Research Foundation (grant no. Ll 1676/2-1 to W.L.).
Ron Henkel is a graduate student at the Heidelberg Institute for Theoretical Studies. His research topic is database and information systems with an application in systems biology.
Robert Hoehndorf is an assistant professor in computer science. His research focuses on the applications of ontologies in biology and biomedicine, with a particular emphasis on integrating and analysing heterogeneous, multimodal data.
Tim Kacprowski is a graduate student at the University of Greifswald. His research tackles different problems in cross-omics integrative bioinformatics. University Medicine Greifswald, Interfaculty Institute for Genetics and Functional Genomics, Department of Functional Genomics.
Christian Knüpfer is a postdoctoral researcher in the Artificial Intelligence Group, University Jena. His research is focused on knowledge representation and its application to different fields ranging from systems biology to medieval studies.
Wolfram Liebermeister is a physicist working in systems biology. In his research on complex biochemical networks, he highlights functional aspects such as variability, information, metabolic control and the economy of cellular resources.
Dagmar Waltemath is a junior research group leader at the University of Rostock. Her research focuses on the development of methods and tools for model management in computational biology.
References
- 1. Le Novère N. Quantitative and logic modelling of molecular and gene networks. Nat Rev Genet 2015;16(3):146–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wolkenhauer O. Why model? Front Physiol 2014;5:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosen R. Life Itself. Columbia University Press, New York, 1991. [Google Scholar]
- 4. Wang RS, Saadatpour A, Albert R.. Boolean modeling in systems biology: an overview of methodology and applications. Phys Biol 2012;9(5):055001.. [DOI] [PubMed] [Google Scholar]
- 5. Hucka M, Finney A, Sauro HM, et al. The Systems Biology Markup Language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 2003;19(4):524–31. [DOI] [PubMed] [Google Scholar]
- 6. Cuellar AA, Lloyd CM, Nielsen PF, et al. An overview of CellML 1.1, a biological model description language. Simulation 2003;79(12):740–7. [Google Scholar]
- 7. Hucka M, Bergmann FT, Keating SM, et al. A profile of today’s SBML-compatible software. In: 2011 IEEE Seventh International Conference on, e-Science Workshops (eScienceW) IEEE, Stockholm, Sweden, 2011. pp. 143–50.
- 8. Degtyarenko K, de Matos P, Ennis M, et al. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res 2008;36(Suppl 1):D344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sansone SA, Rocca-Serra P, Field D, et al. Toward interoperable bioscience data. Nat Genet 2012;44(2):121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waltemath D. Management of simulation studies in computational biology. In: Invited Presentations, Junior Research Groups and Research Highlights at GCB 2015 PeerJ preprints, 2015.
- 11. Yu T, Lloyd CM, Nickerson DP, et al. The physiome model repository 2. Bioinformatics 2011;27(5):743–4. [DOI] [PubMed] [Google Scholar]
- 12. Henkel R, Endler L, Peters A, et al. Ranked retrieval of computational biology models. BMC Bioinformatics 2010;11(1):423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lange M, Henkel R, Müller W, et al. Information retrieval in life sciences: a programmatic survey In: Approaches in Integrative Bioinformatics. Springer, Heidelberg, 2014, 73–109. [Google Scholar]
- 14. Krause F, Uhlendorf J, Lubitz T, et al. Annotation and merging of SBML models with semanticSBML. Bioinformatics 2010;26(3):421–2. [DOI] [PubMed] [Google Scholar]
- 15. Clark C, Kalita J.. A comparison of algorithms for the pairwise alignment of biological networks. Bioinformatics 2014;30(16):2351–9. [DOI] [PubMed] [Google Scholar]
- 16. Tapinos A, Mendes P.. A method for comparing multivariate time series with different dimensions. PloS One 2013;8(2):e54201.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Duda RO, Hart PE, Stork DG.. Pattern Classification, 2nd edn Wiley, New York, 2000. [Google Scholar]
- 18. Le Novère N, Hucka M, Mi H, et al. The systems biology graphical notation. Nat Biotechnol 2009;27(8):735–41. [DOI] [PubMed] [Google Scholar]
- 19. Courtot M, Juty N, Knüpfer C, et al. Controlled vocabularies and semantics in systems biology. Mol Syst Biol 2011;7:543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waltemath D, Adams R, Bergmann FT, et al. Reproducible computational biology experiments with SED- ML - the simulation experiment description markup language. BMC Syst Biol 2011;5(1):198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scharm M,, Wolkenhauer O, Waltemath D.. An algorithm to detect and communicate the differences in computational models describing biological systems. Bioinformatics 2016; 32(4):563–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Saffrey P, Orton R.. Version control of pathway models using XML patches. BMC Syst Biol 2009;3(1):34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miller A, Yu T, Britten R, et al. Revision history aware repositories of computational models of biological systems. BMC Bioinformatics 2011;12(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Apweiler R, Bairoch A, Wu CH, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2004; 32(D1):115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet, 25:25–29, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pesquita C, Faria D, Falcao AO, et al. Semantic similarity in biomedical ontologies. PLoS Comput Biol 2009;5(7):e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schulz M, Krause F, Novère NL, et al. Retrieval, alignment, and clustering of computational models based on semantic annotations. Mol Syst Biol 2011;7(1):512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pabinger S, Snajder R, Hardiman T, et al. MEMOSys 2.0: an update of the bioinformatics database for genome-scale models and genomic data. Database 2014;2014:bau004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wimalaratne SM, Grenon P, Hermjakob H, et al. Biomodels linked dataset. BMC Syst Biol 2014;8(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo X, Liu R, Shriver CD, et al. Assessing semantic similarity measures for the characterization of human regulatory pathways. Bioinformatics 2006;22(8):967–73. [DOI] [PubMed] [Google Scholar]
- 31. Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. CRC Press, Boca Raton, 2006. [Google Scholar]
- 32. Gay S, Soliman S, Fages F.. A graphical method for reducing and relating models in systems biology. Bioinformatics 2010;26(18):i575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Milo R, Shen-Orr S, Itzkovitz S, et al. Network motifs: simple building blocks of complex networks. Science 2002;298(5594):824–7. [DOI] [PubMed] [Google Scholar]
- 34. Alon U. Biological networks: the tinkerer as an engineer. Science 2003;301(5641):1866–7. [DOI] [PubMed] [Google Scholar]
- 35. Bergmann FT, Sauro HM.. Comparing simulation results of SBML capable simulators. Bioinformatics 2008;24(17):1963–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cooper J, Mirams GR, Niederer SA.. High-throughput functional curation of cellular electrophysiology models. Prog Biophys Mol Biol 2011;107(1):11–20. [DOI] [PubMed] [Google Scholar]
- 37. Cooper J, Scharm M, Mirams GR.. The cardiac electrophysiology web lab. Biophys J 2016;110(2):292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tyson JJ. Modeling the cell division cycle: cdc2 and cyclin interactions. Proc Natl Acad Sci 1991;88(16):7328–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoops S, Sahle S, Gauges R, et al. COPASI—a complex pathway simulator. Bioinformatics 2006;22(24):3067–74. [DOI] [PubMed] [Google Scholar]
- 40. Knüpfer C, Beckstein C, Dittrich P, et al. Structure, function, and behaviour of computational models in systems biology. BMC Syst Biol 2013;7(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scharm M, Waltemath D, Mendes P, et al. COMODI: an ontology to characterise differences in versions of computational models in biology. J Biomed Semantics 2016;7:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldbeter A. A minimal cascade model for the mitotic oscillator involving cyclin and cdc2 kinase. Proc Natl Acad Sci USA 1991;88(20):9107–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gao X, Xiao B, Tao D, Li X.. A survey of graph edit distance. Pattern Anal Appl 2010;13(1):113–29. [Google Scholar]
- 44. Chelliah V, Juty N, Ajmera I, et al. Biomodels: ten-year anniversary. Nucleic Acids Res 2015;43(D1):D542–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henkel R, Wolkenhauer O, Waltemath D.. Combining computational models, semantic annotations and simulation experiments in a graph database. Database 2015;2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alm R, Waltemath D, Wolfien M, et al. Annotation-based feature extraction from sets of SBML models. J Biomed Semantics 2015;6(1):20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Henkel R, Lambusch F, Wolkenhauer O, et al. Finding patterns in biochemical reaction networks. PeerJ 2016;4:e1479v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tyson JJ, Novák B.. Functional motifs in biochemical reaction networks. Annu Rev Phys Chem 2010;61:219.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schlatter R, Philippi N, Wangorsch G, et al. Integration of boolean models exemplified on hepatocyte signal transduction. Brief Bioinform 2012;13(3):365–76. [DOI] [PubMed] [Google Scholar]
- 50. Karr JR, Sanghvi JC, Macklin DN, et al. A whole-cell computational model predicts phenotype from genotype. Cell 2012;150(2):389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Thiele I, Swainston N, Fleming RMT, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol 2013;31(5):419–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schulz M, Klipp E, Liebermeister W.. Propagating semantic information in biochemical network models. BMC Bioinformatics 2012;13(1):18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Waltemath D, Henkel R, Hälke R, et al. Improving the reuse of computational models through version control. Bioinformatics 2013;29(6):742–8. [DOI] [PubMed] [Google Scholar]
- 54. Krause F, Schulz M, Swainston N, et al. Sustainable model building: the role of standards and biological semantics In: Methods in Enzymology, Volume 500 of Methods in Systems Biology. Academic Press, Cambridge, US, 2011, 371–95. [DOI] [PubMed] [Google Scholar]
- 55. Waltemath D, Henkel R, Winter F, et al. Reproducibility of model-based results in systems biology In: Systems Biology. Springer, Heidelberg, 2013, 301–320. [Google Scholar]
- 56. Waltemath D, Wolkenhauer O.. How modeling standards, software, and initiatives support reproducibility in systems biology and systems medicine. IEEE Trans Biomed Eng 2016. [DOI] [PubMed] [Google Scholar]
- 57. Berg J, Lässig M.. Local graph alignment and motif search in biological networks. Proc Natl Acad Sci USA 2004;101(41):14689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yan X, Han J Gspan: graph-based substructure pattern mining. In: Proceedings 2002 IEEE International Conference on Data Mining, 2002 (ICDM 2002) IEEE, 2002, pp. 721–4.
- 59. Rosenke C, Waltemath D. How can semantic annotations support the identification of network similarities? In: Proceedings of the 7th International Workshop on Semantic Web Applications and Tools for Life Sciences2014, Berlin. CEUR Workshop Proceedings, Aachen, vol. 2, p. 11. [Google Scholar]
- 60. Jupp S, Malone J, Bolleman J, et al. The EBI RDF platform: linked open data for the life sciences. Bioinformatics 2014;30(9):1338–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tourè V, Mazein A, Waltemath D, et al. STON: exploring biological pathways using the SBGN standard and graph databases. 2016, in press. [DOI] [PMC free article] [PubMed]
- 62. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]