SUMMARY
Mediation analysis is an important tool in the behavioral sciences for investigating the role of intermediate variables that lie in the path between a treatment and an outcome variable. The influence of the intermediate variable on the outcome is often explored using a linear structural equation model (LSEM), with model coefficients interpreted as possible effects. While there has been significant research on the topic, little work has been done when the intermediate variable (mediator) is a high-dimensional vector. In this work, we introduce a novel method for identifying potential mediators in this setting called the directions of mediation (DMs). DMs linearly combine potential mediators into a smaller number of orthogonal components, with components ranked based on the proportion of the LSEM likelihood each accounts for. This method is well suited for cases when many potential mediators are measured. Examples of high-dimensional potential mediators are brain images composed of hundreds of thousands of voxels, genetic variation measured at millions of single nucleotide polymorphisms (SNPs), or vectors of thousands of variables in large-scale epidemiological studies. We demonstrate the method using a functional magnetic resonance imaging study of thermal pain where we are interested in determining which brain locations mediate the relationship between the application of a thermal stimulus and self-reported pain.
Keywords: Directions of mediation; Principal components analysis; fMRI, Mediation analysis; Structural equation models; High-dimensional data
Introduction
Mediation and path analyses have been pervasive in the social and behavioral sciences (e.g., Baron and Kenny, 1986; MacKinnon, 2008; Preacher and Hayes, 2008), and have found widespread use in many applications, including psychology, behavioral science, economics, decision-making, epidemiology, and neuroscience. In the past couple of decades, the topic has also begun to receive a great deal of attention in the statistical literature, particularly in the area of causal inference (e.g., Holland, 1988; Robins and Greenland, 1992; Angrist and others, 1996; Ten Have and others, 2007; Albert, 2008; Jo, 2008; Sobel, 2008; VanderWeele and Vansteelandt, 2009; Imai and others, 2010; Lindquist, 2012; Pearl, 2014). When the effect of a treatment 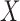 on an outcome
on an outcome 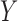 is at least partially directed through an intervening variable
is at least partially directed through an intervening variable 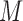 , then
, then 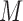 is said to be a mediator. The three-variable path diagram shown in Figure 1 illustrates this relationship. The influence of the intermediate variable on the outcome is frequently ascertained using linear structural equation models (LSEMs), with the model coefficients interpreted as causal effects; see below for discussion of the assumptions under which this interpretation is warranted. Typically, interest centers on parsing the effects of the treatment on the outcome into separable direct and indirect effects, representing the influence of
is said to be a mediator. The three-variable path diagram shown in Figure 1 illustrates this relationship. The influence of the intermediate variable on the outcome is frequently ascertained using linear structural equation models (LSEMs), with the model coefficients interpreted as causal effects; see below for discussion of the assumptions under which this interpretation is warranted. Typically, interest centers on parsing the effects of the treatment on the outcome into separable direct and indirect effects, representing the influence of 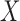 on
on 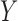 unmediated and mediated by
unmediated and mediated by 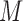 , respectively.
, respectively.
Fig. 1.
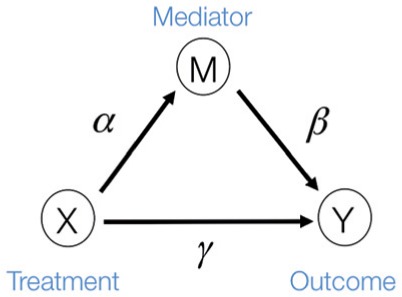
The three-variable path diagram representing the standard mediation framework. The variables corresponding to 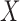 ,
, 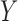 , and
, and  are all scalars, as are the path coefficients
are all scalars, as are the path coefficients 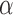 ,
, 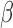 , and
, and 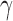 .
.
To date most research in mediation analysis has been devoted to the case of a single mediator, with some attention given to the case of multiple mediators (e.g., Preacher and Hayes, 2008; Albert and Nelson, 2011; VanderWeele and Vansteelandt, 2014; Wang and others, 2013; Imai and Yamamoto, 2013; Daniel and others, 2015). However, high-dimensional mediation has received scarce attention.Recent years have seen a tremendous increase of new applications measuring massive numbers of variables, including brain imaging, genetics, epidemiology, and public health studies. It has therefore become increasingly important to develop methods to deal with mediation in the high-dimensional setting, i.e., when the number of mediators is much larger than the number of observations. Such an extension is the focus of this work.
As a motivating example, consider functional magnetic resonance imaging (fMRI), an imaging modality where the signal of interest is the blood oxygenation level dependent (BOLD) response; a measure of the metabolic demands (i.e., oxygen consumption) of active neurons (Ogawa and others, 1990; Kwong and others, 1992; Lindquist, 2008). In fMRI experiments, a multivariate time series of 3D brain volumes are obtained for each subject, where each volume consists of hundreds of thousands of equally sized volume elements (voxels). A number of previous studies have used fMRI to investigate the relationship between painful heat and self-reported pain (Apkarian and others, 2005; Bushnell and others, 2013). Recently, studies have focused on trial-by-trial modeling of the relationship between the intensity of noxious heat and self-reported pain (Wager and others, 2013; Atlas and others, 2014). In Woo and others (2015), for example, a series of thermal stimuli were applied at various temperatures (ranging from 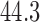 to
to 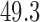
 C in
C in 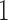
 increments) to the left forearm of each of
increments) to the left forearm of each of 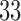 subjects. In response, subjects gave subjective pain ratings at a specific time point following the offset of the stimulus. During the course of the experiment, brain activity in response to the thermal stimuli was measured across the entire brain using fMRI. One of the goals of the study was to search for brain regions whose activity level act as potential mediators of the relationship between temperature and pain rating.
subjects. In response, subjects gave subjective pain ratings at a specific time point following the offset of the stimulus. During the course of the experiment, brain activity in response to the thermal stimuli was measured across the entire brain using fMRI. One of the goals of the study was to search for brain regions whose activity level act as potential mediators of the relationship between temperature and pain rating.
In this context, we are interested in whether the effect of temperature, 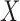 , on reported pain,
, on reported pain, 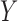 , is mediated by the brain response,
, is mediated by the brain response, 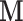 . Here both
. Here both 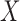 and
and 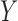 are scalars, while
are scalars, while 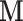 is the estimated brain activity measured over a large number of different voxels/regions. We assume that the values of
is the estimated brain activity measured over a large number of different voxels/regions. We assume that the values of 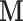 are either parameters or contrasts (linear combinations of parameters) obtained by fitting the general linear model (GLM), where for each subject, the relationship between the stimuli and the BOLD response is analyzed at the voxel level (Lindquist and others, 2012). Standard mediation techniques are applicable to univariate mediators. An early approach to mediation in neuroimaging Caffo and others, 2008 took the route of re-expressing the multivariate images into targeted, simpler, composite summaries on which mediation analysis was performed. In contrast, the identification of univariate mediators on a voxel-wise basis has come to be known as mediation effect parametric mapping (Wager and others, 2008; Wager and others, 2009b; Wager and others, 2009a). This approach, however, ignores the relationship between voxels, and identifies a series of univariate mediators rather than an optimized, multivariate linear combination. A multivariate extension should focus on identifying latent brain components maximally effective as mediators, i.e., those that are simultaneously most predictive of the outcome and predicted by the treatment.
are either parameters or contrasts (linear combinations of parameters) obtained by fitting the general linear model (GLM), where for each subject, the relationship between the stimuli and the BOLD response is analyzed at the voxel level (Lindquist and others, 2012). Standard mediation techniques are applicable to univariate mediators. An early approach to mediation in neuroimaging Caffo and others, 2008 took the route of re-expressing the multivariate images into targeted, simpler, composite summaries on which mediation analysis was performed. In contrast, the identification of univariate mediators on a voxel-wise basis has come to be known as mediation effect parametric mapping (Wager and others, 2008; Wager and others, 2009b; Wager and others, 2009a). This approach, however, ignores the relationship between voxels, and identifies a series of univariate mediators rather than an optimized, multivariate linear combination. A multivariate extension should focus on identifying latent brain components maximally effective as mediators, i.e., those that are simultaneously most predictive of the outcome and predicted by the treatment.
Thus, in this work we consider the same simple three-variable path diagram depicted in Figure 1, with the novel feature that we have a very high-dimensional vector of potential mediators 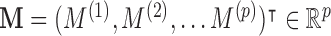 . While an LSEM can be used to estimate mediation effects (defined precisely below), in this setting there are too many mediators to allow reasonable interpretation (unless the model coefficients are highly structured) and there are many more mediators than subjects, precluding estimation using standard procedures. To overcome these problems, a new model, called the directions of mediation (DMs) is developed. DM’s linearly combine activity in different voxels into a smaller number of orthogonal components, with components ranked based upon the proportion of the LSEM likelihood (assuming normally distributed errors) each accounts for. Ideally, the components form a small number of uncorrelated mediators that represent interpretable networks of voxels. The approach shares some similarities with partial least squares (Wold, 1985), which is a dimension reduction approach based on the correlation between a response variable (e.g.,
. While an LSEM can be used to estimate mediation effects (defined precisely below), in this setting there are too many mediators to allow reasonable interpretation (unless the model coefficients are highly structured) and there are many more mediators than subjects, precluding estimation using standard procedures. To overcome these problems, a new model, called the directions of mediation (DMs) is developed. DM’s linearly combine activity in different voxels into a smaller number of orthogonal components, with components ranked based upon the proportion of the LSEM likelihood (assuming normally distributed errors) each accounts for. Ideally, the components form a small number of uncorrelated mediators that represent interpretable networks of voxels. The approach shares some similarities with partial least squares (Wold, 1985), which is a dimension reduction approach based on the correlation between a response variable (e.g., 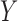 ) and a set of explanatory variables (e.g.,
) and a set of explanatory variables (e.g., 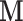 ). In contrast, for DMs the dimension reduction is based on the complete
). In contrast, for DMs the dimension reduction is based on the complete 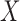 –
–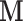 –
–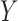 relationship.
relationship.
This article is organized as follows. In Section 2, we define direct and indirect effects for the multivariate mediator setting. In Section 3, we introduce the DMs, and provide an estimation algorithm for estimating the DMs and their associated path coefficients when the mediator is high dimensional. In Section 4, we discuss a method for performing inference on the DMs. Finally, in Sections 5 and 6 the efficacy of the approach is illustrated through simulations and an application to the fMRI study of thermal pain.
2. A mutivariate causal mediation model
Let 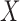 denote an exposure/treatment for a given subject (e.g., thermal pain), and
denote an exposure/treatment for a given subject (e.g., thermal pain), and 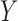 an outcome (e.g., reported pain). Suppose there are multiple mediators
an outcome (e.g., reported pain). Suppose there are multiple mediators 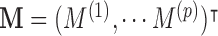 in the path between treatment and outcome; in the fMRI context, the mediators are
in the path between treatment and outcome; in the fMRI context, the mediators are 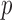 dependent activations over the
dependent activations over the 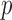 voxels. Here we assume for simplicity that each subject is scanned under one condition.
voxels. Here we assume for simplicity that each subject is scanned under one condition.
Using potential outcomes notation (Rubin, 1974), let 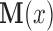 denote the value of the mediators if treatment
denote the value of the mediators if treatment 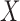 is set to
is set to 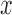 . Similarly, let
. Similarly, let 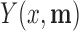 denote the outcome if
denote the outcome if 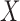 is set to
is set to 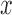 and
and 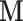 is set to
is set to 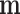 . The controlled unit direct effect of
. The controlled unit direct effect of 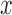 vs.
vs. 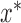 is defined as
is defined as 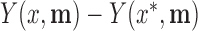 , the natural unit direct effect as
, the natural unit direct effect as 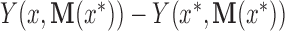 , and the natural unit indirect effect as
, and the natural unit indirect effect as 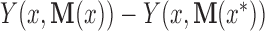 . Note that for these nested counterfactuals to be well defined it must be hypothetically possible to intervene on the mediator without affecting the treatment. We discuss this assumption in the context of fMRI in Section 7.
. Note that for these nested counterfactuals to be well defined it must be hypothetically possible to intervene on the mediator without affecting the treatment. We discuss this assumption in the context of fMRI in Section 7.
The total unit effect is the sum of the natural unit direct and unit indirect effects, i.e.,
| (2.1) |
The direct effect could also be defined as 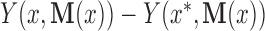 . In general, this would lead to a different decomposition of the total effect; however, as we consider linear models below, this is not of further concern. Suppose the following four assumptions hold for the set of mediators:
. In general, this would lead to a different decomposition of the total effect; however, as we consider linear models below, this is not of further concern. Suppose the following four assumptions hold for the set of mediators:
| (2.2) |
In words, these assumptions imply there is no confounding for the relationship between: (i) treatment 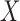 and outcome
and outcome 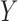 ; (ii) mediators
; (ii) mediators 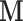 and outcome
and outcome 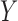 ; (iii) treatment
; (iii) treatment 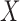 and mediators
and mediators 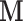 ; and (iv) no confounding for the relationship between mediator and outcome that is affected by the treatment. Together, they are often referred to as sequential ignorability assumptions. See Robins and Richardson (2010) and Pearl (2014) for detailed discussion of these assumptions, and for a critical evaluation of these assumptions in the high-dimensional setting, see Huang and Pan (2015). VanderWeele and Vansteelandt (2014) showed that under (2.2) the average direct and indirect effects are identified from the regression function for the observed data.
; and (iv) no confounding for the relationship between mediator and outcome that is affected by the treatment. Together, they are often referred to as sequential ignorability assumptions. See Robins and Richardson (2010) and Pearl (2014) for detailed discussion of these assumptions, and for a critical evaluation of these assumptions in the high-dimensional setting, see Huang and Pan (2015). VanderWeele and Vansteelandt (2014) showed that under (2.2) the average direct and indirect effects are identified from the regression function for the observed data.
Suppose then (2.2) and the following model for the observed data hold:
| (2.3) |
Note that this model encodes the assumptions of linear relations among treatment, mediators, and outcome and, importantly, the absence of any treatment–mediator interaction in the outcome regression. When the treatment interacts with one or more of the mediators, the LSEM framework considered in this article is not appropriate for mediation analysis (Ogburn, 2012).
The average controlled direct effect, average natural direct effect, and average indirect effect are expressed as follows:
| (2.4) |
| (2.5) |
| (2.6) |
Note the average controlled direct effect and natural direct effect are equivalent whenever there is no treatment-mediator interaction, as is assumed throughout.
When the counterfactuals are well defined and the assumptions in (2.2) hold, the right-hand sides of (2.5) and (2.6) identify causal mediation effects. When one or more of the assumptions in (2.2) fail to hold, or if the counterfactuals are not well defined, the right-hand sides of (2.5) and (2.6) may still be used in exploratory analysis to identify potential mediators. For example, they could identify linear combinations of voxels that correspond to specific brain functions, suggesting mediation through correlates of those brain functions. Throughout, for simplicity, we use “direct effect” and “indirect effect” to refer to the right-hand sides of (2.5) and (2.6), respectively; we are agnostic as to whether these expressions can be interpreted causally or should be taken as exploratory. Similarly, we use “mediator” agnostically to refer to variables that temporally follow treatment and precede outcome and potentially may lie on a causal pathway between them.
We also note that the model only considers the case where the entire vector 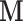 is subject to a single level of intervention (
is subject to a single level of intervention (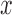 ). More generally, it would be possible to define potential mediators when some elements of
). More generally, it would be possible to define potential mediators when some elements of 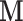 are subject to
are subject to 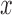 and other elements of
and other elements of 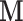 are subject to
are subject to 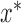 , which could support various decompositions of direct/indirect effects due to pathways acting through different
, which could support various decompositions of direct/indirect effects due to pathways acting through different 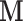 . In the fMRI setting described herein we anticipate that the same elements of
. In the fMRI setting described herein we anticipate that the same elements of 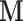 (i.e., brain regions) are subject to both
(i.e., brain regions) are subject to both 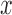 and
and 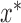 . For example, warm and hot thermal stimuli give rise to activation in the same brain regions, but with different intensities.
. For example, warm and hot thermal stimuli give rise to activation in the same brain regions, but with different intensities.
Fitting the system (2.3) is straightforward if the number of mediators is small. However, the estimates become unstable as 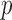 increases, and in fMRI the number of mediators will greatly exceed the sample size. Therefore we seek an orthogonal transformation of the mediators. This both simplifies and stabilizes the parameter estimates in the model, allowing us to estimate the direct and indirect effects using a series of LSEMs, one for each transformed mediator; see Figure 2 for an illustration. The novelty of our approach lies in choosing the transformation so that the transformed mediators are ranked by the proportion of the likelihood of the full LSEM that they account for.This has the benefit of potentially: (i) providing more interpretable mediators (i.e., linear combinations of voxels rather then individual voxels) and (ii) potentially reducing the number of mediators needed to estimate the indirect effect.Finally, it is easier to assess potential treatment by mediator interactions using a lower dimensional summary of the mediator variables than it is doing this directly in the high-dimensional setting.
increases, and in fMRI the number of mediators will greatly exceed the sample size. Therefore we seek an orthogonal transformation of the mediators. This both simplifies and stabilizes the parameter estimates in the model, allowing us to estimate the direct and indirect effects using a series of LSEMs, one for each transformed mediator; see Figure 2 for an illustration. The novelty of our approach lies in choosing the transformation so that the transformed mediators are ranked by the proportion of the likelihood of the full LSEM that they account for.This has the benefit of potentially: (i) providing more interpretable mediators (i.e., linear combinations of voxels rather then individual voxels) and (ii) potentially reducing the number of mediators needed to estimate the indirect effect.Finally, it is easier to assess potential treatment by mediator interactions using a lower dimensional summary of the mediator variables than it is doing this directly in the high-dimensional setting.
Fig. 2.
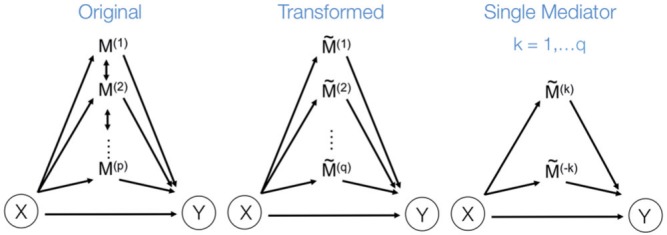
(Left) The three-variable path diagram used to represent multivariate mediation. Here the 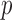 mediators are assumed to be correlated. (Center) A similar path diagram after an orthogonal transformation of the mediators, making the
mediators are assumed to be correlated. (Center) A similar path diagram after an orthogonal transformation of the mediators, making the 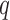 transformed mediators uncorrelated with one another. (Right) Because the mediators are uncorrelated, a series of LSEMs, one for each transformed mediator
transformed mediators uncorrelated with one another. (Right) Because the mediators are uncorrelated, a series of LSEMs, one for each transformed mediator 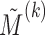 , can be used to estimate direct and indirect effects.
, can be used to estimate direct and indirect effects.
3. Directions of mediation
In this section, we introduce a transformation of the space of mediators, determined by finding linear combinations of the original mediators that (i) are orthogonal and (ii) are chosen to maximize the likelihood of the underlying three-variable SEM. We first formulate the model before introducing an estimation algorithm, and an extension to the case when multiple trials are observed for each subject. We conclude with a discussion regarding estimation for the case when 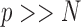 , where
, where 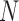 represents the total number of observations.
represents the total number of observations.
3.1. Model formulation
Let 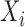 and
and 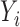 denote univariate variables, and
denote univariate variables, and  , for
, for 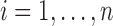 , where
, where 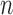 denotes the number of subjects. We denote the full dataset
denotes the number of subjects. We denote the full dataset 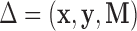 , where
, where 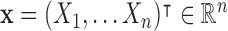 ,
, 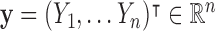 , and
, and 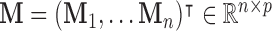 . Now let
. Now let 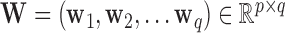 be a linear transformation matrix, where
be a linear transformation matrix, where 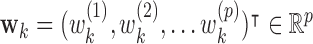 , for
, for 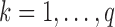 ; and let
; and let 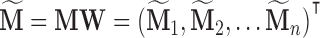 where
where 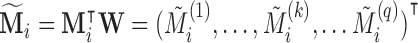 with
with 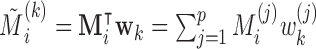 . We assume the relationship between the variables is given by the following LSEM:
. We assume the relationship between the variables is given by the following LSEM:
| (3.1) |
where 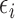 and
and 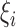 are i.i.d. normal with mean
are i.i.d. normal with mean 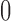 and variances
and variances 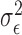 and
and 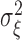 . The parameters of the LSEM can be estimated using linear regression. However, under the additional condition that the new transformed variables
. The parameters of the LSEM can be estimated using linear regression. However, under the additional condition that the new transformed variables 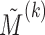 are orthogonal, we can estimate the parameters separately for each
are orthogonal, we can estimate the parameters separately for each 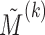 . Thus, for each
. Thus, for each 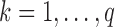 we can fit the following LSEM:
we can fit the following LSEM:
| (3.2) |
where 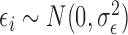 and
and 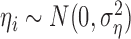 , for
, for 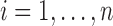 . Using this notation, we can express the average direct and indirect effects defined in (2.5) and (2.6) as
. Using this notation, we can express the average direct and indirect effects defined in (2.5) and (2.6) as 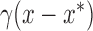 and
and 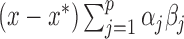 , respectively. Hence, these effects can be estimated either in the original or orthogonalized space. However, for large values of
, respectively. Hence, these effects can be estimated either in the original or orthogonalized space. However, for large values of 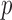 the model expressed in (3.2) is computationally preferable to the one expressed in (2.3).
the model expressed in (3.2) is computationally preferable to the one expressed in (2.3).
Let 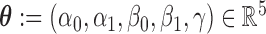 be the parameter vector for the LSEM in (3.2) for
be the parameter vector for the LSEM in (3.2) for 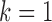 . We seek to simultaneously estimate
. We seek to simultaneously estimate 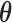 and find the first direction of mediation (DM)
and find the first direction of mediation (DM) 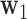 , defined as the coefficient vector of the linear combination of the elements of
, defined as the coefficient vector of the linear combination of the elements of 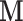 that maximizes the likelihood of the underlying LSEM. In our motivating example,
that maximizes the likelihood of the underlying LSEM. In our motivating example, 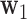 is a linear combination of voxel activations. Thus, similar to principal components analysis (PCA) (Andersen andothers, 1999) or independent components analysis (McKeown and others, 1997) when applied to fMRI data, the weights can be mapped back onto the brain, with the resulting maps interpreted as coherent networks that together act as mediators of the relationship between treatment and outcome. Also like PCA, subsequent directions can be found that maximize the likelihood of the model, conditional on these being orthogonal to the previous directions.
is a linear combination of voxel activations. Thus, similar to principal components analysis (PCA) (Andersen andothers, 1999) or independent components analysis (McKeown and others, 1997) when applied to fMRI data, the weights can be mapped back onto the brain, with the resulting maps interpreted as coherent networks that together act as mediators of the relationship between treatment and outcome. Also like PCA, subsequent directions can be found that maximize the likelihood of the model, conditional on these being orthogonal to the previous directions.
To formalize, let 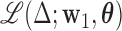 be the joint likelihood of the SEM stated in (3.2). The DMs are defined as follows:
be the joint likelihood of the SEM stated in (3.2). The DMs are defined as follows:
Step 1: The first DM is the vector 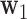 , with norm 1, that maximizes the conditional joint likelihood
, with norm 1, that maximizes the conditional joint likelihood 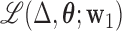 , i.e.,
, i.e.,
subject to
Step 2: The second DM is the vector 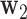 , with norm 1 and orthogonal to
, with norm 1 and orthogonal to 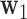 , that maximizes the conditional joint likelihood
, that maximizes the conditional joint likelihood 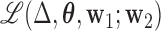 , i.e.,
, i.e.,
subject to
Step k: The 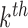 DM is the vector
DM is the vector 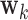 , with norm 1 and orthogonal to
, with norm 1 and orthogonal to 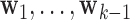 , that maximizes the conditional joint likelihood
, that maximizes the conditional joint likelihood 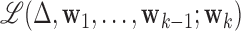 , i.e.,
, i.e.,
subject to
Remark 1 According to the model formulation the signs of the DMs are unidentifiable. A change in sign of 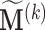 can be offset by a change in sign of both
can be offset by a change in sign of both 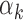 and
and 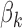 .
.
Remark 2 As the transformed mediators are ranked based upon the proportion of the likelihood of the full LSEM explained, one could potentially limit the number of DMs computed to achieve dimension reduction. However, this could cause the estimate of the indirect effect to be biased.
3.2. Estimation
Here we describe how to estimate the parameters associated with the first DM. Assuming joint normality, the joint 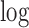 -likelihood function for
-likelihood function for 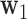 and
and 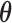 can be expressed as:
can be expressed as:
| (3.3) |
where 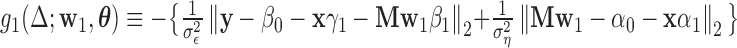 .
.
The goal is to find both the parameters of the LSEM and the first DM that jointly maximize 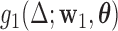 , under the constraint that the
, under the constraint that the 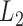 norm of
norm of 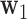 equals 1. Consider the Lagrangian
equals 1. Consider the Lagrangian
The dual problem can be expressed:
where 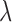 is the Lagrange multiplier. To solve this problem, we propose a method where
is the Lagrange multiplier. To solve this problem, we propose a method where 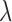 is profiled out by one set of parameters of interest. We establish, under the assumption that the first partial derivatives of the objective function and the constraint function exist, the closed form solution for the path coefficients, the first DM, and
is profiled out by one set of parameters of interest. We establish, under the assumption that the first partial derivatives of the objective function and the constraint function exist, the closed form solution for the path coefficients, the first DM, and 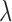 as follows:
as follows:
| (3.4) |
| (3.5) |
| (3.6) |
where 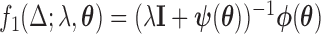 ;
; 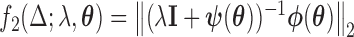 ,
, 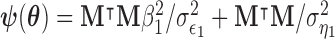 , and
, and 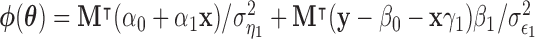 . Using these results we outline an iterative procedure for jointly estimating the first DM and path parameters as described in Algorithm 1. Further, in the Appendices A and B of supplementary material available at Biostatistics online we show that the estimated parameters are consistent and asymptotically normal (see Theorems 1 and 2).
. Using these results we outline an iterative procedure for jointly estimating the first DM and path parameters as described in Algorithm 1. Further, in the Appendices A and B of supplementary material available at Biostatistics online we show that the estimated parameters are consistent and asymptotically normal (see Theorems 1 and 2).
Algorithm 1
First DM
Step 0: Initiate
, denoted
. Step 1: For each
, set:
(3.7)
(3.8)
(3.9) Step 2: Repeat Step 1 until convergence; each time set
.
3.3. Higher order DMs
To estimate higher order DMs, we investigated two alternative approaches. The first uses additional penalty parameters (one for each additional constraint), and the second subtraction and Gram–Schmidt projections. While the former approach is likely to achieve global maxima, the latter is computationally more efficient, and provides a good approximation of higher order DMs; thus we focus on this approach here. Using this approach, estimates of the 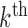 direction of mediation,
direction of mediation, 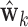 , and the associated path coefficients,
, and the associated path coefficients, 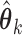 , are obtained by computing:
, are obtained by computing:
subject to
where
| (3.10) |
The performance of the projection approach is evaluated in Section 5.
3.4. Multi-trial model
In our setting, multiple observations (i.e., thermal stimulations and subsequent pain reports) are acquired for each subject. Here we describe a simple extension of the model described in (3.1). Throughout we assume that each subject has 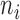 observations, so that
observations, so that 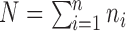 . Let
. Let 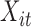 ,
, 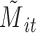 , and
, and 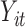 denote subject
denote subject 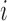 ’s treatment, orthogonalized meditator, and outcome, respectively, on the
’s treatment, orthogonalized meditator, and outcome, respectively, on the 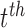 trial. While we ultimately want to express the relationship between these variables using a model that incorporates subject-specific random effects, here we use a simpler formulation that allows us to retain the estimation procedure outlined in the previous sections. We assume the relationship between the variables is given by the following LSEM:
trial. While we ultimately want to express the relationship between these variables using a model that incorporates subject-specific random effects, here we use a simpler formulation that allows us to retain the estimation procedure outlined in the previous sections. We assume the relationship between the variables is given by the following LSEM:
| (3.11) |
where 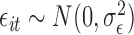 and
and 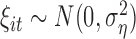 , for
, for 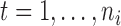 and
and  . Note that if each subject has a single observation, then
. Note that if each subject has a single observation, then 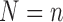 , and this model is equivalent to (3.1). As (3.11) generalizes (3.1), we use it in the continuation.
, and this model is equivalent to (3.1). As (3.11) generalizes (3.1), we use it in the continuation.
3.5. High-dimensional DMs
The estimation procedure described in Section 3.2 works well in the low-dimensional setting, but becomes cumbersome as 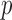 increases. Therefore it is critical to augment it with a matrix decomposition technique. Here we use a generalized version of population value decomposition (PVD) (Caffo and others, 2010; Crainiceanu and others, 2011), which in contrast to singular value decomposition provides population-level information about
increases. Therefore it is critical to augment it with a matrix decomposition technique. Here we use a generalized version of population value decomposition (PVD) (Caffo and others, 2010; Crainiceanu and others, 2011), which in contrast to singular value decomposition provides population-level information about 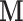 . Throughout we assume that the data for each subject
. Throughout we assume that the data for each subject 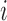 is stored in an
is stored in an 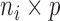 matrix,
matrix, 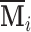 , whose
, whose 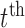 row contains voxel-wise activity for the measurements of the
row contains voxel-wise activity for the measurements of the 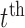 trial for the
trial for the 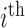 subject. All
subject. All 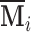 matrices are stacked vertically to form the
matrices are stacked vertically to form the 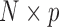 matrix
matrix 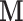 , where
, where 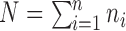 .
.
The generalized PVD (GPVD) of 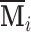 (formally derived in Appendix C of the supplementary material available at Biostatistics online) is given by
(formally derived in Appendix C of the supplementary material available at Biostatistics online) is given by
| (3.12) |
where  is an
is an 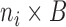 matrix,
matrix, 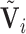 is an
is an 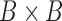 matrix of subject-specific coefficients,
matrix of subject-specific coefficients, 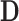 is a
is a 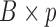 population-specific matrix, and
population-specific matrix, and 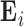 is an
is an 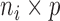 matrix of residuals. Here the value of
matrix of residuals. Here the value of 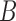 is chosen based upon a criteria such as total variance explained, in a similar manner as in PCA. See supplementary material available at Biostatistics online for details on how to compute these components.
is chosen based upon a criteria such as total variance explained, in a similar manner as in PCA. See supplementary material available at Biostatistics online for details on how to compute these components.
To estimate the DMs, perform GPVD on 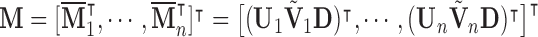 . Next, stack all
. Next, stack all 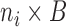 matrices
matrices 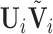 vertically to form an
vertically to form an 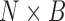 matrix
matrix
| (3.13) |
Let 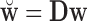 , where
, where 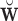 is
is 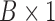 . Now
. Now 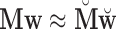 and dim(
and dim(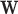 )
) 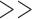 dim(
dim(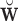 ). Thus, we can use
). Thus, we can use 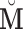 to estimate
to estimate 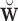 , which is significantly less computationally intensive than using
, which is significantly less computationally intensive than using 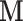 to estimate
to estimate 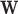 . Since
. Since 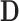 can be obtained via GPVD, we can retrieve the original estimator of
can be obtained via GPVD, we can retrieve the original estimator of 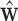 via the generalized inverse, i.e.,
via the generalized inverse, i.e., 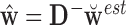 , where
, where 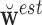 is the estimated
is the estimated 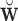 and
and  indicates the generalized inverse.
indicates the generalized inverse.
4. Inference
Here we discuss an approach for using a bootstrap procedure to perform inference. Consider 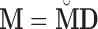 , where
, where 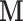 is
is 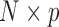 ,
, 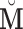 is
is 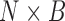 ,
, 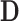 is
is 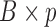 , and
, and 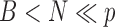 . The bootstrap procedure can be outlined as follows:
. The bootstrap procedure can be outlined as follows:
(1) Create a bootstrap sample
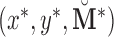 consisting of
consisting of 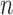 subjects, by randomly resampling subjects with replacement from the data set
subjects, by randomly resampling subjects with replacement from the data set 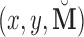 ;
;(2) Estimate the low-dimensional bootstrap DM
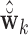 of length
of length 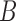 for
for 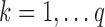 .
.(3) Compute
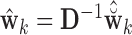 , where
, where 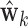 is the high-dimensional bootstrap DM of length
is the high-dimensional bootstrap DM of length 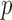 ;
;(4) Repeat steps 1–3
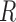 times. Stack values of
times. Stack values of 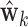 vertically to form the
vertically to form the 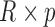 matrix
matrix 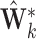 .
.
We seek to use the distribution to test whether specific elements of 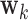 differ from
differ from 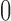 . Note the columns of
. Note the columns of 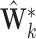 are the bootstrap values of the
are the bootstrap values of the 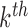 DM corresponding to voxel
DM corresponding to voxel 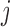 , from which we can form a distribution. There will be two types of distributions: unimodal and bimodal. The occurrence of bimodal distributions is due to the fact that the signs of the DM are not identifiable. To circumvent this we instead focus on obtaining a bootstrap distribution for
, from which we can form a distribution. There will be two types of distributions: unimodal and bimodal. The occurrence of bimodal distributions is due to the fact that the signs of the DM are not identifiable. To circumvent this we instead focus on obtaining a bootstrap distribution for 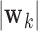 .
.
We begin by performing the bootstrap procedure as outlined above. Next, we sort the bootstrap distributions for all voxels by their median, and choose voxels whose median values lies in either the second or third quartile. We randomly sample among these voxels (in our application we sample 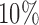 of the voxels whose medians lie in these quartiles), and combine their distribution to create a pseudo-null distribution. Finally, we fit a half-normal distribution to the pseudo-null and use this distribution to estimate a p-value for each element of
of the voxels whose medians lie in these quartiles), and combine their distribution to create a pseudo-null distribution. Finally, we fit a half-normal distribution to the pseudo-null and use this distribution to estimate a p-value for each element of 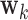 .
.
5. Simulation study
We performed a series of simulation studies to investigate the efficacy of our approach. A description of these simulations and the results are in the Appendix D of supplementary material available at Biostatistics online.
6. An fmri study of thermal pain
The data comes from the fMRI study of thermal pain described in Section 1. Here 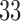 healthy, right-handed participants completed the study (age
healthy, right-handed participants completed the study (age 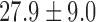 years,
years, 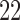 females). All participants provided informed consent, and the Columbia University Institutional Review Board approved the study. The data consists of 7 runs, consisting of between
females). All participants provided informed consent, and the Columbia University Institutional Review Board approved the study. The data consists of 7 runs, consisting of between  separate trials, in which participants experienced and rated the heat stimuli. During each trial, thermal stimulations were delivered to the volar surface of the left inner forearm. Each stimulus lasted
separate trials, in which participants experienced and rated the heat stimuli. During each trial, thermal stimulations were delivered to the volar surface of the left inner forearm. Each stimulus lasted 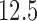 s, with
s, with 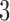 s ramp-up and
s ramp-up and 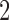 s ramp-down periods and
s ramp-down periods and 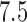 s at the target temperature. Six levels of temperature, ranging from
s at the target temperature. Six levels of temperature, ranging from 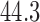 to
to 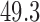
 C in increments of
C in increments of 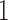
 C, were administered to each participant. Each stimulus was followed by a
C, were administered to each participant. Each stimulus was followed by a 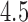 –
–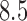 s long pre-rating period, after which participants rated the intensity of the pain on a scale of
s long pre-rating period, after which participants rated the intensity of the pain on a scale of 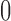 –
–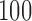 . Each trial concluded with a
. Each trial concluded with a 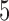 –
–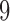 s resting period. For more information about the data acquisition and preprocessing, see Appendix E of supplementary material available at Biostatistics online.
s resting period. For more information about the data acquisition and preprocessing, see Appendix E of supplementary material available at Biostatistics online.
A single trial analysis approach was used by constructing a GLM design matrix with separate regressors for each trial (Rissman and others, 2004). Boxcar regressors, convolved with the canonical hemodynamic response function, were constructed to model periods for the thermal stimulation and rating periods for each trial. Other regressors that were not of direct interest included (i) intercepts for each run; (ii) linear drift across time within each run; (iii) the six estimated head movement parameters (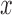 ,
, 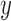 ,
, 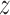 , roll, pitch, and yaw), their mean-centered squares, derivatives, and squared derivative for each run; (iv) indicator vectors for outlier time points; (v) indicator vectors for the first two images in each run; and (vi) signal from white matter and ventricles. Using the results of the GLM analysis, whole-brain maps of activation were computed.
, roll, pitch, and yaw), their mean-centered squares, derivatives, and squared derivative for each run; (iv) indicator vectors for outlier time points; (v) indicator vectors for the first two images in each run; and (vi) signal from white matter and ventricles. Using the results of the GLM analysis, whole-brain maps of activation were computed.
In summary, 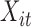 and
and 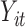 are the temperature level and pain rating, respectively, assigned on trial
are the temperature level and pain rating, respectively, assigned on trial 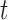 to subject
to subject 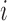 , and
, and 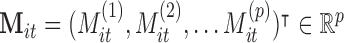 is the whole-brain activation measured over
is the whole-brain activation measured over 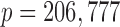 voxels, defined as the regression parameter corresponding to the stimulus in the associated GLM. In addition,
voxels, defined as the regression parameter corresponding to the stimulus in the associated GLM. In addition, 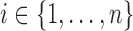 and
and 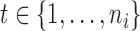 , where
, where 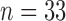 and
and 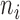 takes subject-specific values between
takes subject-specific values between 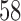 and
and 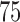 . The data was arranged in a matrix
. The data was arranged in a matrix 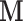 of dimension
of dimension 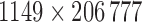 , where each row consists of activation from a single trial on a single subject over
, where each row consists of activation from a single trial on a single subject over 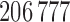 voxels, and each column is voxel specific. The temperature level and reported pain are represented as the vectors
voxels, and each column is voxel specific. The temperature level and reported pain are represented as the vectors 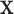 and
and 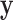 , respectively, both of length
, respectively, both of length 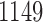 .
.
Each DM corresponding to 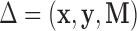 , is a vector of length
, is a vector of length 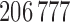 , whose estimation is computationally infeasible without first performing data reduction. Hence, we use the GPVD approach outlined in Section 3.5. We choose
, whose estimation is computationally infeasible without first performing data reduction. Hence, we use the GPVD approach outlined in Section 3.5. We choose 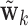 to have dimension
to have dimension 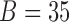 , to ensure that the number of rows of
, to ensure that the number of rows of 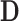 is less than or equal to the minimum number of trials per subject. This value ensures that
is less than or equal to the minimum number of trials per subject. This value ensures that 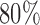 of the total variability of
of the total variability of 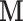 is explained after dimension reduction. The population-specific matrix
is explained after dimension reduction. The population-specific matrix 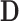 of dimension
of dimension 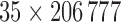 was obtained using GPVD, and the lower dimensional mediation matrix
was obtained using GPVD, and the lower dimensional mediation matrix  of dimension
of dimension 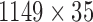 , according to (3.13). The terms
, according to (3.13). The terms 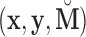 were placed into the algorithm outlined in (3.7)–(3.9), using starting values
were placed into the algorithm outlined in (3.7)–(3.9), using starting values 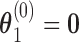 . Finally, values of
. Finally, values of 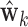 , of length
, of length 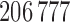 , were computed using
, were computed using 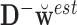 .
.
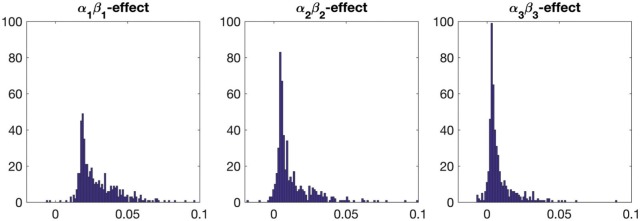
We illustrate the results for the first three DMs. The parameter estimates are 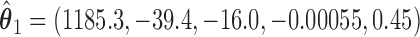 ,
, 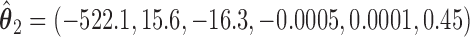 , and
, and 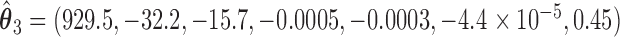 . Figure 3 shows the bootstrap distribution for
. Figure 3 shows the bootstrap distribution for 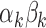 for
for  . Clearly, the center of the distributions move toward
. Clearly, the center of the distributions move toward  for increasing
for increasing 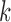 , illustrating their decreasing contribution. However, for each value of
, illustrating their decreasing contribution. However, for each value of 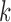 the contribution of
the contribution of 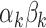 remains significantly different from
remains significantly different from 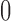 at the
at the 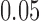 level.
level.
Fig. 3.
Results of the fMRI study of thermal pain. Bootstrap distributions for 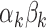 corresponding to the first three DMs.
corresponding to the first three DMs.
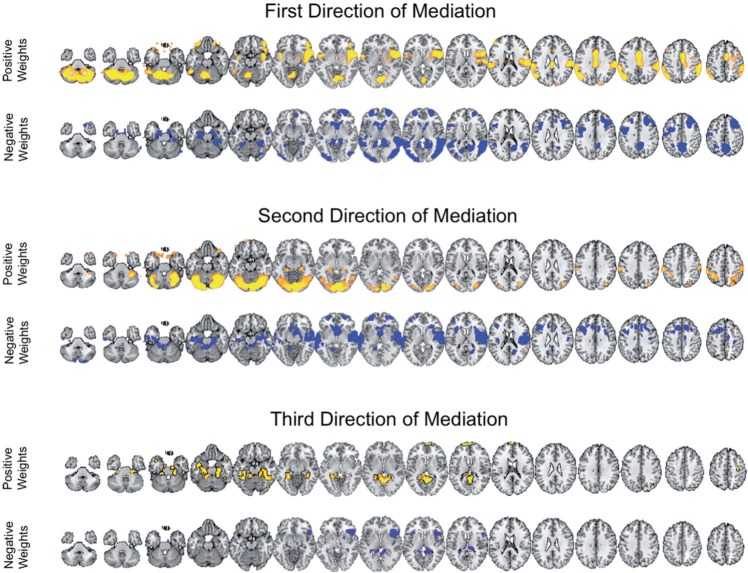
Figure 4 shows the estimated weight maps 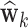 for the first three DMs, thresholded using false discovery rate (FDR) correction with
for the first three DMs, thresholded using false discovery rate (FDR) correction with 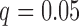 . These maps allow us to visualize latent brain components that are maximally effective as mediators. For each DM, we separate the maps according to whether the voxel-specific value is positive or negative. The maps are consistent with regions typically considered active in pain research, but also reveal some interesting structure that has not been uncovered by previous methods.
. These maps allow us to visualize latent brain components that are maximally effective as mediators. For each DM, we separate the maps according to whether the voxel-specific value is positive or negative. The maps are consistent with regions typically considered active in pain research, but also reveal some interesting structure that has not been uncovered by previous methods.
Fig. 4.
Results of the fMRI study of thermal pain. Weight maps 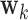 corresponding to the first three DMs, mapped back onto the brain. Significant weights are separated into those with positive and negative values, respectively, for the each DM. All maps are thresholded using FDR correction with
corresponding to the first three DMs, mapped back onto the brain. Significant weights are separated into those with positive and negative values, respectively, for the each DM. All maps are thresholded using FDR correction with 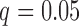 .
.
The first DM shows positive weights on both targets of ascending nociceptive (pain-related) pathways, including the anterior cingulate, mid-insula, posterior insula,parietal operculum/S2, the approximate hand area of S1, and cerebellum. Negative weights were found in areas often anti-correlated with pain, including parts of the lateral prefrontal cortex, parahippocampal cortex, and ventral caudate, and other regions including anterior frontal cortex, temporal cortex, and precuneus. These are associated with distinct classes of functions other than physical pain and are not thought to contain nociceptive neurons, but are still thought to play a role in mediating pain by processing elements of the context in which the pain occurs.
The second DM also contains some nociceptive targets and other, non-nociceptive regions that partially overlap with and are partially distinct from the first direction. This component splits nociceptive regions, with positive weights on S1 and negative weights on the parietal operculum/S2 and amygdala, possibly revealing dynamics of variation among pain processing regions once the first direction of mediation is accounted for. Positive weights are found on visual and superior cerebellar regions and parts of the hippocampus, and negative weights on the nucleus accumbens/ventral striatum and parts of dorsolateral and superior prefrontal cortex. The latter often correlate negatively with pain. Finally, the third DM involves parahippocampal cortex and anterior insula/Ventrolateral prefrontal cortex (VLPFC), both regions related to pain.
7. Discussion
This article addresses the problem of mediation analysis in the high-dimensional setting. The first DM is the linear combination of the elements of a vector of potential mediators that maximizes the likelihood of the underlying three variable SEM. Subsequent directions can be found that maximize the likelihood of the SEM conditional on being orthogonal to previous directions.
The causal interpretation for the parameters of the DM approach rests on strong untestable assumptions, namely sequential ignorability. For example, the assumption 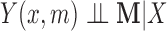 holds if the mediators are randomly assigned to the subjects. However, this is clearly not the case here, and instead, we must assume they behave as if they were. This assumption is unverifiable in practice and ultimately depends on context. In the neuroimaging setting, its validity may differ across brain regions, making causal claims more difficult to access. In practice, this assumption could potentially be weakened by allowing for conditioning on potential confounders. However, no such covariates were available in this particular study. These caveats notwithstanding, we believe the proposed approach still has utility for performing exploratory mediation analysis and detecting regions that potentially mediate the relationship between treatment and outcome, allowing these regions to be explored further in more targeted studies. For further discussion of sequential ignorability assumptions in neuroimaging and the potential to intervene on the mediator without affecting the treatment, see Lindquist and Sobel (2011, 2012).
holds if the mediators are randomly assigned to the subjects. However, this is clearly not the case here, and instead, we must assume they behave as if they were. This assumption is unverifiable in practice and ultimately depends on context. In the neuroimaging setting, its validity may differ across brain regions, making causal claims more difficult to access. In practice, this assumption could potentially be weakened by allowing for conditioning on potential confounders. However, no such covariates were available in this particular study. These caveats notwithstanding, we believe the proposed approach still has utility for performing exploratory mediation analysis and detecting regions that potentially mediate the relationship between treatment and outcome, allowing these regions to be explored further in more targeted studies. For further discussion of sequential ignorability assumptions in neuroimaging and the potential to intervene on the mediator without affecting the treatment, see Lindquist and Sobel (2011, 2012).
The nested counterfactuals defining mediation effects require an assumption that it is possible to intervene on the mediator without affecting the treatment. This assumption is meant to ensure the existence of independent mechanisms affecting treatment and mediator, to rule out, for example, cases where the treatment and the mediator must by necessity cooccur. Transcranial magnetic stimulation (TMS) manipulates the mediator 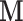 (i.e., brain activity) directly without affecting a treatment such as exposure to heat or pain. Using this technique, one can simulate temporary brain lesions while the subject performs certain tasks. TMS and other similar tools are not technologically advanced enough to allow us to intervene to set
(i.e., brain activity) directly without affecting a treatment such as exposure to heat or pain. Using this technique, one can simulate temporary brain lesions while the subject performs certain tasks. TMS and other similar tools are not technologically advanced enough to allow us to intervene to set 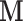 to any user-specified value
to any user-specified value 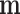 , but it is indeed hypothetically possible to activate or block brain activity on a voxel level. Intervening on a network of voxels is a thornier proposition, but it may be possible to activate a network through direct brain stimulation without affecting treatment.
, but it is indeed hypothetically possible to activate or block brain activity on a voxel level. Intervening on a network of voxels is a thornier proposition, but it may be possible to activate a network through direct brain stimulation without affecting treatment.
When deriving the direct and indirect effect in Section 2, we assumed each subject was scanned under one condition. However, in most fMRI experiments subjects are scanned under multiple conditions, as in our motivating pain data set. Extension of the casual model to this case will allow for single subject studies of mediation in which unit direct effects on the mediators and unit total effects on outcomes are observed. In some instances, the observability of these unit effects can be used to estimate both single subject and population averaged models under weaker and/or alternative conditions than those in (2.2). We leave this extension for future work. In addition, in our motivating example the mediator is brain activation measured with error. Thus, an extension would be to modify the model to deal with systematic errors of measurement in the mediating variable (Sobel and Lindquist, 2014).
One property of the DM framework is that the signs of the estimates are unidentifiable. To address this issue, there are two possible solutions. First, we can use Bayesian methods to apply a sign constraint based on prior knowledge. Second, if the magnitude of the voxel-wise mediation effect is of interest, we can consider a non-negativity constraint. For example, through re-parameterization, as by setting 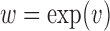 . This can be necessary because, under some circumstances, the coexistence of positive and negative elements of
. This can be necessary because, under some circumstances, the coexistence of positive and negative elements of 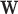 could cancel out potential mediation effects. For example, assume
could cancel out potential mediation effects. For example, assume 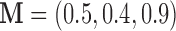 and
and 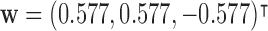 . Then
. Then 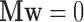 , making the estimate of
, making the estimate of 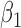 unavailable. It, however, does not necessarily imply the non-existence of a mediation effect.
unavailable. It, however, does not necessarily imply the non-existence of a mediation effect.
In many settings, the response 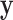 and the mediator
and the mediator 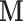 are not necessarily normally distributed, but instead follow some distribution from the exponential family. It can be shown that we can estimate both the DMs and path coefficients under this setting using a type of generalized estimating equation (GEE). Essentially, conditioning on the DM, the path coefficient can be estimated using two sets of GEEs. The DM can then be estimated conditioning on the estimated coefficients.
are not necessarily normally distributed, but instead follow some distribution from the exponential family. It can be shown that we can estimate both the DMs and path coefficients under this setting using a type of generalized estimating equation (GEE). Essentially, conditioning on the DM, the path coefficient can be estimated using two sets of GEEs. The DM can then be estimated conditioning on the estimated coefficients.
In this work, we have assumed that there are no covariates included in the proposed mediation model. However, in observational studies, one often needs to adjust for covariates. In future work, we intend to extend the model and the method to allow for covariates. Finally, while in this work the likelihood is penalized using an L2 norm, in other situations one may require different penalties such as the L1 norm (or some combination of the two). The methods proposed can be extended in these directions, but again we leave this for future work.
Supplementary Material
Supplementary Material
Supplementary material is available at http://biostatistics.oxfordjournals.org. Conflict of Interest: None declared.
Acknowledgments
Conflict of Interest: None declared.
Funding
NIH (R01EB016061, R01DA035484, and P41 EB015909) and NSF (0631637).
References
- Albert J. M. (2008). Mediation analysis via potential outcomes models. Statistics in medicine 27, 1282–1304. [DOI] [PubMed] [Google Scholar]
- Albert J. M. and Nelson S. (2011). Generalized causal mediation analysis. Biometrics 67, 1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen A. H., Gash D. M. and Avison M. J. (1999). Principal component analysis of the dynamic response measured by fMRI: a generalized linear systems framework. Magnetic Resonance Imaging 17, 795–815. [DOI] [PubMed] [Google Scholar]
- Angrist J. D., Imbens G. W. and Rubin D. B. (1996). Identification of causal effects using instrumental variables. Journal of the American Statistical Association 91, 444–455. [Google Scholar]
- Apkarian A. V., Bushnell M. C., Treede R.-D. and Zubieta J.-K. (2005). Human brain mechanisms of pain perception and regulation in health and disease. European Journal of Pain 9, 463–463. [DOI] [PubMed] [Google Scholar]
- Atlas L. Y., Lindquist M. A., Bolger N. and Wager T. D. (2014). Brain mediators of the effects of noxious heat on pain. PAIN® 155, 1632–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron R. M. and Kenny D. A. (1986). The moderator-mediator variable distinction in social psychological research: Conceptual, strategic and statistical considerations. Journal of Personality and Social Psychology 51, 1173–1182. [DOI] [PubMed] [Google Scholar]
- Bushnell M. C., Čeko M. and Low L. A. (2013). Cognitive and emotional control of pain and its disruption in chronic pain. Nature Reviews Neuroscience 14, 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffo B., Chen S., Stewart W., Bolla K., Yousem D., Davatzikos C. and Schwartz B. S. (2008). Are brain volumes based on magnetic resonance imaging mediators of the associations of cumulative lead dose with cognitive function? American journal of epidemiology 167, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caffo B. S., Crainiceanu C. M., Verduzco G., Joel S., Mostofsky S. H., Bassett S. S. and Pekar J. J. (2010). Two-stage decompositions for the analysis of functional connectivity for fmri with application to alzheimer’s disease risk. NeuroImage 51, 1140–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crainiceanu C. M., Caffo B. S., Luo S., Zipunnikov V. M. and Punjabi N. M. (2011). Population value decomposition, a framework for the analysis of image populations. Journal of the American Statistical Association 106, 775–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel R. M., De Stavola B. L., Cousens S. N. and Vansteelandt S. (2015). Causal mediation analysis with multiple mediators. Biometrics 71, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland P. W. (1988). Causal inference, path analysis and recursive structural equation models (with discussion). Sociological Methodology 18, 449–493. [Google Scholar]
- Huang Y.-T. and Pan W.-C. (2015). Hypothesis test of mediation effect in causal mediation model with high-dimensional continuous mediators. Biometrics. [DOI] [PubMed] [Google Scholar]
- Imai K., Keele L. and Tingley D. (2010). A general approach to causal mediation analysis. Psychological methods 15, 309. [DOI] [PubMed] [Google Scholar]
- Imai K. and Yamamoto T. (2013). Identification and sensitivity analysis for multiple causal mechanisms: Revisiting evidence from framing experiments. Political Analysis 21, 141–171. [Google Scholar]
- Jo B. (2008). Causal inference in randomized experiments with mediational processes. Psychological Methods 13, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong K. K., Belliveau J. W., Chesler D. A., Goldberg I. E., Weisskoff R. M., Poncelet B. P., Kennedy D. N., Hoppel B. E., Cohen M. S. and Turner R. (1992). Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proceedings of the National Academy of Sciences 89, 5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M. A. (2012). Functional causal mediation analysis with an application to brain connectivity. Journal of the American Statistical Association 107, 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M. A. and Sobel M. E. (2011). Graphical models, potential outcomes and causal inference: Comment on Ramsey, Spirtes and Glymour. NeuroImage 57, 334–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M. A. and Sobel M. E. (2012). Cloak and DAG: A response to the comments on our comment. NeuroImage 76, 446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M. A., Spicer J., Asllani I. and Wager T. D. (2012). Estimating and testing variance components in a multi-level glm. NeuroImage 59, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist M. A. (2008). The statistical analysis of fMRI data. Statistical Science 23, 439–464. [Google Scholar]
- MacKinnon D. P. (2015). Mediation Analysis. The Encyclopedia of Clinical Psychology. 1–9. [Google Scholar]
- McKeown M. J., Makeig S., Brown G. G., Jung T.-P., Kindermann S.S., Bell A. J. and Sejnowski T. J. (1997). Analysis of fMRI data by blind separation into independent spatial components. Technical Report, DTIC Document. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S., Lee T.-M., Kay A. R. and Tank D. W. (1990). Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proceedings of the National Academy of Sciences 87, 9868–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogburn E. L. (2012). Commentary on “Mediation analysis without sequential ignorability: Using baseline covariates interacted with random assignment as instrumental variables” by Dylan Small. Journal of Statistical Research 46, 105. [PMC free article] [PubMed] [Google Scholar]
- Pearl J. (2014). Interpretation and identification of causal mediation. Psychological methods 19, 459. [DOI] [PubMed] [Google Scholar]
- Preacher K. J. and Hayes A. F. (2008). Asymptotic and resampling strategies for assessing and comparing indirect effects in multiple mediator models. Behavior Research Methods 40, 879–891. [DOI] [PubMed] [Google Scholar]
- Rissman J., Gazzaley A. and D’Esposito M. (2004). Measuring functional connectivity during distinct stages of a cognitive task. Neuroimage 23, 752–763. [DOI] [PubMed] [Google Scholar]
- Robins J. M. and Greenland S. (1992). Identifiability and exchangeability of direct and indirect effects. Epidemiology 3, 143–155. [DOI] [PubMed] [Google Scholar]
- Robins J. M. and Richardson T. S. (2010). Alternative graphical causal models and the identification of direct effects. In: Shrout P. (editor), Causality and Psychopathology: Finding the Determinants of Disorders and Their Cures. Oxford University Press, pp. 103–158. [Google Scholar]
- Rubin D. B. (1974). Estimating causal effects of treatment in randomized and nonrandomized studies. Journal of Educational Psychology 66, 688–701. [Google Scholar]
- Sobel M. E. (2008). Identification of causal parameters in randomized studies with mediating variables. Journal of Educational and Behavioral Statistics 33, 230–251. [Google Scholar]
- Sobel M. E. and Lindquist M. A. (2014). Causal inference for fmri time series data with systematic errors of measurement in a balanced on/off study of social evaluative threat. Journal of the American Statistical Association 109, 967–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Have T. R., Joffe M. M., Lynch K. G., Brown G. K., Maisto S.A. and Beck A. T. (2007). Causal mediation analyses with rank preserving models. Biometrics 63, 926–934. [DOI] [PubMed] [Google Scholar]
- VanderWeele T. and Vansteelandt S. (2009). Conceptual issues concerning mediation, interventions and composition. Statistics and its Interface 2, 457–468. [Google Scholar]
- VanderWeele T. and Vansteelandt S. (2014). Mediation analysis with multiple mediators. Epidemiologic methods 2, 95–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager T. D., Atlas L. Y., Lindquist M. A., Roy M., Woo C.-W. and Kross E. (2013). An fmri-based neurologic signature of physical pain. New England Journal of Medicine 368, 1388–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager T. D., Davidson M. L., Hughes B. L., Lindquist M. A. and Ochsner K.N. (2008). Prefrontal-subcortical pathways mediating successful emotion regulation. Neuron 59, 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager T. D., van Ast V., Davidson M. L., Lindquist M. A. and Ochsner K. N. (2009a). Brain mediators of cardiovascular responses to social threat, Part II: Prefrontal subcortical pathways and relationship with anxiety. NeuroImage 47, 836–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager T. D., Waugh C. E., Lindquist M. A., Noll D. C., Fredrickson B. L. and Taylor S. F. (2009b). Brain mediators of cardiovascular responses to social threat, Part I: Reciprocal dorsal and ventral sub-regions of the medial prefrontal cortex and heart-rate reactivity. NeuroImage 47, 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Nelson S. and Albert J. M. (2013). Estimation of causal mediation effects for a dichotomous outcome in multiple-mediator models using the mediation formula. Statistics in medicine 32, 4211–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold H. (2006). Partial Least Squares. Encyclopedia of Statistical Sciences. 9. [Google Scholar]
- Woo C. W., Roy M., Buhle J. T. and Wager T. D. (2015). Distinct brain systems mediate the effects of nociceptive input and self-regulation on pain. PLoS Biology 13, e1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.