Summary
Integration of proteomic and metabolomic data from early stage lung adenocarcinoma tissues identified increased glycosylation and glutaminolysis, elevated Nrf2 activation, increased NAD-salvaging pathways and elevated polyamine biosynthesis linked to differential regulation of one-carbon metabolism, all important elements of early carcinogenesis.
Abstract
Lung cancer is the leading cause of cancer mortality in the United States with non-small cell lung cancer adenocarcinoma being the most common histological type. Early perturbations in cellular metabolism are a hallmark of cancer, but the extent of these changes in early stage lung adenocarcinoma remains largely unknown. In the current study, an integrated metabolomics and proteomics approach was utilized to characterize the biochemical and molecular alterations between malignant and matched control tissue from 27 subjects diagnosed with early stage lung adenocarcinoma. Differential analysis identified 71 metabolites and 1102 proteins that delineated tumor from control tissue. Integrated results indicated four major metabolic changes in early stage adenocarcinoma (1): increased glycosylation and glutaminolysis (2); elevated Nrf2 activation (3); increase in nicotinic and nicotinamide salvaging pathways and (4) elevated polyamine biosynthesis linked to differential regulation of the s-adenosylmethionine/nicotinamide methyl-donor pathway. Genomic data from publicly available databases were included to strengthen proteomic findings. Our findings provide insight into the biochemical and molecular biological reprogramming that may accompany early stage lung tumorigenesis and highlight potential therapeutic targets.
Introduction
Lung cancer continues to be the leading cause of cancer-related deaths for men and women in the United States and worldwide (1). While clinical studies show that low dose spiral computerized tomography can reduce lung cancer mortality by 20%, its use has been hindered by high false positive rates (1) largely because of difficulty distinguishing between benign and malignant solid pulmonary nodules (2), which leads to over-treatment, anxiety, and use of invasive procedures. Although false positive rates can be reduced with Positron Emission Tomography - Computed Tomography (PET-CT), a more comprehensive understanding of the underlying biochemical and molecular perturbations that accompany tumorigenesis is needed. Such knowledge would be instrumental in elucidating ‘lung cancer’ signatures in addition to identifying molecular drivers of tumorigenesis, candidate targets of intervention and to aid with diagnosis by identifying candidate biomarkers for clinical use.
Early perturbations in cellular metabolism are a hallmark of cancer (3). Recent advances in mass spectrometry have enabled comprehensive profiling of lipids, carbohydrates, amino acids, organic acids and nucleotides allowing for global evaluation of metabolic alterations that occur in a variety of biological matrices (4,5). Metabolomics has enabled new insights into the pathology of cancer and revealed new biomarkers potentially useful for diagnosis and prognosis (6). However, to-date, investigations of lung cancer have been limited and studies that have looked at blood plasma are limited by small sample sizes with mixed histologies (7–10). Recently, we compared the metabolome of early stage (stage IA/IB) non-small cell lung cancer (NSCLC) adenocarcinoma and matched control tissue and identified metabolomic evidence of decreased glucose levels, changes in cellular redox status, elevations in nucleotide metabolites, reduced purine salvage, increased de novo purine synthesis and increased protein glycosylation (5). While such knowledge is instrumental for understanding the underlying biochemical alterations that accompany tumorigenesis, it may still be insufficient to completely resolve complex biological systems or pathologies. Therefore, the combination of multiple ‘omic’ platforms is of considerable value and presents a more accurate representation of the organismal phenotype under a pathological state. In this context, given the direct relationship between metabolites as substrate and products of their related enzymes, the integration of proteomics and metabolomics serves as a particularly powerful approach to study lung tumorigenesis.
In the current investigation, we combined metabolomics and proteomics data utilizing a ‘systems biology’ approach to characterize alterations at the biochemical and molecular level of lung adenocarcinoma tissue compared to matched non-malignant tissue (control) in a subset of subjects (n = 27) from our initial metabolomics (5) using described previously strategies (11). We further coupled these findings with gene expression data from the Gene Expression Omnibus (GEO) database. We hypothesize that identification of cancer-induced cellular and tissue-level biochemical and molecular changes offers a more robust method for identification of candidate diagnostic markers and targets of therapeutic intervention and will improve our understanding of the changes that accompany adenocarcinoma tumorigenesis.
Materials and methods
Sample acquisition
De-identified malignant and adjacent non-malignant (control) lung tissues were harvested in the operating room from patients having resection for NSCLC, none of whom received preoperative treatment. The matching control lung tissue was always taken from areas 8–10 cm removed from the cancer bed. Two to three tissue pieces were aliquoted into 1.5 ml Nunc vials in the operating room after resection of the cancer, and frozen in liquid nitrogen within 30 s. Each vial was barcoded, and stored at −80°C. Specimens were annotated for diagnosis (including stage), age, gender, race, histology and smoking status (pack-years). For this study, the following criteria was used: (a) current or former smokers, (b) adenocarcinoma histology, (c) pathological stage IA or IB and (d) signed the IRB consent form.
Metabolomics analysis
The MiniX database (5) was used as a Laboratory Information Management System and for sample randomization prior to all analytical procedures. Sample identifications were kept blinded during the entire metabolomics analysis to minimize bias. Sample preparation and metabolomics analyses were described previously (5). Mass spectrometry metabolomics data have been deposited to the Metabolomics Workbench (http://www. metabolomicsworkbench.org) with dataset identifier ST000619.
Proteomic analysis of tissues
Paired tissue samples from 27 current or former smokers with early stage (IA/IB) NSCLC adenocarcinoma were assessed for alterations in the metabolome and proteome (Table 1). The mean age and smoking packs per year for subjects were 71 and 35, respectively, with more females than males (Table 1). Tissue samples were minced into two to three pieces and rinsed with cold PBS. One hundred microliters of HB buffer [0.25 M sucrose, 20 mM Hepes-KOH, containing protease inhibitors (Roche)] was added to each sample, followed by homogenization for 3 min (Bullet Blender Storm, Wisbiomed) as described previously (12). N-glycans were released, also described previously (12), and ethanol precipitated tissue samples were used for proteomic analysis. Protein pellets were solubilized in 100 µL of 6 M urea in 50 mM ammonium bicarbonate. Dithiothreitol (200 mM) was added (final concentration of 5 mM), and samples were incubated for 30 min at 37°C, followed by addition of 20 mM iodoacetamide (IAA) (final concentration of 15mM), with incubation for 30 min (RT), and then 20 µl 200 mM DTT added to quench the iodoacetamide reaction. Lys-C/trypsin (Promega, Madison, WI, USA) was next added (1:25 enzyme:protein ratio), incubated (4 h at 37°C), then diluted to <1 M urea with 50 mM ammonium bicarbonate, and digested overnight at 37°C. Samples were desalted (C18 Macro Spin columns, Nest Group) and lyophilized. Samples were dissolved in 2% ACN/0.1% TFA for LC MS/MS analysis
Table 1.
Patient characteristics
| Subjects, N | 27 |
|---|---|
| By stage (NSCLC adenocarcinoma) | |
| IA, N (%) | 15 (55.6%) |
| IB, N (%) | 12 (44.4%) |
| Gender (M/F) | (10/17) |
| Age (mean ± SD) | 71 ± 9 |
| Smoking packs per year (mean ± SD) | 35 ± 27 |
LC-MS/MS analysis
Samples were randomized into eight blocks of four normal and four tumor samples to keep identification of samples unknown during LC-MS/MS analysis. Three technical replicates were acquired for each sample resulting in 162 total LC-MS/MS analyses. LC separation was done on a Waters Nano Acquity UHPLC (Waters Corporation, Milford, MA, USA) with a Proxeon nanospray source. Mass spectra was collected on an Orbitrap Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) in a data-dependent mode with one MS precursor scan followed by 15 MS/MS scans as described previously (13). Detailed information on instrument parameters and mass spectra collection is provided in the Supplemental Materials and Methods. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (14) via the PRIDE partner repository with the dataset identifier PXD002612.
Protein identification
Protein identification was performed similarly to Zhang et al. (15). Additional details are provided in Supplemental Materials and Methods.
Statistical analysis
Detailed information regarding statistical analysis is provided in Supplemental Materials and Methods.
Proteomic (n = 3026) and metabolomic (n = 462) variables were merged for matched control and adenocarcinoma tissue for 27 patients. Mixed effects models were calculated to identify differentially regulated proteins and metabolites between adenocarcinoma and control tissues. Models were fit to measure variables given patient age, gender, pack-years of smoking history and cancer status with patient identifiers included as a random factor to account for the correlation of measurements from the same patient. A chi-squared test was used to assess the significance of metabolic differences through comparison of the full model to a reduced model not including a cancer term. The significance levels (i.e. P-values) were adjusted for multiple hypothesis testing according to Benjamini and Hochberg at a false discovery rate (FDR) of 5% (abbreviated pFDR < 0.05).
Gene expression analysis
Transcriptomic profiling data of 59 NSCLC adenocarcinomas (78% stage I) and 59 adjacent non-malignant tissue from Selamat et al. (16) was downloaded from the GEO database (GEO accession number GSE32863). The experiment was conducted using Illumina HumanWG-6 v3.0 expression BeadChip platform (16). Detailed information of the patient characteristics is provided elsewhere (16). Differential gene expression analysis between lung adenocarcinoma and control tissues was performed with an online tool GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) that wraps around two R packages: GEOquery and Linear Models for Microarray Analysis. The GEOquery parsed the GEO data into R and differentially expressed genes were identified by Linear Models for Microarray Analysis. The raw P-values were adjusted with Benjamini and Hochberg at a FDR of 0.05.
Pathway over-representation analysis
Pathway over-representation analysis was performed using Integrated Molecular Pathway Level Analysis (17). Only protein IDs and metabolites with a raw P-value < 0.05 were considered. Detailed information regarding filtering of redundant over-represented pathways is provided in the Supplemental Materials and Methods.
Network analysis
Thirty-eight significantly different metabolites (pFDR < 0.05) and 1102 significantly different proteins (raw P-value < 0.05) comparing adenocarcinoma with non-malignant tissues were used separately to query the integrated networks of metabolites, proteins and genes with Grinn, an R-based software tool for omic data integration (11). Grinn collects biochemical relationships from publicly available databases (e.g. KEGG, Reactome and HMDB), which include information such as associations between enzymatic proteins and metabolites, and gene–protein encoding pairs. Networks were generated in Cytoscape. The networks were mapped with the relative changes of metabolites, proteins and genes levels between lung malignant and non-malignant tissues. The purpose of systems biology software is to help synthesize data and improve presentation of results in order to identify concurrent and related alterations leading to important clues about the biological connection between specific changes in metabolites, which will lead to further studies demonstrating a causal link.
Results
Comparison of physical and biochemical characteristics
Metabolomics analysis identified a total of 462 compounds, 83 with known annotations. Mixed effects models were used to identify 71 significantly different metabolites between control and adenocarcinoma tissues after adjusting for the FDR (pFDR < 0.05) (Supplementary Table S1, available at Carcinogenesis Online). Proteomics analysis yielded 3026 protein groups. Mixed effects models were also used to identify 1102 significantly different proteins (raw P-value < 0.05) between control and adenocarcinoma tissues; however, no protein remained significantly following FDR adjustment (Supplementary Table S1, available at Carcinogenesis Online). Gene expression data was obtained as described in the materials and methods section and data is provided in Supplementary Table S1, available at Carcinogenesis Online.
Pathway over-representation analysis of metabolic and proteomic alterations
Integrated pathway over-representation analysis of metabolomic and proteomic data was used to identify significantly perturbed pathways in adenocarcinoma relative to control. Of the top 10 over-represented pathways, 5 were related to altered metabolism highlighted by glucose metabolism, Warburg Effect and gluconeogenesis (Table 2 and Supplementary Table S2, available at Carcinogenesis Online).
Table 2.
Top 10 pathways determined from metabolomic and proteomic data results
| Rank | Pathway | No. of overlapping genes | No. of genes in pathway | p-Value | Q-Value | No. of overlapping metabolites |
|---|---|---|---|---|---|---|
| 1 | Metabolism | 216 | 1427 (1485) | 8.51E-23 | 1.66E-19 | 39 |
| 2 | Cytoplasmic ribosomal proteins | 44 | 88 (88) | 1.62E-25 | 6.32E-22 | 0 |
| 3 | Glucose metabolism | 32 | 67 (70) | 3.95E-18 | 2.21E-15 | 5 |
| 4 | Superpathway of conversion of glucose to acetyl CoA and entry into the TCA cycle | 28 | 48 (52) | 4.42E-19 | 5.33E-16 | 3 |
| 5 | Warburg effect | 27 | 45 (45) | 6.82E-19 | 5.33E-16 | 4 |
| 6 | Ribosome––Homo sapiens (human) | 46 | 134 (135) | 2.31E-18 | 1.50E-15 | 0 |
| 7 | Translation | 59 | 205 (217) | 6.21E-19 | 5.33E-16 | 0 |
| 8 | SRP-dependent cotranslational protein targeting to membrane | 47 | 154 (164) | 2.01E-16 | 5.24E-14 | 0 |
| 9 | Eukaryotic translation elongation | 46 | 138 (148) | 8.77E-18 | 4.28E-15 | 0 |
| 10 | Gluconeogenesis | 19 | 32 (33) | 1.50E-13 | 2.34E-11 | 5 |
Adenocarcinoma exhibit increased glycosylation and glutamine utilization
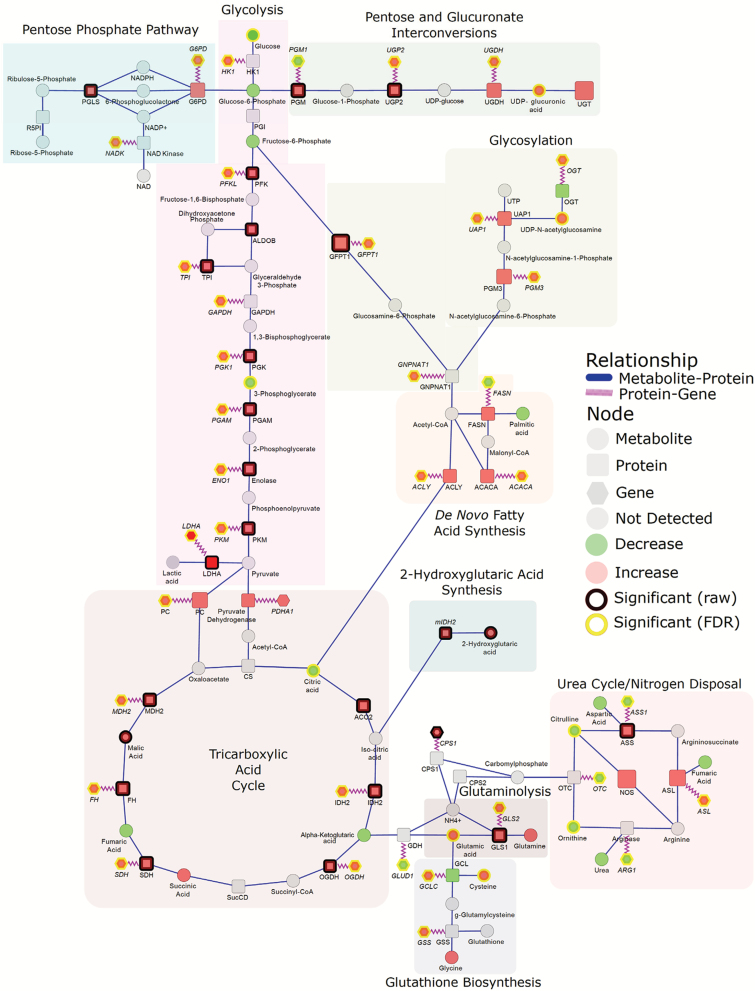
Analysis of the metabolome revealed significant reductions in glucose and the glycolytic intermediate 3-phosphoglycerate whereas virtually all enzymes related to the glycolytic cascade were elevated (Figure 1). Downstream metabolites in the glycolytic cascade serve as important precursors for other proximal metabolic pathways. Specifically, glucose-6-phosphate serves as a branching point between the glycolytic pathway, pentose phosphate pathway (PPP) and pentose and glucuronate interconversion pathways whereas fructose-6-phosphate serves as a precursor for the generation of glucosamine-6-phosphate, an important metabolite involved in glycosylation (Figure 1). Our analysis indicated adenocarcinoma-associated elevations in enzymes involved in both the PPP and pentose and glucuronate interconversion pathways (Figure 1). PPP-related enzymes were glucose-6-phosphate dehydrogenase and 6-phosphogluconolactose whereas pentose and glucuronate interconversions-related enzymes were phosphoglucomutase, UDP-glucose pyrophosphorylase 2, UDP-glucose-6 dehydrogenase and UDP-glucuronosyltransferase (Figure 1). Transcriptomic data corroborated adenocarcinoma-associated elevations in glucose-6-phosphate dehydrogenase, UDP-glucose pyrophosphorylase 2 and UDP-glucose-6 dehydrogenase; however, phosphoglucomutase 1 was decreased. UDP-glucuronic acid, an intermediate in the pentose and glucuronate interconversion pathways, was also significantly elevated in adenocarcinoma compared to control (Figure 1). Additionally, numerous enzymes related to glycosylation including glutamine-fructose-6-phosphate transaminase 1, phosphoglucomutase 3 and UDP-N-acetylglucosamine pyrophosphorylase were increased in adenocarcinoma compared to control; however, only glutamine-fructose-6-phosphate transaminase 1 was statistically significant (Figure 1 and Supplementary Table S1, available at Carcinogenesis Online). The adenocarcinoma-dependent elevations in these enzymes were corroborated by the elevations in the encoding genes, GFPT1, PGM3 and UAP1 in addition to glucosamine-phosphate N-acetyltransferase 1 (GNPNAT1) and O-linked N-acetylglucosamine (GlcNAc) transferase (OGT) (Figure 1). UDP-N-acetylglucosamine, the end-product of the hexosamine pathway and principal substrate for O-linked OGTs, was also significantly elevated in adenocarcinoma compared to control tissue (Figure 1). Also detected were increases in fucosyltransferases FUT6 (alpha 1–3) and FUT8 (alpha 1–6) protein levels in adenocarcinomas, which support increased fucosylation in tumorigenesis. FUT8 is responsible for core fucosylation in N-linked glycans and FUT6 is involved in the biosynthesis of the E-selectin ligand, sialyl-Lewis X, through catalyzing transfer of fucose from GDP-beta-fucose to alpha-2,3 sialylated substrates. FUT6 was not detected in control tissues (Supplementary Table S1, available at Carcinogenesis Online). Also detected was FUCA1 (fucosidase, alpha-L- 1, tissue), which selectively removes fucose from N-linked glycoproteins, showing a potential balancing of fucosylation in adenocarcinoma.
Figure 1.
Integrated networks of lung adenocarcinoma outlining biochemical relationships between metabolites, proteins and genes related to glycolysis and proximal biosynthetic pathways. Metabolites, proteins and genes are represented by circle, square and hexagonal, respectively. Node size indicates absolute magnitude of change. Node color shows the direction of changes comparing lung tumor and control tissues (red: increased in adenocarcinoma; green: decreased). Gray nodes represent features that were not detected. Significance is determined by yellow borders (pFDR < 0.05) or black borders (raw P < 0.05). Blue edges connect metabolites and proteins based on their enzymatic association. Purple edges link a protein and its encoding gene. Shaded areas represent the different biosynthetic pathways.
All tricarboxylic acid cycle (TCA)-related enzymes were elevated in adenocarcinoma relative to control (Figure 1). No measured TCA-related metabolites were found to differ between control and adenocarcinoma except for citrate, which was significantly reduced (pFDR < 0.05) in adenocarcinoma (Figure 1). Glutamine, like glucose, serves as a primary carbon donor for the TCA cycle via entry through glutamate-to- α-ketoglutarate. SLC1A5, the amino-acid transporter that mediates uptake of glutamine, was significantly elevated in adenocarcinoma relative to control in the independent dataset (Supplementary Table S1, available at Carcinogenesis Online). Whereas glutamine was not significantly different between adenocarcinoma and control, glutamate was significantly elevated (Figure 1). Glutaminase, the enzyme that mediates deamination of glutamine to glutamate, and SLC25A22, the mitochondrial transporter that mediates the transfer of glutamate across the inner mitochondrial membrane, were elevated 6-fold, and 125-fold in adenocarcinoma compared to control, respectively (Figure 1 and Supplementary Table S1, available at Carcinogenesis Online). SLC25A22 was also found to be significantly elevated in adenocarcinoma relative to control tissue (Supplementary Table S1, available at Carcinogenesis Online).
Adenocarcinoma display heightened antioxidant defense mechanisms and xenobiotic metabolizing pathways
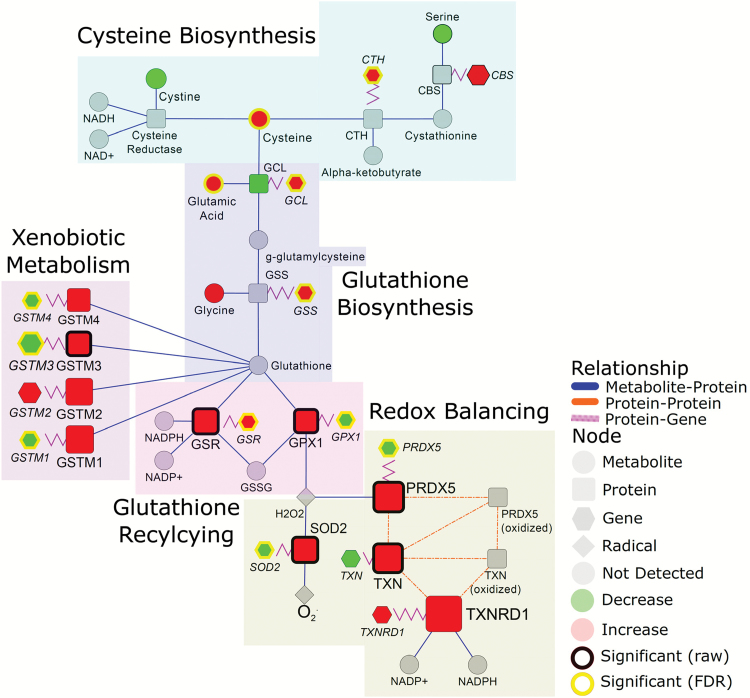
Cysteine and glutamate, important components in glutathione synthesis, and alpha-tocopherol, a lipophilic antioxidant, were significantly elevated 1.6-, 1.3- and 2.2-fold in adenocarcinoma compared to control tissue, respectively, (Figure 2 and Supplementary Table S1, available at Carcinogenesis Online). Cystine, the oxidized dimer of cysteine, tended to be reduced in adenocarcinoma compared to control, albeit not statistically significant (raw P-value: 0.057) (Figure 2). SLC7A11, the sodium-dependent cystine-glutamate antiporter that facilitates cystine uptake, was significantly elevated in adenocarcinoma relative to control tissue (Supplementary Table S1, available at Carcinogenesis Online). While we did not detect cystathionine nor the enzymes in the transsulfuration pathway, transcriptomic data on adenocarcinoma tissue indicated significant increases in cystathionine gamma-lyase (CTH) suggesting that the increase in adenocarcinoma-associated cysteine may be a consequence of both cystine reduction and increased flux into the transsulfuration pathway (Figure 2).
Figure 2.
Integrated networks of lung adenocarcinoma outlining biochemical pathways related to cysteine synthesis, glutathione synthesis and recycling and redox quenching. Metabolites, proteins and genes are represented by circle, square and hexagonal, respectively. Node size indicates absolute magnitude of change. Node color shows the direction of changes comparing lung tumor and control tissues (red: increased in adenocarcinoma; green: decreased). Gray nodes represent features that were not detected. Significance is determined by yellow borders (pFDR < 0.05) or black borders (raw P < 0.05). Blue edges connect metabolites and proteins based on their enzymatic association. Purple edges link a protein and its encoding gene. Shaded areas represent the different biosynthetic pathways.
In association with increases in cysteine and glutamate, numerous enzymes related to glutathione biosynthesis, glutathione recycling, xenobiotic metabolism and redox balancing were elevated in adenocarcinomas relative to control tissue. These findings were highlighted by adenocarcinoma-dependent elevations in mitochondrial superoxide dismutase, thioredoxin, peroxiredoxin, glutathionine-s-transferases, glutathionine peroxidase and glutathionine reductase (Figure 2; data analysis in Supplementary Table S1, available at Carcinogenesis Online).
NAD metabolism in early stage lung adenocarcinoma
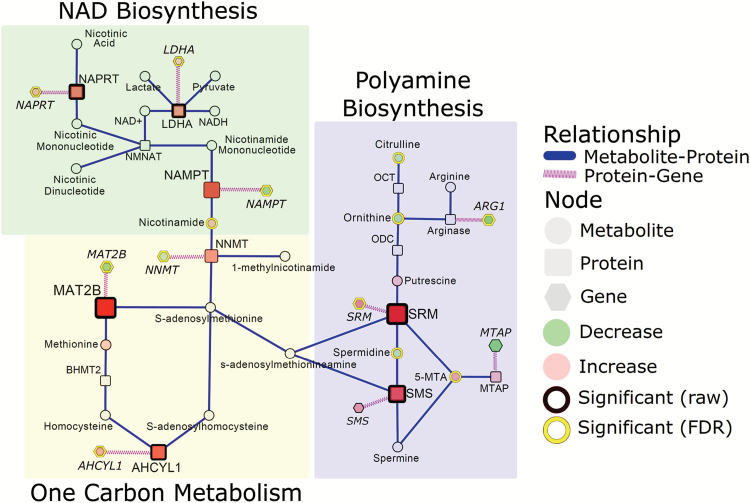
Nicotinamide dinucleotide adenine (NAD+) is an important oxidizing agent vital to metabolizing pathways such as glycolysis and the TCA cycle. NAD+ can be produced de novo via tryptophan catabolism or through the nicotinic and nicotinamide salvaging pathways. Our analysis of the metabolome and proteome found significant increases in nicotinamide, a precursor for NAD+, and nicotinate phosphoribosyltransferase (NAPRT) in adenocarcinoma relative to control (Figure 3). The encoding gene NAPRT was also found to be significantly elevated in adenocarcinomas relative to control in the independent dataset (Figure 3). Nicotinamide phosphoribosyltransferase (NAMPT), the rate-limiting enzymes involved in NAD+ salvaging, and nicotinamide N-methyltransferase (NNMT) were elevated in adenocarcinoma compared to control (Figure 3). However, the encoding genes, NNMT and NAMPT, were significantly reduced in adenocarcinomas compared to control in the independent dataset (Figure 3).
Figure 3.
Integrated networks of lung adenocarcinoma depicting the relationship between NAD synthesis, one carbon metabolism and polyamine biosynthesis. Metabolites, proteins and genes are represented by circle, square and hexagonal respectively. Node size indicates absolute magnitude of change. Node color shows the direction of changes comparing lung tumor and control tissues (red: increased in adenocarcinoma; green: decreased). Gray nodes represent features that were not detected. Significance is determined by yellow borders (pFDR < 0.05) or black borders (raw P < 0.05). Blue edges connect metabolites and proteins based on their enzymatic association. Purple edges link a protein and its encoding gene. Shaded areas represent the different biosynthetic pathways.
Adenocarcinoma tissue exhibits altered one-carbon metabolism and polyamine biosynthesis
In addition to serving as a substrate for NAD+ biosynthesis nicotinamide also participates in one carbon metabolism, acting as a precursor for 1-methylnicotinamide (not detected). NNMT, the enzyme that mediates the transfer of the methyl group from s-adenosylmethionine (SAM) to nicotinamide to generate 1-methylnicotinamide, was elevated in adenocarcinoma relative to control (Figure 3). Consistently, our analysis of the proteome indicated significant adenocarcinoma-dependent elevations in methionine adenosyltransferase II beta (MAT2B) and adenosylhomocysteinase-like 1 (AHCYL1) (Figure 3). Only AHCYL1 was found to be elevated, whereas MAT2B was reduced in adenocarcinoma compared to control (Figure 3). SAM also serves as a precursor for s-adenosylmethionineamine (dSAM). dSAM provides the propylamine groups to putrescine and spermidine to generate spermidine and spermine, respectively, with a concomitant buildup of the byproduct 5-methylthioadenosine (5-MTA) (Figure 3). Spermidine was significantly reduced in adenocarcinoma; whereas spermidine synthase and spermine synthase are elevated at both the protein and mRNA level (Figure 3). Moreover, 5-MTA was elevated in adenocarcinoma relative to control (Figure 3). Putrescine is generated through the decarboxylation of ornithine by ornithine decarboxylase (ODC). Although ornithine decarboxylase and OTC were not detected in this study, ornithine and citrulline were significantly reduced in adenocarcinomas compared to control (Figure 3).
Alterations in nucleotide metabolism characterized by elevations in 5,6-dihydrouracil in adenocarcinoma tissue
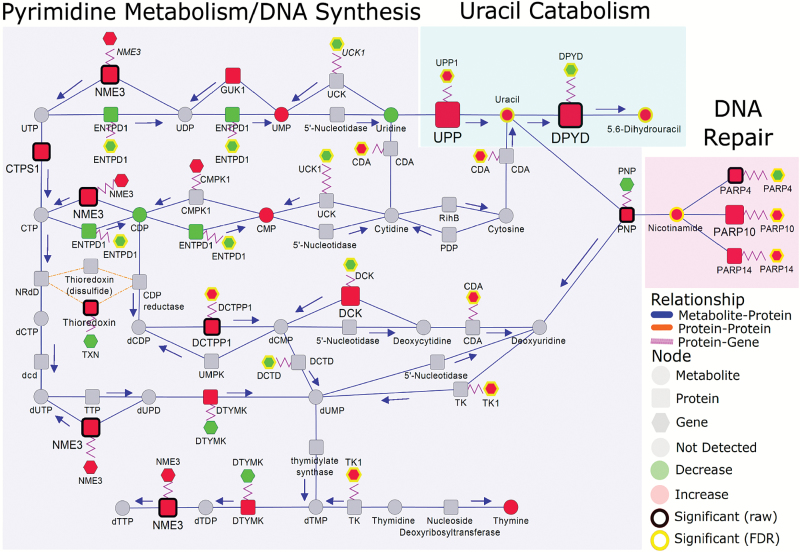
Many proteins involved in de novo pyrimidine synthesis were up-regulated in adenocarcinoma compared to control (Figure 4). Of interest is the 2.7-fold increase in adenocarcinoma-associated 5,6-dihydrouracil, a catabolic product of uracil that was also elevated in tumor relative to control (Figure 4 and Supplementary Table S1, available at Carcinogenesis Online). The catabolism of uracil to 5,6-dihydrouracil is mediated by dihydropyrimidine dehydrogenase (DPYD), which was increased 23-fold in adenocarcinoma (Figure 4 and Supplementary Table S1, available at Carcinogenesis Online). Additionally, uridine phosphorylase, the enzyme mediating the catabolism of uridine to uracil, and its encoding gene, UPP1, were also significantly elevated in adenocarcinoma compared to control (Figure 4).
Figure 4.
Integrated networks illustrating relationship between metabolites, proteins and genes related to pyrimidine metabolism, uracil catabolism and DNA repair. Metabolites, proteins and genes are represented by circle, square and hexagonal respectively. Node size indicates absolute magnitude of change. Node color shows the direction of changes comparing lung tumor and control tissues (red: increased in adenocarcinoma; green: decreased). Gray nodes represent features that were not detected. Significance is determined by yellow borders (pFDR < 0.05) or black borders (raw P < 0.05). Blue edges connect metabolites and proteins based on their enzymatic association. Purple edges link a protein and its encoding gene. Shaded areas represent the different biosynthetic pathways. Arrows indicate the direction of the respective enzymatic reaction.
Noteworthy, many nicotinamide-dependent poly(ADP)ribose polymerases (PARPs), important components in DNA repair, were elevated in adenocarcinoma for which PARP10 and PARP14 also indicated significant elevations at the mRNA level (Figure 4).
Discussion
The present study integrates proteomic and metabolomic data with gene expression information in order to characterize biochemical and molecular changes potentially associated with lung tumorigenesis. Specific focus was placed on NSCLC adenocarcinoma, since adenocarcinoma is the most common histological type of lung cancer (18), thereby minimizing bias from mixed histologies or tumor stage. Evaluation of early stage (Stage IA/IB) adenocarcinomas allows us to capture early events of tumorigenesis, important for early detection. Metabolites are the final output of all cellular activity and is the most sensitive gauge of physiological activity, especially what might be biologically occurring during early tumorigenesis. Because of the dynamic nature of connected metabolomic and proteomic changes and to better understand these changes in early stage lung adenocarcinoma, which are difficult to study in human tissues and we can only surmise from this investigation, more examination into this topic is needed. Future studies investigating flux information is warranted to better understand directionality of fluxes, e.g. to understand the utilization of such compounds as glutamate or glucose (19,20) in lung cancer tissues, as well as the magnitude of pathway activities, e.g. for the TCA cycle where metabolite concentrations appear to remain stable even when enzyme abundances were up-regulated. These studies are likely better addressed in animal models where the process of lung carcinogenesis is more controlled than in human studies.
The preferential use of glucose for aerobic glycolysis and lactic acid fermentation rather than oxidative phosphorylation, also referred to as the Warburg effect, which is a well-documented phenomenon that occurs in various malignancies (21). Our analysis indicated that adenocarcinoma tissue displayed reductions in glucose and elevations in virtually all proteins involved in the glycolytic cascade consistent with increased glycolysis. However, our analysis implicates that the adenocarcinoma-associated increase in glycolysis is not purely to increase pyruvate and lactate, but to also increase the bioavailability of down-stream intermediates that are used as precursors for other proximal biosynthetic pathways such the PPP (22) and glycosylation (23). Our integrative analysis clearly shows that adenocarcinomas exhibit increased machinery involved in both the PPP and glycosylation (Figure 1). We have already reported that adenocarcinoma exhibit differential N-glycosylation patterns relative to control (12). O-GlcNacylation of G6PD and PFK1 has been shown to promote the PPP and tumor growth (23,24), suggesting a potential interplay between post-translational modifications of proteins by glycosylation and tumor metabolism. The functional implications of altered glycosylation patterns remain to be determined.
Interestingly, TCA-related proteins but not metabolites were elevated in cancer. TCA-associate metabolites were largely unchanged with the exception of citrate, which was significantly reduced in adenocarcinoma. Glutamine, like glucose, can serve as a central carbon donor for the TCA cycle via entry through its catabolism to glutamate and subsequently α-ketoglutarate. While glutamine was unchanged, glutamate was significantly elevated in adenocarcinoma relative to control. Malignancies have been shown to have a strong reliance on glutamine (25). The lack of change in glutamine may, therefore, be attributed increased uptake of glutamine from the tumor microenvironment thereby providing steady-state levels. This is supported by the observation that SLC1A5, the amino-acid transporter that facilitates glutamine uptake (26), was significantly elevated in adenocarcinoma compared to control (Supplementary Table S1, available at Carcinogenesis Online). The conversion of glutamine to glutamate is mediated by glutaminase, which was also significantly elevated in adenocarcinoma relative to control. Previous studies have demonstrated that glutamine serves as a primary carbon donor for oxidative phosphorylation via glutaminolysis in NSCLC (27). In addition, one of the main products, malic acid, serves as substrate for malic enzyme that produces additional NADPH that is needed for both biosynthesis and reducing oxidized glutathione in ROS defense (19). Therefore, the reduction in citrate and evidence of increased glutamine deamination via GLS and mitochondrial glutamate transporters (SLC25A22 and SLC25A18) suggests an increased utilization of glutamine as a substrate for oxidative phosphorylation and NADPH production rather than glucose, especially since increased glucose utilization is needed for glycolysis, PPP and glycosylation rather than for oxidative phosphorylation. The alternative, use of glutamate via citrate lyase for production of acetyl-CoA, would not only likely lead to higher citrate levels, but may be restricted to tumors with oxygen deficiencies (20) which are far less likely for early stage adenocarcinomas.
Previous studies have demonstrated the dependence of NSCLC cancer cells to GLS1, namely the GAC splice variant in cells with high rates of glutaminolysis and dependence on glutamine for cell growth (28). Studies in triple-negative breast cancer have shown that GLS1 inhibition by CB-839, an oral non-competitive selective inhibitor of GAC and KGA splice variants of GLS1, resulted in significant anti-tumor activity in both a patient-derived xenograft (PDX) model and in JIMT-1 implanted CB.17 SCID mouse model (29). Therefore, targeting GLS1 may serve as a viable therapeutic target in NSCLC adenocarcinoma; however, further studies will fully determine the benefit of GLS inhibition in NSCLC adenocarcinoma, an aspect that cannot be addressed in our current study.
Reactive oxygen species are common by-products of cellular reactions and bioenergetics pathways mentioned above. Thus, to compensate and prevent excessive buildup of potentially toxic radicals, intricate defense mechanisms must be in place. Nrf2 is master regulator of the antioxidant response and xenobiotic metabolism (30). Nrf2 is constantly ubiquitinated by KEAP1 resulting in its rapid degradation in the proteasome (30). KEAP1 is frequently mutated in NSCLC adenocarcinoma, resulting in the liberation and nuclear translocation of Nrf2 and activation of its target genes (31). Although mutational status was not assessed on these samples, many Nrf2 regulated enzymes, particularly those related to glutathione synthesis and recycling, were up-regulated in adenocarcinoma compared to control. These changes were met with similar elevations in cysteine and glutamate, integral components in glutathione biosynthesis. DeNicola and colleagues indicated previously that Nrf2 regulates serine biosynthesis in subsets of NSCLC cancers harboring KEAP1 mutations by controlling expression of serine/glycine biosynthesis enzyme genes PHGDH, PSAT1 and SHMT2 to support glutathione and nucleotide production (32). In our analysis, only SHMT2 was found to be elevated in adenocarcinoma compared to control (Supplementary Table S1, available at Carcinogenesis Online). Interestingly, we observed adenocarcinoma-associated reductions in cystine, the oxidized dimer of cysteine. Uptake of cystine is facilitated by SLC7A11, the amino-acid antiporter that mediates the glutamate-cystine exchange. Cystine can be reduced to increase intracellular cysteine availability and, subsequently, regulate glutathione pools (33). Although SLC7A11 was not detected in our proteomic data, transcriptomic data indicated significant increases in SLC7A11 in adenocarcinoma compared to control tissue (Supplementary Table S1, available at Carcinogenesis Online). Thus, it is possible that the elevation in cysteine is derived from the reduction of cystine and/or from serine through the transulfuration pathway. Despite this, our data clearly demonstrate increased antioxidant defense mechanisms and elevated Nrf2 activation.
Bioenergetic pathways, such as glycolysis and the TCA cycle, and glutathione recycling are dependent on the availability of reducing equivalents that mediate energy transfer. Furthermore, NAD+ serves as an important modulator of redox metabolism. Targeting NAD+ pools has become an attractive target for therapeutic intervention (34). Our analysis of the metabolome indicated adenocarcinoma-associated increase in nicotinamide, a precursor for NAD+ synthesis. Nicotinamide serves as the substrate for NAMPT, the rate-limiting enzyme in the NAD+ salvaging pathway (34). Although NAMPT was not significantly altered (P: 0.059), it tended to be elevated in lung adenocarcinoma. However, NAD+ can also be generated through the alternative nicotinate salvaging pathway for which NAPRT is the rate-limiting enzyme (35), or through de novo synthesis from tryptophan catabolism. De novo synthesis of NAD+ from tryptophan catabolism is likely to be minimal since cancer cells tend to lack at least one enzyme in the kynurenine pathway (36). Our analysis indicated significant adenocarcinoma-dependent elevations in NAPRT for which the encoding gene, NAPRT, was also elevated in the independent transcriptomic dataset, suggesting that adenocarcinoma may have preferential use of nicotinate for NAD+ biosynthesis rather than nicotinamide. This is an important consideration given that much focus has been given to the inhibition of NAMPT but not NAPRT (37,38). Recently, O’Brien and colleagues demonstrated that NAMPT inhibition with the oral NAMPT-inhibitor GNE-617 inhibited NAD+ generation by greater than 98% and reduced tumor volume in NAPRT-deficient PC3 and HT-1080 xenograft models (39). However, co-administration of nicotinic acid markedly abrogated the growth-inhibitor effects of GNE-617 (39). These findings were conserved in patient-derived xenograft models of SAO-737 sarcoma and STO-399 gastric cancer (39). This would suggest a negative role of nicotinic acid in antagonizing the beneficial effects of NAMPT inhibition in cancerous cells.
The nicotinamide pathway is also intricately linked to the polyamine biosynthesis pathway and one-carbon metabolism via controlling the levels of available methyl donors through SAM pools (40). Polyamine biosynthesis is known to be deregulated in various malignancies (41). This study indicates significant adenocarcinoma-associated reductions in ornithine, citrulline and spermidine, whereas 5-MTA was significantly elevated. 5-MTA, through the action of 5′-deoxy-5′-methylthioadenosine phosphorylase (MTAP), is involved in SAM salvage, purine salvage and spermidine synthesis (42). MTAP activity is known to be reduced in a wide variety of tumor types including NSCLC (43). It has been shown previously that increased levels of 5-MTA inhibit MTAP activity leading to decreased levels of polyamines in NSCLC (44). Based on only metabolomic evidence (5) the reduction in spermidine was thought to be due to inhibition of the polyamine pathway via increased 5-MTA. However, additional integrated proteomic data shows that spermidine synthase and spermine synthase were both significantly elevated in adenocarcinoma, whereas MTAP was no different. spermidine synthase and SMS facilitate the transfer of the propylamine group from dSAM, derived from SAM (42), to produce spermidine and spermine from putrescine and spermidine, respectively, resulting in 5-MTA as a by-product (42). Although methionine was not different between cancer and control tissue, MAT2B, the regulatory enzyme responsible for SAM production from methionine, was up-regulated in adenocarcinoma tissue. Thus, the adenocarcinoma-associated reduction in spermidine and elevation in 5-MTA are more likely a consequence of increased polyamine biosynthesis rather than suppression. Increased flux into the polyamine biosynthetic pathway would also explain the reductions in ornithine and citrulline. Ornithine serves as a precursor for both putrescine and citrulline via ornithine decarboxylase and ornithine transcarbamylase, respectively (42). The coordinated reduction in both ornithine and citrulline and evidence of increased polyamine biosynthesis would indicate that ornithine is being diverted into the polyamine pathway. Importantly, spermidine and spermine (not detected) can undergo acetylation to ultimately derive at the metabolic end-product N1,N12-diacetylspermine. We have identified previously N1,N12-diacetylspermine as a novel pre-diagnostic marker of lung cancer (45). Our results suggest an increase in polyamine biosynthesis and provides a direct link to our previously identified diagnostic marker, N1,N12-diacetylspermine (45).
The biological versatility of nicotinamide is also connected to regulation of DNA repair, principally through its importance as a cofactor for nicotinamide-dependent PARPs. Our analysis indicated adenocarcinoma elevations in numerous PARPs for which mRNA expressions of PARP10 and PARP14 were also significantly elevated in the independent dataset. PARP10 has been shown to interact with PCNA, a master regulator of DNA replication and S-phase-coupled repair, to promote stability and to protect against DNA alterations (46), whereas PARP14 has been linked to promoting the aerobic glycolysis (47). The elevation in PARPs coincided with a general up-regulation of machinery involved in pyrimidine synthesis (Figure 4). Interestingly, adenocarcinomas exhibited increased uracil catabolism relative to control tissue. In particular, uracil and 5,6-dihydrouracil were significantly elevated in adenocarcinomas compared to control. These changes were equally met with adenocarcinoma-dependent elevations in UPP1 and DPYD, the enzymes that mediate the catabolism of uridine to uracil and 5,6-dihydrouracil, respectively. The functional relevance of 5,6-dihydrouracil remains to be determined; however, its elevation implies elevated DPYD activity that is known to be increased in lung adenocarcinoma compared to control tissue (48). The degree of patients’ DPYD activity has been related to improved efficacy of cytotoxic effects from common postoperative adjuvant therapy for NSCLC anti-cancer drug, 5-fluorouracil and its derivatives (48,49). DPYD expression has also been shown to be induced upon EMT induction and be necessary for cells to acquire mesenchymal characteristics (50), a reliance that may be mediated through 5,6-dihydrouracil. The functional relevance of 5,6-dihydrouracil accumulation in NSCLC adenocarcinoma warrants further investigation.
In conclusion, our findings provide considerable insight into biochemical and molecular perturbations that accompany tumorigenesis. In particular, cancer-associated changes in NSCLC adenocarcinoma tissue were highlighted by four major metabolic changes (1) increased glycosylation and glutaminolysis (2), elevated Nrf2 activation (3), increase in both nicotinic and nicotinamide salvaging pathways and (4) elevated polyamine biosynthesis linked to differential regulation of the SAM/nicotinamide methyl-donor pathway. These findings are consistent with those of other lung cancer tissue studies and may highlight potential candidate targets for therapeutic interventions.
Authors Contributions
JFF––wrote manuscript, interpreted data and generated figures/tables. DDG––wrote manuscript, interpreted data and performed statistical analyses. KW––generated figures. BCF––generated metabolomic data. MRS––generated proteomic data. WNR––provided knowledge and expertise about lung cancer, added in study design and critically reviewed manuscript. DRG––provided knowledge and expertise about lung cancer, provided funding and critically reviewed manuscript. BSP––generated proteomic data, provided expertise and knowledge of proteomic findings, aided in study design and critically reviewed manuscript. OF––generated metabolomic data, provided expertise and knowledge of metabolomic findings, aided in study design and critically reviewed manuscript. HP––provided funding and samples, provided knowledge and expertise, aided in study design and critically reviewed manuscript. SM––conceived study design, provided funding, provided knowledge and expertise, helped write the manuscript and critically reviewed manuscript.
Ethics approval and consent to participate
Lung cancer (malignant) and control frozen tissues were obtained from the NYU Biorepository (Harvey Pass, MD) following an approved NYU institution IRB protocol with patient consent (Study Title: NYU Lung Cancer Biomarker Center, Study# i8896, Jun-Chieh Tsay, MD, PI).
Funding
LUNGevity Foundation and Thomas G. Labrecque Foundation [201118739 (SM)]; National Institute of Health [NIH 5U01CA111295-06 (HP)]; National Institutes of Health, West Coast Metabolomics Center [U24 DK097154 (OF)]; National Cancer Institute [NCI U01CA086137 (WNR)] and private donations (DRG).
Supplementary Material
Acknowledgements
We acknowledge the effort of Carol Stroble for tissue sample preparation, N-glycan release and protein precipitation.
Conflict of Interest Statement: The authors declare no conflicts of interest.
Abbreviations
- DPYD
dihydropyrimidine dehydrogenase
- FDR
false discovery rate
- LDCT
low dose spiral computerized tomography
- NAD
nicotinamide dinucleotide adenine
- NAMPT
nicotinamide phosphoribosyltransferase
- NAPRT
nicotinate phosphoribosyltransferase
- NSCLC
non-small cell lung cancer
- PARPs
poly(ADP)ribose polymerases
- PPP
pentose phosphate pathway
- SAM
s-adenosylmethionine
- TCA
tricarboxylic acid cycle
- 5-MTA
5-methylthioadenosine
References
- 1. National Lung Screening Trial Research, T et al. (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl. J. Med., 365, 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Harzheim D., et al. (2015) The solitary pulmonary nodule. Respiration, 90, 160–172. [DOI] [PubMed] [Google Scholar]
- 3. Ward P.S., et al. (2012) Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell, 21, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fahrmann J.F., et al. (2015) Investigation of metabolomic blood biomarkers for detection of adenocarcinoma lung cancer. Cancer Epidemiol Biomarkers Prev., 24, 1716–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wikoff W.R., et al. (2015) Metabolomic markers of altered nucleotide metabolism in early stage adenocarcinoma. Cancer Prev Res (Phila), 8, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spratlin J.L., et al. (2009) Clinical applications of metabolomics in oncology: a review. Clin. Cancer Res., 15, 431–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lokhov P.G., et al. (2013) Blood plasma metabolites and the risk of developing lung cancer in Russia. Eur. J. Cancer Prev., 22, 335–341. [DOI] [PubMed] [Google Scholar]
- 8. Rocha C.M., et al. (2011) Metabolic signatures of lung cancer in biofluids: NMR-based metabonomics of blood plasma. J. Proteome Res., 10, 4314–4324. [DOI] [PubMed] [Google Scholar]
- 9. Hori S., et al. (2011) A metabolomic approach to lung cancer. Lung Cancer, 74, 284–292. [DOI] [PubMed] [Google Scholar]
- 10. Wen T., et al. (2013) Exploratory investigation of plasma metabolomics in human lung adenocarcinoma. Mol. Biosyst., 9, 2370–2378. [DOI] [PubMed] [Google Scholar]
- 11. Wanichthanarak K., et al. (2015) Genomic, proteomic, and metabolomic data integration strategies. Biomark Insights, 10, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ruhaak L.R., et al. (2015) Differential N-Glycosylation patterns in lung adenocarcinoma tissue. J. Proteome Res, 14, 4538–4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grapov D., et al. (2015) The human colostrum whey proteome is altered in gestational diabetes mellitus. J. Proteome Res., 14, 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vizcaino J.A., et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol., 32, 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang B., et al. (2014) Proteogenomic characterization of human colon and rectal cancer. Nature, 513, 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Selamat S.A., et al. (2012) Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res., 22, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamburov A., et al. (2011) Integrated pathway-level analysis of transcriptomics and metabolomics data with IMPaLA. Bioinformatics, 27, 2917–2918. [DOI] [PubMed] [Google Scholar]
- 18. Breathnach O.S., et al. (2001) Twenty-two years of phase III trials for patients with advanced non-small-cell lung cancer: sobering results. J. Clin. Oncol., 19, 1734–1742. [DOI] [PubMed] [Google Scholar]
- 19. Fan J., et al. (2014) Quantitative flux analysis reveals folate-dependent NADPH production. Nature, 510, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Metallo C.M., et al. (2012) Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature, 481, 380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vander Heiden M.G., et al. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science, 324, 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Patra K.C., et al. (2014) The pentose phosphate pathway and cancer. Trends Biochem. Sci., 39, 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yi W., et al. (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science, 337, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rao X., et al. (2015) O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nat. Commun., 6, 8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mohamed A., et al. (2014) Altered glutamine metabolism and therapeutic opportunities for lung cancer. Clin. Lung Cancer, 15, 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Geldermalsen M., et al. (2016) ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene, 35, 3201–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sellers K., et al. (2015) Pyruvate carboxylase is critical for non-small-cell lung cancer proliferation. J. Clin. Invest., 125, 687–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van den Heuvel A.P., et al. (2012) Analysis of glutamine dependency in non-small cell lung cancer: GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol. Ther., 13, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gross M.I., et al. (2014) Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther., 13, 890–901. [DOI] [PubMed] [Google Scholar]
- 30. Taguchi K., et al. (2011) Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells, 16, 123–140. [DOI] [PubMed] [Google Scholar]
- 31. Singh A., et al. (2006) Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med., 3, e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeNicola G.M., et al. (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet., 47, 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang Y., et al. (2005) Cystine-glutamate transporter SLC7A11 in cancer chemosensitivity and chemoresistance. Cancer Res., 65, 7446–7454. [DOI] [PubMed] [Google Scholar]
- 34. Chiarugi A., et al. (2012) The NAD metabolome—a key determinant of cancer cell biology. Nat. Rev. Cancer, 12, 741–752. [DOI] [PubMed] [Google Scholar]
- 35. Hara N., et al. (2007) Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J. Biol. Chem., 282, 24574–24582. [DOI] [PubMed] [Google Scholar]
- 36. Xiao Y., et al. (2013) Dependence of tumor cell lines and patient-derived tumors on the NAD salvage pathway renders them sensitive to NAMPT inhibition with GNE-618. Neoplasia, 15, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sampath D., et al. (2015) Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) as a therapeutic strategy in cancer. Pharmacol. Ther., 151, 16–31. [DOI] [PubMed] [Google Scholar]
- 38. Okumura S., et al. (2012) Nicotinamide phosphoribosyltransferase: a potent therapeutic target in non-small cell lung cancer with epidermal growth factor receptor-gene mutation. J. Thorac. Oncol., 7, 49–56. [DOI] [PubMed] [Google Scholar]
- 39. O’Brien T., et al. (2013) Supplementation of nicotinic acid with NAMPT inhibitors results in loss of in vivo efficacy in NAPRT1-deficient tumor models. Neoplasia, 15, 1314–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sperber H., et al. (2015) The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition. Nat. Cell Biol., 17, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gerner E.W., et al. (2004) Polyamines and cancer: old molecules, new understanding. Nat. Rev. Cancer, 4, 781–792. [DOI] [PubMed] [Google Scholar]
- 42. Sauter M., et al. (2013) Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J., 451, 145–154. [DOI] [PubMed] [Google Scholar]
- 43. Watanabe F., et al. (2009) Immunohistochemical diagnosis of methylthioadenosine phosphorylase (MTAP) deficiency in non-small cell lung carcinoma. Lung Cancer, 63, 39–44. [DOI] [PubMed] [Google Scholar]
- 44. Basu I., et al. (2011) Growth and metastases of human lung cancer are inhibited in mouse xenografts by a transition state analogue of 5′-methylthioadenosine phosphorylase. J. Biol. Chem., 286, 4902–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wikoff W.R., et al. (2015) Diacetylspermine is a novel prediagnostic serum biomarker for non-small-cell lung cancer and has additive performance with pro-surfactant protein B. J. Clin. Oncol., 33, 3880–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nicolae C.M., et al. (2014) The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J. Biol. Chem., 289, 13627–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iansante V., et al. (2015) PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat. Commun., 6, 7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Miyoshi T., et al. (2005) Thymidylate synthase and dihydropyrimidine dehydrogenase in non-small cell lung cancer: relationship between mRNA expression and activity. Anticancer Res., 25, 923–930. [PubMed] [Google Scholar]
- 49. Shintani Y., et al. (2011) Low dihydropyrimidine dehydrogenase correlates with prolonged survival in patients with lung adenocarcinoma treated with 5-fluorouracil. Anticancer Res., 31, 4665–4671. [PubMed] [Google Scholar]
- 50. Shaul Y.D., et al. (2014) Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell, 158, 1094–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.