Summary
Single-institutional methylome data from 1007 samples revealed that genes encoding transcription factors involved in development and differentiation are commonly methylated in multiple organ cancers. Disruption of the differentiated state may be a common feature of DNA methylation alterations during carcinogenesis.
Abstract
The aim of this study was to clarify the significance of DNA methylation alterations shared by cancers derived from multiple organs. We analyzed single-institutional methylome data by single-CpG-resolution Infinium assay for 1007 samples of non-cancerous tissue (N) and corresponding cancerous tissue (T) obtained from lung, stomach, kidney, breast and liver. Principal component analysis revealed that N samples of each organ showed distinct DNA methylation profiles, DNA methylation profiles of N samples of each organ being inherited by the corresponding T samples and DNA methylation profiles of T samples being more similar to those of N samples in the same organ than those of T samples in other organs. In contrast to such organ and/or carcinogenetic factor-specificity of DNA methylation profiles, when compared with the corresponding N samples, 231 genes commonly showed DNA hypermethylation in T samples in four or more organs. Gene ontology enrichment analysis showed that such commonly methylated genes were enriched among “transcriptional factors” participating in development and/or differentiation, which reportedly show bivalent histone modification in embryonic stem cells. Pyrosequencing and quantitative reverse transcription-PCR revealed an inverse correlation between DNA methylation levels and mRNA expression levels of representative commonly methylated genes, such as ALX1, ATP8A2, CR1 and EFCAB1, in tissue samples. These data suggest that disruption of the differentiated state of precancerous cells via alterations of expression, independent of differences in organs and/or carcinogenetic factors, may be a common feature of DNA methylation alterations during carcinogenesis in multiple organs.
Introduction
As well as genetic alterations, epigenetic changes have also been shown to play a key role in human carcinogenesis (1,2). Aberrant DNA methylation is one of the most important epigenetic alterations resulting in chromosomal instability and silencing of tumor-related genes (3–5). We and other groups have indicated that aberrant DNA methylation participates even in early and precancerous stages in different organs exposed to various carcinogenetic factors such as cigarette smoking, chronic inflammation and persistent infection with viruses and other pathogenic microorganisms (6–10). In addition, aberrant DNA methylation is frequently associated with tumor aggressiveness and poorer patient outcome, and can be employed clinically as a prognostic marker (11–16). Thus DNA methylation profiling is generally considered to reflect the biological and clinicopathological heterogeneity of cancers in various organs. However, most previous studies in this field have been conducted using only a single type of organ, and only a limited number of papers have directly compared the DNA methylation profiles of multiple organ cancers (17–19). In particular, little is known about DNA methylation alterations during carcinogenesis shared by various organs.
Recent data from large-scale cancer genomic databases, such as The Cancer Genome Atlas (https://tcga-data.nci.nih.gov/tcga/), International Cancer Genome Consortium (https://icgc.org) and International Human Epigenome Consortium (http://epigenomesportal.ca/ihec/) have allowed detailed genetic and epigenetic characterization of cancers derived from various organs (20–22). However, meta-analysis using such databases has certain drawbacks due to heterogeneity in the quality of samples, the criteria used for clinical diagnosis and the technical platforms employed. In contrast, our group has performed methylome analysis of 1007 quality samples of non-cancerous tissue (N) and corresponding cancerous tissue (T) obtained from five organs collected by experienced pathologists, then stored in liquid nitrogen until analysis and histologically diagnosed by such pathologists at a single institution, the National Cancer Center, Tokyo, Japan. Moreover, single-CpG resolution, highly quantitative Infinium assay has been applied to methylome analysis of these 1007 specimens at a single laboratory. This consistency of sample quality, diagnostic criteria and technical platforms is considerably advantageous for avoiding inter-observer and inter-experimenter bias, providing an overall view of DNA methylation profiles of cancers arising in multiple organs.
Here, we analyzed our own methylome data for 1007 tissue samples of lung, stomach, kidney, breast and liver. In order to further clarify the significance of DNA methylation alterations during carcinogenesis, abnormalities shared by cancers of multiple organs were identified.
Materials and methods
Patients and tissue samples
We employed 1007 tissue samples (498 samples of N and 509 samples of T) obtained surgically from five organs: 166 paired N and T samples of lung tissue resected from 166 patients with primary lung adenocarcinomas, 109 N samples of gastric mucosa and 105 T samples from 109 patients with primary gastric adenocarcinomas, 104 paired N and T samples of nephrectomy specimens from 104 patients with primary clear cell renal cell carcinomas, 83 N samples and 97 T samples of mastectomy specimens from 97 patients with carcinomas of the breast (74 patients with invasive ductal carcinoma of no special type, 7 patients with carcinoma with apocrine differentiation, 6 patients with ductal carcinoma in situ, 4 patients with invasive lobular carcinoma, 2 patients with metaplastic carcinoma, 2 patients with mucinous carcinoma, 1 patient with invasive micropapillary carcinoma and 1 patient with acinic cell carcinoma) and 36 N samples and 37 T samples of hepatectomy specimens from 37 patients with hepatocellular carcinomas. The age, sex and clinicopathological backgrounds of these 513 patients are summarized in Supplementary Table 1, available at Carcinogenesis Online. These patients did not receive preoperative treatment (except for one patient with breast cancer who received preoperative chemotherapy) and underwent surgical resection at the National Cancer Center Hospital, Tokyo, Japan. Histological diagnosis was made in accordance with the World Health Organization classification (23–27), and with the Tumor-Node Metastasis classification (28).
All tissue specimens kept in liquid nitrogen were provided by the National Cancer Center Biobank, Tokyo, Japan. This study was approved by the Ethics Committee of the National Cancer Center, Tokyo, Japan, and performed in accordance with the Declaration of Helsinki. All patients included in this study provided written informed consent for the use of their materials.
Infinium assay
High-molecular weight DNA was extracted from fresh frozen tissue samples using phenol–chloroform, followed by dialysis. Genome-wide CpG methylation profiling was performed on lung, stomach and kidney samples using the Infinium HumanMethylation27 BeadChip (Illumina, San Diego, CA), and on breast and liver samples using the Infinium HumanMethylation450 BeadChip (Illumina) (29). Five-hundred-nanogram samples of DNA were subjected to bisulfite conversion using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). After hybridization, the specifically hybridized DNA was fluorescence-labeled by a single-base extension reaction and detected using a BeadScan or iScan reader (Illumina) in accordance with the manufacturer’s protocol. The data were then assembled using GenomeStudio methylation software (Illumina). At each CpG site, the ratio of the fluorescence signal was measured using a methylated probe relative to the sum of the methylated and unmethylated probes, i.e. the so-called β-value, which ranges from 0.00 to 1.00, reflecting the methylation level of an individual CpG site.
Pyrosequencing
DNA methylation levels for the ALX1, ATP8A2, CR1, EFCAB1, MMP26 and PNOC genes were measured by pyrosequencing. The polymerase chain reaction (PCR) and sequencing primers were designed using Pyrosequencing Assay Design Software ver.1.0 (QIAGEN, Hilden, Germany). To overcome any PCR bias, we optimized the PCR conditions as described previously (13). One CpG site, the same as the original Infinium probe CpG site, or two to six CpG sites included in or located close to the CpG islands, which included or were close to the original Infinium probe CpG sites, were evaluated using pyrosequencing. When multiple CpG sites were evaluated by pyrosequencing, the average DNA methylation levels of the sites were calculated. Each of the primer sequences and PCR conditions are given in Supplementary Table 2, available at Carcinogenesis Online. Positional relationships among CpG sites evaluated using pyrosequencing, the original Infinium probe CpG sites, and the nearest CpG islands are illustrated schematically in the footnote of Supplementary Table 2, available at Carcinogenesis Online. The PCR product was generated from bisulfite-treated DNA and subsequently captured on streptavidin-coated beads. Quantitative sequencing was performed on a PyroMark Q24 (QIAGEN) using the Pyro Gold Reagents (QIAGEN) in accordance with the manufacturer’s protocol.
Real-time quantitative reverse transcription (RT)-PCR analysis
Using TRIzol reagent (Life Technologies, Carlsbad, CA), total RNA was isolated from 19 paired N and T samples from 19 patients with gastric adenocarcinomas, 21 paired N and T samples from 21 patients with clear cell renal cell carcinomas, and 20 paired N and T samples from 20 patients with hepatocellular carcinomas, additional tissue specimens having been available after DNA extraction. (Because of insufficient sample volume, 10 paired samples obtained from patients whose samples were not subjected to Infinum assay were also included in the RT-PCR analysis as well as in the pyrosequening.) cDNA was reverse-transcribed from total RNA using random primers and Superscript III RNase H–Reverse Transcriptase (Life Technologies). Levels of expression of mRNA for the ATP8A2, ALX1, CR1, EFCAB1, MMP26 and PNOC genes were analyzed using custom TaqMan Expression Assays on the 7500 Fast Real-Time PCR System (Life Technologies) employing the relative standard curve method in accordance with the manufacturer’s protocol. The probes and PCR primer sets employed are summarized in Supplementary Table 3, available at Carcinogenesis Online. Experiments were performed in triplicate, and the mean value for the three experiments was used as the CT value. All CT values were normalized to that of GAPDH in the same sample.
Gene ontology (GO) enrichment analysis
In order to reveal the function of proteins encoded by genes showing DNA methylation alterations shared by multiple organ cancers and to reveal the biological processes in which such proteins participate, GO enrichment analysis was conducted using the GeneGO MetaCore™ software (version 6.7) (Thomson Reuters, NY).
Statistics
DNA methylation profiles of N and T samples from each organ were analyzed using principal component analysis (PCA). Infinium probes showing significant differences in DNA methylation levels between N and T samples were defined in terms of the false discovery rate (FDR) of q = 0.01 for Welch’s t test and more than 0.1 or less than −0.1 of the ΔβT-N value. Differences in DNA methylation levels assessed by pyrosequencing and mRNA expression levels assessed by real-time quantitative RT-PCR analysis between N and T samples were examined by Mann–Whitney U test at a significance level of P < 0.05. All statistical analyses were performed using programming language R.
Results
Infinium assay of tissue specimens
About 25979 probes were shared between the Infinium HumanMethylation27 BeadChip and the Infinium HumanMethylation450 BeadChip. Good accordance of the β-values of the shared probes (average correlation coefficient: 0.984) was confirmed in both N and T representative kidney samples (Supplementary Figure 1, available at Carcinogenesis Online). Among 25979 shared probes, all 964 probes on chromosomes X and Y were excluded from further analysis to avoid any gender-specific methylation bias. In addition, the call proportions (P < 0.01 for detection of signals above the background) for the 13 probes shown in Supplementary Table 4, available at Carcinogenesis Online, in all 1007 tissue samples were less than 90%. Since such a low proportion may have been attributable to polymorphism at the probe CpG sites, these 13 probes were excluded from the present assay. Thus, 25 002 probes on autosomal CpG sites were used for further analyses.
DNA methylation levels (β values) for samples of the lung, stomach, kidney and breast have been deposited in the Integrative Disease Omics Database (http://gemdbj.ncc.go.jp/omic/). The DNA methylation status of lung, stomach and kidney for some of the probes has been detailed separately in our previous papers that were not intended to give such an overall view of multiple organ cancers (8–10,14,30).
DNA methylation profiles of N and T samples from various organs
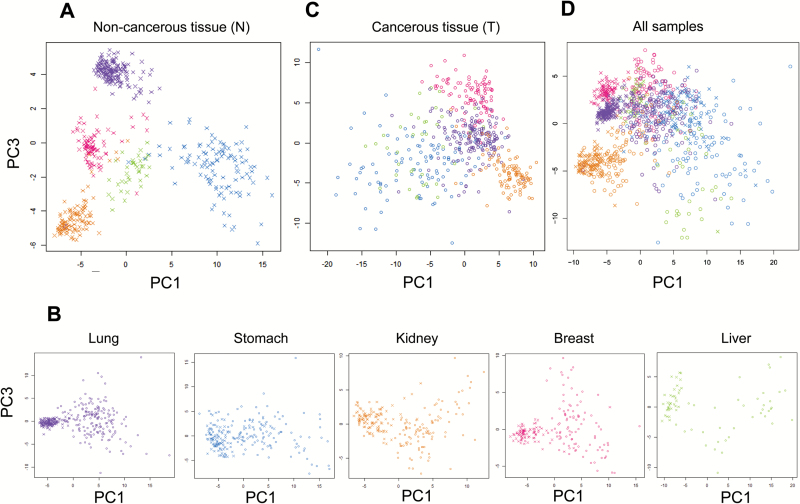
PCA revealed that N samples from each organ had a distinct DNA methylation profile that differed from DNA methylation profiles of N samples from other organs (Figure 1A). N samples from the stomach and liver showed obvious intra-organ heterogeneity, whereas those from the lung, kidney and breast did not (Figure 1A). PCA also showed that the DNA methylation profiles of T samples differed from these of N samples in each organ, although some N samples showed DNA methylation profiles similar to those of T samples from the same organ (Figure 1B). DNA methylation profiles of T samples from each specific organ differed from those of T samples from other organs; T samples shown in Figure 1C showed more obvious intra-organ heterogeneity than N samples shown in Figure 1A. PCA using all 1007 samples revealed that the DNA methylation profiles of T samples bore a closer resemblance to those of N samples from the same organ than to T samples from other organs (Figure 1D).
Figure 1.
Principal component analysis (PCA) based on DNA methylation profiles obtained by Infinium assay of samples of lung (purple), stomach (blue), kidney (yellow), breast (pink) and liver (green). (A) PCA for samples of non-cancerous tissue (N) from all of the five organs examined. Each sample is shown as an x-mark. (B) PCAs for samples of N (x-mark) and the corresponding cancerous tissue (T, clear circles) from each organ. (C) PCAs for samples of T (clear circles) from all of the five organs examined. (D) PCAs for all samples of N (x-marks) and T (clear circles) from all of the five organs examined.
Comparison of DNA methylation alterations in T samples and N samples from various organs
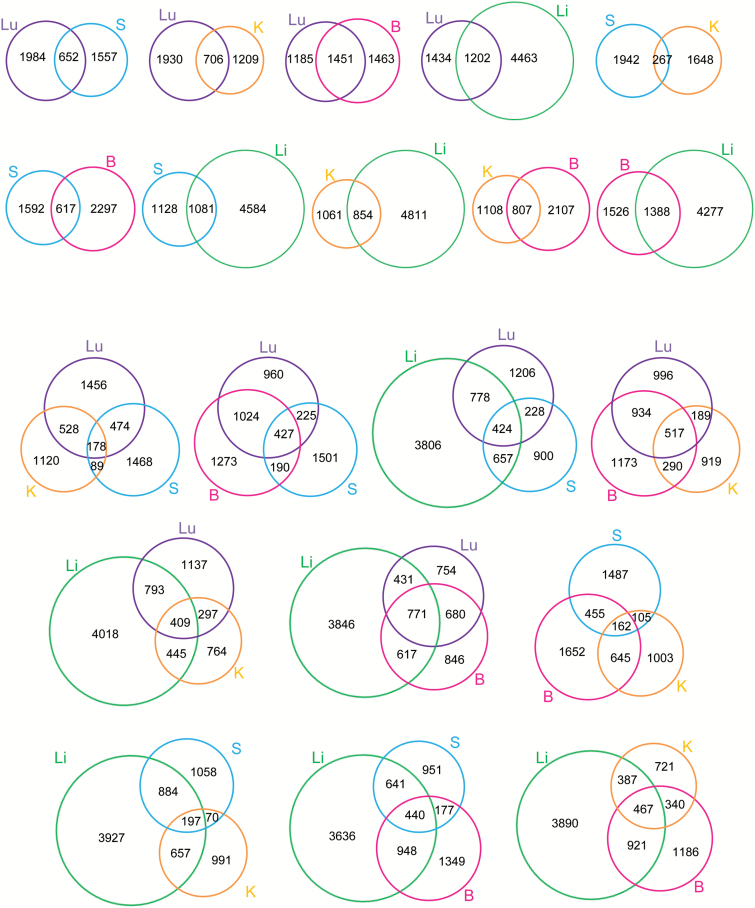
We identified 2636 probes that were aberrantly methylated in lung adenocarcinomas in comparison with the corresponding N samples [Welch’s t test (FDR, q = 0.01) and more than 0.1 or less than −0.1 of the ΔβT-N value]. Similarly, we found 2209, 1915, 2914 and 5665 probes that were aberrantly methylated in gastric adenocarcinomas, clear cell renal cell carcinomas, carcinomas of the breast and hepatocellular carcinomas, respectively, in comparison with the corresponding N samples. The numbers of aberrantly methylated probes shared by 2–5 cancers are summarized in Supplementary Table 5, available at Carcinogenesis Online, and shown using Venn diagrams in Figure 2. When we examined pairs of organs, 35.4% (1451/4099) of aberrantly methylated probes were shared between lung adenocarcinomas (2636 probes) and breast cancers (2914 probes), whereas only 6.9% (267/3857) of them overlapped between clear cell renal cell carcinomas (1915 probes) and gastric adenocarcinomas (2209 probes). Similar tendencies were again observed when we compared DNA methylation alterations among sets of three organs; Figure 2 shows that the majority of DNA methylation alterations were diversely distributed among multiple organs.
Figure 2.
Venn diagrams showing the numbers of probes that were aberrantly methylated in cancerous tissue (T) samples relative to corresponding samples of non-cancerous tissue (N) from the lung (Lu), stomach (S), kidney (K), breast (B) and liver (Li).
Aberrantly methylated genes shared by cancers of multiple organs
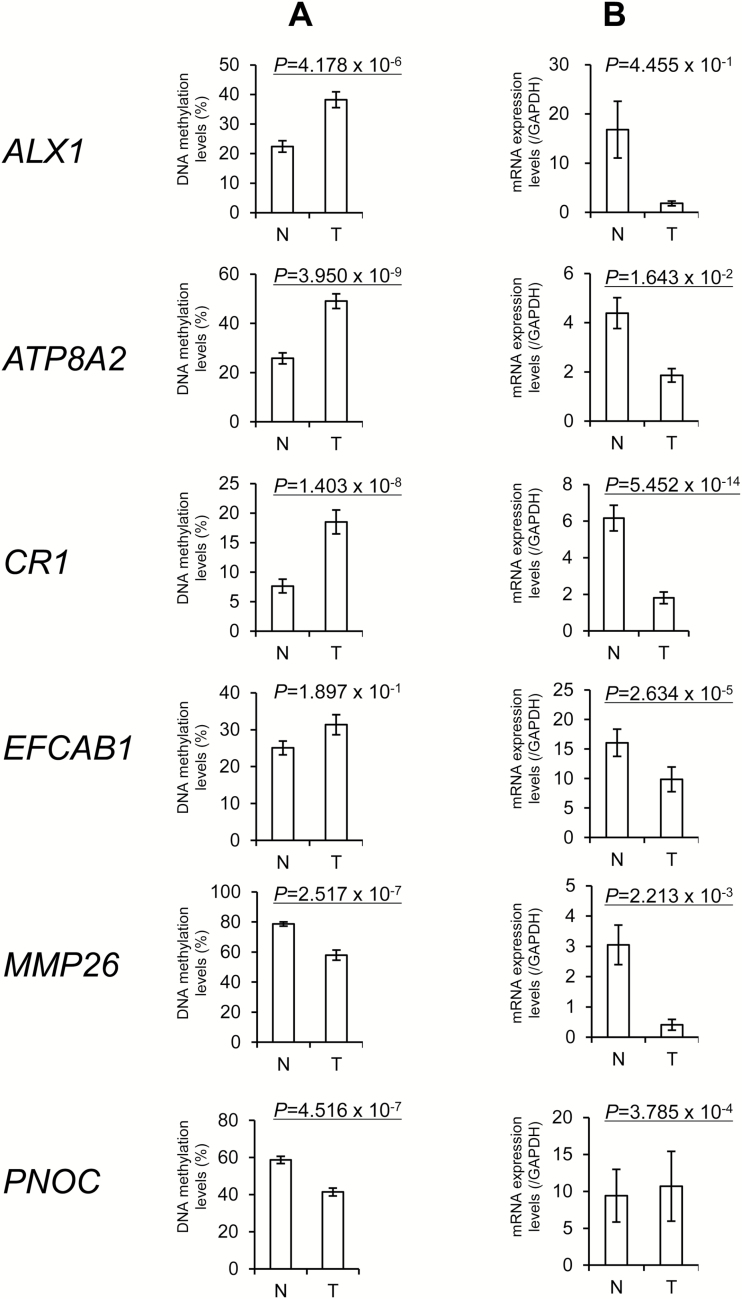
Next, we focused on DNA methylation alterations commonly observed among multiple organ cancers. Thirty-nine probes designed for 33 genes and 69 probes designed for 66 genes showed DNA hypermethylation and DNA hypomethylation, respectively, in T samples from all of the five organs examined, in comparison with the corresponding N samples (Supplementary Table 5, available at Carcinogenesis Online). In order to confirm that such DNA hypermethylation commonly observed in all five organ cancers actually results in gene silencing, DNA methylation levels of representative genes (ALX1, ATP8A2, CR1 and EFCAB1) were verified by pyrosequencing, and their mRNA expression levels were evaluated by real-time quantitative RT-PCR analysis. When we examined 60 paired N and T samples (19, 21 and 20 paired samples from the stomach, kidney and liver, respectively) for which total RNA was available, the DNA methylation levels of the four genes examined were higher in T samples than in N samples, although DNA hypermethylation of the EFCAB1 gene did not reach statistically significant levels, perhaps because the number of samples was insufficient (Figure 3). Levels of mRNA expression for all four genes showing DNA hypermethylation were lower in T samples than in N samples (Figure 3). These results suggested that DNA hypermethylation of these genes may result in a reduction of mRNA expression in tissue samples. On the other hand, DNA hypomethylation of the MMP26 gene did not result in an elevation of mRNA expression in T samples relative to N samples (Figure 3).
Figure 3.
Inverse correlation between DNA methylation levels and mRNA expression levels for representative genes commonly methylated in all five of the organs examined. DNA methylation levels (A) and mRNA expression levels (B) for the ALX1, ATP8A2, CR1, EFCAB1, MMP26 and PNOC genes genes in 60 paired samples of non-cancerous tissue (N) and corresponding cancerous tissue (T) from patients with gastric adenocarcinomas (n = 19), clear cell renal cell carcinomas (n = 21) and hepatocellular carcinomas (n = 20) were determined by pyrosequencing and quantitative real-time reverse transcription-PCR analysis, respectively. P values of < 0.05 are underlined.
GO enrichment analysis using aberrantly methylated genes shared by cancers of multiple organs
Two hundred thirty-one genes (281 probes) commonly showing DNA hypermethylation in T samples from any four organs or all five organs examined (as summarized in Supplementary Table 6, available at Carcinogenesis Online) were evaluated for protein function by enrichment analysis using the MetaCore software, and compared with the protein function distribution of genes within the GeneGo databases. Genes showing DNA hypermethylation shared by cancers of multiple organs were clearly overrepresented by “transcriptional factors” (P = 5.003 × 10–35), being 8.569 times more abundant than expected (Table 1). In addition, such genes were significantly enriched among “receptors” (P = 2.137 × 10–5), “ligands” (P = 6.684 × 10–3) and “kinases” (P = 2.558 × 10–2) (Table 1). Two hundred fifty-eight genes (307 probes) commonly showing DNA hypomethylation in T samples from any four organs or all of the five organs examined were significantly enriched among “receptors” (P = 1.460 × 10–25), “ligands” (P = 1.237 × 10–5) and “proteases” (P = 4.400 × 10–5) (Table 1).
Table 1.
Gene ontology enrichment analysis by protein function using MetaCore software
| Protein class | r | N | R | N | Expected | Ratio | P |
|---|---|---|---|---|---|---|---|
| (A) 231 genes commonly showing DNA hypermethylation in T samples from any four, or all five of the organs examined | |||||||
| Transcription factors | 55 | 230 | 1201 | 43 037 | 6.418 | 8.569 | 5.003 × 10 –35 |
| Receptors | 23 | 230 | 1615 | 43 037 | 8.631 | 2.665 | 2.137 × 10 –5 |
| Ligands | 8 | 230 | 513 | 43 037 | 2.742 | 2.918 | 6.684 × 10 –3 |
| Kinases | 8 | 230 | 656 | 43 037 | 3.506 | 2.282 | 2.558 × 10 –2 |
| Enzymes | 21 | 230 | 2776 | 43 037 | 14.84 | 1.416 | 6.901 × 10–2 |
| Phosphatases | 1 | 230 | 229 | 43 037 | 1.224 | 0.8171 | 6.536 × 10–1 |
| Proteases | 2 | 230 | 580 | 43 037 | 3.100 | 0.6452 | 3.990 × 10–1 |
| Other | 115 | 230 | 35 519 | 43 037 | 189.8 | 0.6058 | 1.363 × 10–29 |
| (B) 258 genes commonly showing DNA hypomethylation in T samples from any four, or all five of the organs examined | |||||||
| Receptors | 54 | 252 | 1615 | 43 037 | 9.457 | 5.710 | 1.460 × 10 –25 |
| Ligands | 13 | 252 | 513 | 43 037 | 3.004 | 4.328 | 1.237 × 10 –5 |
| Proteases | 13 | 252 | 580 | 43 037 | 3.396 | 3.828 | 4.400 × 10 –5 |
| Kinases | 4 | 252 | 656 | 43 037 | 3.841 | 1.041 | 5.367 × 10–1 |
| Enzymes | 13 | 252 | 2776 | 43 037 | 16.25 | 0.7998 | 2.451 × 10–1 |
| Transcription factors | 5 | 252 | 1201 | 43 037 | 7.032 | 0.7110 | 2.922 × 10–1 |
| Other | 150 | 252 | 35 519 | 43 037 | 208 | 0.7212 | 6.533 × 10–18 |
r, number of objects from the present data set for a given protein class; n, total number of objects from the present data set; R, number of background objects from the database for a given class; N, total number of background objects from the database; Expected, mean value for hypergeometric distribution (n × R/43037); Ratio, the ratio of actual/expected. If the ratio is more than 1, P < 0.05 mean significant enrichment and are underlined.
All statistically significant GO biological processes (P < 0.05) are listed in Supplementary Table 7, available at Carcinogenesis Online. The majority of such processes, in which the hypermethylated genes were enriched, were related to organ development and/or cell differentiation (Table 2A), whereas the hypomethylated genes were, if anything, enriched in immune-related processes (Table 2B). Since enrichment was prominent, we focused on genes that commonly showed DNA hypermethylation in cancers of multiple organs, were annotated as GO protein class “transcriptional factors” (P = 5.003 × 10–35), and were involved in the top GO biological process in Table 2A, “multicellular organismal development” (GO: 0007275) (P = 9.259 × 10–30) (Table 3). Among the 49 “transcriptional factors” listed in Table 3, 45 (92%) were encoded by genes reportedly showing bivalent histone modification, consisting of histone H3 lysine 4 trimethylation (H3K4me3) and H3K27me3 in embryonic stem (ES) cells (31–33) (Table 3). In addition, expression levels of many of these genes are reportedly regulated by DNA methylation (Table 3).
Table 2.
Top 40 statistically significant biological processes revealed by Gene Ontology enrichment analysis using MetaCore software
| Biological process | P |
|---|---|
| (A) Top 20 biological processes enriched by 231 genes commonly showing DNA hypermethylation in T samples from any four, or all five of the organs examined | |
| Multicellular organismal development | 9.259 × 10–30 |
| System development | 3.543 × 10–28 |
| Organ development | 5.112 × 10–26 |
| Anatomical structure development | 5.424 × 10–26 |
| Single-organism developmental process | 8.390 × 10–26 |
| Developmental process | 1.017 × 10–25 |
| Regulation of transcription from RNA polymerase II promoter | 1.545 × 10–25 |
| Single-multicellular organism process | 7.922 × 10–24 |
| Multicellular organismal process | 1.646 × 10–22 |
| Regulation of cell differentiation | 1.902 × 10–22 |
| Cell differentiation | 6.829 × 10–22 |
| Nervous system development | 2.639 × 10–21 |
| Cellular developmental process | 8.912 × 10–21 |
| Regulation of nucleobase-containing compound metabolic process | 1.573 × 10–20 |
| Regulation of nitrogen compound metabolic process | 2.012 × 10–20 |
| Regulation of multicellular organismal development | 2.728 × 10–20 |
| Regulation of cellular biosynthetic process | 5.341 × 10–20 |
| Regulation of developmental process | 8.224 × 10–20 |
| Anatomical structure morphogenesis | 1.422 × 10–19 |
| Regulation of biosynthetic process | 1.925 × 10–19 |
| (B) Top 20 biological processes enriched by 258 genes commonly showing DNA hypomethylation in T samples from any four, or all five of the organs examined | |
| Immune response | 6.621 × 10–16 |
| Immune system process | 6.420 × 10–15 |
| Defense response | 6.877 × 10–15 |
| Keratinization | 2.070 × 10–12 |
| Response to stimulus | 2.633 × 10–12 |
| Defense response to bacterium | 2.435 × 10–11 |
| Regulation of immune system process | 2.943 × 10–11 |
| Innate immune response | 6.144 × 10–11 |
| Defense response to other organism | 2.514 × 10–10 |
| Response to stress | 4.862 × 10–10 |
| Regulation of biological quality | 1.004 × 10–9 |
| Striated muscle contraction | 2.143 × 10–9 |
| Keratinocyte differentiation | 2.234 × 10–9 |
| Response to external stimulus | 3.049 × 10–9 |
| Single-multicellular organism process | 5.854 × 10–9 |
| Regulation of blood pressure | 9.479 × 10–9 |
| Regulation of cytokine production | 1.220 × 10–8 |
| Humoral immune response | 1.376 × 10–8 |
| Multicellular organismal process | 1.605 × 10–8 |
| Cell adhesion | 2.339 × 10–8 |
Table 3.
Genes that commonly showed DNA hypermethylation in T samples from any four, or all five of the organs examined, and which are annotated gene ontology (GO) protein class “transcriptional factors” and involved in the GO biological process “multicellular organismal development”
| Gene symbol | Gene ID | Infinium probe ID | Organsa | Bivalent modificationb | Referencesc | Gene regiond | Regulatory regione |
|---|---|---|---|---|---|---|---|
| ALX1 | 8092 | cg02409351 | Lu, S, K, B, Li | Y | — | TSS200 | P |
| ALX4 | 60529 | cg26365854 | Li, Lu, S, B | Y | Y (R1) | GB/1st Int | |
| ASCL2 | 430 | cg06263495 | Lu, K, B, Li | Y | Y (R2) | GB/5’UTR/1st Ex | |
| BARHL2 | 343472 | cg17241310 | Lu, K, B, Li | Y | — | TSS200 | OC |
| DLX5 | 1749 | cg27016494; cg00503840; cg13462129 | Lu, S, K, Li | Y | Y (R3) | GB | TFBS |
| EBF1 | 1879 | cg05056120 | Lu, S, B, Li | Y | — | TSS1500 | P |
| EGR2 | 1959 | cg19355190 | Lu, S, B, Li | Y | — | GB/5’UTR | P |
| EMX2 | 2018 | cg19358493 | Lu, S, B, Li | Y | Y (R4) | TSS1500 | |
| EOMES | 8320 | cg15540820 | Lu, S, B, Li | Y | — | TSS1500 | |
| FOXD3 | 27022 | cg22815110 | Lu, K, B, Li | N | Y (R5) | GB/1st Ex | P |
| FOXE3 | 2301 | cg18815943 | Lu, K, B, Li | Y | — | GB/1st Ex | TFBS |
| FOXG1 | 2290 | cg10300684 | Lu, S, B, Li | Y | — | GB/5’UTR/1st Ex | |
| GBX2 | 2637 | cg23095584 | Lu, K, B, Li | Y | — | TSS1500 | |
| GCM2 | 9247 | cg02844545 | Lu, K, B, Li | Y | — | GB/5’UTR/1st Ex | TFBS |
| GFI1 | 2672 | cg22341104 | Lu, S, B, Li | Y | Y (R6) | GB/5’UTR/1st Int | P |
| HAND2 | 9464 | cg01580681 | Lu, S, K, B, Li | Y | Y (R7) | GB/1st Ex | P |
| HOXA7 | 3204 | cg23432345; cg26511321 | Lu, S, B, Li; Lu, K, B, Li | Y | Y (R8) | GB/1st Ex; TSS1500 | P |
| HOXA9 | 3205 | cg01354473; cg01381846; cg07778029 | Lu, S, B, Li | Y | Y (R9) | GB/1st Ex; GB/1st Ex; GB/5’UTR/1st Ex | P |
| HOXB4 | 3214 | cg08089301; cg21460081 | Lu, S, B, Li | Y | Y (R10) | GB/1st Ex; TSS1500 | P |
| HOXB6 | 3216 | cg18878432 | Lu, S, K, B | Y | — | TSS1500 | |
| HOXD12 | 3238 | cg03874199 | Lu, S, K, B | Y | — | TSS200 | |
| HOXD9 | 3235 | cg14991487 | Lu, S, B, Li | Y | Y (R11) | TSS200 | P |
| IRF4 | 3662 | cg12741420 | Lu, S, B, Li | Y | Y (R12) | GB/5’UTR/1st Int | P |
| MKX | 283078 | cg02161900 | Lu, S, B, Li | Y | — | TSS1500 | |
| MSC | 9242 | cg23710218 | Lu, S, B, Li | Y | — | GB/5’UTR/1st Ex | P |
| MYOD1 | 4654 | cg18555440 | Lu, K, B, Li | Y | — | GB/1st Ex | CTCFBS |
| NKX3-2 | 579 | cg20073553 | Lu, S, B, Li | Y | — | GB/1st Int | |
| NKX6-2 | 14912 | cg08441806; cg09260089 | Lu, K, B, Li | Y | — | GB/1st Ex; TSS1500 | |
| NR2E1 | 7101 | cg03958979 | Lu, K, B, Li | Y | — | TSS1500 | TFBS |
| PAX7 | 5081 | cg07536847 | Lu, S, B, Li | Y | — | TSS1500 | |
| POU3F3 | 5455 | cg20291049 | Lu, S, B, Li | Y | — | GB/1st Ex | |
| POU4F1 | 5457 | cg08097882 | Lu, S, K, B, Li | Y | Y (R13) | GB/5’UTR/1st Ex | P |
| POU4F2 | 5458 | cg24199834 | Lu, S, K, B, Li | Y | Y (R14) | GB/5’UTR/1st Ex | TFBS |
| PRDM14 | 63978 | cg01295203 | Lu, S, B, Li | N | Y (R15) | TSS1500 | TFBS |
| RAX | 30062 | cg19576304 | Lu, S, K, B | Y | — | GB/1st Int | TFBS |
| RUNX3 | 864 | cg00117172; cg24019564 | Lu, S, K, B; S, K, B, Li | Y | Y (R16) | TSS1500 | |
| SALL3 | 27164 | cg15191648 | Lu, K, B, Li | Y | Y (R17) | TSS200 | |
| SALL4 | 57167 | cg06303238 | Lu, K, B, Li | N | Y (R18) | GB/5’UTR/1st Ex | P |
| SIX6 | 4990 | cg19456540 | Lu, S, B, Li | Y | — | GB/1st Ex | TFBS |
| SOX11 | 6664 | cg20008332 | Lu, S, K, Li | N | Y (R19) | GB/1st Ex | P |
| SOX14 | 8403 | cg16428251 | Lu, K, B, Li | Y | — | GB/5’UTR/1st Ex | OC |
| SOX17 | 64321 | cg02919422 | Lu, S, K, B, Li | Y | Y (R20) | GB/5’UTR/1st Ex | P |
| T | 6862 | cg17188046 | K, Lu, S, B | Y | Y (R21) | TSS200 | |
| TBX20 | 57057 | cg02008154 | Lu, S, K, B, Li | Y | Y (10) | GB/5’UTR/1st Ex | P |
| TLX3 | 30012 | cg25720804; cg25942450 | Lu, K, B, Li | Y | Y (R22) | GB/1st Ex; TSS200 | P |
| TP73 | 7161 | cg04391111 | Lu, S, B, Li | Y | Y (R23) | TSS1500 | P |
| TWIST1 | 7291 | cg20052718; cg26312150 | Lu, S, K, Li | Y | Y (R24) | GB/1st Ex | P |
| WT1 | 7490 | cg04456238; cg06516124; cg12006284; cg13301003 | Lu, S, K, B | Y | Y (R25) | GB; GB/1st Int; GB;GB | OC |
| ZIC1 | 7545 | cg05073035; cg14456683 | Lu, K, B, Li | Y | Y (R26) | GB/5’UTR/1st Ex; TSS200 | P |
Organs in which T samples showed DNA hypermethylation relative to N samples. B, breast cancer; K, kidney cancer; Li, liver cancer; Lu, lung cancer; S, stomach cancer.
Y, bivalent histone modification consisting of H3K4me3 and H3K27me3 in embryonic stem cells were reported in References 31, 32 and/or 33; N, not reported in these references.
Y, restored expression in cell lines by treatment with a demethylating agent such as 5′-aza-2′-deoxycitidine, and/or inverse correlation between DNA methylation levels and mRNA expression levels in cell lines and/or tissue samples have been reported in previous studies; -, not reported in previous studies. References R1–R26 reporting such findings for each gene are summarized in Supplementary Table 8, available at Carcinogenesis Online.
Probe CpG sites were annotated as the region from 200 bp upstream of the transcription start site (TSS) to 1500 bp upstream of it (TSS1500), the region from TSS to 200 bp upstream of TSS (TSS200), the 5’ untranslated region (UTR), the first exon (1st EX), the first intron (1st Int), the gene body (GB) and the 3′UTR based on the UCSC Genome Browser (https://genome.ucsc.edu/).
Probe CpG sites were annotated as gene promotor (P), transcription factor binding site (TFBS), CTCF binding site (CTCFBS) and open chromatin region (OC) based on the Ensembl genome browser 86 (http://asia.ensembl.org/index.html).
Discussion
Here we have presented an overall view of data derived from methylome analyses of 1007 paired N and T samples obtained from lung, stomach, kidney, breast and liver, which were collected at a single hospital and analyzed in a single laboratory. Our hospital is among those that collect the largest number of patients with cancers of various organs in Japan. Pathological diagnosis for each patient is assigned simultaneously to both experienced pathologists specializing in cancers of specific organs and general pathologists. The uniform quality of the samples and the use of a single technical platform enabled us to perform multiple organ analysis more accurately than a meta-analysis employing data sets that had been diagnosed and analyzed separately at different institutions and deposited in various databases.
Our PCA analyses revealed that N samples from each organ showed distinct DNA methylation profiles differing from those of N samples from other organs (Figure 1). Two possibilities can be considered to account for these findings: (a) Epigenetic information generally shows heterogeneity depending on individual cell lineages, tissues and organs (34,35), and differences in the DNA methylation profiles of the present N samples (Figure 1A) may reflect such organ-specificity. (b) We and other groups have shown that N samples obtained from patients with cancers may already be at the precancerous stages with DNA methylation alterations (6–10). The present N samples may already have been affected by carcinogenetic background factors, such as cigarette smoking or chronic obstructive pulmonary disorder in the lung (9,10).
In order to confirm that N samples may already be at the precancerous stage with DNA methylation alterations (6–10) [i.e. DNA methylation field defects (36)], previously obtained Infinium data for 36 normal (control [C]) samples of lung tissue obtained from patients without any lung tumor (9,10), 29 normal C samples of renal cortex tissue obtained from patients without any renal tumor (14), and 36 normal C samples of liver tissue obtained from patients without hepatitis virus infection, chronic hepatitis, liver cirrhosis or any liver tumor were included in the present analysis. We identified 3292, 978 and 9314 probes that were aberrantly methylated in N samples relative to C samples (Welch’s t test [FDR, q = 0.01]) from the lung, kidney and liver, respectively (Supplementary Table 9, available at Carcinogenesis Online), indicating that the DNA methylation field defects had been established in the N samples of the present cohort.
Figure 1A shows differences in DNA methylation profiles at the precancerous stage that reflect differences in carcinogenetic background among various organs. In particular, heterogeneity of the type, severity and history of carcinogenetic factors, such as H.pylori infection and chronic atrophic gastritis in the stomach (8) and hepatitis B or C virus infection (16) and non-alcoholic steatohepatitis in the liver (37), may result in more obvious intra-organ heterogeneity in N samples taken from these organs (Figure 1A): 423 and 3642 genes, for which DNA methylation levels in N samples from patients with gastric adenocarcinomas were significantly correlated with H.pylori infection and intestinal metaplasia (intestinal metaplasia reflects the long history of H.pylori infection, subsequent chronic active gastritis and atrophic gastritis), respectively, are summarized in Supplementary Table 10, available at Carcinogenesis Online.
Analysis of the DNA methylation profiles of N and T samples taken from each individual organ (Figure 1B) showed differences between the two sample types, indicating that DNA methylation alterations are associated with progression from the precancerous N stage to the T stage. On the other hand, since we have reported that DNA methylation profiles at the precancerous stage are inherited by cancers established in the same patient (8–10,14–16), it is feasible that some of the N samples would have shown DNA methylation profiles similar to those of T samples (Figure 1B).
In addition to carcinogenetic factors that might disturb DNA methylation profiles even in N samples, numerous alterations such as chromosomal instability or mutations of genes encoding epigenetic regulators (38–40) would further disturb the DNA methylation status in T samples. This might explain why intra-organ heterogeneity of T samples in Figure 1C was more obvious than that of N samples in Figure 1A. Intra-organ heterogeneity of DNA methylation profiles in T samples may reflect and/or determine the heterogeneity of clinicopathological aggressiveness of each tumor in individual patients. When we focused on various clinicopathological parameters, DNA methylation levels in T samples for the 396, 251, 2532, 1189 and 1475 probes were significantly correlated with pathological TNM stages in patients with lung adenocarcinomas, gastric adenocarcinomas, clear cell renal cell carcinomas, breast cancers and hepatocellular carcinomas, respectively, indicating that such DNA methylation abnormalities participate in malignant progression (Supplementary Table 11, available at Carcinogenesis Online). DNA methylation levels in T samples for the 1331, 62 and 761 probes were significantly correlated with smoking history, H.pylori infection and viral status (hepatitis B virus positive or hepatitis C virus positive) in patients with lung adenocarcinomas, gastric adenocarcinomas, and hepatocellular carcinomas, respectively, indicating that such carcinogenetic factors can induce distinct DNA methylation profiles in cancers (Supplementary Table 11, available at Carcinogenesis Online).
The present single-institutional multiple organ analysis clearly showed that DNA methylation profiles of T samples were more similar to those of N samples in the same organ than to T samples in other organs (Figure 1D). Again, two possibilities can be considered to explain this: (i) Organ-specific DNA methylation profiles may be shared by N and T samples. (ii) The impacts of different carcinogenetic factors in each organ may overwhelm the common characteristics shared by cancers of multiple organs.
Next, we identified the probes in which the DNA methylation status of T samples was significantly altered relative to that in N samples (Supplementary Table 5, available at Carcinogenesis Online): The number of probes showing DNA methylation alterations in T samples (FDR, q = 0.01 and more than 0.1 or less than −0.1 of ΔβT-N) was relatively small for the kidney, but large for the liver. Although so-called “overall DNA hypomethylation and regional DNA hypermethylation” (i.e. a larger number of probes showing DNA hypomethylation) (1,2) was observed for gastric adenocarcinomas, clear cell renal cell carcinomas and hepatocellular carcinomas, the number of probes showing DNA hypermethylation in T samples from the lung and breast was larger than that of probes showing DNA hypomethylation, in accordance with previously published reports of methylome analysis for lung adenocarcinomas (41).
As shown in Figure 2, only 6.9% (267/3857, between gastric adenocarcinomas and clear cell renal cell carcinomas) to 35.4% (1451/4099, between lung adenocarcinomas and breast cancers) of aberrantly methylated probes were shared between cancers of any two organs, indicating that the majority of DNA methylation alterations were diverse among multiple organs, possibly reflecting differences in carcinogenetic background factors (Figure 2, Supplementary Table 5, available at Carcinogenesis Online).
In contrast to such diversity of DNA methylation alterations, a small number of probes commonly showed DNA methylation alterations in cancers of multiple organs. We considered that analysis of such commonly shared DNA methylation alterations might shed light on common processes occurring at an early stage of carcinogenesis in all organs, which are not affected by differences in carcinogenetic factors. We then identified the genes commonly showing aberrant DNA methylation in cancers of multiple organs (Supplementary Table 6, available at Carcinogenesis Online).
Gastric adenocarcinomas were divided into two groups: one group showed DNA methylation aberrations of a larger number of the probes shared by the five organ cancers and listed in Supplementary Table 6, available at Carcinogenesis Online, whereas the other group showed DNA methylation aberrations of a smaller number of the probes (biphasic distribution in Supplementary Figure 2A, available at Carcinogenesis Online). However, the majority of lung adenocarcinomas, clear cell renal cell carcinomas, breast cancers and hepatocellular carcinomas showed DNA methylation aberrations on many probes shared by the five organ cancers and did not show such a biphasic distribution (Supplementary Figure 2A, available at Carcinogenesis Online). Although the cancer-free and overall survival rates of patients with gastric adenocarcinomas harboring DNA methylation aberrations on a larger number of the probes were not different from those of patients harboring them on a smaller number of the probes, accumulation of DNA methylation aberrations was significantly correlated with poorer prognosis of patients with lung adenocarcinomas and clear cell renal cell carcinomas (Supplementary Figure 2B, available at Carcinogenesis Online).
GO enrichment analysis clearly showed that DNA hypermethylation shared by cancers of multiple organs was enriched among “transcriptional factors” participating in early development and/or cell differentiation (Tables 1 and 2). In fact, the majority of genes included among “transcriptional factors” (P = 5.003 × 10–35, Table 1) and the top pathway, “multicellular organismal development” (GO: 0007275) (P = 9.259 × 10–30, Table 2), are so-called bivalent genes that paradoxically show a permissive histone mark (H3K4me3) and a repressive mark (H3K27me3) in ES cells (31–33) (Table 3). It is well known that bivalent histone modification is resolved during ES cell differentiation, and that distinct DNA methylation profiles of genes participating in development are established in terminally differentiated cells, such as epithelial cells, in adult organs (42). Although DNA methylation alterations of bivalent genes have already been reported in cancers (43), the prominent enrichment evident among multiple organs revealed by the present large single-institutional analysis suggests the universal significance of such alterations of genes involved in development and differentiation during carcinogenesis.
As shown in Supplementary Table 12, available at Carcinogenesis Online, DNA methylation alterations occurred in N samples in comparison with C samples even on the multiple genes listed in Table 3. Moreover, Jonckheere–Terpstra trend test revealed that the DNA methylation alterations in N samples in comparison with C samples were inherited by or strengthened in T samples for all the genes listed in Table 3, indicating that DNA methylation alterations of these genes was actually associated with cancer development, even when DNA methylation field defects were taken into consideration (Supplementary Table 12, available at Carcinogenesis Online). DNA methylation alterations listed in Table 3 may not be passenger changes but in fact an essential event during carcinogenesis, since their incidence was commonly high in cancers of various organs and such alterations were prominently accumulated in specific biological processes.
Moreover, our pyrosequencing and quantitative RT-PCR of representative genes, such as ALX1 and ATP8A2 reportedly involved in development (44–46) and ALX1 and CR1 reportedly involved in carcinogenesis (47,48), revealed an inverse correlation between the DNA methylation and mRNA expression levels of such genes. In addition, analyses of the correlation between DNA methylation and mRNA expression levels for all four genes were performed using data for 2,025 samples of N and T deposited in the TCGA database (445 242, 317 790 and 231 samples of the lung, stomach, kidney, breast and liver). A significant inverse correlation for each gene was observed in one to five organs (Supplementary Figure 3, available at Carcinogenesis Online). These facts indicate that aberrant DNA methylation of commonly methylated genes would actually result in silencing of such genes listed in Supplementary Table 6, available at Carcinogenesis Online. In addition, many of the commonly methylated genes are reportedly silenced due to DNA hypermethylation (Table 3). Thus the available data suggest that DNA methylation alterations shared by cancers of multiple organs may commonly disrupt the differentiated status of epithelial cells, from which various cancers originate, via alteration of gene expression levels.
Examination of clinicopathological impact revealed that reduced expression of ATP8A2 was significantly correlated with parameters reflecting tumor aggressiveness, such as lymphatic invasion and blood vessel invasion in gastric adenocarcinomas, and higher histological grade in clear cell renal cell carcinomas (Supplementary Table 13, available at Carcinogenesis Online), indicating the possibility that reduced ATP8A2 expression is crucial even for malignant progression.
On the other hand, DNA hypomethylation of the MMP26 gene did not result in an elevation of mRNA expression (Figure 3). Next, analysis of the correlation between DNA methylation and mRNA expression levels was performed for all 108 probes (69 and 39 probes showing DNA hypomethylation and DNA hypermethylation, respectively) shared by cancers of five organs using data deposited in the TCGA database for the 2025 samples, as shown in Supplementary Figure 3, available at Carcinogenesis Online. We found an inverse correlation between the DNA methylation and mRNA expression levels of 28 (45.2%) of 62 genes showing DNA hypomethylation in the present study and deposited in the public database, whereas an inverse correlation between 29 (93.5%) of 31 genes showing DNA hypermethylation was observed in one or more organs, suggesting that DNA hypermethylation of the probes shared by cancers of these five organs showed a more obvious tendency to result in aberrant expression in comparison with DNA hypomethylation.
Both genetic and epigenetic alterations are generally considered to play a critical role in multistage carcinogenesis. In addition, we and other groups have shown that, at the precancerous stage, DNA methylation alterations frequently precede genetic alterations (1,4,10,49). Recently, through introduction of the APC, KRAS, SMAD4, TP53 and PIK3CA genes into cultured organoids derived from normal intestinal epithelial cells, Matano et al. have shown that driver mutations are essential for growth, but alone are insufficient for malignant progression to tumors with metastatic ability, molecular lesions other than driver mutations being necessary (50). Common and early participation of DNA methylation alterations during carcinogenesis in various organs may disrupt the differentiated state of precancerous cells, thus resulting in an abnormally differentiated and/or de-differentiated state that may predispose cells to transformation induced by subsequent driver mutations.
Funding
This study was supported by the Program for Promotion of Fundamental Studies in Health Sciences (10–42) of the National Institute of Biomedical Innovation (NiBio) and partially supported by the Advanced Research & Development Programs for Medical Innovation (AMED-CREST, 15gm0510003h0405), Emerging/Re-emerging Infectious Diseases Project of Japan (15fk0210034h0001) and Practical Research for Innovative Cancer Control (15ck0106023h0002) from the (Japan Agency for Medical Research and Development AMED) and Grants-in-Aid for Scientific Research (C) (16K08720) from the Japan Society for the Promotion of Science (JSPS). The National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund (26-A-1).
Conflict of Interest Statement: None declared.
Supplementary Material
Abbreviations
- C
control
- ES
embryonic stem
- FDR
false discovery rate
- GO
gene ontology
- N
non-cancerous tissue
- PCA
principal component analysis
- RT
reverse transcription
- T
cancerous tissue
References
- 1. You J.S., et al. (2012) Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell, 22, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baylin S.B., et al. (2011) A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer, 11, 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahuja N., et al. (2016) Epigenetic therapeutics: a new weapon in the war against cancer. Annu. Rev. Med., 67, 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kanai Y. (2010) Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci., 101, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Arai E., et al. (2010) DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics, 2, 467–481. [DOI] [PubMed] [Google Scholar]
- 6. Klebaner D., et al. (2016) X chromosome-wide analysis identifies DNA methylation sites influenced by cigarette smoking. Clin. Epigenetics, 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Planello A.C., et al. (2016) Pre-neoplastic epigenetic disruption of transcriptional enhancers in chronic inflammation. Oncotarget, 7, 15772–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamanoi K., et al. (2015) Epigenetic clustering of gastric carcinomas based on DNA methylation profiles at the precancerous stage: its correlation with tumor aggressiveness and patient outcome. Carcinogenesis, 36, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato T., et al. (2014) Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: Its correlation with smoking history and chronic obstructive pulmonary disease. Int. J. Cancer, 135, 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato T., et al. (2013) DNA methylation profiles at precancerous stages associated with recurrence of lung adenocarcinoma. PLoS One, 8, e59444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Soto J., et al. (2016) The impact of next-generation sequencing on the DNA methylation-based translational cancer research. Transl. Res., 169, 1–18.e1. [DOI] [PubMed] [Google Scholar]
- 12. Tian Y., et al. (2014) Prognostication of patients with clear cell renal cell carcinomas based on quantification of DNA methylation levels of CpG island methylator phenotype marker genes. BMC Cancer, 14, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nagashio R., et al. (2011) Carcinogenetic risk estimation based on quantification of DNA methylation levels in liver tissue at the precancerous stage. Int. J. Cancer, 129, 1170–1179. [DOI] [PubMed] [Google Scholar]
- 14. Arai E., et al. (2012) Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis, 33, 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arai E., et al. (2009) Genome-wide DNA methylation profiles in both precancerous conditions and clear cell renal cell carcinomas are correlated with malignant potential and patient outcome. Carcinogenesis, 30, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arai E., et al. (2009) Genome-wide DNA methylation profiles in liver tissue at the precancerous stage and in hepatocellular carcinoma. Int. J. Cancer, 125, 2854–2862. [DOI] [PubMed] [Google Scholar]
- 17. Gross A.M., et al. (2015) Analysis of matched tumor and normal profiles reveals common transcriptional and epigenetic signals shared across cancer types. PLoS One, 10, e0142618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sánchez-Vega F., et al. (2015) Pan-cancer stratification of solid human epithelial tumors and cancer cell lines reveals commonalities and tissue-specific features of the CpG island methylator phenotype. Epigenetics Chromatin, 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J., et al. (2016) Pan-cancer methylation and expression profiling of adenocarcinomas revealed epigenetic silencing in the WNT signaling pathway. Neoplasma, 63, 208–214. [DOI] [PubMed] [Google Scholar]
- 20. Noushmehr H., et al. (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell, 17, 510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brocks D., et al. (2014) Intratumor DNA methylation heterogeneity reflects clonal evolution in aggressive prostate cancer. Cell Rep., 8, 798–806. [DOI] [PubMed] [Google Scholar]
- 22. Kanai Y., et al. (2014) Multilayer-omics analyses of human cancers: exploration of biomarkers and drug targets based on the activities of the International Human Epigenome Consortium. Front. Genet., 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flejou J. F., et al. (2009) Gastric Carcinoma. World Health Organization classification of tumours. Pathology and Genetics of Tumours of the Digestive System. IARC Press, Lyon. [Google Scholar]
- 24. Eble J.N., et al. (2004) Renal Cell Carcinoma. World Health Organization classification of tumours. Pathology and Genetics of Tumuors of the Urinary System and Male Genital Organs. IARC Press, Lyon. [Google Scholar]
- 25. Colby T.V., et al. (2011) Adenocarcinoma. World Health Organization classification of tumours. Pathology and genetics of tumours of the lung, pleura, thymus and heart. IARC Press, Lyon. [Google Scholar]
- 26. Ellis I.O., et al. (2011) World Health Organization Classification of Tumours of the Breast. IARC Press, Lyon. [Google Scholar]
- 27. Hirohashi S., et al. (2009) Hepatocellular carcinoma. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of the Digestive System. IARC Press, Lyon. [Google Scholar]
- 28. Sobin LH., et al. (2010) International Union Against Cancer (UICC) TNM Classification of Malignant Tumours, 7 ed Wiley-Liss, New York. [Google Scholar]
- 29. Bibikova M., et al. (2009) Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics, 1, 177–200. [DOI] [PubMed] [Google Scholar]
- 30. Arai E., et al. (2014) Multilayer-omics analysis of renal cell carcinoma, including the whole exome, methylome and transcriptome. Int. J. Cancer, 135, 1330–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao X.D., et al. (2007) Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell, 1, 286–298. [DOI] [PubMed] [Google Scholar]
- 32. Pan G., et al. (2007) Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell, 1, 299–312. [DOI] [PubMed] [Google Scholar]
- 33. Ku M., et al. (2008) Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet., 4, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rakyan V.K., et al. (2008) An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs). Genome Res., 18, 1518–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Irizarry R.A., et al. (2009) The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet., 41, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Teschendorff A.E., et al. (2016) DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nat. Commun., 7, 10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J.H., et al. (2014) Epigenetic mechanisms underlying the link between non-alcoholic fatty liver diseases and nutrition. Nutrients, 6, 3303–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee J.J., et al. (2015) Targeted next-generation sequencing reveals high frequency of mutations in epigenetic regulators across treatment-naïve patient melanomas. Clin. Epigenetics, 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huether R., et al. (2014) The landscape of somatic mutations in epigenetic regulators across 1,000 paediatric cancer genomes. Nat. Commun., 5, 3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Plass C., et al. (2013) Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat. Rev. Genet., 14, 765–780. [DOI] [PubMed] [Google Scholar]
- 41. Selamat S.A., et al. (2012) Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res., 22, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bernstein B.E., et al. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell, 125, 315–326. [DOI] [PubMed] [Google Scholar]
- 43. Ohm J.E., et al. (2007) A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat. Genet., 39, 237–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uz E., et al. (2010) Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am. J. Hum. Genet., 86, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gordon D.F., et al. (1996) Human Cart-1: structural organization, chromosomal localization, and functional analysis of a cartilage-specific homeodomain cDNA. DNA Cell Biol., 15, 531–541. [DOI] [PubMed] [Google Scholar]
- 46. Onat O.E., et al. (2013) Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur. J. Hum. Genet., 21, 281–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yuan H., et al. (2013) ALX1 induces snail expression to promote epithelial-to-mesenchymal transition and invasion of ovarian cancer cells. Cancer Res., 73, 1581–1590. [DOI] [PubMed] [Google Scholar]
- 48. Maurya S.K., et al. (2012) Genetic aberrations in gallbladder cancer. Surg. Oncol., 21, 37–43. [DOI] [PubMed] [Google Scholar]
- 49. Sawan C., et al. (2008) Epigenetic drivers and genetic passengers on the road to cancer. Mutat. Res., 642, 1–13. [DOI] [PubMed] [Google Scholar]
- 50. Matano M., et al. (2015) Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med., 21, 256–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.