Abstract
Assessing the functional impacts of somatic mutations in cancer genomes is critical for both identifying driver mutations and developing molecular targeted therapies. Currently, it remains a fundamental challenge to distinguish the patterns through which mutations execute their biological effects and to infer biological mechanisms underlying these patterns. To this end, we systematically studied the association between somatic mutations in protein-coding regions and expression profiles, which represents an indirect measurement of impacts. We defined mutation features (mutation type, cluster and status) and built linear regression models to assess mutation associations with mRNA expression and protein expression. Our results presented a comprehensive landscape of the associations between mutation features and expression profile in multiple cancer types, including 62 genes showing mutation type associated expression changes, 21 genes showing mutation cluster associations and 51 genes showing mutation status associations. We revealed four characteristics of the patterns that mutations impact on expression. First, we showed that mutation type (truncation versus amino acid-altering mutations) was the most important determinant of expression levels. Second, we detected mutation clusters in well-studied oncogenes that were associated with gene expression. Third, we found both similarities and differences in association patterns existed within and across cancer types. Fourth, although many of the observed associations stay stable at both mRNA and protein expression levels, there are also novel associations uniquely observed at the protein level, which warrant future investigation. Taken together, our findings provided implications for cancer driver gene prioritization and insights into the functional consequences of somatic mutations.
Keywords: somatic mutation, mutation cluster, mutation type, cancer gene, gene expression
Introduction
Cancer is a genetic disease where genomic abnormalities directly or indirectly alter gene expression, protein activities and signaling pathways that ultimately contribute to cell proliferation and survival [1, 2]. An important task of cancer research is to identify driver mutations that confer growth advantage [3]. To date, many computational approaches have been developed to prioritize driver mutations using a variety of characteristics of somatic mutations, such as evolutionary conservation information of the mutation sites in multiple species [4], sequence context [5], occurrence frequency [6] and impacts of mutations on transcriptome [7], among others. With the accumulation of multidimensional omics data, studies on mutation impact are drawing increasing attention through integrative approaches [8]. Some examples include the study of the overall expression of downstream genes of the candidate mutation in molecular networks [7], the connection between genomic mutations and transcriptomic changes [9] or the overall impacts on pathways [10]. Although all these methods were demonstrated as effective, it remains critical to distinguish the patterns how mutations exert their impacts and to link these patterns with possible underlying biological interpretation. For example, some mutations introduce premature stop codon and lead to reduced dosage of mRNA transcripts, and some affect protein activities through changing amino acid sequences. To address this challenge, we approached mutation impacts by assessing whether a mutation impacts its residing gene before it expands its impacts, if any, on the expression of its neighborhood genes in the transcriptome, pathways or networks.
Mapping the associations between mutations and their residing genes presents several analytical challenges. First, the mRNA dosage, and in turn the protein concentration, in cancer cells are regulated at multiple levels with different mechanisms, involving regulatory variants [11], promoter methylation, copy number variation (CNV) and copy-neutral loss of heterozygosity (LOH), among others. In particular, for single nucleotide variants (SNVs) and short insertions and deletions (indels) detected in only cancer cells (i.e. somatic mutations), their association with gene expression is not fully understood yet. Second, the biological interpretation for how somatic mutations impact its residing genes is diverse. mRNAs carrying premature stop codon, which can be introduced by truncation mutations (NS), are typically eliminated by the process called nonsense-mediated mRNA decay (NMD), and thus, both the concentration of mRNA transcripts and protein products would be decreased owing to NS [12]. However, if a stop-gain mutation occurs outside of the NMD-target regions, e.g. within 50 bp from the 3′ end of the transcripts [13], mRNAs carrying such mutations may escape NMD. Missense mutations (MS), which comprise another major group of mutations in cancer, typically change the amino acid sequences of proteins and affect the activities of proteins in which they are located (e.g. catalytic efficiency [14, 15], receptor activity [16], phosphorylation [17]). Previous studies have shown that MS could impact other genes’ expression or activities in the same pathways [18]; however, their association with the dosage of their residing genes remains unexplored. Third, for the purpose of identifying driver mutations, lack of an association between a candidate mutation and its coding genes does not necessarily rule out the possibility of the mutation being a driver but may also imply that other mechanisms could exist through which mutations execute their impacts. For example, mutations in DNA repair genes mainly lead to accumulation of somatic mutations in cancer cells, and such impacts may not be well captured through association studies with transcriptomic data.
In this work, we systematically studied the functional footprints of somatic mutations obtained via whole-exome sequencing (WES) in 12 cancer types using the data generated by The Cancer Genome Atlas (TCGA) project [19]. We focused on protein-coding mutations. To overcome the limitation of many low-frequency mutations in the data set, we defined three mutation features and grouped the mutations accordingly; these features are mutation type (MS versus NS), mutation cluster (mutations clustered in a short region, in domains or in 3D space) and mutation status [mutated versus wild type (WT)]. Grouping mutations according to these features provides a natural way of distinguishing the patterns through which mutations impact expression profile, as mutations of the same group are assumed to have similar mechanisms. We asked whether these features of somatic mutations are associated with transcriptional levels (measured by RNA-sequencing, mRNA level) and posttranscriptional modifications [measured by the reverse phase protein array (RPPA) platform, protein level]. In particular, we asked for each gene, whether it is differentially expressed in its mutant samples compared with its WT samples and if the answer is yes, whether such an association is distinguishable among different mutation clusters or different mutation types. Our results showed that among the three features, mutation type had the strongest influence on transcriptional level, especially in tumor suppressor genes by NS. Mutation clusters were detected in well-studied genes, as expected, and yet, they were only found in a small number of genes. Both similarities and differences were observed with these mutation clusters within and across cancer types, such as the same clusters recurrent in multiple cancer types (e.g. KRAS), different clusters in the same genes found in different cancer types (e.g. EGFR) and consistent or inconsistent directions with transcriptional levels. Examination of mutation features with protein expression levels, which represent mutations’ ultimate impacts on cellular signaling processes, confirmed some of the associations detected at the mRNA level and also revealed novel associations that were uniquely observed at the protein level such as ARID1A, BAP1, CTNNB1 and ERBB3. In summary, this study unraveled the effects of somatic mutation features on mRNA and protein expression, providing implications in the design of cancer gene prioritization.
Results
Overview of the data and mutation features
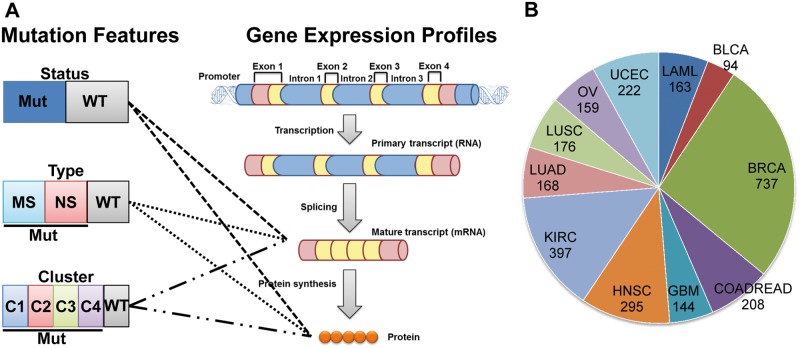
Multi-domain omics data sets from TCGA were downloaded for 12 cancer types [19]: acute myeloid leukemia (conventionally called AML; we followed the abbreviation in the original publication of LAML [19]), bladder urothelial carcinoma (BLCA), breast adenocarcinoma (BRCA), colon and rectal carcinoma (COADREAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous carcinoma (OVCA) and uterine corpus endometrial carcinoma (UCEC). Unless stated specifically, for each cancer type, we obtained somatic mutations, CNV, copy-neutral LOH (GBM and OVCA), methylation, tumor purity (excluding LAML), mRNA expression and protein expression (excluding LAML). For LAML, there is no available RPPA data, so we only performed the association test using mRNA expression data. For OVCA, there is only one significant gene, TP53. Accordingly, we did not discuss further details because TP53 was also detected in other cancer types.
To identify the eligible genes for our follow-up analyses, we first required genes mutated in a sufficient number of samples, i.e. a minimum of five samples in each respective feature group. We defined three types of mutation features: mutation type, mutation cluster and mutation status (Figure 1). For mutation type, we categorized all SNVs and small indels with amino acid changes into MS (including missense SNVs and in-frame indels) and NS (including nonsense SNVs, frame-shift indels and splice site SNVs and splice site indels). We used WT (no SNV or indel in the prospective gene) for comparison. We required the genes eligible for mutation type analysis as those that had at least one mutation type (MS or NS mutations) occurred in at least five samples. As shown in Supplementary Table S1, the number of eligible genes for mutation type analysis varied and could be several hundred in each cancer type. For mutation clusters, we grouped those that were located no more than 5 amino acids (AAs) apart from each other in their protein sequences. The distance of 5 AAs was defined arbitrarily. We explored other distance units, such as 7 AAs and 10 AAs, and found minor difference in the eligible genes (Supplementary Table S2). Thus, we chose 5 AAs as the threshold throughout this work. Using other definitions of mutation clusters is also optional [20]. Surprisingly, the number of eligible genes for mutation cluster analysis was quite moderate in each cancer type, ranging between 3 and 17 (Supplementary Table S1). This observation was unexpected for two reasons. First, numerous previous studies have reported highly recurrently mutated genes in various cancer types [21, 22], and we initially expected that there would have been more genes eligible for the cluster analysis. Second, our way of defining mutation clusters includes mutation hotspots (e.g. the R132 mutation in IDH1 and the G12 mutation in KRAS), which are frequently reported in oncogenes. Accordingly, we would have expected more genes eligible for our mutation cluster analysis. We thus explored the potential reasons that prevented recurrent genes from the inclusion in our clusters. We found that most of the recurrent genes had their mutations scattered across their protein sequences and failed to form any mutation clusters within a short distance of several AAs (e.g. 5 AAs, 7 AAs or 10 AAs, Supplementary Table S2). In this work, we only focused on genes with a high frequency of clustered mutations; thus, many of the recurrent genes that had non-clustering mutations were excluded in our analysis. Finally, for mutation status, we defined two groups, mutated or WT. Because we were more interested in testing whether the impacts of mutations on expression were mediated through mutation types or mutation clusters, we required eligible genes for the mutation status test as those that were eligible for at least one of the mutation type tests and mutation cluster tests (Figure 1).
Figure 1.
Illustration of association studies of mutation features with expression in human cancer. (A) Schematic description of the three mutation features and analysis steps. We defined mutation types, mutation clusters and mutation statuses as three independent mutation features. For each gene, its WT samples remained the same in all three features, whereas its mutated samples (Mut status) were further separated as NS or MS in the feature of mutation type, or categorized into multiple cluster groups (C1, C2, C3 and C4 in the feature of mutation cluster). For gene expression, we considered mRNA expression measured by RNA sequencing data and protein expression measured by the RPPA platform. (B) Pie chart shows the number of samples used in each cancer that had somatic mutation, RNA sequencing and copy number variation data in each sample. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
Furthermore, we used two additional ways to cluster mutations. First, we proposed to cluster mutations according to their locations in protein domains based on the PFAM database [23]. For each gene, we grouped the samples whose mutations occurred in the same protein domain and compared the mRNA expression of the gene in these samples with that in WT samples. For genes with multiple domains, we tested each domain independently. This strategy increased the number of eligible genes for the association test (Supplementary Table S2). Second, we proposed to cluster mutations according to their space locations in 3D protein structures (herein, named ‘space clustering’). Specifically, we extracted the residues reported in Kamburov et al. [24] that were closely located in the Protein Data Bank (PDB) chains (Euclidean distance < 7Å). For each gene, we tested whether its MS belonging to these close residues showed any association with gene expression.
Mutation features associated with mRNA expression
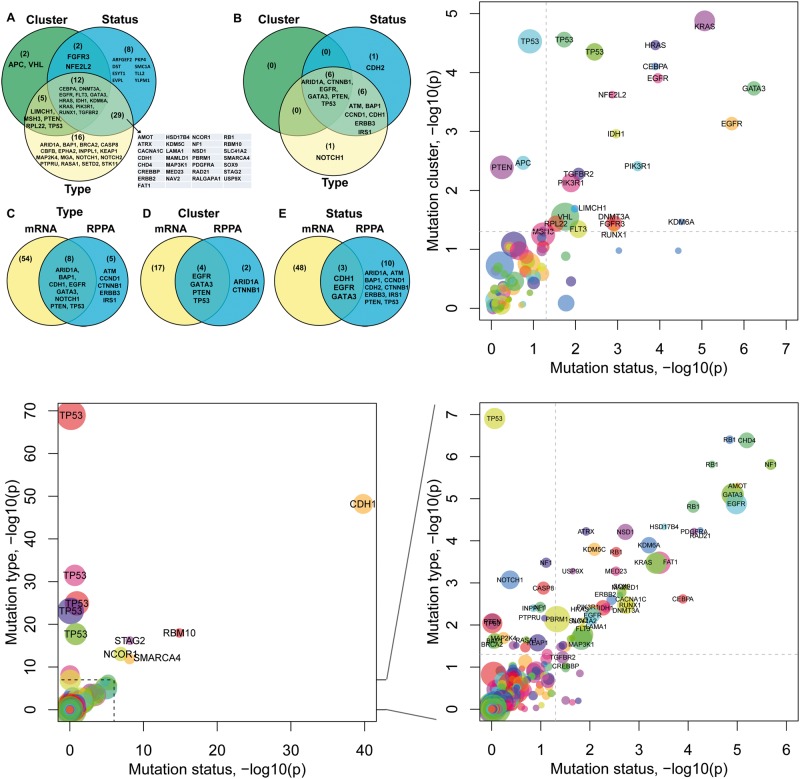
We aimed to learn the relationship between features of somatic mutations and measured expression levels by building multivariate linear regression (MLR) models in which mutations predict mRNA expression or protein expression. With MLR and assistance from Wilcoxon rank sum test for each particular feature group (e.g. a mutation type, a mutation cluster or a mutation status) versus the WT group samples, we conducted a systematic association test for each gene between its mutation features and its transcriptional changes obtained from RNA-sequencing in each of the 12 cancer types. Using false discovery rate (FDR) <0.05, we found 62 genes (76 times considering different cancer types) that had at least one mutation type significantly associated with their transcriptional changes, 21 genes (25 times) that had at least one cluster of mutations associated with their transcriptional changes and 51 genes (55 times) that were associated with mutation status (i.e. differentially expressed in mutated samples and the respective WT samples) (Figure 2). We, respectively, referred these three sets of genes as mutation type-associated genes (Supplementary Figure S1), cluster-associated genes (Supplementary Figure S2) and status-associated genes (Supplementary Figure S3).
Figure 2.
Overview of mutational impacts on gene expression. (A and B) Venn diagrams showing genes associated with each mutation feature based on their mRNA expression (A) and protein expression (B). (C–E) Comparison of genes associated with mutation types (C), mutation clusters (D) or mutation statuses (E) at the mRNA level and at the protein level, respectively. (F) Plot of the impact of mutation clusters versus the impact of mutation statuses. Each dot represents a gene in a cancer type. Node color indicates cancer type, e.g. TP53 was found in multiple cancer types. The x-axis and y-axis show the negative log10 P-values for each gene obtained from the regression models in which the mutation status of a gene predicts its mRNA expression (x-axis) or its mutation cluster predicts its mRNA expression (y-axis), respectively. The dash line indicates where P = 0.05. (G–H) Plots of the impact of mutation types versus the impact of mutation statuses (G), and its zoom-in view (H). The x-axis shows the negative log10 P-value of the regression model fitted for each gene in which the gene’s mutation status predicts its mRNA expression. The y-axis shows the negative log10 P-value obtained using a gene’s mutation type to predict its mRNA expression. P-values shown are after multiple testing correction. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
In all three tests, the WT samples, which were used as the reference group, remained the same for each gene. The mutated samples in the status test were the combination of NS and MS samples in the type test, and also the combination of samples with all mutation clusters (Figure 1). Thus, the type and cluster tests could be considered as a nested form of the status test with mutated samples being further categorized, and a comparison of the three sets genes would help reveal genes with specific association relationships with their mutations. As shown in Figure 2, 12 genes were shared by all three gene sets, i.e. their mRNA expression levels were associated simultaneously with mutation types, mutation clusters and mutation statuses (Figure 2A). Eight genes were only associated with mutation statuses but were neither associated with mutation types nor with mutation clusters, indicating that although their mRNA expression differed in mutated samples versus WT samples, such effects were not attributable to any particular mutation type or mutation cluster. We manually checked each of these eight genes and found no particular pattern, and thus, we chose not to follow them further. Sixteen genes were associated only with mutation types but not with mutation statuses. Examples of such genes include BAP1, EPHA2, KEAP1, NOTCH1, NOTCH2, SETD2 and STK11. One possible explanation to their failure in the status test is that these genes might be primarily impacted by one type of the mutations, e.g. NS, whereas the other type had no detectable association with their expression changes, and inclusion of all mutated samples as one group (i.e. mutation status test) weakened the potential association in the MLR analysis. Finally, two genes, APC and VHL, were associated with mutation clusters but not with mutation statuses, indicating that only certain clusters of mutations, but not all mutations, of these genes were correlated with mRNA expression changes. Put together, the observation of genes that were only associated with particular feature groups of mutations but not all mutations highlighted the importance to examine the effects of mutation features independently.
Two alternative ways to cluster mutations are protein domains and residue space locations. When grouping mutations by their locations in domains, we found 19 genes (27 times) that had one or more domains significantly associated with mRNA expression changes (FDR < 0.05, Supplementary Table S4). Nine of these genes were detected in our mutation cluster test aforementioned. When we grouped MS, which were closely located in space, we found 13 genes (19 times) that showed nominal association with mRNA expression changes (Supplementary Table S5). Because of the limited numbers of eligible genes, here we used nominal P-values instead of the adjusted values. A comparison of the significant genes identified by mutation clusters, protein domains and residue space locations was shown in Supplementary Figure S5. The overlapping genes by all three tests included several well-established cancer genes (e.g. EGFR, FGFR3, HRAS, KRAS, PIK3R1 and TP53), whereas the genes uniquely identified as associated with protein domain (CDH1, CHD4, IDH2, MAP3K1, MED23, MED24, RB1, SMC1A and TMCC3) or space locations (EP300, KEAP1 and TLR4) were also worth further investigation.
Assessment of potential confounding effects
As has been widely reported, cancer samples frequently acquire genetic and epigenetic alterations, which in turn influence gene expression through various mechanisms. To validate the associations that we observed between mutation features and gene expression, we assessed other potential confounding factors including CNV, methylation status, tumor purity (excluding LAML) and copy-neutral LOH (GBM).
Effects of CNV
To control the potential confounding effects by CNV on gene expression levels, we conducted three types of analyses to fully assess the impact of CNVs in our results. First, we used CNV as a covariate for all our association tests for mutation type and mutation status. Applying this model, we found that for the 55 significant events in mutation status test, except one gene that was removed owing to lack of CNV data, all the remaining 54 events were nominally significant and 46 of them remained significant at FDR < 0.05. For the 23 significant events (MS) and 55 significant events (NS) in the mutation-type test, all remained nominally significant and after multiple testing correction, 21 (91.3%) remained significant for MS and 38 (69.1%) for NS.
Second, we restricted the association test in the subset of samples with the most prevalent copy number status. For the majority of genes, their most prevalent status is copy number neutral; for a few number of genes, the most prevalent status is copy number gain or copy number loss. We made this adjustment, instead of using copy number neutral for all genes, in order that the largest subset of samples could be analyzed. This restricted test reduced sample size and, thus, many genes were no longer eligible for the test. Applying this model, we found that for the 55 significant events in mutation status test, after removing one gene with no CNV data, 43 (79.6%) of them remained nominally significant and 28 (51.9%) were significant at FDR < 0.05. For the 23 significant events for MS in mutation-type test, 18 were eligible for the test (requiring the most prevalent CNV status occurred in ≥8 samples), all remained nominally significant and after multiple testing correction, 14 (77.8%) remained significant. Similarly, for the 55 significant events for NS, 39 were eligible for the controlled test and all were nominally significant, including 38 (97.4%) significant ones after multiple testing correction.
Third, we used linear regression to obtain the residual expression of copy number status and used the residual expression for the association test. For the 54 significant events in mutation status test excluding the one with missing CNV, all remained nominally significant and 45 (83.3%) stayed significant after multiple testing correction. For the mutation cluster test, 24 of the 25 (96%) significant events remained significant (FDR < 0.05). For the 23 significant events (MS) and 55 significant events (NS) in mutation type test, 21 (91.3%) remained significant for MS and 54 (98.2%) for NS (FDR < 0.05). Collectively, these results suggested that the majority of our findings (>80%) on mutation feature association were also significant after accounting for CNV, indicating that these associations with mutation features were beyond the impact of CNVs.
Effects of methylation
Similar to our analysis strategy for CNV, we conducted two types of analyses to control the effect of methylation. First, we used methylation status as a covariate in all our association tests. Second, we used linear regression to obtain the residual expression of methylation status and used the residual expression in the association test. As shown in Supplementary Table S6, both strategies showed that methylation status had generally minor impact on our association results. Our initial screen showed that the majority of genes eligible for the association tests did not have methylation data. Because the methylation data set we used only selected 2203 probes that were methylated in a sufficient number of samples (>10% of any of the tumor types or 50% of any of the well-defined subtypes), genes with no methylation data in this data set indicated that the methylation statuses of these genes showed no substantial variation across samples. More specifically, for those genes from our list of significant genes that had methylation data in the 2203 probes, they remained significant after we controlled the effects by methylation status (Supplementary Table S6).
Control for tumor purity
Tumor cellularity and purity are confounding factors that might influence gene expression changes. We conducted the association analyses by controlling the tumor purity for the cancer types excluding LAML. As a result, for the significant genes that we detected to be associated with their mutation features, the majority of them remained significant after adjusting tumor purity: 43/49 = 87.8% in mutation status test, 20/21 = 95.2% in mutation cluster test, 17/21 = 81.0% in mutation-type test for MS and 46/52 = 88.5% in mutation-type test for NS, respectively. For the remaining genes that were no longer in the significant gene lists after adjusting tumor purity, we found that they were not eligible for the test because of the decreased small sample size. Taken together, we consider tumor purity had minor effects on our association results.
Control for copy-neutral LOH
Similarly, for copy-neutral LOH status that may have an impact on gene expression, we included LOH status as a covariate and conducted the association tests. We performed this analysis in GBM only because this is the only cancer type having a reasonable number of samples with LOH data. There were other data sets for 11 cancer types [22], but they were not made publicly available when this study was conducted. We did not observe dramatic changes in our detected association results. The significant genes still remained significant after adjusting for LOH status.
Mutation features associated with protein expression
To assess the association between mutation features and protein expression levels measured by the RPPA platform, we performed the MLR association test on the genes that are available with RPPA data. By using FDR < 0.05, we found 15 antibodies for 13 genes that had at least one mutation type significantly associated with their protein expression changes, 8 antibodies for 6 genes that had at least one cluster of mutations associated with their protein expression change and 15 antibodies for 13 genes that had differential protein expression levels in mutated samples versus WT samples (Figure 2). Some genes whose mRNA expression levels were significantly associated with mutation features were successfully validated at protein levels, including BAP1 (antibody: Bap1 − c − 4) and CDH1 (E − Cadherin) in mutation type and status tests, EGFR (EGFR, EGFR_pY1068, and EGFR_pY1173) in all three tests, NOTCH1 (Notch1) in mutation-type test and PTEN (PTEN) and TP53 (p53) in mutation cluster and type tests.
We next studied the genes that were not consistently identified at the mRNA and protein expression levels with details. Notably, the RPPA platform for protein expression levels only provided measurements for 187 antibodies targeting ∼150 genes. Thus, many genes that were detected at the mRNA level were not available for protein expression test, partially explaining the dramatic decrease of the number of associated genes from the mRNA level to the protein level. Furthermore, there are often posttranscriptional regulation mechanisms that place additional layers of control on mRNA and protein expression, such as microRNA regulation, but were not examined in our work.
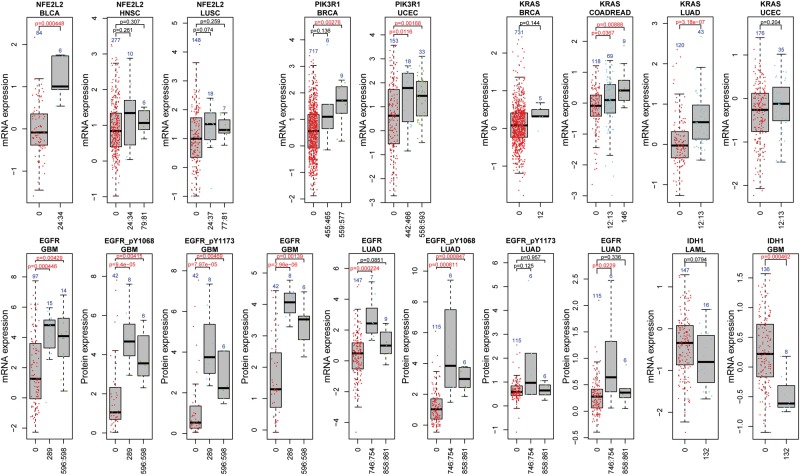
On the contrary, for genes that were only detected at the protein level but were missed by the mRNA expression tests, we sought for the potential explanations with particular interests. There were 5 genes in the mutation type test, 2 genes in the cluster test and 10 genes in the status test, which were uniquely significant at the protein level (Figure 1). We use ARID1A and CTNNB1 as examples to illustrate their mRNA and protein expression changes. ARID1A (AT-rich interactive domain 1A) encodes a member of the SWI/SNF family, which regulates gene transcription through altering chromatic structures. ARID1A has been recently identified as a tumor suppressor gene whose deficiency has been found in a broad spectrum of cancer types [25, 26]. As shown in Figure 3, samples with NS mutations of ARID1A turned to be substantially significant at the protein level compared with the mRNA level in BLCA (pmRNA = 0.18, pRPPA = 4.29 × 10−4, Wilcoxon test, nominal P-value), BRCA (pmRNA = 0.16, pRPPA = 3.61 × 10−3), KIRC (pmRNA = 0.38, pRPPA = 0.02) and UCEC (pmRNA = 0.02, pRPPA = 5.11 × 10−9). As for mutation clusters, ARID1A had a cluster covering the amino acids between the 1324th and 1335th positions in its protein sequence that occurred in seven samples with decreased protein expression levels in UCEC (pmRNA = 0.06, pRPPA = 0.02), including three samples with nonsense SNVs and four with splice site SNVs. We inferred that the association between the observed ARID1A protein expression decrease and this cluster was attributable to the NS mutation type as discussed above, because all the seven samples had NS mutations. The gene CTNNB1 had only noticeable changes in UCEC and, surprisingly, it was the MS mutation type that showed decreased expression level (pmRNA = 0.10, pRPPA = 2.74 × 10−5; Supplementary Figure S4). The significant cluster in CTNNB1 was located between the 32th and 41th positions. No significant difference was observed in samples with mutations in that cluster compared with CTNNB1 WT samples at the mRNA expression level (pmRNA = 0.10); however, significant decrease was observed at the protein level (beta-catenin, pRPPA = 4.27 × 10−5, Supplementary Figure S4).
Figure 3.
Comparison of ARID1A expression level in each mutation feature group. (A) Mutation cluster plots on mRNA and protein expression. The x-axis shows the cluster index, where x = 0 indicates the wild type (WT) group and x = 27 indicates the 27th mutation cluster around 1324th–1335th amino acids. (B) Mutation type plots in eight cancers. The top panel showed ARID1A mRNA expression versus its three mutation types: NS, MS and WT. The bottom panel showed its protein expression versus the three mutation types. The P-values were calculated by Wilcoxon rank sum test between the NS group and the WT group. (C) Mutation status plots. Similarly to (B), the top and bottom panel showed ARID1A mRNA and protein expression, respectively, versus its mutation status. The P-values shown were obtained based on Wilcoxon rank sum test comparing the mutated group (Mut) and the WT group. In (B) and (C), we showed all eight cancer types in which ARID1A was eligible for the corresponding feature association test and highlighted those that were nominally significant (nominal P < 0.05). Sample size was labeled for each feature group. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
We found more similar genes to ARID1A and CTNNB1, such as ATM, CTNND1, ERBB3 and IRS1, which were uniquely associated with mutation types at the protein expression level but not at the mRNA expression level, and ATM, BAP1, CCND1, ERBB3, IRS1 and PTEN, which were uniquely associated with mutation statuses at the protein expression. Notably, for ERBB3, there are two antibodies showing the association: HER3 and HER3_pY1289. ERBB3 was not detected with differential expression at the mRNA level for its MS mutations (pmRNA = 0.82) but showed increased protein expression by one of its antibody HER3_pY1289 (pRPPA = 0.01), but not by the other antibody HER3 (pRPPA = 0.73). A full list of these genes is presented in Supplementary Figure S4.
Oncogenic genes and tumor suppressor genes are associated with different mutation features
Tumor suppressor genes have been reported to be frequently disrupted by NS, leading to the onset and progression of cancer [27], whereas proto-oncogenes are activated through multiple mechanisms in cancer [28]. We compared the gene sets obtained by each mutation feature test with tumor suppressor genes and oncogenes, respectively (Supplementary Table S3). The strongest overrepresentation was observed with mutation-type-associated genes, in which 45 (45/62 = 72.6%) were tumor suppressor genes. In particular, genes whose expression levels were specially associated with the truncation mutation type (n = 55, truncation-associated genes) formed the majority of the overlapping tumor suppressor genes (n = 41, 74.5%). On the contrary, MS-associated genes had comparable proportion of tumor suppressor genes (n = 6) and oncogenes (n = 8). Although expected, these results provide a systematic evaluation of the effects of NS on gene expression in multiple cancer types.
The tumor suppressor genes identified in our list of truncation-associated genes included RB1, TP53, NF1, PTEN, NOTCH1 and NOTCH2. Except NOTCH1/2 genes that were only detected in HNSC, all the other genes were recurrently decreased by NS in multiple cancer types. In addition to tumor suppressor genes, another major functional group of genes that were associated with NS were chromatin modification genes, such as ATRX, BAP1, CHD4, CREBBP, DNMT3A, IDH1, KDM5C, KDM6A, NSD1, PBRM1, SETD2 and SMARCA4, indicating that these genes may also function in cancer through inactivation mechanisms. Finally, MSH3, a DNA repair gene whose loss of function leads to increased mutation rates [29], was also found to be significantly associated with NS mutations.
Similarities and differences in association patterns among mutation clusters
The 21 genes with functional mutation clusters included well-studied cancer genes (Table 1). Three of them were recurrent in multiple cancer types: EGFR, PIK3R1 and TP53. The recurrent genes increased only by two when we relaxed FDR from 0.05 to 0.2; these additional genes were KRAS and PIK3CA. We choose to discuss several representative genes below (Figure 4).
Table 1.
Significant genes with mutation cluster associations
| Cancer type | Gene | Cluster index | Position (AA) a | Number of samples | Wilcoxon b |
Regression-based test
c
|
||
|---|---|---|---|---|---|---|---|---|
| P | Model p | t value | Pr(>|t|) | |||||
| LAML | CEBPA | 7 | 300::310 | 6 | 1.14 × 10−4 | 8.75 × 10−6 | 4.599 | 8.75 × 10−6 |
| LAML | DNMT3A | 17 | 882 | 21 | 0.018 | 7.98 × 10−3 | −2.691 | 7.98 × 10−3 |
| LAML | RUNX1 | 6 | 171:174 | 6 | 0.042 | 0.019 | 1.994 | 0.048 |
| LAML | FLT3 | 2 | 593::604 | 28 | 0.041 | 0.020 | 2.180 | 0.031 |
| LAML | FLT3 | 4 | 835:839 | 12 | 0.035 | 0.020 | 2.072 | 0.040 |
| BLCA | NFE2L2 | 1 | 24::34 | 6 | 4.48 × 10−4 | 3.96 × 10−5 | 4.328 | 3.96 × 10−5 |
| BLCA | KDM6A | 10 | 555 | 5 | 0.035 | 0.013 | −2.557 | 0.013 |
| BLCA | FGFR3 | 4 | 248:249 | 5 | 3.26 × 10−3 | 0.017 | 2.430 | 0.017 |
| BRCA | GATA3 | 4 | 329::335 | 10 | 7.37 × 10−5 | 1.86 × 10−5 | 2.914 | 3.67 × 10−3 |
| BRCA | GATA3 | 5 | 345::365 | 10 | 1.45 × 10−3 | 1.86 × 10−5 | 2.038 | 0.042 |
| BRCA | GATA3 | 7 | 395::410 | 17 | 7.33 × 10−5 | 1.86 × 10−5 | 2.806 | 5.16 × 10−3 |
| BRCA | GATA3 | 8 | 419::445 | 16 | 1.23 × 10−3 | 1.86 × 10−5 | 2.766 | 5.82 × 10−3 |
| BRCA | PIK3R1 | 3 | 455::465 | 6 | 0.136 | 7.74 × 10−4 | 2.045 | 0.041 |
| BRCA | PIK3R1 | 5 | 559::577 | 9 | 2.76 × 10−3 | 7.74 × 10−4 | 3.228 | 1.30 × 10−3 |
| COADREAD | TP53 | 9 | 194:195:196 | 6 | 1.04 × 10−3 | 1.63 × 10−6 | −4.423 | 1.74 × 10−5 |
| COADREAD | TP53 | 10 | 207:212:213 | 6 | 2.18 × 10−3 | 1.63 × 10−6 | −4.514 | 1.19 × 10−5 |
| COADREAD | APC | 3 | 213:216 | 11 | 8.10 × 10−5 | 4.00 × 10−4 | −4.318 | 3.13 × 10−5 |
| COADREAD | APC | 16 | 564:567 | 6 | 0.093 | 4.00 × 10−4 | −2.688 | 8.14 × 10−3 |
| COADREAD | TGFBR2 | 1 | 125 | 13 | 1.28 × 10−3 | 9.29 × 10−4 | −3.362 | 9.29 × 10−4 |
| COADREAD | MSH3 | 2 | 381 | 9 | 0.017 | 0.012 | −2.543 | 0.012 |
| GBM | EGFR | 9 | 289 | 15 | 4.46 × 10−4 | 1.84 × 10−4 | 3.490 | 6.73 × 10−4 |
| GBM | EGFR | 14 | 596:598 | 14 | 4.29 × 10−3 | 1.84 × 10−4 | 2.691 | 8.13 × 10−3 |
| GBM | IDH1 | 1 | 132 | 8 | 4.62 × 10−4 | 5.46 × 10−4 | −3.537 | 5.46 × 10−4 |
| HNSC | TP53 | 7 | 98::110 | 9 | 0.05 | 3.28 × 10−6 | −2.509 | 0.013 |
| HNSC | TP53 | 13 | 265::286 | 42 | 0.264 | 3.28 × 10−6 | 2.120 | 0.035 |
| HNSC | TP53 | 14 | 292::306 | 10 | 6.78 × 10−5 | 3.28 × 10−6 | −4.595 | 6.64 × 10−6 |
| HNSC | HRAS | 1 | 11:12:13 | 9 | 1.09 × 10−3 | 7.61 × 10−6 | 4.558 | 7.61 × 10−6 |
| KIRC | VHL | 2 | 39::45 | 5 | 0.814 | 9.33 × 10−3 | −2.143 | 0.033 |
| KIRC | VHL | 3 | 60::137 | 123 | 0.019 | 9.33 × 10−3 | 2.278 | 0.023 |
| KIRC | VHL | 4 | 143::189 | 62 | 0.053 | 9.33 × 10−3 | 2.430 | 0.016 |
| LUAD | KRAS | 1 | 12:13 | 43 | 3.18 × 10−7 | 2.21 × 10−6 | 4.911 | 2.21 × 10−6 |
| LUAD | EGFR | 4 | 746:751:754 | 7 | 2.24 × 10−4 | 4.16 × 10−5 | 4.436 | 1.70 × 10−5 |
| LUSC | TP53 | 10 | 151::163 | 19 | 9.77 × 10−4 | 1.11 × 10−5 | 3.559 | 5.05 × 10−4 |
| LUSC | TP53 | 11 | 172::183 | 13 | 4.56 × 10−4 | 1.11 × 10−5 | 3.910 | 1.42 × 10−4 |
| LUSC | TP53 | 12 | 193:195:196 | 6 | 0.044 | 1.11 × 10−5 | 2.158 | 0.033 |
| LUSC | TP53 | 14 | 234::252 | 24 | 4.62 × 10−5 | 1.11 × 10−5 | 4.433 | 1.84 × 10−5 |
| LUSC | TP53 | 15 | 259::287 | 27 | 2.63 × 10−3 | 1.11 × 10−5 | 3.458 | 7.16 × 10−4 |
| UCEC | PTEN | 12 | 165::182 | 8 | 0.050 | 2.39 × 10−4 | 1.981 | 0.049 |
| UCEC | PTEN | 15 | 233:237:240 | 12 | 8.48 × 10−4 | 2.39 × 10−4 | −3.678 | 3.04 × 10−4 |
| UCEC | PTEN | 20 | 335::344 | 8 | 1.25 × 10−3 | 2.39 × 10−4 | 3.252 | 1.35 × 10−3 |
| UCEC | PIK3R1 | 8 | 442::466 | 18 | 0.012 | 8.75 × 10−4 | 2.493 | 0.013 |
| UCEC | PIK3R1 | 12 | 558:593 | 33 | 1.68 × 10−3 | 8.75 × 10−4 | 3.108 | 2.16 × 10−3 |
| UCEC | LIMCH1 | 3 | 421 | 6 | 6.25 × 10−3 | 3.62 × 10−3 | −2.943 | 3.62 × 10−3 |
| UCEC | RPL22 | 1 | 16 | 23 | 2.19 × 10−3 | 8.61 × 10−3 | −2.652 | 8.61 × 10−3 |
aTwo continuous colons (::) indicate that there are multiple positions with mutations. AA: amino acid position.
bWilcoxon test was performed in the comparison of the samples having a single cluster with the wild type samples.
cRegression-based test was performed using all eligible clusters.
Figure 4.
Comparison of mutation clusters of the five genes in different cancer types. We showed similarities and differences in the impacts of mutation clusters of five representative genes in different cancer types: NEF2L2, PIK3R1, KRAS, EGFR and IDH1. X-axis indicates the amino acid positions of each cluster. For a fair comparison, we chose all cancer types in which the gene was eligible for the cluster test (e.g. the mutation cluster occurred in ≥5 samples), regardless of the association being significant or not. For EGFR, we showed its different mutation clusters in different cancer types and their association with mRNA or protein expression. Three antibodies for EGFR, i.e. EGFR, EGFR_pY1068 and EGFR_pY1173, were shown for the relationship between mutation clusters and the protein expression. For the other four genes, we showed the same or overlapping clusters associated with consistent trend of mRNA expression changes in multiple cancer types. Sample size and P-value were added on each plot. The P-values shown were obtained from Wilcoxon rank sum test. For the ease of illustration, we presented nominal P-values. A colour version of this figure is available at BIB online: https://academic.oup.com/bib.
NFE2L2
The gene NFE2L2 encodes nuclear factor-erythroid 2 p45-related factor 2 (also called Nrf2). NFE2L2 is a transcription factor that regulates many genes responsible for oxidative stress and chemical detoxification. NFE2L2 contains seven domains. Among them, the Neh2 domain contains two binding motifs, DLG (29th–31th codons) and ETGE (79th–82th codons), which interact with KEAP1 (Kelch-like ECH-associated protein 1) [30]. The interaction between NFE2L2 and KEAP1 negatively regulates the expression of NFE2L2. In normal conditions, KEAP1 binds to the DLG and ETGE motifs of NFE2L2 and brings NFE2L2 to KEAP1-CUL3-E3 ubiquitin ligase complex, leading to ubiquitination and subsequent degradation of NFE2L2 [31]. In cases of stress, NFE2L2 is stabilized and function to regulate the expression of many cytoprotective genes [31, 32]. Constitutive activation of NFE2L2 in cancer cells may confer cancer cells the ability to survive against drugs [33]. In our work, we observed that NFE2L2 had two recurrent mutation clusters, one around amino acids at the 24th–34th positions (cluster 1), which overlaps with the DLG motif, and the other around the 80th amino acid (cluster 2), which overlaps with the ETGE motif. Both clusters were within the KEAP1 binding domain [30]. Samples with mutation cluster 1 had noticeable trend of increase in NFE2L2 mRNA expression changes in three cancer types (BLCA, HNSC and LUSC), although only in BLCA the difference in expression changes was significant (pBLCA = 4.48 × 10−4). The second mutation cluster around the 80th position occurred in HNSC and LUSC, but the impact by the statistical test was not significant, although with an observed trend of increase in expression. We hypothesized that these mutations blocked the NFE2L2–KEAP1 binding and allowed NFE2L2 to escape KEAP1-mediated degradation. Concordantly, we observed increased mRNA expression of NFE2L2 in samples with clusters 1 and 2 mutations. Future work with larger sample sizes will warrant clarification of the impact of the mutation cluster around the 80th position in NFE2L2.
PIK3R1
PIK3R1, together with PIK3CA, are key players in the phosphatidylinositol 3-kinase signaling pathway. The gene PIK3R1 encodes p85α, a regulatory unit important for the stability of p110α encoded by PIK3CA. The longest transcript of PIK3R1 has 724 amino acids, including multiple domains [34]. In our analysis, we found PIK3R1 had two mutation clusters around the 455th–465th positions (cluster 1) and the 558th–593th positions (cluster 2), respectively. Both clusters were within the critical inter-SH2 (iSH2) domain required for the inhibition of p110α activity. Mutations in the iSH2 domain of PIK3R1 may lead to increased catalytic activity of p110α. As shown in Figure 4, cluster 1 was significantly associated with gene expression in UCEC (pmRNA = 0.01), whereas cluster 2 was significantly associated with gene expression changes in both BRCA (pmRNA = 2.76 × 10−3) and UCEC (pmRNA = 1.68 × 10−3). We also observed the same mutation clusters in GBM, but could not test their association with gene expression owing to insufficient sample size in gene expression data.
EGFR
EGFR is a well-studied oncogene in many cancers. We found completely different clusters of EGFR in GBM and in LUAD (Figure 4). The two clusters in GBM surrounded the 289th position and the 596th–598th positions, respectively. The two clusters in LUAD, however, were distributed around the 746th–754th and 858th–861th positions. In GBM, samples with either mutation cluster of EGFR had significantly higher gene expression compared with EGFR WT samples (pmRNA = 4.46 × 10−4 for the 289th position, pmRNA = 4.29 × 10−3 for the 596th–598th positions). Both clusters in GBM co-occurred with EGFR amplifications. To evaluate the impact of CNV status in the cluster test, we conducted the analysis only in the subset of GBM samples with EGFR amplification. However, the association was no longer significant in either cluster. Given this observation, we could not determine whether the mutation–cluster association was owing to the SNV clusters or copy number gain, or a combined effect of both events. In LUAD, the cluster around the 746th–750th positions, which mainly involved a deletion of 5 AAs (15 bp), showed significantly increased expression (pmRNA = 2.24 × 10−4), whereas the cluster around the 858th–861th positions failed to reach a statistical significance albeit with the same trend of increase in gene expression (pmRNA = 0.09). The complete different clusters found in the same gene EGFR but in different cancer types further highlighted the importance of comparative studies of somatic mutations and reflects the strong heterogeneity in cancer.
Another two genes, IDH1 and KRAS, were also found with recurrent clusters in multiple cancer types. IDH1 had a hotspot at the 132th codon, residing in the substrate binding sites, in both LAML and GBM, with an observed decrease at mRNA expression level (pmRNA = 0.08 in LAML and pmRNA = 4.62 × 10−4 in GBM). The cluster in KRAS was found at the 12th–13th positions, with significantly increased mRNA expression in LUAD (n = 43, pmRNA = 3.18 × 10−7) and COADREAD (n = 69, pmRNA = 0.04, nominal P-value), and similar but not significant trend in BRCA (n = 5, pmRNA = 0.14) or UCEC (n = 35, pmRNA = 0.20). Collectively, these results indicated that both similarities and differences of mutation features existed in the same gene across multiple cancer types, providing implications for future studies.
Discussion
In this work, we present an association study of mutation features with mRNA and protein expression levels in multiple cancer types. We discovered 72 genes whose mutation types (62), mutation clusters (21) or mutation statuses (51) were associated with their mRNA expression levels. We found 14 genes whose protein expression levels were associated with their mutation features, including 13 with mutation types, 6 with mutation clusters and 13 with mutation statuses. Eight genes were replicated at both the mRNA and the protein expression level, whereas there were also genes that were uniquely detected at the protein level but not the mRNA level. As for each mutation feature, our analysis indicated that tumor suppressor genes were sensitive to NS. Finally, genes whose mRNA expression changes with their mutation clusters were discovered, illustrating that both recurrent clusters and unique clusters in the same gene may be associated with their expression in different cancer types.
The association results we reported are likely reliable, as we have controlled multiple potential confounding factors in our analyses, such as CNV, methylation status, tumor purity and copy-neutral LOH. These genetic or epigenetic events all had been reported in previous studies to impact gene expression in certain cancer types and certain genes. These factors could be the true driving sources that cause expression changes. Therefore, controlling for the potential effects of these factors was required to clarify whether the associations we detected might have biases. The majority of our association results remained significant after controlling the effects by CNV, methylation status, tumor purity or copy-neutral LOH, supporting that these associations are likely true between mutation features and expression. However, there are still limitations in our association analyses. For example, owing to the lack of data, we could not take into consideration of the status of heterozygous variants. It could occur when the mutant allele was on the un-transcribed strand or isoform(s). Such mutations were likely passenger mutations, as they do not have impact on gene expression. We may exclude these mutations in future work when such information can be extracted from the original data sets. Our work is also limited to the alteration types that we could collect. In addition to the alternation types that we used, which commonly affect gene expression, there are other alternation types that may impact gene expression such as internal tandem duplication [35], but they are not often readily available to use. Considering the complicated and multiple levels of regulations on gene expression, caution should be taken when interpreting the data.
We studied two major types of mutations: NS and MS. For NS, as most mRNA transcripts carrying these mutations undergo NMD, we observed decreased expression level at both the mRNA and protein levels. Exceptions were observed in a few genes, which could escape NMD and express normally at the mRNA level but would have generated truncated protein products [36]. Such genes only have detectable decrease expression at the protein level. For MS, some of them formed clusters that were associated with increasing expression levels. Mutation clusters could take multiple forms, such as sequence clusters, protein domains and space clusters in 3D structures. Although MS mainly change amino acid sequences and are not expected with expression changes, one possible explanation is that these mutations might confer growth advantage to cancer cells and were positively selected when their residing genes showed increased expression levels. This was further supported by the similar association relationships for the same mutation clusters with mRNA expression across multiple cancer types, such as NFE2L2, KRAS and PIK3R1. Taken together, these observations highlighted that different patterns of mutation impacts are associated with different biological mechanisms and understanding of these patterns can be benefited by integration of both mRNA and protein expression data.
A main challenge on the association analysis between somatic mutations and mRNA and protein expression is to distinguish causal effects from reactive effects. For the mutation-type associations, the decreased expression associated with NS in many genes was supported by recent studies. When the mutations leading to premature stop codons are located in NMD-target regions, which is generally considered as 50 bp upstream of the last exon–exon junction in a transcript, these variants could trigger NMD-mediated degradation and result in a measurable decrease of mRNA expression [12, 13]. Exceptions could occur when a nonsense mutation escapes NMD (e.g. located outside of NMD-target regions) or a stop codon read-through occurs. For MS, interpretations and mechanisms remained elusive on how they are linked to differential expression changes. Our preliminary speculation is that the associations observed across cancer types are likely to reflect the positive selection on these mutations and their associated expression change, which might lead to an advantage in cell growth that is related to cancer development. Overall, caution should be taken when interpreting our results, and future experimental validation is required to warrant mechanistic insights.
The reported results provide insights into studies of driver mutations and genes and may potentially help the designs of integrative analyses in future translational studies. Integrative approaches are believed to offer advantages over mutation analysis alone by combining multidimensional genomics data such as genetic annotations, transcriptomic profile and protein expression [37]. With multi dimensional data becoming available for large-scale samples, methodologies are rapidly being developed to take advantage of comprehensive and complementary information by integrating multi-omics data [3, 7, 8, 38, 39]. However, caution should be taken when detecting driver mutations based on the transcriptional consequences of somatic mutations. In our results, although we observed many somatic mutations, most of them were not linked to any expression changes of their residing genes. These genes included, as expected, many passenger mutations (such as TTN), and also genes whose impacts are not predictable through expression dosage but protein activity changes. To measure the impacts of the latter, examination of their neighborhood genes should be considered such as those in the same signaling pathways or those closely located in molecular networks.
We introduced three ways of clustering somatic mutations for their association study: cluster by linear amino acid sequence, cluster by protein domain and cluster by space clustering in 3D protein structures. Each approach has its own strength but also the weakness in discovering genes with unique mutation features. Although we mainly reported and discussed the genes identified by linear amino acid distance, as shown in Supplementary Figure S5, there were nine genes uniquely associated with protein domains and three genes uniquely associated with space clustering, which deserved further investigation too. Most of these genes had been studied in previous work and some were functionally correlated. For example, among the six domain-associated genes identified in BRCA (CDH1, PIK3CA, MED23, MAP3K1, GATA3 and TMCC3, Supplementary Table S4), a recent study reported that some of these genes formed mutually exclusive gene sets (MEGS) [40]. One reported MEGS module included TP53 (identified by all three clustering methods), CDH1 (unique to domain cluster), GATA3 (unique to domain cluster) and MAP3K1 (unique to domain cluster). The second MEGS module included PIK3CA (identified by domain and space cluster but not linear sequence cluster), TP53 and GATA3. And the third MEGS module contained TP53, GATA3, FOXA1 and MED23 (unique to domain cluster). More importantly, we found a number of kinase domains associated with expression changes, such as ‘PI3-kinase family, p85-binding domain (PF02192)’ in PIK3CA, ‘Protein kinase domain (PF00069)’ in MAP3K1 and ‘Tyrosine kinase (PF07714)’ in EGFR (Supplementary Table S4).
The numbers of feature-associated genes were not as high as would have been expected, considering that regulatory variants had been predicted as prevalent [41]. There are a few possible explanations. First, we focused on somatic mutations obtained from WES and tested those that resulted in amino acid changes, excluding synonymous SNVs in coding regions. Mutations in noncoding regions, such as promoter, untranslated regions and intronic or intergenic regions, were not tested either. Second, unlike regulatory variants, the majority of somatic mutations with amino acid changes in coding regions may not function through regulation of gene expression. Rather, they could change protein confirmation, obstruct protein–protein interactions and activate/deactivate kinase activities. Third, it is worth noting that among the small number of eligible genes for mutation cluster test, only a few of them were associated with gene expression changes. Our step to define the eligible genes had already preselected genes with mutation clusters, which included many well-studied cancer genes. In previous studies, these eligible genes would have been reported as having hotspot mutations and predicted as candidate driver genes. Here, we showed that many such mutation clusters were not associated with expression changes, implying that their impact on cancer might be executed by other ways, rather than changing the expression of their residing genes. It is likely that the consequences of somatic mutations on the transcriptome level may not be restricted to the gene itself but its participating pathways.
Conclusion
We systematically examined the associations between mutation features and mRNA/protein expression levels in multiple cancer types, aiming to address the functional consequence of somatic coding mutations in terms of their expression and protein products. Our analyses revealed that mutation type was the most important determinant of expression level and mutation clusters could be detected in well-studied oncogenes that were associated with gene expression. We found both similarities and differences in association patterns existed within and across cancer types. In summary, our results suggested that mutation features were important factors in somatic mutation data analyses for their functional consequences.
Materials and methods
TCGA data acquisition and process
SNV and indel reannotations
We downloaded SNV and indel data from the synapse Web site (syn1729383), which is listed as the data deposition for the pan-cancer study in [17]. The downloaded SNVs and indels were organized in the MAF format. We annotated them using the tool Oncotator (v1.5.1.0) [42]. Reference transcripts were downloaded from GENCODE (hg19). For each protein-coding gene, we use its longest transcript for annotation. For some cancer types, there are hyper-mutated samples with extremely large size of SNVs and indels. We removed those with >1000 nonsilent SNVs and indels from our analysis.
CNV data
CNVs were downloaded from http://cbio.mskcc.org/cancergenomics/pancan_tcga/. CNVs were originally obtained using the tool GISTIC [43]. The downloaded data included five levels to represent different CNV status, i.e. deletion (CN = −2), copy loss (CN = −1), neutral, copy gain (CN = 1) and amplification (CN = 2). In our analysis, we simplified the group as copy number loss (CN < 0), copy number neutral (CN = 0) and copy number gain (CN > 0).
We conducted three types of analyses to fully assess the impact of CNVs in our results. The first two were similar to a stratification test and were sensitive to the number of samples in each group, and thus, they were applied to only mutation type and mutation status analysis but not mutation cluster analysis. The third method was applied to all three association tests. First, we used CNV as a covariate for all our association tests. Specifically, for each gene, we constructed the MLR model as: , where is the mRNA expression of the gene, represents the mutation status vector ( for the ith sample) or mutation type vector , and represents the CNV vector . Note is a factorized vector where values in represent independent groups, rather than ordinal numbers. We choose the P-value for to determine whether the corresponding mutation feature is significantly associated with gene expression levels. Second, we restricted the association test in the subset of samples with the most prevalent copy number status. Third, we used linear regression to obtain the residual expression of copy number status: . Then, we used the residual expression, , for the association test. This model does not stratify CNV status, and so, it is applicable to all genes with CNV data.
Methylation data
We downloaded methylation data from UCSC Cancer Genome Browser (28 January 2015). There were two platforms used by TCGA to measure methylation levels: Infinium HumanMethylation 27K (HM27K) and Infinium HumanMethylation 450K (HM450K). We chose the TCGA PANCAN AWG (PanCancer Analysis of Whole Genomes) dichotomized DNA methylation data across 12 TCGA cohorts in the PANCAN12 study (file name: TCGA_PANCAN12_hMethyl-2015-01-28.tgz). This data set had been preprocessed by performing a moderate probe-design-dependent platform normalization to remove systematic platform bias [44]. The file contains a merged data set on 25 978 probes shared by the HM27K and HM450K platforms. A detailed description of the preprocessing procedure can be found elsewhere [44]. Briefly, the major filtering criteria included removing probes with a standard deviation of >0.05 (to control batch and platform effects) and removing probes that showed methylation (median β value > 0.2) in any of the 12 matched normal tissue types. The data set we downloaded included probes with dichotomized β values at 0.3. Samples with a β value ≥ 0.3 were designated methylated and samples with a β value < 0.3 were designated unmethylated. The original working group of PANCAN AWG had selected the 2203 probes that were methylated in >10% of any of the tumor types or 50% of any of the well-defined subtypes for clustering. Thus, genes not tagged by these probes indicated that their methylation statuses did not change substantially across cancer types or subtypes.
Tumor purity
We obtained the tumor purity predictions for TCGA samples from the work by Yoshihara et al. (2013). In the original work, the authors described a method to calculate stromal and immune scores to predict the level of infiltrating stromal and immune cells, from which they inferred tumor purity in tumor tissue. We downloaded the tumor purity score for each sample from the supplementary materials of Yoshihara et al. (2013) and included tumor purity as a covariate in our MLR model. This analysis applied to all cancer types excluding LAML.
Copy-neutral LOH
We searched in the TCGA database for copy-neutral LOH data. The LOH status was determined by Hudson Alpha Institute for Biotechnology using Human 1MDuo SNP chip as part of the TCGA initiative. We systematically searched data for the 12 cancer types used in this study. There were only three cancers that had LOH data: GBM, LUSC and OVCA. For LUSC, there were only 23 samples with available LOH data. For OVCA, we found only one gene that was significant in our association tests, and we did not discuss this cancer type in most of our analysis. We then excluded OVCA as well as from the LOH analysis. Thus, only GBM was eligible for the LOH analysis. We preprocessed the LOH data following the works in [45, 46]. Specifically, we considered only the LOH events with log2 values >0.075. We included LOH status as a covariate and constructed the MLR model as follows: .
RNA sequencing data
Normalized mRNA expression data from RNA sequencing (HiSeq V2) was downloaded from UCSC Cancer Genome Browser (28 January 2015) [47]. Two layers of filters based on gene expression were applied to remove low expression genes. The first filter eliminated genes with low variability across patients (var < 0.1), as measured by the variance per gene. The second filter eliminated genes that had static expression levels in more than half of all patients, i.e. genes whose minimum and median expression levels remained the same across samples. Genes remained after these filters were considered as expressed in the corresponding cancer type.
Protein expression data
Normalized protein expression data measured by the RPPA were downloaded from The Cancer Proteome Atlas [48]. The RPPA platform is an antibody-based screening tool that measures ∼150 proteins (187 antibodies) and their posttranscriptional (e.g. phosphorylation) levels. The data we downloaded had been normalized using the ‘replicates-based normalization (RBN)’ method across 11 cancer types (no RPPA data for LAML) [49].
Statistical tests
We examined three mutation features that likely impact expression changes. Mutation type is a categorical parameter with three statuses: MS, NS and WT. Mutation cluster is a factor with multiple levels. Mutation status is described as a categorical parameter with two groups, mutated or not. We grouped mutations that are located with no more than 5 AAs away from each other. Genes with insufficient representations in any cluster grouping or type grouping (with a threshold of five samples) were excluded. MLR was performed for each feature independently, using mutation features as the predictive parameter and expression values as the responsive parameter. Genes that are significantly impacted by any feature (evaluated by the model P-value) were collected. To avoid potential false discoveries owing to data inflation, we further examined each mutation cluster or mutation type for these genes using Wilcoxon rank sum test by comparing the group of samples with the feature of interest and the WT samples. Only those with a significant mutation cluster or mutation type were kept. Multiple testing correction [50] was conducted in each cancer type.
Key Points
We presented a comprehensive landscape of the associations between mutation features and expression profile in multiple cancer types, including 62 genes showing mutation type associated with expression changes, 21 genes showing mutation cluster associations and 51 genes showing mutation status associations.
Mutation type (truncation versus amino acid-altering mutations) was found as the most important determinant of expression levels.
Mutation clusters were detected in well-studied oncogenes that were associated with gene expression.
We found both similarities and differences in association patterns existed within and across cancer types.
Although many of the observed associations stay stable at both mRNA and protein expression levels, there were also novel associations uniquely observed at the protein level, which warrant future investigations.
Supplementary Data
Supplementary data are available online at http://bib.oxfordjournals.org/.
Supplementary Material
Acknowledgments
The authors thank Vanderbilt Advanced Computing Center for Research & Education (ACCRE) for providing computing resources and support. The results shown here are in part based on data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Funding
This work was partially supported by National Institutes of Health grants (R01LM011177, P30CA068485, P50CA095103 and P50CA098131), Ingram Professorship Funds (to Z.Z.), Breast Cancer SPORE pilot grant (to Z.Z.), an American Cancer Society Institutional Research Grant pilot project (#IRG-58-009-55, to P.J.) and a Center for Quantitative Sciences (CQS) pilot grant (to P.J.). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript
Peilin Jia is currently an assistant professor in the School of Biomedical Informatics at The University of Texas Health Science Center at Houston.
Zhongming Zhao is a professor in the School of Biomedical Informatics at The University of Texas Health Science Center at Houston and a professor in the Department of Biomedical Informatics at Vanderbilt University Medical Center.
References
- 1. Hanahan D, Weinberg RA.. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 2. Watson IR, Takahashi K, Futreal PA, et al. Emerging patterns of somatic mutations in cancer. Nat Rev Genet 2013;14:703–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Raphael BJ, Dobson JR, Oesper L, et al. Identifying driver mutations in sequenced cancer genomes: computational approaches to enable precision medicine. Genome Med 2014;6:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reva B, Antipin Y, Sander C.. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 2011;39:e118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hou JP, Ma J.. DawnRank: discovering personalized driver genes in cancer. Genome Med 2014;6:56.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gonzalez-Perez A, Mustonen V, Reva B, et al. Computational approaches to identify functional genetic variants in cancer genomes. Nat Methods 2013;10:723–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paull EO, Carlin DE, Niepel M, et al. Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE). Bioinformatics 2013;29:2757–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ng S, Collisson EA, Sokolov A, et al. PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics 2012;28:i640–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Q, Seo JH, Stranger B, et al. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 2013;152:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Noensie EN, Dietz HC.. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol 2001;19:434–9. [DOI] [PubMed] [Google Scholar]
- 13. Nagy E, Maquat LE.. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci 1998;23:198–9. [DOI] [PubMed] [Google Scholar]
- 14. Chen J, Chen H, Zhu T, et al. Asp120Asn mutation impairs the catalytic activity of NDM-1 metallo-beta-lactamase: experimental and computational study. Phys Chem Chem Phys 2014;16:6709–16. [DOI] [PubMed] [Google Scholar]
- 15. Lane A, Segura-Cabrera A, Komurov K.. A comparative survey of functional footprints of EGFR pathway mutations in human cancers. Oncogene 2014;33:5078–89. [DOI] [PubMed] [Google Scholar]
- 16. Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med 2005;2:e313.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Reimand J, Bader GD.. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol Syst Biol 2013;9:637.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ding J, McConechy MK, Horlings HM, et al. Systematic analysis of somatic mutations impacting gene expression in 12 tumour types. Nat Commun 2015;6:8554.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature 2013;502:333–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tamborero D, Gonzalez-Perez A, Lopez-Bigas N.. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics 2013;29:2238–44. [DOI] [PubMed] [Google Scholar]
- 21. Melton C, Reuter JA, Spacek DV, et al. Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet 2015;47:710–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet 2013;45:1134–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 2016;44:D279–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamburov A, Lawrence MS, Polak P, et al. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci USA 2015;112:E5486–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SY, Kim DW, Lee HS, et al. Loss of AT-rich interactive domain 1A expression in gastrointestinal malignancies. Oncology 2015;88:234–40. [DOI] [PubMed] [Google Scholar]
- 26. Chou A, Toon CW, Clarkson A, et al. Loss of ARID1A expression in colorectal carcinoma is strongly associated with mismatch repair deficiency. Hum Pathol 2014;45:1697–703. [DOI] [PubMed] [Google Scholar]
- 27. Payne SR, Kemp CJ.. Tumor suppressor genetics. Carcinogenesis 2005;26:2031–45. [DOI] [PubMed] [Google Scholar]
- 28. Levine AJ, Puzio-Kuter AM.. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010;330:1340–4. [DOI] [PubMed] [Google Scholar]
- 29. The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayes JD, Dinkova-Kostova AT.. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 2014;39:199–218. [DOI] [PubMed] [Google Scholar]
- 31. Jaramillo MC, Zhang DD.. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 2013;27:2179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh A, Misra V, Thimmulappa RK, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med 2006;3:e420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ji L, Li H, Gao P, et al. Nrf2 pathway regulates multidrug-resistance-associated protein 1 in small cell lung cancer. PLoS One 2013;8:e63404.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ross RL, Burns JE, Taylor CF, et al. Identification of mutations in distinct regions of p85 alpha in urothelial cancer. PLoS One 2013;8:e84411.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spencer DH, Abel HJ, Lockwood CM, et al. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn 2013;15:81–93. [DOI] [PubMed] [Google Scholar]
- 36. Bordeira-Carrico R, Pego AP, Santos M, et al. Cancer syndromes and therapy by stop-codon readthrough. Trends Mol Med 2012;18:667–78. [DOI] [PubMed] [Google Scholar]
- 37. Khurana E, Fu Y, Chen J, et al. Interpretation of genomic variants using a unified biological network approach. PLoS Comput Biol 2013;9:e1002886.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shen R, Mo Q, Schultz N, et al. Integrative subtype discovery in glioblastoma using iCluster. PLoS One 2012;7:e35236.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bashashati A, Haffari G, Ding J, et al. DriverNet: uncovering the impact of somatic driver mutations on transcriptional networks in cancer. Genome Biol 2012;13:R124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hua X, Hyland PL, Huang J, et al. MEGSA: a powerful and flexible framework for analyzing mutual exclusivity of tumor mutations. Am J Hum Genet 2016;98:442–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ongen H, Andersen CL, Bramsen JB, et al. Putative cis-regulatory drivers in colorectal cancer. Nature 2014;512:87–90. [DOI] [PubMed] [Google Scholar]
- 42. Ramos AH, Lichtenstein L, Gupta M, et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015;36:E2423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mermel CH, Schumacher SE, Hill B, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol 2011;12:R41.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. UCSC Cancer Genome Browser. https://genome-cancer.ucsc.edu/.
- 45. Pedersen BS, De S.. Loss of heterozygosity preferentially occurs in early replicating regions in cancer genomes. Nucleic Acids Res 2013;41:7615–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pedersen BS, Konstantinopoulos PA, Spillman MA, et al. Copy neutral loss of heterozygosity is more frequent in older ovarian cancer patients. Genes Chromosomes Cancer 2013;52:794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cline MS, Craft B, Swatloski T, et al. Exploring TCGA pan-cancer data at the UCSC Cancer Genomics Browser. Sci Rep 2013;3:2652.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li J, Lu Y, Akbani R, et al. TCPA: a resource for cancer functional proteomics data. Nat Methods 2013;10:1046–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Akbani R, Ng PK, Werner HM. et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun 2014;5:3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Benjamini Y, Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.