This review highlights the achievements of the late Professor Alan Clarke in developing the first knockout mice and sets his work in the general context of the scientific field he contributed to, namely using mouse models to understand cancer biology.
Abstract
Over the past 20 years, huge advances have been made in modelling human diseases such as cancer using genetically modified mice. Accurate in vivo models are essential to examine the complex interaction between cancer cells, surrounding stromal cells, tumour-associated inflammatory cells, fibroblast and blood vessels, and to recapitulate all the steps involved in metastasis. Elucidating these interactions in vitro has inherent limitations, and thus animal models are a powerful tool to enable researchers to gain insight into the complex interactions between signalling pathways and different cells types. This review will focus on how advances in in vivo models have shed light on many aspects of cancer biology including the identification of oncogenes, tumour suppressors and stem cells, epigenetics, cell death and context dependent cell signalling.
Introduction
The discovery of mutated genes in tumours spawned a whole new field of science, dedicated to understanding the link between genetics and cancer. In 1953, Carl Nordling hypothesized that cancer was caused by the accumulation of mutations over time (1), and this theory was further supported by analysis of retinoblastoma patients by Alfred Knudson in 1971. Knudson observed that inherited retinoblastoma developed in both eyes of children, whilst sporadic retinoblastoma developed in older patients and usually only in one eye (2). He correctly hypothesized that retinoblastoma in young patients was due to inheritance of a single mutation, later identified to be in the Retinoblastoma-1 (RB1) gene, however pathology would only occur if a second mutation occurred in a somatic cell.
Nordling and Knudson did not have the genetic tools to functionally demonstrate that their theories were correct, but researchers were working hard to develop the ability to introduce a mutation into a gene and thus study its requirement in development and disease. Sir Martin Evans took the first steps towards this goal in 1981, by developing a method to culture pluripotent stem cells from mouse embryos (3). This was the precursor which subsequently allowed Oliver Smithies and Mario Capecchi to modify cultured embryonic stem cells using homologous recombination to specifically target the HPRT gene (4,5). Two years later this technology was used to correct the mutant HPRT gene and generate the first targeted, genetically modified mouse which passed on the modified gene to its progeny via the germ line (6).
Rb1 was the first tumour suppressor knocked out in mice using gene targeting and was published by three groups in the same issue of Nature in 1992 (7–9). However, these mice did not develop retinoblastoma until compound mutant mice were generated which had a mutation in Rb1 and its family member p107 (10). It has since been observed that Rb1 mutations deregulate the cell cycle in several different cancers and that Rb1 interacts with other tumour suppressor genes such as p53 (11).
From these pioneering works, which resulted in the Nobel Prize in 2007 for Sir Martin Evans, Oliver Smithies and Mario Capecchi, researchers have been given the tools to study the function of genes in vivo and these tools has subsequently developed into more sophisticated and precise ways of manipulating genes to yield fundamental advances in many fields of biology. Among these discoveries have been the generation of increasingly accurate mouse models of disease, the identification of stem cells in various tissues and genetic evidence of the interaction between different gene products. This review will focus on some of the extraordinary advances in the fields of cell signalling (and particularly Wnt signalling), apoptosis and stem cells in the intestine, and how these findings have increased our understanding of intestinal cancer, and led to novel therapeutic strategies.
The origins of mouse models of epithelial cancer
In a complementary approach to the gene targeting techniques described above, several groups were investigating the genetic events that resulted from exposure to carcinogens (reviewed in ref. (12). In 1983, two groups identified that tumours induced by experimentally applied carcinogens were due to an activating mutation to the Harvey-ras (H-ras) oncogene (13,14). These types of experiments were among the first to demonstrate that carcinogens could mutate specific genes to transform cells. Subsequently, William Dove’s treated mice with ethylnitrosourea, with the aim of identifying new tumour suppressor genes. They identified a pedigree of mice that developed multiple intestinal neoplasias throughout the length of the intestinal tract with 100% penetrance, and named this gene Min (Multiple intestinal neoplasia) (15). Although originally it was not known which gene was responsible for the tumourigenesis subsequent work showed the gene mutated was the Adenomatous Polyposis coli (APC) gene. This gene is the most commonly mutated gene in sporadic human colorectal cancer (CRC) and people who are germline heterozygous for APC, familial adenomatous polyposis patients are strongly predisposed to CRC. Thus, this first mouse model of intestinal cancer would enable researchers to investigate the molecular events of intestinal tumourigenesis and growth in vivo. Moreover, from a very early stage these mice were used to investigate potential chemopreventative agents such as aspirin (16) which have subsequently proved to have activity in humans (17).
The ability to direct mutations into a selected gene in embryonic stem cells and generate mice from these cells has provided great insights into developmental biology and disease, specific examples of which are provided below. In addition to deletion of parts of genes, homologous recombination made it possible to introduce the exact mutations found in human disease, which could sometimes hyperactive the gene product rather than delete it. However, there are some limitations to what can be achieved using this approach. For example, about 15% of all knockout mice are embryonic lethal and therefore any role for the mutated gene in adults cannot be assessed. To overcome this, scientists began looking for ways to inducibly delete genes in adult somatic cells of mice.
An important step towards this goal was the generation of transgenic mice. Superseding the generation of knockout mice, five groups independently demonstrated that they were able to successfully microinject oocytes with DNA constructs, which were then implanted into pseudo-pregnant recipient mice, to produce transgenic mice in which the introduced DNA was transmitted to progeny and retained expression (18–22). This type of transgenic mouse differs from the classical knockouts to mutate a specific gene, in that the introduced DNA is randomly integrated into the host DNA, but this technology has had an equally important impact on the study of genes in vivo. Indeed Jerry Adams and Suzanne Cory generated the first oncomouse by fusing an immunoglobulin enhancer (Eµ) to the Myc gene. These mice developed pre-B-cell and mature B-cell lymphomas and supported the hypothesis that the Ig-Myc translocations observed in patients were malignant events (23). Transgenic mice also provided the genetic tools for the ability to conditionally delete a gene in a specific tissue in adult somatic cells when used in combination with knockout mice.
Inducible manipulation of genes in vivo
Temporal regulation of eukaryotic promoters to manipulate gene expression proved unsuccessful. However, in 1992 Manfred Gossen and Hermann Bujard generated a tetracycline-controlled transactivator (tTA) by fusing the tet repressor element of Escherichia coli to the activating domain of virion protein 16 of herpes simplex virus. Importantly, they then demonstrated that this construct worked in mammalian cells in vitro (24), and transgenic mice (25). This technology was used to demonstrate an essential role for mutant H-Ras in tumour maintenance (26), and was more recently refined to allow temporal expression of shRNAs, which is proving to be a powerful research tool (27).
The most widely used approach to conditionally manipulate genes in vivo is the Cre-LoxP (Causes recombination-Locus of crossover P1) system. Cre is a site-specific DNA recombinase used by bacteriophage P1 during viral genome replication and is not expressed in mammals. It recognizes specific 34bp palindromic sequences of DNA, termed LoxP sites, and recombines the DNA between two LoxP sites, thus deleting it. In 1988, Sauer and Henderson demonstrated this system was able to work in mammalian cells, with the advantage that no mammalian protein is capable of recognising the lox sites (28), and in 1994 a T-cell-specific deletion of the DNA polymerase β gene was achieved in vivo (29).
In addition to deletion of specific areas of DNA, Cre-Lox technology also allows for conditional activation of genes, by inserting a lox flanked stop codon into a gene. The subsequent Cre-mediated removal of the stop codon then permits read-through and transcription of the gene (30). Regulation of Cre recombinase activity is most commonly achieved by use of a tissue-specific promoter, allowing spatial control of Cre expression. More recent versions of this technology have incorporated a secondary, temporal control of recombinase activity by fusing the Cre enzyme to a modified oestrogen receptor, allowing for activation of Cre only after administration of tamoxifen (TM). Several groups have demonstrated tissue-specific manipulation of gene activity by using transgenic Cre-ERTM mice (31–33), which was superseded by an improved version called Cre-ERT2 (34).
These types of experiments use a combination of transgenic mice to generate the Cre-expressing line, and knockin mice to generate the Lox-flanked alleles, thus bringing together the two technologies to enable the ability to conditionally manipulate genes. More recently combining Cre-Lox technology with another system that allows conditional activation/deletions of alleles independently of TM exposure (recombinase flippase [FLP] flippase recognition target [FRT]), has allowed questions on tumour maintenance and/or the importance of stromal genes to be investigated as now tumours can be driven by one system and genes deleted by the other (35). Although the Cre-Lox system allows tissue specific, inducible manipulation of gene expression, presently the introduced genetic change cannot be reversed to interrogate questions on tumour maintenance. This however was achieved by refining of the tetracycline system, in which the tTA protein can only operate to activate transcription in the presence or absence of tetracycline, or a tetracycline derivative such as doxycycline. Thus, in the tet-Off system, tTA binds to a tetracycline-responsive promoter element and activates transcription of a downstream target gene, which is blocked by the presence of doxycycline. In the tet-On system, the reverse tTA (rtTA) fusion protein can only recognize the tetracycline-responsive promoter element and activate transcription of the target gene in the presence of doxycycline. As the binding of tTA or rtTA to the tetracycline-responsive promoter element does not alter the genetic material itself this is a reversible mechanism of gene manipulation (36) which has now been used in mouse models for several decades. This system has allowed key oncogenes such as MYC and KRAS to be switched on and off and the mechanism of tumour regression investigated. With the development of conditional gene modification researchers now had very powerful tools in which they could investigate molecular cell biology in vivo, and the chapters below will highlight some of the important discoverers in intestinal biology and cancer using these technologies.
Using mouse models to study intestinal carcinogenesis
Interaction of oncogenes and tumour suppressor genes in intestinal cancer
As mentioned above, two years after William Dove’s group had generated the Min mouse, the same group published that the gene mutated was homologous to a gene called APC, which was mutated in patients with familial adenomatous polyposis and sporadic intestinal tumours (37). Familial adenomatous polyposis patients are born with a mutation in one allele of APC, and during adulthood a somatic mutation of the remaining wild-type allele triggers tumorigenesis, thus supporting the Knudsonian ‘two-hit’ hypothesis. At this early stage of the research, the function of the APC gene was unknown, however, during a screen for binding partners, β-catenin was identified as an interactor of APC (38). β-catenin had already been identified as the mammalian homologue to the Drosophila gene armadillo, a component of the Wingless (Wnt) signalling pathway (39), and comprehensive in vitro experiments using a panel of APC mutant proteins demonstrated that cells with mutant APC had activated β-catenin and the Wnt pathway (40,41). Thus, the link had been made between mutations in Apc and aberrant Wnt signalling in colon cancer. Apc was one of the first genes to be conditionally deleted in the adult intestine. Shibata and colleagues proved loss of Apc was sufficient to drive tumourigenesis when they delivered AdCre to the colon of mice and observed colonic polyp formation (42). Acute deletion throughout the intestine happened some years after this following the development of efficient inducible intestinal cre recombinases; AhCre (43,44) and VillinCreER (45). Loss of both copies of Apc led to a rapid phenotype now termed a ‘crypt progenitor-like phenotype’ with rapid proliferation and perturbed differentiation (44,45). Importantly, these experiments showed a rapid translocation of β-catenin to the nucleus, and activation of Wnt signalling target genes immediately following Apc loss, thus supporting the model that Apc loss, and deregulation of Wnt signalling, is one of the earliest events during tumour initiation. This was supported by experiments in which a mutant form of β-catenin, which could not be phosphorylated and targeted for degradation, was expressed under the control of the cytokeratin 19 gene to drive expression in the intestine. These mice also developed intestinal tumours, although more readily in the small intestine than the colon, demonstrating that deregulated Wnt signalling can lead to tumourigenesis (46).
The idea that tumourigenesis was a multistep process had been postulated in 1958 (47) and was further refined by Weinberg (48) and Vogelstein (49). The classical model for the sequential order of mutations in colon cancer proposed mutations in Apc as the initial trigger, followed by mutations in the same cell for K-ras, Tgf-β signalling and p53 (50). Compound mutants now enabled researchers to functionally investigate this and establish which oncogenes or tumour suppressors co-operated with one another. Work from the Taketo lab was the first to show that ApcMin/+ mice with an additional heterozygous mutation to the Tgf-β signal transducer Smad4 developed invasive intestinal tumours, thus demonstrating that oncogenes and tumour suppressors co-operate to promote tumour progression in vivo (51).
The use of conditional alleles allowed more accurate models of cancer to be generated, thus avoiding the embryonic lethality of systemic mutations, which is well illustrated by a series of papers from Alan Clarke’s group. They showed that knock-in of the activating mutation of K-ras (LSL-KRASG12V) in the intestine did not induce tumourigenesis, but rather accelerated tumour growth and progression after Apc had been deleted (52). Interestingly, expression of a different mutant form of Kras in the intestine, K-rasG12D, induced tumourigenesis (53). However, only a single tumour was observed in the small intestine of 25% of mice aged at least a year, suggesting that additional mutations are required in concert with Kras to initiate tumourigenesis. Serrated tumours did develop in the colon of 50% of K-rasG12D; Ink4a/Arf-/- mice, which allowed K-ras-induced senescence to be overcome (53). This highlights the use of mice in identifying the context-specific nature of oncogenes as in contrast to the intestine, the expression of mutant KRAS in the lung rapidly induces multiple adenomas (54). An elegant example of how the advent of very specific cre recombinases allowed researchers to elucidate the cell of origin of intestinal cancer comes from work examining the function of the tumour suppressor phosphatase and tensin homolog (Pten). Deletion of Pten specifically in the intestinal epithelium using VillinCreER did not induce tumourigenesis, but compound mutant mice in which Apc and Pten were co-deleted resulted in invasive adenocarcinoma (55). In contrast, another group observed rapid intestinal tumourigenesis when Pten was deleted from the intestine, but this group used the Mx1Cre which drives recombination in the epithelium and the stroma, prompting suggestions that stromal loss of Pten was important (56). These data strongly suggest that stromal deletion of Pten may give rise to tumours within the intestine, a situation similar to what has been shown with Lkb1 (57). However, when mice with epithelial specific deletion of Pten were aged (using AhCre to drive recombination), they developed intestinal tumours that were histologically consistent with juvenile polyps observed in Cowden Syndrome, with expanded mesenchymal compartments and immune infiltrates, which progressed to become invasive neoplasias (58). These data suggest that Pten deletion alone can trigger intestinal tumourigenesis, albeit with extended latency, and that Pten deficient epithelial cells can affect the underlying stromal cells. However, potentially the most efficient system to form tumours driven by Pten loss is when there is loss in both the tumour and the surrounding mesenchyme (56). This may be similar to the situation in breast cancer where the Leone group have shown elegantly that Pten deletion in fibroblasts can drive aggressive breast cancers in mice that much more closely resemble that of humans (59).
These experiments also suggest that the classic sequential order of mutations is not always required to induce tumourigenesis. This was illustrated in further experiments from Alan Clarke’s group designed to elucidate the co-operation between Apc, Pten and Kras during intestinal tumourigenesis. Compound mutant mice in which Apc and Pten were deleted concurrently with an activated KrasG12D mutation in the intestinal epithelium resulted in invasive adenocarcinoma (60). Surprisingly, a tumourigenic synergy was also revealed between Pten and K-ras, as mice with deleted Pten and mutant K-ras, but wild-type for Apc, also developed intestinal tumours (60). These lesions were markedly different from the triple mutant tumours however, and instead displayed a spectrum of serrated lesions, which progressed to invasive adenocarcinoma and metastasized to the pancreas, liver and lungs in 41% of mice. Pten and K-Ras both activate PI3K, and indeed serrated tumours in the double mutant Pten K-ras mice displayed increased PI3K signalling (60). This led to further studies investigating PI3K in colon cancer which revealed that deletion of Apc concurrently with expression of the PI3K activating mutation found in humans, Pik3caH1047R, resulted in invasive intestinal tumours in mice (61).
p53 (or tumour protein 53, TP53), was among the first of the mouse knockouts (62), and developed multiple tumour types predominantly sarcomas and lymphomas (63). These studies demonstrated it was a ‘bona fide’ tumour suppressor gene. It has since been identified as the gene most frequently mutated in all types of cancers, with an estimated 50% of tumours possessing a p53 mutation (64). Interestingly, p53-/- mice did not develop intestinal tumours, however ApcMin/+p53-/- mice had increased tumour initiation with an increase in invasive tumours (65), suggesting that p53 mutations are more important during later stages of intestinal cancer. One problem with these studies were that as the p53 knockout mouse rapidly succumbed to lymphoma it was difficult to unravel the precise impact on intestinal tumourigenesis. Human intestinal tumours also frequently harbour a specific mutation to p53 (R175H), which was found to possess different properties to Wt p53 and also to deletion of p53. Indeed, when Apc was deleted in the intestine with co-expression of p53R172H (the mouse version of the human mutation) using AhCre, all mice now developed invasive adenocarcinoma (66). Similarly, deletion of exons 2–10 of Tp53 specifically in mouse intestinal enterocytes using VillinCre did not induce tumourigenesis until mice were treated with the carcinogen azoxymethane which causes activating mutations in β-catenin (and hence high wnt pathway) (67). These tumours were invasive, and formed in the colon of azoxymethane (due to colonic metabolism of azoxymethane). This overcomes a potential problem of the ApcMin mice in recapitulating human cancer, the preponderance of small intestinal tumours though also suggests a similar molecular mechanism regulates tumourigenesis in the small intestine and colon (co-operation between deregulated Wnt and p53), and highlights the usefulness of carcinogenic models to gain further insight into intestinal cancer. Furthermore, the development of colonic specific Cre mice such as Cdx2Cre has enabled researchers to better mimic human disease, in which tumours develop predominantly in the colon (68), allowing the study of the interaction between colon tumours and the commensal microbiome which is most abundant and diverse in the colon. Indeed deletion of Apc inhibits differentiation of mucus secreting cells (44) which has been shown to result in defects in the barrier function of the colonic tumours observed in Cdx2CreERT;Apcflox/flox mice (67) to allow invasion of microbial species (69). This in turn has been shown to induce NF-κB activation and increase cyctokine expression to fuel colonic tumour growth (69), which is enhanced in tumours with mutant p53 (70).
Of all the cell signalling pathways, the transforming growth factor-β (TGF-β) pathway is one where the potential tumour suppressive and promoting pathways needed to be tested in vivo to elucidate the functional relevance in cancer. Both SMAD4 and TGFBR2 are commonly mutated or inhibited in CRC, though there are compelling data to suggest upregulation of TGF-β ligand particularly is stromal fibroblasts may drive by tumour progression. Deletion of the TGF-β receptor Tgfbr2 specifically in the intestinal epithelium using VillinCre did not induce the formation of tumours, however, co-deletion of Apc and Tgfbr2 in the intestine promoted the progression of adenomas (observed after Apc deletion alone) to adenocarcinomas (71). However, the same group later showed that Apc mutations are not always required to reveal the oncogenic capacity of deregulated Tgf-β signalling, as mice with mutant Kras (KrasG12D) and deletion of Tgfbr2 specifically in the intestine also developed intestinal tumours with ~15% of them becoming invasive and metastasising to the lymph nodes and/or lungs (72). This is of interest to human CRC as many tumours that have mismatch repair deficiency (approximately 20%) do not mutate APC and have mutations in TGFBR2 (due to a microsatellite repeat which is commonly mutated)
TGF-β has very recently also been identified as a regulator of the serrated neoplasia pathway in the intestine (73). Colon cancer is currently classified into four consensus molecular subtypes according to several biological parameters including gene expression and molecular aberrations (74). Work from JP Mademas lab showed that sessile serrated adenoma organoids carrying BRAFV600E mutations respond differently from tubular adenoma organoids when exposed to TGF-β, and acquired a mesenchymal phenotype, characteristic of the CMS4 group which has a very poor clinical prognosis (73). Indeed work from Eduard Batlle’s group suggests that these mesenchymal expression patterns in serrated tumours are due to TGF-β ligand secretion from the stroma, and these may confer the poor prognosis of this subtype (75), suggesting that TGFβ inhibition in this group may be a therapeutic strategy (75). Metastatic genetically engineered mouse models of CRC are now required more than ever so these treatments can be modelled in an immunocompetent setting.
One question that remains from alternative models of colon cancer that lack Apc mutation is whether they activate Wnt signalling during tumour progression. Allan Bradleys group observed that intestinal tumours developed in VillinCre;BrafV637E mice, with Wnt signaling deregulation observed in very early high-grade dysplasia (76). Sequencing revealed that several Wnt regulator genes were mutated in these Braf driven lesions, including Apc, suggesting Wnt signalling is important during early dysplasia progression (76). In support of this model, most intestinal tumour organoids that are generated without Apc mutations are able to grow in a Wnt and R-Spondin (Wnt agonist) independent manner, strongly suggestive of mutations somewhere in the Wnt pathway. Thus, it is likely that invasive tumours observed in long latency mouse models of cancer have acquired additional mutations to genes which drive progression such as Wnt regulators. The continued importance of aberrant Wnt signalling in intestinal cancer has recently been shown in a study from Scott Lowes’ lab in which they used a doxycycline-regulated shRNA which enabled them to initially inhibit Apc and then conditionally re-express it in vivo. Using this system they induced regression, via differentiation of colonic adenocarcinomas upon re-expression of Apc, even in tumours harbouring oncogenic mutations to Kras and p53 (77).
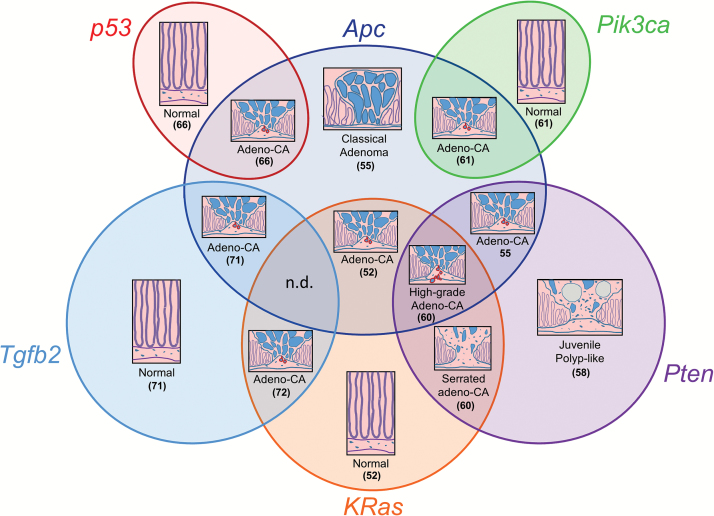
Together, the experiments above have given us many novels insights into the molecular events that regulate tumour initiation, growth and progression, and provide a roadmap from normal tissue to invasive tumour via the oncogenes and tumour suppressor genes involved (Figure 1). This basic understanding of the mechanisms driving cancer is critical to identify therapeutic targets and strategies. For a more detailed review of the development of mouse models of intestinal carcinogenesis, see our recent review (78).
Figure 1.
Venn diagram illustrating in vivo co-operation between mutations in the mouse intestine as determined by the phenotype of compound mutant mice. Adeno-CA, Adenocarcinoma. n.d., Not determined. Numbers indicate publication of phenotype as listed in the references.
Using mouse models to assess the importance of apoptosis to carcinogenesis
Mouse models have allowed the study of fundamental processes involved in carcinogenesis. In a landmark paper in 1982, David Vaux investigated the role of bcl-2, which had been previously identified in patients with follicular lymphoma as being translocated into the immunoglobulin heavy chain locus. He discovered that bcl2 did not promote proliferation, but rather prevented the death of cells deprived of growth factor, and hypothesized that genes regulating cell survival such as bcl-2 are important during transformation (79). This discovery of a new type of oncogene that inhibits cell death triggered huge interest into how cell death is regulated and how it might be exploited to treat cancer (80).
The differences between the phenotype of Rb1-/- mice and that of humans with Rb1 mutations questioned the validity of using mouse models of human disease. However, subsequent studies to reveal the reason for these differences led to substantial increases in our understanding of the molecular events behind genotype and phenotype, and inadvertently, that evasion of apoptosis (cell death) was a hallmark of cancer.
Although originally implicated as a regulator of cell cycle following DNA damage (81,82), it was also discovered that p53 regulated cell death (83,84). Indeed, this was able to help explain one of the molecular differences between mice and humans with regards to Rb1, as it was elegantly demonstrated in compound genetic mutant mice that Rb1-/- cells were deleted in the murine retina via p53-dependent apoptosis, thus explaining why Rb1-/- mice did not develop retinoblastoma (85).
p53 is also required for intestinal crypt cells to induce apoptosis after exposed to gamma irradiation in vivo (86), via a mechanism that requires Myc to activate p53 via Mdm2 (87) However, studies in p53 knockout mice showed that there was not simple correlations between loss of apoptosis, clonogenicity and carcinogenesis (88). For example, loss of p53 sensitizes mice to radiation despite blocking the apoptosis response, and increased p53 in this context can protect mice from radiation-induced pathologies. Remarkably the reason for this is not understood (89,90).
Separate studies from the groups of Tyler Jacks and Gerard Evan demonstrated that genetically restoring p53 function resulted in regression of lymphomas and sarcomas in mouse models, thus providing proof of principal that reactivation of p53 function could be therapeutically attractive for treating cancer (91,92) As one of the first genes to be knocked out in mice, p53 has been one of the most widely studied and has been implicated in multiple cell functions including apoptosis, proliferation, invasion, metabolism and stemness, and interacts with other oncogenes and tumour suppressors (93). Consequently, huge efforts had been devoted to generate or identify compounds that could restore wild-type activity to mutated p53, and of these APR-246 (PRIMA-1met) (94) was the first to be used in clinical trials (95), and is currently in phase Ib/II trials for patients with high-grade serous ovarian cancer (96).
The gp130/Jak/Stat signalling pathway is also frequently deregulated in many cancers, including those of the gastrointestinal tract, and thus has attracted research interest into the therapeutic benefit of targeting this pathway (97). One of the first experiments using transgenic mice to elucidate the role of this pathway in cancer identified that deletion of Stat3 in the mammary gland blocked apoptosis, thus implicating this pathway as a regulator of apoptosis in vivo for the first time (98). More recent studies have identified a similar role for Stat3 to regulate cell survival in intestinal tumourigenesis (99), highlighting the gp130/Jak/Stat pathway as an attractive target for therapeutic intervention for gastrointestinal cancer (100,101).
The ability to therapeutically induce apoptosis in cancer cells was very challenging, due in part to the observation that mutant p53 prevented the induction of pro-apoptotic BH3-only proteins (102). BH3-only proteins induce apoptosis by inhibiting Bcl2 family proteins, and thus an ingenious way to by-pass this problem was to mimic the function of BH3-only proteins which was achieved by Feik, Rosenberg and co-workers who developed a BH3-only memetic which had potent cell killing properties (103). In 2016, the Food and Drug Administration approved the use of a BH3-only memetic (Venclexta [venetoclax]) for the treatment of patients with chronic lymphocytic leukemia. This was the culmination of 28 years of research, after the original identification of Bcl2 as a regulator of cell death (80), and highlights the importance of basic research and how understanding molecular cell biology in vivo, can lead directly to translational outcomes to improve patient care.
Using mouse models to assess the functional importance of alterations in methylation to carcinogenesis
Epigenetic changes such as methylation act to regulate gene expression whereby methylation of regulatory CpG island shores, up to 2 kb away from gene promoter regions, can inhibit transcription (104). The CpG islands of tumour suppressor gene such as p16 (105) and indeed APC (106) in breast cancer are often highly methylated and this correlates with low expression. Global methylation was also observed to be frequently reduced (hypomethylated) in tumours (107) and thus an early question in the field was to define if this change in methylation was a cause or consequence of transformation. The ApcMin/+ mouse served as a useful mouse model to answer this question, with compound mutants in which the DNA methyltransferase gene Dnmt1 was also mutated resulted in marked decrease in intestinal tumour formation (108,109). These data suggested that methylation was an important regulator of tumourigenesis and not merely a non-functional outcome of the transformation itself.
Several subsequent studies have supported this model by investigating the consequences of disrupting methylation in mouse models of intestinal cancer. The methyl binding domain (Mbd) proteins, which can recognize methylated CpGs and recruit transcriptional repressors, have also been shown to influence tumourigenesis, as ApcMin/+Mbd2-/- mice developed significantly less tumours than their ApcMin/+ littermates (110). Similarly, genetic deletion of the DNA methylation-dependant transcriptional repressor Kaiso also reduced tumourigenesis in Kaiso deficient ApcMin/+ mice (111). It was thus speculated that epigenetic gene silencing that contributes to the cancer phenotype. In support of this hypothesis, transcriptome analysis of the intestine of Apc deleted Mbd2-/- mice revealed an increase in the transcriptional repressor Lect2, which was subsequently shown to be a novel Wnt pathway inhibitor (112). This provided a mechanism by which deficiency of methylation attenuates Wnt signalling to reduce intestinal tumourigenesis (112). Indeed, genome-wide analysis of promoter DNA methylation in several different cancers, including colon cancer, identified that Wnt pathway repressors are frequently regulated by epigenetic mechanisms (113). However, the precise role of Wnt target genes and methylation in colon cancer may not be as simple as first predicted as some Wnt target genes associated with the cancer stem cell (CSC) signature such as Lgr5, Ascl2 and Axin2 become silenced by CpG methylation during tumour progression, and can predict for recurrence-free survival (114).
It should be noted that not all Mbd proteins act in a similar fashion to that of Mbd2 described above. For example, ApcMin/+; Mbd4-/- mice actually had an increased susceptibility to develop intestinal tumours associated with more CpG to TpG mutations in the Apc gene (115). In addition to its role as a thymine glycosylate, Mbd4 has also been shown to interact with the DNA mismatch repair protein Mlh1 (116), which is able to mediate apoptosis in the intestinal epithelium and potentially increase the survival of transformed cells (117).
Hundreds of genes have altered methylation status in colon cancer and elucidating exactly which of these are important during each stage of colon cancer is of continued interest in the field. One of the challenges in studying methylation has been establishing a causative link between methylation and gene regulation at a single locus. For example, the genetic studies to investigate epigenetic regulators such as the methyl binding domain proteins result in expression changes to many genes, but cannot distinguish which of the changes is important for any observed phenotype without additional compound mutations. The development of CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated 9) technology (described in further detail in the ‘Organoids and CRSPR/Cas9 gene editing’ section below) allows researchers to fuse epigenetic regulatory domains such as the acetyletransferase p300 (118) or histone demethylase LSD1 (119) onto mutant Cas9 proteins which do not cleave DNA, and thus it is now possible to direct methylation, or demethylation to any locus using specific gRNA. Not only is this a powerful research tool, but it also presents a potential clinical tool to modify pathological methylation of tumour suppressor genes or oncogenes in the future.
As methylation is considered a hallmark of cancer, another potentially powerful use of understanding methylation changes is its use as a biomarker. In recent years, advances in technology have enabled comprehensive genome wide methylome analysis of tumours and matched normal tissue, and panels of markers have been identified which could help detect early-stage colon cancer when it is much easier to treat (reviewed in ref. (120)). Non-invasive methods of screening for methylation biomarkers are also being developed using more accessible samples including blood, urine and faeces (120).
In addition to its use as a biomarker, methylated DNA is also being analysed to help predict which patients will respond to certain treatments. For example, treatment with an anti-epidermal growth factor receptor (EGFR) antibody is restricted to patients without mutations in KRAS (121), and methylation associated suppression of PTEN has also been shown to predict for a lack of benefit from these drugs (122). Thus, the information gained from the original studies to understand the role of methylation in cancer could lead to methylation biomarkers for detection and drug response in the clinic and potentially novel screening strategies.
Using mouse models to define intestinal stem cells
Identification and isolation of the intestinal stem cell (ISC) has long been considered key to understanding tumourigenesis of the intestine, since they were thought to be the only epithelial cell type which persists long enough to accumulate the multiple mutations required for transformation. However, the location and identity of ISCs has been a topic of intense research and debate. The most popular model as the putative location of ISCs was originally proposed in 1965 (123), and further supported by work from Chris Potten and colleagues, as just above the Paneth cells at +4 from the base of the crypts (124). In contrast a second model proposed by Chen and Leblond in 1974, after they observed a population of slender cells wedged between Paneth cells at the base of the crypts which divided once a day called crypt base columnar cells (CBCs) (125). Rudimentary lineage tracing showed that 3H-Thymidine retaining cells were originally restricted to CBCs but subsequently, radioactive differentiated cells were observed, thus suggesting CBCs could give rise to differentiated cells, and that these cells, and not the +4 cells were the ISCs (125).
In 2002, Hans Clevers group demonstrated that Wnt signalling was required for the development of intestinal crypts, and thus hypothesized Wnt signalling targets may mark the ISC (126). In one of the first experiments using cDNA microarrays to assay expression of large sets of genes, they analysed data from colon cancer cells with and without active Wnt signalling and mapped these genes back to the intestine using in situ hybridization to see where deregulated genes were expressed in normal crypts (127). One of the genes upregulated in their array was leucine-rich-repeat-containing G-protein-coupled receptor 5, Lgr5 or Gpr49. Nick Barker generated an Lgr5EGFP- CreERT2 knockin mouse in which EGFP was driven from the endogenous promoter of Lgr5, and Cre could be induced upon injection of TM under control of the endogenous Lgr5 regulatory region. He observed green fluorescent protein expression in the CBCs, and was then able to demonstrate that these cells gave rise to all the differentiated cells of the intestinal epithelium in long term experiments in which the Lgr5EGFP-CreERT2 mouse was crossed to the ROSA26LACZLSL mouse to allow genuine lineage tracing in vivo (128). This landmark paper transformed the field of stem cell biology and although several other stem cell markers for the +4 position have been proposed, including Bmi-1, Lrig1, HopX and mTERT, Lgr5 remains a robust and convincing stem cell marker in intestinal crypts, although debate still continues especially regarding populations of reserve stem cells and the inherent plasticity and interconversion of stem cells (reviewed in ref. (129)).
Developing transgenic mice and identifying ISC markers has allowed researchers to gain huge insight into the function of this important cell population. An area of great debate in the field was the nature of the turn-over and population dynamics of ISCs in their crypt base niche. There were two models that researchers considered most likely; firstly, that of asymmetrical division, whereby a stem cell gives rise to another stem cell and a transit amplifying cell, and secondly, stochastic, neutral drift, in which stem cells divide symmetrically and their fates are dependent on environmental factors such as their location in the niche relative to Wnt expressing Paneth cells. In 2010, two independent studies using fate mapping experiments demonstrated that neutral drift was the more likely of the two models. The Clevers group used the Lgr5Cre R26-Confetti mouse in fate mapping experiments to demonstrate that neutral drift was the more likely of the two models (130), whilst the Winton group used very low deletion of a single reporter (in ISC marker agnostic approach) to reach the same conclusion (131). Both groups also suggest similar patterns in adenoma however as yet no-one has experimentally examined the nature of stem cells in carcinomas (132). From elegant modelling studies in human tumours, a big bang model has been proposed where selection occurs early but once tumours are large neutral drift best explains the evolution of tumours (133).
Given the rapid turnover of the intestine, every 4–5 days, which is driven by continuous and rapid proliferation of cells in the transit amplifying zone, just above the crypt base, ‘reserve’ ISC or label retaining cells must be able to escape these processes. Paneth cells, which secrete antimicrobial peptides and enzymes such as defensin and lysozyme, are located at the base of the crypt between the stem cells and therefore seem to escape this upward migration and reside within the intestine for many weeks (134). In 2013 the Winton group solved this long-standing question using H2B-YFP to identify long-term, label retaining cells. They found that these slow cycling cells were located in the stem cell niche and had a combined secretory and stem cell signature, and over time differentiated into Paneth cells (134). Furthermore, label retaining cells were able to function as a reserve pool of stem cells as they were able to contribute to the stem cell pool after crypts were damaged chemically or by irradiation (134–136).
A population of reserve stem cells is also proposed at the +4 position, and marked by one of several different markers including Bmi-1 (137), Lrig1 (138) and Hoxp (139). For example, ablation of Lgr5+ cells does not perturb intestinal homeostasis (137,140), due to additional cells expressing Bmi-1 (137) or Hoxp (139) having the capacity to compensate and subsequently re-express Lgr5. Taken together, these experiments demonstrate a high level of plasticity whereby a population of functional adult stem cells can be maintained in situations of stress. This plasticity in the intestinal epithelium has most likely evolved to enable it to cope with the harsh physical and chemical stresses encountered in the gut. For a more detailed review of this area see the recent publication by Snippert and Vermuelen (136).
Stem cells, Wnt signalling and intestinal regeneration
Mouse models have also allowed the investigation of the intestines remarkable capacity to regenerate following injury. If it was not for this regenerative function it is unlikely that anyone would survive radiotherapy. The canonical Wnt signalling pathway is activated during intestinal regeneration as assayed by the presence of nuclear β-catenin and upregulation of Wnt target genes including c-Myc (141). Indeed c-Myc was demonstrated to be required for efficient intestinal regeneration, since epithelial deletion of c-Myc prior to irradiation resulted in a marked reduction in the number of regenerating crypts via a mechanism involving focal adhesion kinase and Akt/mTOR (141). C-Myc was also shown to be critical for stem cell function in the intestinal epithelium, as deletion specifically in this tissue using AhCre; c-Mycfl/fl mice triggered rapid repopulation of the epithelium in a process similar to regeneration, but differing in that the epithelium is not denuded prior to the event but rather is triggered via a deleterious event in the stem cells (in this case deletion of c-Myc). This process of repopulation is another mechanism to assist the intestinal epithelium to cope with its harsh environment, and was also observed upon deletion of other critical genes including Stat3 (142), Chk1 (143) and Brg1 (144).
Lgr5 + cells have been demonstrated to be essential for intestinal regeneration as ablation of Lgr5+ cells using Lgr5DTR mice treated with diphtheria toxin completely blocked regeneration following irradiation (140). The importance of Wnt signalling during regeneration was functionally demonstrated when it was shown that the Wnt receptor Fzd7 was required for efficient regeneration (145). Fzd7 was also demonstrated to be the predominant Wnt receptor transmitting Wnt signalling in the Lgr5+ ISCs since deletion was deleterious and triggered repopulation (145).
The source and requirement for Wnt ligands in the intestine during homeostasis and regeneration is still an area of controversy. Hans Clevers’ lab showed that Paneth cells were required for efficient establishment of mini-gut organoid cultures, and that they secreted various growth factors including Wnt3 (146). The Wnt agonist R-Spondin 1 is also required in the culture medium to culture organoids (146). In support of the model whereby Paneth cells supply Wnt ligand for intestinal regeneration, Alan Clarke’s group showed that the capacity of the intestinal epithelium to repopulate after deletion of β-catenin was severely compromised in the absence of functional Paneth cells resulting in loss of intestinal epithelial integrity (147). This supported a previous study from Ricardo Fodde’s group showing that Paneth cells lose their differentiated identity after irradiation and can proliferate to contribute to regeneration (135). Interestingly, inflammation induced by treating mice with 3% dextran sodium sulfate was recently shown to promote the secretion of secreted phospholipases A2 (sPLA2s) in Paneth cells which increased Wnt signalling to regulate stem cell function (148). This could provide a possible mechanism for Paneth cells positively regulating regeneration/inflammation in the intestine. In contrast to these results, a study from Frederic de Sauvages group observed a robust regenerative response in the intestine of Paneth cell-deficient Gfi1KI/KI mice, suggesting Paneth cells are dispensable for regeneration (140).
Surprisingly, there was no detectable phenotype when Wnt3 was deleted from the intestine in vivo, however, cultured organoids did require Paneth cell-derived Wnt3, suggesting that a mesenchymal source of Wnt could compensate for the loss of epithelial Wnt3 (149). Indeed, Wnt2b was able to activate Wnt signalling and could restore the growth of intestinal organoids in which Wnt secretion was perturbed (149,150). Mice lacking Wnt2b have normal intestinal homeostasis (151), suggesting that mesenchymal Wnt2b acts as a safeguard to ensure Wnt ligands can be delivered to the ISCs should Paneth cell-derived Wnt3 be compromised in situations that require the intestine to regenerate. This is supported by the observation that Fzd7, the Wnt receptor transmitting Wnt signalling in Lgr5+ ISCs can bind Wnt3 and Wnt2b with equal affinity (145). Recently, it has been proposed that Wnt ligands can be secreted in extracellular vesicles by macrophages during intestinal regeneration (152), which may also suggest that under different stimuli Wnt secretion from different cells can regulate either homeostasis (low Wnt levels expressed from epithelium allowing normal differentiation) or regeneration in the intestine (higher Wnt levels expressed from macrophages to drive hyperproliferation and repair).
The biology of the colonic epithelium is slightly different to that of the small intestine, as the colonic epithelium does not contain Paneth cells. However, a subset of C-Kit+ goblet cells has been identified that are able to support Lgr5+ stem cells by secreting Notch and epidermal growth factor ligands (153). As organoids cultured from the colonic epithelium also require Wnt signalling to grow (150), this suggests a more pronounced dependence on mesenchymal Wnts in the colon, which is supported by the observation that no Wnt genes are reported to be expressed in the colon epithelium (149).
The studies above illustrate the importance of Wnt signalling during regeneration and homeostasis of the intestine but it is important to recognize that Wnt signalling does not function in isolation, but rather interacts with other signalling pathways that are rate limiting for Wnt-induced phenotypes including gp130/Jak/Stat (99 100), Robo1/Slit (154,155), EGFR/Yap (156), Notch (157), mTORC1 (158) and Rac1 (159). These studies and many others highlight the huge complexity and interaction between multiple signalling pathways in the intestine.
The cell of origin and CSCs
The cell of origin for intestinal cancer was unknown at the time when Lgr5 was discovered, and two models were the subject of great debate; the bottom-up model which proposed the cell of origin at the base of the crypts (160), and the top-down model which proposed the transformation towards the luminal surface and spreads down (161). Using the Lgr5CreERT2 mouse to delete Apc specifically in Lgr5+ ISCs, Barker and colleagues observed rapid, widespread tumour initiation throughout the intestinal tract (162). Importantly, they also observed that deletion of Apc in differentiated enterocytes (using low levels of recombination in AhCre Apcfl/fl mice) caused the formation of small lesions that rarely went on to form tumours (or if they did it was at long latency), thus demonstrating that Lgr5+ stem cells were indeed efficient cells of origin for intestinal cancer (162). This paper seemed to have settled the debate in favour of the bottom-up model, however, two papers then discovered that transformation could occur in Lgr5-negative cells outside of the crypt base stem cell niche, ether in cells with Apc and Kras mutations (163), or those expressing the bone morphogenic protein antagonist Gremlin (164). More recently, the development of a new Cre (carbonic anhydrase), which does not induce in colonic stem cells but only in more differentiated cells, confirmed these results in the colon (165). Whilst Apc loss alone was unable to cause tumours (only small lesions), if both Apc and Kras were mutated mice then developed tumours with a “top down morphology” (165). These papers demonstrate that the CBC is not the exclusive cell of origin for all subtypes of CRC, and that the top-down model is also valid. However, it should be remembered that in human cancer it would be very rare that three alleles would be lost simultaneously, so the top down model would require the first APC mutation in a stem cell to repopulate the crypt (for a more thorough review of this area, see ref. (166)). It is possible that multiple mutations could occur in the same cell at the same time, in situations of high cellular stress such as inflammation or during massive chromosomal rearrangements (133). Indeed the ‘big bang’ model predicts that driver mutations are all present from the beginning of tumourigenesis, however they may not necessarily have been acquired by the same cell at the same time, but rather are acquired rapidly in a cell after the first mutation has occurred which then results in a large clonal expansion (133).
The discovery of Lgr5 as an ISC marker also helped contribute to the continuing debate regarding CSCs. CSCs are hypothesized to be a small population of cells that reside within the tumour and are responsible for the growth of the tumour, and thus a very attractive target for therapeutic intervention. Experiments investigating CSCs using transplantation of sorted cell populations in immunodeficient mice are compromised by the culture procedure the cells must undergo during the assay. This may skew the outcomes towards a cell population which is inadvertently selected for by the process itself (167). Therefore, it was important to investigate stem cell potential in an autotochronous model without transplantation. This was achieved using the R26-Confetti mouse, which can express one of four random colours upon recombination. It was demonstrated that the progeny of transformed cells could be lineage traced in the tumours of Lgr5Cre; Apcfl/fl; R26-Confetti mice as patches of single colour, demonstrating that each patch of the tumour (colour) had originated from a single cell (168). Importantly, the R26-confetti mouse retains two of its four colours in the confetti allele after recombination, thus allowing for re-tracing upon a second injection of TM, which subsequently showed that only small populations of cells recombined to generate a second colour, demonstrating that the rare Lgr5+ CSCs give rise to the other cell types present in the tumour (168). It should be noted this study showed that a population of cells was acting as stem cells in adenomas but not their functional relevance. Furthermore, depletion of Lgr5+ cells in the intestine with diphtheria toxin did not prevent the formation of hyperplastic crypts when Apc was deleted, suggesting that Lgr5+ cells are not required for crypt hyperplasia (140). However, the diphtheria toxin depletion of Lgr5+ cells in an established intestinal tumour of, for example a Min mouse, has not been published to date and thus the definite role of Lgr5+ CSCs has yet to be fully established.
The experiments above suggest that targeting the cell of origin or CSCs could be an attractive strategy for treating colon cancer. Indeed co-deletion of Apc and either Brahma-related gene 1 (Brg1) (169) or one allele of Stat3 (100) specifically in intestinal Lgr5+ cells, significantly reduced tumourigenesis. Interestingly, the mechanism for these two similar phenotype outcomes was different, as Brg1 deficiency attenuated Wnt signalling (169), whilst reduced Stat3 induced senescence to reduce tumour growth and initiation even though Wnt signalling remained high (100).
Organoids and CRSPR/Cas9 gene editing
Using genetically engineered mouse models has allowed researchers to gain huge insight into the signalling pathways and cell biology required for homeostasis, stem cell function and cancer in the intestine. This knowledge enabled Hans Clevers’ group to develop a robust technique for the long-term 3D culture of intestinal organoids, which shared many of the physiological characteristics of the in vivo epithelium (146,170). This very close resemblance to the primary tissue from which they are derived gives organoids a distinct advantage over 2D cultured cell lines and to date culture conditions have been published for 15 different types of tissue from mice and humans (171).
Importantly, cancer organoids have also been established from humans thus providing a physiologically accurate, expandable supply of ex vivo tissue from patients. This type of approach is particularly useful to gain rapid knowledge about the transcriptome and genetic and epigenetic mutations of a patient’s tumour and also provides the ability to perform high-throughput screens of drug combinations to determine the most effective treatment for each individual.
Another exciting potential use for organoids is in regenerative medicine, in which diseased tissue can be transplanted with healthy tissue. Indeed, engraftment of orthotopically-transplanted organoids has been used to repair damaged tissue in vivo in the intestine (172), pancreas (173) and liver (174). CRISPR/Cas9 is the latest technology to be developed to target and manipulate specific sequences of DNA (175) or RNA (176), which allows rapid, cheap replacement of mutated genes. This represents considerable promise for regenerative medicine in which mutant genes can be replace in patient-derived cultured organoids, thus generating healthy tissue which can be expanded for replacing diseased tissue (171).
CRISPR/Cas9 was discovered as a mechanism of immunity in bacteria in which the CRISPR transcript, crRNA, guides the endonuclease Cas9 to cleave unwanted, invading DNA from a virus or plasmid (175). A non-coding trans-activating crRNA (tracrRNA) was found to be required to facilitate crRNA to associate with Cas9 and cleave DNA (177). It was subsequently shown that by fusing the crRNA and the tracrRNA together, a single guide RNA (sgRNA) could be constructed that contained the specific targeting sequence that was able to associate with Cas9 to facilitate site-directed gene editing (178). Thus, two components are required to target a gene; Cas9, and a sgRNA comprised of a gene target sequence of 23 base pair target sequence fused to a tracrRNA. When these are introduced into a cell, or indeed a fertilized zygote to generate mutant mice, they form a complex, which is guided to the gene of interest by the 23 base pair sequence, and introduce a double strand break in the DNA. If disruption of the gene of interest is the goal, then the non-homologous end joining can be allowed to proceed until an error occurs, leaving indels, and subsequent disruption of the gene. The introduction of new sequences of DNA is also possible with the addition of a donor DNA template during the process, thus allowing much more complex gene editing (175). Further utilizations of CRIPR/Cas9 technology are currently being developed at a rapid pace including introduction of epigenetic silencing (118) and even conditional Cas9 (mutated to prevent DNA cleavage)-mediated gene silencing, and editing, dependant on exposure to blue light (179,180) or doxycycline (181).
CRISPR/Cas9 was recently employed to demonstrate the multistep mechanism of intestinal tumour progression in human intestinal organoids, in which combinations of mutations in APC, KRAS, SMAD4 and P53 enabled transformed organoids to grow without the addition of stem cell niche factors (182). These organoids displayed several characteristics of intestinal tumours including chromosomal instability and aneuploidy, and became invasive when xenotransplanted into immunocompromised mice, thus demonstrating the considerable power of combining these two technologies to provide a fast and cheap way of generating a model of colon cancer in human cells.
Tumours contain hundreds of mutant genes, and developing accurate animal models of disease will probably require that at least three to five of these genes is mutated. Achieving this using the traditional methods of generating mouse mutants will be very expensive and time consuming. However the use of CRISPR/Cas9 can significantly speed up this process, as multiple mutations can be introduced into a single fertilized zygote and therefore the first generation of mice are already compound mutants, thus alleviating expensive, timely cross-breeding to generate the desired genotypes. For a more comprehensive review of CRISPR/Cas9 technology please see (175).
The expandable nature of organoids provides a large supply of tissue for use in regenerative medicine, and high throughput assays for drug discovery and drug combination screens. However, as organoids do not contain any stroma or immune components, there are limitations to what can be accurately modelled in this system to date (171). This is particularly frustrating with regards to the current advances being made with immune checkpoint inhibitors as they cannot be tested in organoids. Similarly, patient-derived xenografts are a very useful platform to study the response of fresh human tumour tissue to novel therapies, especially in cancers that are very heterogeneous such as breast, ovarian and gastric cancer. However, patient-derived xenografts are performed in immunocompromised mice, which again prevents analysis of the host immune system during therapy. Animal models therefore remain the most accurate way of simulating disease, allowing analysis from tumour initiation through to metastasis, and with the advent of CRISPR/Cas9 to allow cheap generation of compound mutants, including inducible compound mutants, the use of animal models of disease is entering a new and exciting phase. Indeed, very recent work has used CRISPR/Cas to mutate Apc, Kras and Trp53 in colonic organoids which developed invasive tumours after orthotopic, syngeneic transplantation into the mouse colon using a modified colonoscope, suggesting the next stage of in vivo mouse models has begun (183,184).
Conclusion
Animal models of disease, particularly intestinal cancer, offer significant advantages over the in vitro and ex vivo technologies that have been developed and refined in the last decade some of which are described above. Recent research has begun to uncover the intimate relationship between the biology of the intestine and its resident commensal microbiome, suggesting it also plays an important role during colon cancer. Animal models have again been employed to gain insight into this interaction which is more challenging to accurately model in vitro. Thus, with the development of mouse models of colon cancer that are immune-proficient, develop invasive tumours in the correct anatomical site as the human disease (colon) and contain a commensal microbiome, researchers have developed very accurate pre-clinical models of colon cancer.
The massive increase in our understanding of cancer biology in the past 20 years could not have been achieved without the ground-breaking discoveries and advances in technology accomplished by some of the talented scientists mentioned above. This review has described the events that led to the first animal models of disease, the first conditional gene manipulation in vivo, the use of these models to elucidate cancer biology, and finally the exciting developments in utilising CRISPR/Cas9 for cheaper, faster and potentially more complex gene editing in vivo. Inserting new DNA using CRISPR/Cas9 still utilizes the homologous recombination strategies discovered by the pioneers of generating mutant mice, and thus the skills and techniques they developed, for example to design targeting vectors, are still relevant and critical today.
Of those involved in these field-changing advances, we would like to highlight the contributions made by the late Professor Alan Clarke who was a pioneer in the very early use of genetic engineering to make mutant mice, and was involved in the very first successful gene-targeted Hprt mutant mouse, and first author on the subsequent Nature publications generating the p53 and Rb1 mutants. Alan was also at the forefront of the use of these mice to help broaden our understanding of disease and the biology of several organs, and was the first to identify the regulation of apoptosis by p53, demonstrated that loss of Apc activated Wnt signalling to trigger intestinal tumourigenesis, showed the role of methylation regulating genes during intestinal cancer and characterized the genetic co-operation between many oncogenes and tumour suppressor genes in vivo. Alan’s contribution to the field of cancer biology can thus be considered in the highest echelons of scientists past and present, and his findings will continue to help shape the field as it progresses.
Funding
T.J.P. is funded by a Wellcome Trust ISSF Fellowship, and a Capital Medical University/Cardiff University Fellowship. V.M.D. is funded by a European Cancer Stem Cell Research institute fellowship and O.J.S. is funded by a Cancer Research UK programme grant (A21139) and ERC investigator award ‘ColonCan’ (311301).
Acknowledgements
This review is dedicated to our friend and colleague Professor Alan Clarke, whose scientific brilliance continues to inspire us even after his tragic death at the end of 2015, and whom we miss greatly. Thank you to the Fellows at the European Cancer Stem Cell Research Institute for their helpful discussions regarding the generation of the figure in this review.
Conflict of Interest Statement: None declared.
Abbreviations
- APC
adenomatous Polyposis coli
- CBC
crypt base columnar cell
- CRC
colorectal cancer
- CSC
cancer stem cell
- EGFR
epidermal growth factor receptor
- ISC
intestinal stem cell
- Mbd
methyl binding domain
- TGF-β
transforming growth factor-β
- TM
tamoxifen
References
- 1. Nordling C.O. (1953) A new theory on cancer-inducing mechanism. Br. J. Cancer, 7, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Knudson A.G., Jr (1971) Mutation and cancer: statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U. S. A., 68, 820–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evans M.J., et al. (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature, 292, 154–156. [DOI] [PubMed] [Google Scholar]
- 4. Thomas K.R., et al. (1987) Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell, 51, 503–512. [DOI] [PubMed] [Google Scholar]
- 5. Doetschman T., et al. (1987) Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature, 330, 576–578. [DOI] [PubMed] [Google Scholar]
- 6. Thompson S., et al. (1989) Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell, 56, 313–321. [DOI] [PubMed] [Google Scholar]
- 7. Clarke A.R., et al. (1992) Requirement for a functional Rb-1 gene in murine development. Nature, 359, 328–330. [DOI] [PubMed] [Google Scholar]
- 8. Jacks T., et al. (1992) Effects of an Rb mutation in the mouse. Nature, 359, 295–300. [DOI] [PubMed] [Google Scholar]
- 9. Lee E.Y., et al. (1992) Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature, 359, 288–294. [DOI] [PubMed] [Google Scholar]
- 10. Zhang J., et al. (2004) The first knockout mouse model of retinoblastoma. Cell Cycle, 3, 952–959. [PubMed] [Google Scholar]
- 11. Chen J., et al. (1992) T-antigen mutant activities in vivo: roles of p53 and pRB binding in tumorigenesis of the choroid plexus. Oncogene, 7, 1167–1175. [PubMed] [Google Scholar]
- 12. Loeb L.A., et al. (2008) Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res., 68, 6863–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balmain A., et al. (1983) Mouse skin carcinomas induced in vivo by chemical carcinogens have a transforming Harvey-ras oncogene. Nature, 303, 72–74. [DOI] [PubMed] [Google Scholar]
- 14. Sukumar S., et al. (1983) Induction of mammary carcinomas in rats by nitroso-methylurea involves malignant activation of H-ras-1 locus by single point mutations. Nature, 306, 658–661. [DOI] [PubMed] [Google Scholar]
- 15. Moser A.R., et al. (1990) A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science, 247, 322–324. [DOI] [PubMed] [Google Scholar]
- 16. Mahmoud N.N., et al. (1998) Aspirin prevents tumors in a murine model of familial adenomatous polyposis. Surgery, 124, 225–231. [PubMed] [Google Scholar]
- 17. Burn J., et al. ; CAPP2 Investigators. (2011) Long-term effect of aspirin on cancer risk in carriers of hereditary colorectal cancer: an analysis from the CAPP2 randomised controlled trial. Lancet, 378, 2081–2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gordon J.W., et al. (1980) Genetic transformation of mouse embryos by microinjection of purified DNA. Proc. Natl. Acad. Sci. U. S. A., 77, 7380–7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brinster R.L., et al. (1981) Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell, 27(1 Pt 2), 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costantini F., et al. (1981) Introduction of a rabbit beta-globin gene into the mouse germ line. Nature, 294, 92–94. [DOI] [PubMed] [Google Scholar]
- 21. Harbers K., et al. (1981) Microinjection of cloned retroviral genomes into mouse zygotes: integration and expression in the animal. Nature, 293, 540–542. [DOI] [PubMed] [Google Scholar]
- 22. Wagner T.E., et al. (1981) Microinjection of a rabbit beta-globin gene into zygotes and its subsequent expression in adult mice and their offspring. Proc. Natl. Acad. Sci. U. S. A., 78, 6376–6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Adams J.M., et al. (1985) The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature, 318, 533–538. [DOI] [PubMed] [Google Scholar]
- 24. Gossen M., et al. (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. U. S. A., 89, 5547–5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Furth P.A., et al. (1994) Temporal control of gene expression in transgenic mice by a tetracycline-responsive promoter. Proc. Natl. Acad. Sci. U. S. A., 91, 9302–9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chin L., et al. (1999) Essential role for oncogenic Ras in tumour maintenance. Nature, 400, 468–472. [DOI] [PubMed] [Google Scholar]
- 27. Zuber J., et al. (2011) Toolkit for evaluating genes required for proliferation and survival using tetracycline-regulated RNAi. Nat. Biotechnol., 29, 79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sauer B., et al. (1988) Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. U. S. A., 85, 5166–5170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gu H., et al. (1994) Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science, 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 30. Hayashi S., et al. (2002) Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol., 244, 305–318. [DOI] [PubMed] [Google Scholar]
- 31. Brocard J., et al. (1997) Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc. Natl. Acad. Sci. U. S. A., 94, 14559–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwenk F., et al. (1998) Temporally and spatially regulated somatic mutagenesis in mice. Nucleic Acids Res., 26, 1427–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Indra A.K., et al. (1999) Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res., 27, 4324–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Feil R., et al. (1997) Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun., 237, 752–757. [DOI] [PubMed] [Google Scholar]
- 35. Schönhuber N., et al. (2014) A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat. Med., 20, 1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Urlinger S., et al. (2000) Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. U. S. A., 97, 7963–7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Su L.K., et al. (1992) Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science, 256, 668–670. [DOI] [PubMed] [Google Scholar]
- 38. Rubinfeld B., et al. (1993) Association of the APC gene product with beta-catenin. Science, 262, 1731–1734. [DOI] [PubMed] [Google Scholar]
- 39. Peifer M., et al. (1990) The segment polarity gene armadillo encodes a functionally modular protein that is the Drosophila homolog of human plakoglobin. Cell, 63, 1167–1176. [DOI] [PubMed] [Google Scholar]
- 40. Morin P.J., et al. (1997) Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science, 275, 1787–1790. [DOI] [PubMed] [Google Scholar]
- 41. Korinek V., et al. (1997) Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science, 275, 1784–1787. [DOI] [PubMed] [Google Scholar]
- 42. Shibata H., et al. (1997) Rapid colorectal adenoma formation initiated by conditional targeting of the Apc gene. Science, 278, 120–123. [DOI] [PubMed] [Google Scholar]
- 43. Ireland H., et al. (2004) Inducible Cre-mediated control of gene expression in the murine gastrointestinal tract: effect of loss of beta-catenin. Gastroenterology, 126, 1236–1246. [DOI] [PubMed] [Google Scholar]
- 44. Sansom O.J., et al. (2004) Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev., 18, 1385–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Andreu P., et al. (2005) Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development, 132, 1443–1451. [DOI] [PubMed] [Google Scholar]
- 46. Harada N., et al. (2000) Development of an automated synthesis apparatus for L-[3-11C] labeled aromatic amino acids. Appl. Radiat. Isot., 52, 845–850. [DOI] [PubMed] [Google Scholar]
- 47. Foulds L. (1958) The natural history of cancer. J. Chronic Dis., 8, 2–37. [DOI] [PubMed] [Google Scholar]
- 48. Weinberg R.A. (1989) Oncogenes, antioncogenes, and the molecular bases of multistep carcinogenesis. Cancer Res., 49, 3713–3721. [PubMed] [Google Scholar]
- 49. Vogelstein B., et al. (1988) Genetic alterations during colorectal-tumor development. N. Engl. J. Med., 319, 525–532. [DOI] [PubMed] [Google Scholar]
- 50. TCGA. (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takaku K., et al. (1998) Intestinal tumorigenesis in compound mutant mice of both Dpc4 (Smad4) and Apc genes. Cell, 92, 645–656. [DOI] [PubMed] [Google Scholar]
- 52. Sansom O.J., et al. (2006) Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc. Natl. Acad. Sci. U. S. A., 103, 14122–14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bennecke M., et al. (2010) Ink4a/Arf and oncogene-induced senescence prevent tumor progression during alternative colorectal tumorigenesis. Cancer Cell, 18, 135–146. [DOI] [PubMed] [Google Scholar]
- 54. Johnson L., et al. (2001) Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature, 410, 1111–1116. [DOI] [PubMed] [Google Scholar]
- 55. Marsh V., et al. (2008) Epithelial Pten is dispensable for intestinal homeostasis but suppresses adenoma development and progression after Apc mutation. Nat. Genet., 40, 1436–1444. [DOI] [PubMed] [Google Scholar]
- 56. He X.C., et al. (2007) PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat. Genet., 39, 189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Katajisto P., et al. (2008) LKB1 signaling in mesenchymal cells required for suppression of gastrointestinal polyposis. Nat. Genet., 40, 455–459. [DOI] [PubMed] [Google Scholar]
- 58. Marsh Durban V., et al. (2014) Epithelial-specific loss of PTEN results in colorectal juvenile polyp formation and invasive cancer. Am. J. Pathol., 184, 86–91. [DOI] [PubMed] [Google Scholar]
- 59. Trimboli A.J., et al. (2009) Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature, 461, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Davies E.J., et al. (2014) PTEN loss and KRAS activation leads to the formation of serrated adenomas and metastatic carcinoma in the mouse intestine. J. Pathol., 233, 27–38. [DOI] [PubMed] [Google Scholar]
- 61. Hare L.M., et al. (2014) Physiological expression of the PI3K-activating mutation Pik3ca(H1047R) combines with Apc loss to promote development of invasive intestinal adenocarcinomas in mice. Biochem. J., 458, 251–258. [DOI] [PubMed] [Google Scholar]
- 62. Donehower L.A., et al. (1992) Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 356, 215–221. [DOI] [PubMed] [Google Scholar]
- 63. Jacks T., et al. (1994) Tumor spectrum analysis in p53-mutant mice. Curr. Biol., 4, 1–7. [DOI] [PubMed] [Google Scholar]
- 64. Kandoth C., et al. (2013) Mutational landscape and significance across 12 major cancer types. Nature, 502, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Halberg R.B., et al. (2000) Tumorigenesis in the multiple intestinal neoplasia mouse: redundancy of negative regulators and specificity of modifiers. Proc. Natl. Acad. Sci. U. S. A., 97, 3461–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Muller P.A., et al. (2009) Mutant p53 drives invasion by promoting integrin recycling. Cell, 139, 1327–1341. [DOI] [PubMed] [Google Scholar]
- 67. Schwitalla S., et al. (2013) Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell, 23, 93–106. [DOI] [PubMed] [Google Scholar]
- 68. Hinoi T., et al. (2007) Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res., 67, 9721–9730. [DOI] [PubMed] [Google Scholar]
- 69. Grivennikov S.I., et al. (2012) Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature, 491, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cooks T., et al. (2013) Mutant p53 prolongs NF-κB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer Cell, 23, 634–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Muñoz N.M., et al. (2006) Transforming growth factor beta receptor type II inactivation induces the malignant transformation of intestinal neoplasms initiated by Apc mutation. Cancer Res., 66, 9837–9844. [DOI] [PubMed] [Google Scholar]
- 72. Trobridge P., et al. (2009) TGF-beta receptor inactivation and mutant Kras induce intestinal neoplasms in mice via a beta-catenin-independent pathway. Gastroenterology, 136, 1680–8.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fessler E., et al. (2016) TGFβ signaling directs serrated adenomas to the mesenchymal colorectal cancer subtype. EMBO Mol. Med., 8, 745–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Guinney J., et al. (2015) The consensus molecular subtypes of colorectal cancer. Nat. Med., 21, 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Calon A., et al. (2015) Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat. Genet., 47, 320–329. [DOI] [PubMed] [Google Scholar]
- 76. Rad R., et al. (2013) A genetic progression model of Braf(V600E)-induced intestinal tumorigenesis reveals targets for therapeutic intervention. Cancer Cell, 24, 15–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dow L.E., et al. (2015) Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell, 161, 1539–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jackstadt R., et al. (2016) Mouse models of intestinal cancer. J. Pathol., 238, 141–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vaux D.L., et al. (1988) Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature, 335, 440–442. [DOI] [PubMed] [Google Scholar]
- 80. Delbridge A.R., et al. (2016) Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer, 16, 99–109. [DOI] [PubMed] [Google Scholar]
- 81. Kastan M.B., et al. (1991) Participation of p53 protein in the cellular response to DNA damage. Cancer Res., 51(23 Pt 1), 6304–6311. [PubMed] [Google Scholar]
- 82. Kastan M.B., et al. (1992) A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 71, 587–597. [DOI] [PubMed] [Google Scholar]
- 83. Clarke A.R., et al. (1993) Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature, 362, 849–852. [DOI] [PubMed] [Google Scholar]
- 84. Lowe S.W., et al. (1993) p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature, 362, 847–849. [DOI] [PubMed] [Google Scholar]
- 85. Morgenbesser S.D., et al. (1994) p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature, 371, 72–74. [DOI] [PubMed] [Google Scholar]
- 86. Clarke A.R., et al. (1994) p53 dependence of early apoptotic and proliferative responses within the mouse intestinal epithelium following gamma-irradiation. Oncogene, 9, 1767–1773. [PubMed] [Google Scholar]
- 87. Phesse T.J., et al. (2014) Endogenous c-Myc is essential for p53-induced apoptosis in response to DNA damage in vivo. Cell Death Differ., 21, 956–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sansom O.J., et al. (2002) The ability to engage enterocyte apoptosis does not predict long-term crypt survival in p53 and Msh2 deficient mice. Oncogene, 21, 5934–5939. [DOI] [PubMed] [Google Scholar]
- 89. Christophorou M.A., et al. (2006) The pathological response to DNA damage does not contribute to p53-mediated tumour suppression. Nature, 443, 214–217. [DOI] [PubMed] [Google Scholar]
- 90. Kirsch D.G., et al. (2010) p53 controls radiation-induced gastrointestinal syndrome in mice independent of apoptosis. Science, 327, 593–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ventura A., et al. (2007) Restoration of p53 function leads to tumour regression in vivo. Nature, 445, 661–665. [DOI] [PubMed] [Google Scholar]
- 92. Martins C.P., et al. (2006) Modeling the therapeutic efficacy of p53 restoration in tumors. Cell, 127, 1323–1334. [DOI] [PubMed] [Google Scholar]
- 93. Kruiswijk F., et al. (2015) p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol., 16, 393–405. [DOI] [PubMed] [Google Scholar]
- 94. Bykov V.J., et al. (2002) Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med., 8, 282–288. [DOI] [PubMed] [Google Scholar]
- 95. Lehmann S., et al. (2012) Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J. Clin. Oncol., 30, 3633–3639. [DOI] [PubMed] [Google Scholar]
- 96. Bykov V.J., et al. (2016) Targeting of mutant p53 and the cellular redox balance by APR-246 as a strategy for efficient cancer therapy. Front. Oncol., 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Jones S.A., et al. (2011) Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J. Clin. Invest., 121, 3375–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Chapman R.S., et al. (1999) Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev., 13, 2604–2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bollrath J., et al. (2009) gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell, 15, 91–102. [DOI] [PubMed] [Google Scholar]
- 100. Phesse T.J., et al. (2014) Partial inhibition of gp130-Jak-Stat3 signaling prevents Wnt-β-catenin-mediated intestinal tumor growth and regeneration. Sci. Signal., 7, ra92. [DOI] [PubMed] [Google Scholar]
- 101. Stuart E., et al. (2014) Therapeutic inhibition of Jak activity inhibits progression of gastrointestinal tumors in mice. Mol. Cancer Ther., 13, 468–474. [DOI] [PubMed] [Google Scholar]
- 102. Villunger A., et al. (2003) p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science, 302, 1036–1038. [DOI] [PubMed] [Google Scholar]
- 103. Oltersdorf T., et al. (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature, 435, 677–681. [DOI] [PubMed] [Google Scholar]
- 104. Irizarry R.A., et al. (2009) The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet., 41, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Herman J.G., et al. (1995) Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res., 55, 4525–4530. [PubMed] [Google Scholar]
- 106. Jin Z., et al. (2001) Adenomatous polyposis coli (APC) gene promoter hypermethylation in primary breast cancers. Br. J. Cancer, 85, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Feinberg A.P., et al. (1988) Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res., 48, 1159–1161. [PubMed] [Google Scholar]
- 108. Eads C.A., et al. (2002) Complete genetic suppression of polyp formation and reduction of CpG-island hypermethylation in Apc(Min/+) Dnmt1-hypomorphic Mice. Cancer Res., 62, 1296–1299. [PubMed] [Google Scholar]
- 109. Laird P.W., et al. (1995) Suppression of intestinal neoplasia by DNA hypomethylation. Cell, 81, 197–205. [DOI] [PubMed] [Google Scholar]
- 110. Sansom O.J., et al. (2003) Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat. Genet., 34, 145–147. [DOI] [PubMed] [Google Scholar]
- 111. Prokhortchouk A., et al. (2006) Kaiso-deficient mice show resistance to intestinal cancer. Mol. Cell. Biol., 26, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Phesse T.J., et al. (2008) Deficiency of Mbd2 attenuates Wnt signaling. Mol. Cell. Biol., 28, 6094–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Suzuki H., et al. (2004) Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet., 36, 417–422. [DOI] [PubMed] [Google Scholar]
- 114. de Sousa E.M.F., et al. (2011) Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell, 9, 476–485. [DOI] [PubMed] [Google Scholar]
- 115. Millar C.B., et al. (2002) Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science, 297, 403–405. [DOI] [PubMed] [Google Scholar]
- 116. Bellacosa A., et al. (1999) MED1, a novel human methyl-CpG-binding endonuclease, interacts with DNA mismatch repair protein MLH1. Proc. Natl. Acad. Sci. U. S. A., 96, 3969–3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Sansom O.J., et al. (2003) MBD4 deficiency reduces the apoptotic response to DNA-damaging agents in the murine small intestine. Oncogene, 22, 7130–7136. [DOI] [PubMed] [Google Scholar]
- 118. Hilton I.B., et al. (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol., 33, 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kearns N.A., et al. (2015) Functional annotation of native enhancers with a Cas9-histone demethylase fusion. Nat. Methods, 12, 401–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lam K., et al. (2016) DNA methylation based biomarkers in colorectal cancer: A systematic review. Biochim. Biophys. Acta, 1866, 106–120. [DOI] [PubMed] [Google Scholar]
- 121. Allegra C.J., et al. (2009) American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J. Clin. Oncol., 27, 2091–2096. [DOI] [PubMed] [Google Scholar]
- 122. Sood A., et al. (2012) PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin. Colorectal Cancer, 11, 143–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Cairnie A.B., et al. (1965) Cell proliferation studies in the intestinal epithelium of the rat. I. Determination of the kinetic parameters. Exp. Cell Res., 39, 528–538. [DOI] [PubMed] [Google Scholar]
- 124. Potten C.S. (1977) Extreme sensitivity of some intestinal crypt cells to X and gamma irradiation. Nature, 269, 518–521. [DOI] [PubMed] [Google Scholar]
- 125. Cheng H., et al. (1974) Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am. J. Anat., 141, 537–561. [DOI] [PubMed] [Google Scholar]
- 126. Korinek V., et al. (1998) Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet., 19, 379–383. [DOI] [PubMed] [Google Scholar]
- 127. Gregorieff A., et al. (2005) Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology, 129, 626–638. [DOI] [PubMed] [Google Scholar]
- 128. Barker N., et al. (2007) Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature, 449, 1003–1007. [DOI] [PubMed] [Google Scholar]
- 129. Barker N. (2014) Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat. Rev. Mol. Cell Biol., 15, 19–33. [DOI] [PubMed] [Google Scholar]
- 130. Snippert H.J., et al. (2010) Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell, 143, 134–144. [DOI] [PubMed] [Google Scholar]
- 131. Lopez-Garcia C., et al. (2010) Intestinal stem cell replacement follows a pattern of neutral drift. Science, 330, 822–825. [DOI] [PubMed] [Google Scholar]
- 132. Kozar S., et al. (2013) Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell, 13, 626–633. [DOI] [PubMed] [Google Scholar]
- 133. Sottoriva A., et al. (2015) A Big Bang model of human colorectal tumor growth. Nat. Genet., 47, 209–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Buczacki S.J., et al. (2013) Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature, 495, 65–69. [DOI] [PubMed] [Google Scholar]
- 135. Roth S., et al. (2012) Paneth cells in intestinal homeostasis and tissue injury. PLoS One, 7, e38965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Vermeulen L., et al. (2014) Stem cell dynamics in homeostasis and cancer of the intestine. Nat. Rev. Cancer, 14, 468–480. [DOI] [PubMed] [Google Scholar]
- 137. Tian H., et al. (2011) A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature, 478, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Powell A.E., et al. (2012) The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell, 149, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Takeda N., et al. (2011) Interconversion between intestinal stem cell populations in distinct niches. Science, 334, 1420–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Metcalfe C., et al. (2014) Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell, 14, 149–159. [DOI] [PubMed] [Google Scholar]
- 141. Ashton G.H., et al. (2010) Focal adhesion kinase is required for intestinal regeneration and tumorigenesis downstream of Wnt/c-Myc signaling. Dev. Cell, 19, 259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Matthews J.R., et al. (2011) Absolute requirement for STAT3 function in small-intestine crypt stem cell survival. Cell Death Differ., 18, 1934–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Greenow K.R., et al. (2009) Chk1 deficiency in the mouse small intestine results in p53-independent crypt death and subsequent intestinal compensation. Oncogene, 28, 1443–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Holik A.Z., et al. (2013) Brg1 is required for stem cell maintenance in the murine intestinal epithelium in a tissue-specific manner. Stem Cells, 31, 2457–2466. [DOI] [PubMed] [Google Scholar]
- 145. Flanagan D.J., et al. (2015) Frizzled7 functions as a Wnt receptor in intestinal epithelial Lgr5(+) stem cells. Stem Cell Reports, 4, 759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Sato T., et al. (2011) Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature, 469, 415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Parry L., et al. (2013) Evidence for a crucial role of paneth cells in mediating the intestinal response to injury. Stem Cells, 31, 776–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Schewe M., et al. (2016) Secreted Phospholipases A2 Are Intestinal Stem Cell Niche Factors with Distinct Roles in Homeostasis, Inflammation, and Cancer. Cell Stem Cell, 19, 38–51. [DOI] [PubMed] [Google Scholar]
- 149. Farin H.F., et al. (2012) Redundant sources of Wnt regulate intestinal stem cells and promote formation of Paneth cells. Gastroenterology, 143, 1518–1529.e7. [DOI] [PubMed] [Google Scholar]
- 150. Valenta T., et al. (2016) Wnt Ligands Secreted by Subepithelial Mesenchymal Cells Are Essential for the Survival of Intestinal Stem Cells and Gut Homeostasis. Cell Rep., 15, 911–918. [DOI] [PubMed] [Google Scholar]
- 151. Goss A.M., et al. (2009) Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell, 17, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Saha S., et al. (2016) Macrophage-derived extracellular vesicle-packaged WNTs rescue intestinal stem cells and enhance survival after radiation injury. Nat. Commun., 7, 13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Rothenberg M.E., et al. (2012) Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology, 142, 1195–1205.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Zhou W.J., et al. (2013) Induction of intestinal stem cells by R-spondin 1 and Slit2 augments chemoradioprotection. Nature, 501, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Phesse T.J., et al. (2013) Responding to R-spondin: slit2 potentiates intestinal regeneration. Cell Stem Cell, 13, 512–514. [DOI] [PubMed] [Google Scholar]
- 156. Gregorieff A., et al. (2015) Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature, 526, 715–718. [DOI] [PubMed] [Google Scholar]
- 157. Tian H., et al. (2015) Opposing activities of Notch and Wnt signaling regulate intestinal stem cells and gut homeostasis. Cell Rep., 11, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Faller W.J., et al. (2015) mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature, 517, 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Myant K.B., et al. (2013) ROS production and NF-κB activation triggered by RAC1 facilitate WNT-driven intestinal stem cell proliferation and colorectal cancer initiation. Cell Stem Cell, 12, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Preston S.L., et al. (2003) Bottom-up histogenesis of colorectal adenomas: origin in the monocryptal adenoma and initial expansion by crypt fission. Cancer Res., 63, 3819–3825. [PubMed] [Google Scholar]
- 161. Shih I.M., et al. (2001) Top-down morphogenesis of colorectal tumors. Proc. Natl. Acad. Sci. U. S. A., 98, 2640–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Barker N., et al. (2009) Crypt stem cells as the cells-of-origin of intestinal cancer. Nature, 457, 608–611. [DOI] [PubMed] [Google Scholar]
- 163. Schwitalla S., et al. (2013) Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell, 152, 25–38. [DOI] [PubMed] [Google Scholar]
- 164. Davis H., et al. (2015) Aberrant epithelial GREM1 expression initiates colonic tumorigenesis from cells outside the stem cell niche. Nat. Med., 21, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Tetteh P.W., et al. (2016) Generation of an inducible colon-specific Cre enzyme mouse line for colon cancer research. Proc. Natl. Acad. Sci. U. S. A., 113, 11859–11864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Mills J.C., et al. (2015) Reserve stem cells: Differentiated cells reprogram to fuel repair, metaplasia, and neoplasia in the adult gastrointestinal tract. Sci. Signal., 8, re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Clevers H. (2011) The cancer stem cell: premises, promises and challenges. Nat. Med., 17, 313–319. [DOI] [PubMed] [Google Scholar]
- 168. Schepers A.G., et al. (2012) Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science, 337, 730–735. [DOI] [PubMed] [Google Scholar]
- 169. Holik A.Z., et al. (2014) Brg1 loss attenuates aberrant wnt-signalling and prevents wnt-dependent tumourigenesis in the murine small intestine. PLoS Genet., 10, e1004453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Sato T., et al. (2009) Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature, 459, 262–265. [DOI] [PubMed] [Google Scholar]
- 171. Fatehullah A., et al. (2016) Organoids as an in vitro model of human development and disease. Nat. Cell Biol., 18, 246–254. [DOI] [PubMed] [Google Scholar]
- 172. Yui S., et al. (2012) Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5⁺ stem cell. Nat. Med., 18, 618–623. [DOI] [PubMed] [Google Scholar]
- 173. Huch M., et al. (2013) Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J., 32, 2708–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Huch M., et al. (2015) Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell, 160, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Wright A.V., et al. (2016) Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell, 164, 29–44. [DOI] [PubMed] [Google Scholar]
- 176. Abudayyeh O.O., et al. (2016) C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science, 353, aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Deltcheva E., et al. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature, 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Jinek M., et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Polstein L.R., et al. (2015) A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat. Chem. Biol., 11, 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Nihongaki Y., et al. (2015) Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat. Biotechnol., 33, 755–760. [DOI] [PubMed] [Google Scholar]
- 181. Dow L.E., et al. (2015) Inducible in vivo genome editing with CRISPR-Cas9. Nat. Biotechnol., 33, 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Drost J., et al. (2015) Sequential cancer mutations in cultured human intestinal stem cells. Nature, 521, 43–47. [DOI] [PubMed] [Google Scholar]
- 183. Roper J., et al. (2017) In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol., 35, 569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Fumagalli A., et al. (2017) Genetic dissection of colorectal cancer progression by orthotopic transplantation of engineered cancer organoids. Proc. Natl. Acad. Sci. U. S. A., 114, E2357–E2364. [DOI] [PMC free article] [PubMed] [Google Scholar]