Although MUC1 is believed to be an oncoprotein, this study uncovers cell-type specific functions of MUC1 in the tumor microenvironment. Our results add complexity to the roles of MUC1 in cancer and help to develop intervention approaches against lung cancer.
Abstract
Mucin 1 (MUC1) is a tumor antigen that is aberrantly overexpressed in various cancers, including lung cancer. Our previous in vitro studies showed that MUC1 facilitates carcinogen-induced EGFR activation and transformation in human lung bronchial epithelial cells (HBECs), which along with other reports suggests an oncogenic property for MUC1 in lung cancer. However, direct evidence for the role of MUC1 in lung carcinogenesis is lacking. In this study, we used the 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK)-induced A/J mouse lung tumor model to investigate the effect of whole-body Muc1 knockout (KO) on carcinogen-induced lung carcinogenesis. Surprisingly, lung tumor multiplicity was significantly increased in Muc1 KO compared to wild-type (WT) mice. The EGFR/AKT pathway was unexpectedly activated, and expression of the EGFR ligand epiregulin (EREG) was increased in the lung tissues of the Muc1 KO compared to the WT mice. EREG stimulated proliferation and protected against cigarette smoke extract (CSE)-induced cytotoxicity in in vitro cultured human bronchial epithelial cells. Additionally, we determined that MUC1 was expressed in human fibroblast cell lines where it suppressed CSE-induced EREG production. Further, suppression of MUC1 cellular activity with GO-201 enhanced EREG production in lung cancer cells, which in turn protected cancer cells from GO-201-induced cell death. Moreover, an inverse association between MUC1 and EREG was detected in human lung cancer, and EREG expression was inversely associated with patient survival. Together, these results support a promiscuous role of MUC1 in lung cancer development that may be related to cell-type specific functions of MUC1 in the tumor microenvironment, and MUC1 deficiency in fibroblasts and malignant cells results in increased EREG production that activates the EGFR pathway for lung carcinogenesis.
Introduction
MUC1 (MUC1 in humans and Muc1 in animals) is a mucin-like glycosylated protein expressed on the apical membrane surface of bronchial epithelial cells and plays an important role in the resolution of inflammation during respiratory tract infection (1). MUC1 expression is elevated in various cancers including lung cancer (2–4), where apical polarity of MUC1 is lost. MUC1 overexpression is correlated with poor survival in lung cancer patients (5). Because MUC1 is considered as a tumor antigen, it has been used as an immunotherapy target (6,7). MUC1 interacts with a variety of cellular partners that contribute to malignancy and chemotherapy resistance in cancer cells, and thus is proposed to function as an oncoprotein (8,9). Thus, directly targeting MUC1 has been a focus for cancer therapy (8). Our previous studies demonstrated that chronic exposure of cigarette smoke (CS) resulted in increased MUC1 expression in mouse airway epithelial cells (10). In addition to expression in lung epithelial cells, MUC1 is also expressed in macrophages where it potentiates CSE-induced tumor necrosis factor α (TNF-α) secretion, which may contribute to an inflammatory microenvironment that, in turn, facilitates MUC1 expression in epithelial cells (10). While chronic pulmonary inflammation causes sustained MUC1 expression in lung epithelial cells, MUC1 is also likely involved in inflammation-associated cancer development (11). Additionally, MUC1 contributes to cell transformation induced by the CS carcinogen derivative benzo[a]pyrene diol epoxide (BPDE) by potentiating epidermal growth factor receptor (EGFR)-mediated cellular signaling (12). All these findings, primarily obtained from in vitro studies, suggest an oncogenic role for MUC1 in lung carcinogenesis. However, direct in vivo evidence for MUC1 in lung cancer development is still lacking.
Among the MUC1-modulated pathways, the EGFR pathway is particularly interesting because it is a major driving force for lung carcinogenesis (13). EGFR mutation and aberrant activation are frequently found in human lung cancer (13). Lung cancer cells acquire dependence on EGFR activity for survival, substantiating the use of EGFR inhibitors for lung cancer therapy (13). There are seven EGFR ligands that include epidermal growth factor (EGF), transforming growth factor α (TGFα), epiregulin (EREG), heparin-binding EGF-like growth factor (HBEGF), amphiregulin (AREG), betacellulin (BTC) and epigen (EPGN) (14). While EGF and TGFα have been extensively studied for EGFR signaling, EREG has emerged as an important EGFR activator for tumor promotion based on studies with human lung cancer tissues and an experimental murine lung carcinogenesis model (15,16). Whether MUC1 regulates EGFR ligands during carcinogenesis has not been addressed.
When activated by its ligands, EGFR dimerizes and autophosphorylates its C-terminal tyrosine kinase domain in the cytoplasm to initiate a signaling cascade (14,17). The two major downstream pathways of EGFR signaling, ERK and Akt, are important signals for EGFR-mediated oncogenesis. While the underlying mechanisms by which CS carcinogens activate EGFR in lung epithelial cells have not been clearly elucidated, our recent in vitro studies suggest that MUC1 facilitates carcinogen-induced HBEC transformation partly through stabilization of the activated form of EGFR (12). Therefore, MUC1 in epithelial cells may play an oncogenic role for lung carcinogenesis. MUC1 expressed in pulmonary macrophages may also promote lung cancer development (10). However, whether MUC1 is expressed in other pulmonary stromal cells and its role in lung carcinogenesis is unknown.
The purpose of the current study was to investigate the role of MUC1 in lung carcinogenesis in vivo. An unexpected lung tumor-promoting effect of Muc1 was detected in whole-body Muc1 knockout (KO) mice. Mechanism exploration revealed that MUC1 suppressed the production of EGFR ligand EREG in fibroblasts and malignant cells. The results imply a promiscuous role of MUC1 in lung cancer development that may be related to cell-type specific functions of MUC1 in the tumor microenvironment, in which MUC1 deficiency in fibroblasts and malignant cells results in increased EREG production that activates the EGFR pathway to compensate for MUC1 loss, promoting lung carcinogenesis.
Materials and methods
Reagents
Carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) was purchased from ChemSyn Chemical (Concord, Canada). The primary antibody against Mucin 1 (GP1.4) was purchased from Santa Cruz Biotechnology. Antibodies against EREG and MUC1 Ab-5, a hamster monoclonal antibody that recognizes the MUC1 CT domain, were obtained from Thermo Scientific. The β-Actin antibody and MUC1 inhibitor GO-201 were from Sigma-Aldrich. The antibody against phospho-EGFR (Y1068) was purchased from Abcam. Antibodies against ERK, and phospho-ERK (Y185/187), and phospho-Akt (Ser 473) were from Invitrogen. Recombinant human EREG and antibodies against EGFR and Akt were from Cell Signaling Technology. Small interfering RNA (siRNA; SiGenome SMARTpool) for MUC1, EREG, and negative control siRNA were purchased from Dharmacon. Recombinant human IL-6, TNF-α, and IL-1β were purchased from eBioscience (San Diego, CA). Mouse cytokine array C2000 and mouse EREG ELISA kit were purchased from Raybiotech.Inc. Human EREG ELISA kit was from Biomatik. Cigarette smoke extract (CSE) was prepared as described previously (10).
NNK-induced lung tumorigenesis in A/J mice
MUC1 knockout mice with C57BL/6 background, a kind gift from Dr Sandra J Gendler from Mayo Clinic (Scottsdale, AZ) (18), were backcrossed to A/J mice for nine generations to generate MUC1−/− with A/J background. Homologous knockout of Muc1 (Muc1−/−) were confirmed by genotyping (see below). Muc1−/− and WT A/J mice (female, 4–6 weeks old) were randomly placed into the experimental or control groups (n = 15/group). The experimental groups received 3 injections (i.p.) of NNK (50 mg/kg) over a week, while the control mice received an equivalent volume of PBS. Thirty and forty-five weeks after NNK treatment, 15 mice from each group were euthanized. The left lungs were fixed in 4% buffered paraformaldehyde and used for pathological analysis. The right lung tissues were collected, stored at −80°C for RNA extraction and molecular analysis. The experimental design and group information for the animal study are shown in Figure 1A and B, respectively. The tumors on the formalin-fixed left lungs that were visible were enumerated before the lung tissues were embedded in paraffin and stained with hematoxylin and eosin (H&E) for histological examination. Pulmonary lesions were classified as hyperplasia or neoplasia (adenoma or carcinoma) after H&E staining, and examined by a pathologist. The multiplicity was calculated as the average number of lesions in the left lung of each group.
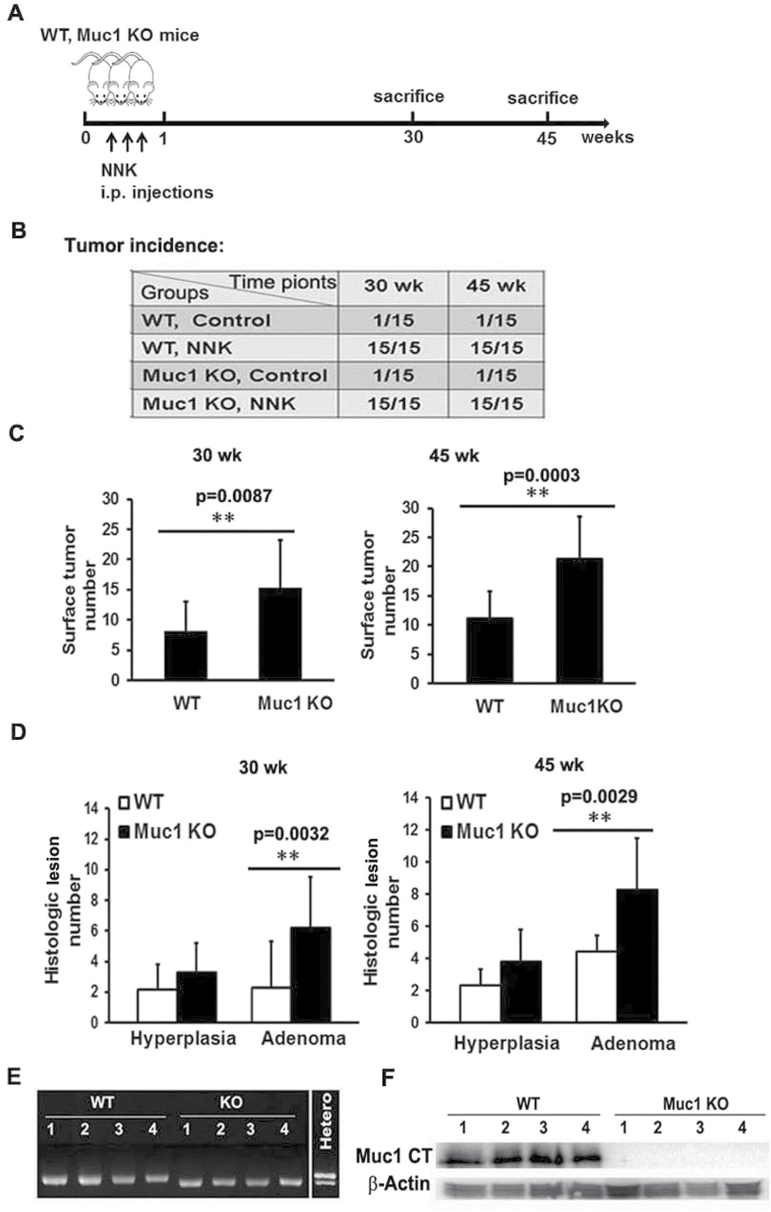
Figure 1.
Whole-body Muc1 KO increased lung tumor multiplicity in NNK-treated A/J mice. (A) Experimental design of the animal study: Muc1−/− and WT A/J mice were treated with 3 doses of NNK (50 mg/kg, i.p.) in the first week. Mice were euthanized at 30 and 45 weeks following NNK treatment. (B) Tumor incidence in both control and experimental groups in the Muc1−/− and WT mice at 30 and 45 weeks after NNK treatment. A total of 15 mice were used in each group. (C) The number of tumors on formalin-fixed left lungs at 30 and 45 weeks were determined by counting with the naked eye. Data shown are mean ± S.D; **P < 0.01. (D) Histological quantification of lung lesions at 30 and 45 weeks after NNK treatment were examined by H&E staining of paraffin-embedded left lung sections from both the Muc1 −/− and WT mice. Pulmonary lesions were classified as hyperplasia or neoplasia (adenoma or carcinoma). Data shown are mean ± S.D; **P < 0.01. (E and F) Confirmation of mouse strain by RT-PCR and Western blot. Genomic DNA and total protein samples were isolated from lung tissues of four randomly selected mice in both the WT mice group and the Muc1 −/− group. Genomic DNA sample from a heterozygous mouse served as a control for PCR assay. Muc1 protein in lung tissues was detected by Western blot with antibody MUC1 Ab-5 recognizing the C-terminal domain of MUC1 (MUC1-CT). β-Actin was detected as an input control.
Genotyping of Muc1 KO mice
Genomic DNA isolated from lung tissues of four randomly selected mice in the WT mice group and Muc1 KO mice group was digested with alkaline lysis reagent and neutralized with 40 mM Tris-HCl (pH = 5). Genomic DNA samples were subjected to a PCR assay with following primers as reported with some modification (18): WT and mutant (MT) shared forward: 5′-ACC TCA CAC ACG GAG CGC CAG; MT reverse: 5′-TTC TGG TGC CGG AAA CCA GGC; WT reverse: 5′-cga att cct cga gcg aat tcc tcg agT CCC CCC TGG CAC ATA CTG GG-3′. The 18 random nucleotides shown in lower case were added to the 5′ end of the WT reverse primer to enable a PCR reaction with mixed WT/MT primers in a single tube to give rise to a longer product for WT mice (288 bp) that can be distinguished from that of the KO mice (261 bp) on agarose gels. The PCR reaction condition was 40 cycles of 95°C, 60 s; 62°C, 30 s; and 72°C, 60 s. The amplified PCR products were resolved on 3% agarose gels with 0.5 μg/ml ethidium bromide, visualized and photographed.
Cell culture and transfection
The human lung cancer cell line A549, human fetal lung fibroblast line HFL-1, and human adult lung fibroblast line Hs888Lu were purchased from American Type Culture Collection (ATCC, Manassas, VA). A549 and HFL-1 cells were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 1 mM of glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. Hs888Lu cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. These cell lines were used within 6 months of receipt from ATCC. The immortalized human bronchial epithelial cells, HBEC-14, were generously provided by Drs. Shay and Minna, Southwestern Medical Center, Dallas, TX (19). HBEC-14 cells were maintained in Keratinocyte serum free medium (K-SFM) (Invitrogen), supplemented with 5 μg/L of human recombinant EGF and 50 mg/L of bovine pituitary extract in plates coated with FNC coating mix (Athena ES). HFL-1, Hs888Lu and A549 cells were authenticated by ATCC before purchase. The HBEC-14 cell line has not been authenticated. For transfection of siRNA, the cells were seeded in 12-well plates at about 60–70% confluence, transfected with siRNA with INTERFERin™ siRNA transfection reagent (Polyplus-transfection) according to manufacturer’s instructions.
Western blot
Cells were washed twice with cold PBS, collected, and lysed with M2 buffer (20 mM Tris-HCl, pH 7.6, 0.5% Nonidet P-40, 250 mM NaCl, 3 mM EDTA, 3 mM EGTA, 2 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 20 mM β-glycerophosphate, 1 mM sodium vanadate and 1 μg/ml leupeptin). Equal amounts of proteins were separated by 12 or 15% SDS-polyacrylamide gels and then transferred to PVDF membranes. After incubating the membranes with the primary and secondary antibodies, the proteins on the membranes were detected by enhanced chemiluminescence (ECL, Millipore). The intensity of the individual bands was quantified by densitometry (ImageJ) and normalized to the corresponding loading control bands. Fold changes were calculated with the control taken as 1.
Cytokine array assay
Normal lung tissues from a control WT mouse and a control Muc1 KO mouse were each homogenized in M2 buffer. Two hundred and fifty micrograms of the extracted protein from each sample was used for cytokine array assay (RayBiotech) according to the manufacturer’s instructions.
Detecting EREG by ELISA
Normal lung tissues from control WT and Muc1 KO mice and tumor tissues from NNK-treated WT and Muc1 KO mice, seven per group for a total of 28 mice, were used to determine EREG concentration with a mouse EREG ELISA kit from Raybiotech. For collection of lung tumor samples from NNK-treated mice, lung tumors and normal lung tissues were separated under a microscope using disposable scalpels. Proteins were extracted from these samples and 10 µg of protein from each sample were subjected to the ELISA assay by following the manufacturer’s instructions. For detection of EREG in fibroblasts, HFL-1 cells were plated in 12-well plates at 70–80% confluence overnight. Cells were then transfected with 20 nM MUC1 siRNA for 24 h, and were maintained in fresh RPMI 1640 without serum before treated as described in the figure legends. Twenty-four hours later, the culture medium was collected and used to determine the concentration of EREG with a human EREG ELISA kit by following the manufacturer’s instructions. For determining GO-201-induced EREG secretion, A549 cells were seeded in a 12-well plate at 70–80% confluence overnight in RPMI 1640 with 0.5% FBS, before the cells were treated with GO-201 for 8 h. After treatment, the culture media were collected, and EREG concentration was measured by ELISA.
Cell viability and proliferation assay
Cell viability was assessed using a 3-(4,5-dimethylthiazolyl-2-)-2,5-diphenyltetrazolium bromide (MTT) cell proliferation assay as described previously (20). Briefly, A549 or HBEC-14 cells were seeded in 48-well plates at 50~60% confluence overnight, and then treated as indicated in the figure legends. After treatment, cells were incubated in MTT for 2–3 h. The percentage of viable cells was calculated using the following formula: Cell viability (%) = (Absorbance of treated sample/Absorbance of control) × 100. For proliferation assay, HBEC-14 cells were seeded in 24-well plates at a density of 2 × 104 cells/well in blank K-SFM, cultured overnight (day 0), and treated with 20 ng/ml recombinant human EREG for different time periods. The viable cells were measured by MTT assay and expressed as relative cell numbers to the control cells (day 0).
Reverse transcription-polymerase chain reaction (RT-PCR)
HFL-1 cells were treated with CSE (20 μg/ml total particulate matter [TPM]) for different time points and total RNA was extracted from each sample using TRIzol® Reagent (Life technology). Two micrograms of RNA were used as a template for cDNA synthesis with a reverse transcription kit (Promega). An equal volume of cDNA product was subjected to PCR analysis. The following primers were used in the PCR reactions: MUC1, forward primer 5′- ACAATTGACTCTGGCCTTCCG-3′ and reverse primer 5′- TGGGTTTGTGTAAGAGAGGCT-3′; β-actin, forward primer: 5′- CCAGCCTTCCTTCCTGGGCAT-3′ and reverse primer 5′-AGGAGCAATGATCT. The PCR reaction condition for MUC1 was 32 cycles of 95°C, 45 s; 57°C, 45 s; and 72°C, 45 s. For β-actin, the PCR cycles were 23, and the repeated condition was 95°C, 45 s; 55°C, 45 s; and 72°C, 45 s. The amplified PCR products were resolved on 2% agarose gels with 0.5 μg/ml ethidium bromide, visualized and photographed.
Analysis of expression relationship between MUC1 and EREG with The Cancer Genome Atlas (TCGA) database
Level 3 transcriptome data measured by RNA-seq and patient clinical data (e.g. demographics, tumor stage, and prognosis) for lung adenocarcinomas (n = 462) and paired normal lungs (n = 109) were downloaded from The Cancer Genome Atlas (TCGA) project. Correlation between EREG and MUC1 expression in lung adenocarcinomas was analyzed using Spearman correlation analysis. Logistic regression was used to assess the association between gene expression and tumor stage (stages 3 and 4 versus stages 1 and 2). The effect of EREG and MUC1 expression on overall survival was assessed using the Cox regression model with adjustment for important covariates. The Kaplan–Meier estimator was also used to estimate the survival function between high versus low gene expression. All these analyses were conducted using SAS 9.4.
Statistics
All data are expressed as mean ± S.D. Statistical significance was examined by two-way analysis of variance. In all analyses, P < 0.05 was considered statistically significant.
Results
Whole-body Muc1 KO increased lung tumor multiplicity in NNK-treated A/J mice
The effect of Muc1 KO on lung carcinogenesis was examined with an established protocol using the NNK-induced lung cancer model in the A/J mouse (Figure 1A), which progressively generates benign and malignant tumors in mouse lungs (59). All mice (100%) in the WT and Muc1 KO groups developed lung lesions by 30 weeks following NNK treatment, while only one mouse in the control groups of each phenotype had a spontaneous lung lesion (Figure 1B). Similar results were obtained at 45 weeks (Figure 1B). Surprisingly, a significantly higher number of lung surface tumors were found in the Muc1 KO mice compared to that of the WT mice at both 30 and 45 weeks after NNK treatment, which was confirmed by histological analysis of left lung sections. The number of histological lesions (mainly hyperplasias and adenomas) was higher in the Muc1 KO mice than the WT mice at both 30 and 45 weeks after NNK treatment (Figure 1C and D and Supplementary Figure S1). The validity of the Muc1 KO mice was confirmed by genotyping, RT-PCR and Western blot using lung tissues from the mice (Figure 1E and F and data not shown). No adenocarcinoma was seen in any group. These results contradict the proposed oncogenic role of MUC1, while suggesting a tumor-suppressing effect.
Activation of the EGFR/Akt pathway in the lung and tumor tissues from Muc1 KO mice
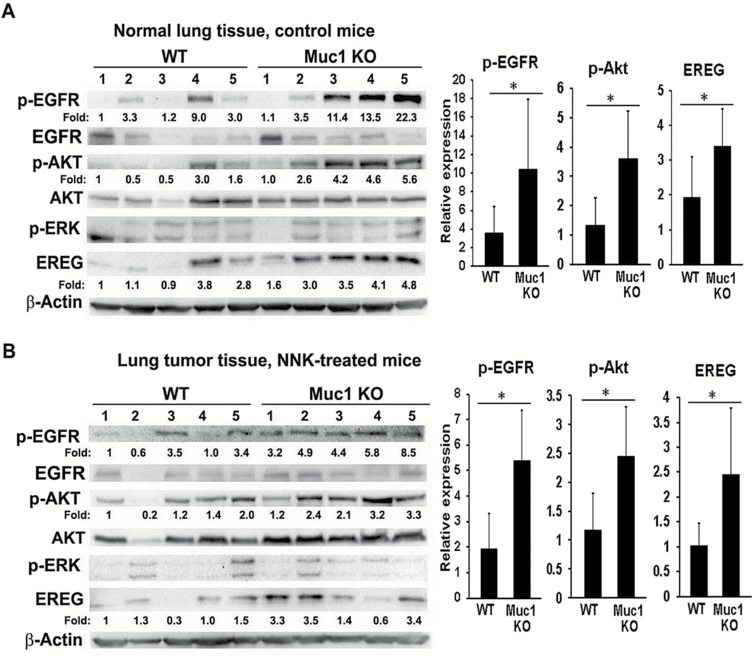
Activation of the EGFR signaling pathway is a driving force of lung carcinogenesis, and our previous study showed that MUC1 is involved in carcinogen-induced EGFR activation in HBECs (10,12). Thus, we examined the status of EGFR pathway in lung and tumor tissues from the Muc1 KO mice. EGFR and Akt were activated in normal lung and tumor tissues from the Muc1 KO mice compared to tissues from WT mice (Figure 2A and B). In contrast, the activity of ERK was not changed in Muc1 KO mice versus WT mice. These results suggest that the activated EGFR/Akt pathway in the Muc1 KO mice may be involved in the increased tumor formation after NNK treatment.
Figure 2.
The EGFR/Akt pathway was activated in both the lung and tumor tissues in the Muc1 KO mice. Normal lung tissue (A) and tumor tissues (B) from five randomly selected mice in both the WT mice group and the Muc1 KO group were subjected to Western blot assay. The expression of phospho-EGFR (Y1068), -Akt (Ser 473), and -ERK (Y185/187) was determined. The expression of EREG, total EGFR, and Akt was also detected by Western blot. β-Actin was detected as an input control. The intensity of the individual bands was quantified and normalized to the corresponding input control bands. Relative expression of each protein was statistically analyzed and shown to the right of its respective panel. *P < 0.05.
Increased EREG production in the lung of Muc1 KO mice
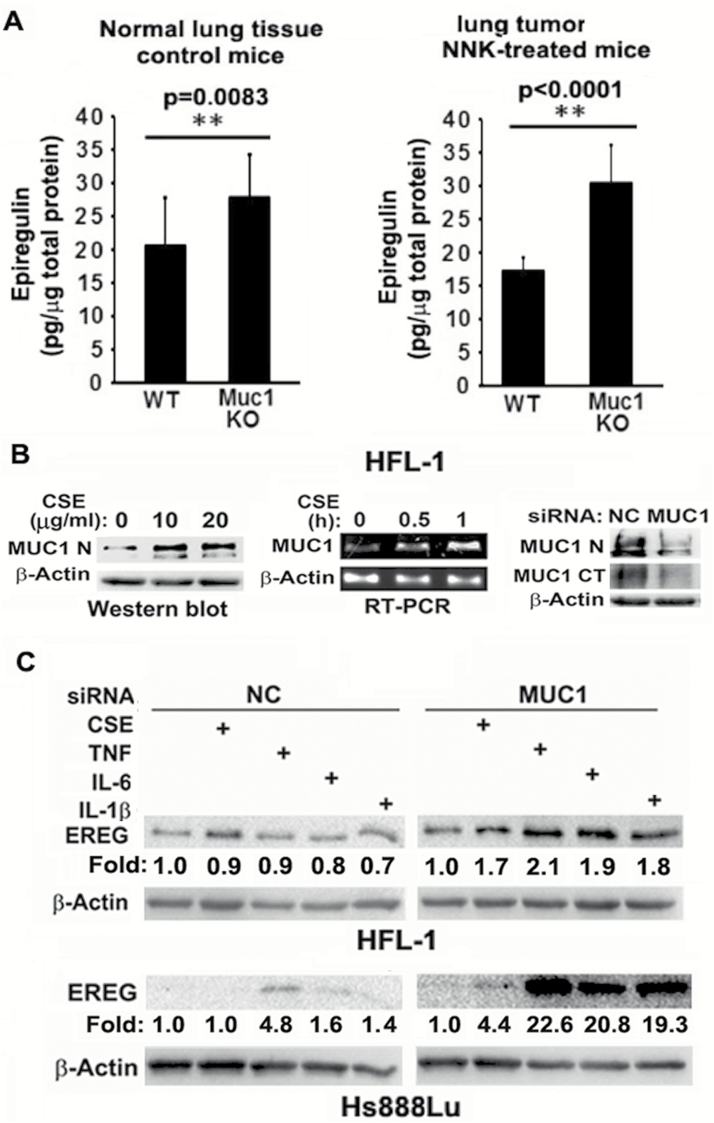
To explore the underlying mechanism by which EGFR/Akt pathway activation occurs in Muc1 KO mice, we performed a cytokine/growth factor array assay using the normal lung tissues from a WT and Muc1 KO mouse. Among the protein factors that had elevated expression in the Muc1 KO mouse when compared with the WT mouse, the EGFR ligand EREG had the highest increase (Supplementary Figure S2). This result was confirmed by an ELISA assay showing that EREG production was increased in normal lung tissues and lung tumors from the Muc1 KO mice when compared with those from the WT mice (Figure 3A). Increased EREG expression in normal lung and tumor tissues from the Muc1 KO mice compared to that from the WT mice was also detected by Western blot (Figure 2A and B). The concomitant activation of the EGFR/Akt pathway and increased EREG production suggest that EREG may be involved in increased lung tumor formation through activation of the EGFR/Akt pathway in Muc1 KO mice.
Figure 3.
MUC1 is expressed and contributes to EREG expression in fibroblasts. (A) Normal lung tissues from seven control WT and seven Muc1 KO mice and tumor tissues from seven NNK-treated WT and seven Muc1 KO mice were used to determine EREG concentration by ELISA assay. Ten micrograms of total protein from each sample was loaded to each well. Data shown are mean ± S.D; **P < 0.01. (B) Left, HFL-1 cells were treated with CSE (0, 10 and 20 µg/ml TPM) overnight. MUC1 expression was detected by Western blot. β-Actin was detected as an input control. Middle, HFL-1 cells were treated with CSE (20 µg/ml TPM) for the indicated time points. MUC1 mRNA level was detected by RT-PCR. β-Actin was detected as an input control. Right, HFL-1 cells were transfected with MUC1 siRNA or negative control siRNA for 48 h. MUC1 expression was detected by Western blot with antibody Muc1 GP1.4 against the extracellular domain (MUC1-N) and antibody MUC1 Ab-5 recognizing the C-terminal domain (MUC1-CT). β-Actin was detected as an input control. (C) HFL-1 and Hs888Lu cells were transfected with MUC1 siRNA or negative control siRNA for 24 h, before the cells were treated with CSE (20 µg/ml TPM), TNF-α (10 ng/ml), IL-6 (10 ng/ml) and IL-1β (10 ng/ml) overnight. EREG expression was detected by Western blot. β-Actin was detected as an input control. The intensity of the individual bands was quantified and normalized to the corresponding input control bands. Fold changes were calculated with the control taken as 1.
MUC1 is expressed and suppresses EREG production in fibroblasts
Because EREG was initially identified in mouse fibroblasts (21), and tumor-associated fibroblasts are a main source of EREG production (22,23), we next focused on EREG production in fibroblasts. We determined that EREG was produced in human fibroblast cell lines HFL-1 and Hs888Lu (Figure 3C). Meanwhile, we detected MUC1 expression, which was robustly induced by CS extract (CSE), in these fibroblast cells by both RT-PCR and Western blot. CSE containing cigarette smoke carcinogens was used to simulate cigarette smoke. Notably, MUC1 expression was detected by Western blot with antibodies against either the extracellular domain (MUC1-N) or the C-terminal domain (MUC1-CT, Figure 3B). The specificity of MUC1 detection was confirmed with a MUC1 siRNA, which specifically blocked MUC1 expression (Figure 3B). MUC1 was expressed in HFL-1 and HS888Lu cells at a comparable protein level to that in HBEC2 (Supplementary Figure S3A), while being at a higher mRNA level than in HBEC2 (Supplementary Figure S3B). The difference in mRNA and protein levels may be due to differences in post-transcriptional regulation in different cell types. Consistently, MUC1 is detectable by immunohistochemistry in fibroblasts in human lung cancer tissues and CS-exposed mouse lung, although much weaker than in cancer cells (Supplementary Figure S4 and data not shown). Altogether, these results confirmed that MUC1 is expressed in fibroblasts. We then examined if MUC1 plays a role in EREG expression in fibroblasts. MUC1 was knocked down before induction of EREG expression with CSE and TNF-α, IL-6, and IL-1β in HFL-1 and Hs888Lu cells, and the expression of EREG was detected by Western blot. CSE, TNF-α, IL-6 and IL-1β were used as inducers of EREG expression because they were reported as EREG regulators or involved in pulmonary inflammation for carcinogenesis (22,24,25). Although CSE, TNF-α, IL-6 and IL-1β barely induced EREG expression in the control cells transfected with NC siRNA, MUC1 knockdown substantially potentiated EREG expression induced by each of the stimuli, despite the extend of response to each stimulus varied between the two cell lines (Figure 4D). These results suggest a novel role for MUC1 in suppressing EREG expression in fibroblasts, which may be involved in regulation of lung carcinogenesis.
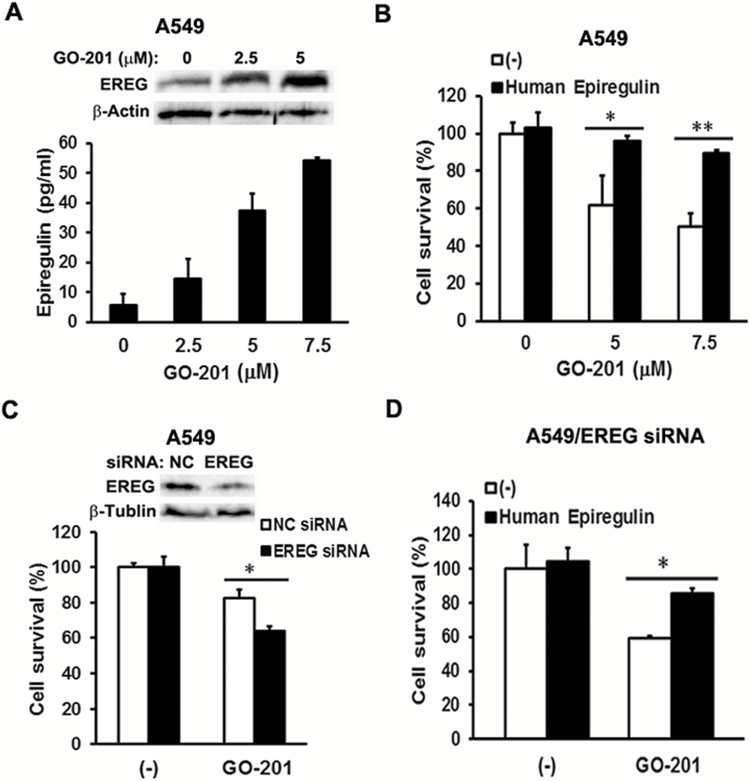
Figure 4.
EREG protects human lung cancer cells from MUC1 inhibitor GO-201 induced cell death. (A) Upper, A549 cells were treated with MUC1 inhibitor GO-201 (0, 2.5 and 5 µM) for 24 h. EREG expression was detected by Western blot. β-Actin was detected as an input control. Lower, A549 cells were cultured in 0.5% FBS containing RPMI medium overnight, before the cells were treated with GO-201 (0, 2.5, 5 and 7.5 µM) for 8 h. After treatment, conditioned media were collected for detection of EREG production by ELISA assay. (B) A549 cells were cultured in 0.5% FBS containing RPMI medium overnight, before the cells were exposed to human EREG (20 ng/ml) for 24 h. After EREG treatment, the cells were treated with GO-201 (0, 5 and 7.5 µM) for 48 h. Cell viability was detected by MTT assay. Data shown are mean ± S.D; **P < 0.01, *P < 0.05. (C) A549 cells were cultured in 10% FBS containing RPMI medium overnight, before the cells were transfected with EREG siRNA or negative control siRNA for 24 h. The cells were then maintained in 0.5% FBS containing RPMI medium for another 24 h. After that, the A549/NC siRNA and A549/EREG siRNA cells were treated with GO-201 (5 µM) for 48 h. Cell viability was detected by MTT assay. Data shown are mean ± S.D; *P < 0.05. (D) The A549/EREG siRNA cells were pre-treated with human EREG (20 ng/ml) for 24 h, before they were treated with GO-201 (5 µM) for 48 h. Cell viability was detected by MTT assay. Data shown are mean ± S.D; *P < 0.05.
EREG promotes HBEC cell proliferation and protects against carcinogen-induced cell death
The oncogenic role of EREG is directly associated with its ability to promote cell proliferation and survival of carcinogen-treated HBECs, which is critical for cell transformation (12,26). Thus, we examined the effects of EREG on bronchial epithelial cell proliferation and survival against the cytotoxicity of CSE. Compared to the control, the number of cells that were supplemented with human EREG exhibited a significant increase, which was in an EREG dose-dependent manner (Supplementary Figure S5A and B). EREG also protected HBEC cells from CSE-induced cytotoxicity (Supplementary Figure S5C). These results suggest EREG potentiates cell survival and proliferation in bronchial epithelial cells during cigarette smoking, which may contribute to lung cancer development.
EREG protects human lung cancer cells from death induced by the MUC1 activity inhibitor GO-201
To further explore the functional relevance of EREG in MUC1-mediated carcinogenesis, we investigated if EREG is involved in MUC1-mediated survival in lung cancer cells. A549 cells were treated with the MUC1 activity inhibitor GO-201 that was shown to kill cancer cells via growth arrest and necrotic death (8,27). GO-201 is a short cell-permeable peptide fragment of the MUC1 cytoplasmic domain that suppresses MUC1-mediated cellular signaling by interrupting the interaction between the MUC1 CT domain and its cellular partners (8,27–29). EREG expression and secretion were induced by GO-201 in A549 cells in a dose-dependent manner (Figure 4A), suggesting that EREG expression and secretion were suppressed by MUC1 in lung cancer cells. Recombinant human EREG effectively attenuated GO-201′s inhibitory effect on cell viability (Figure 4B). Knockdown of EREG expression remarkably sensitized GO-201-induced cytotoxicity (Figure 4C), which was attenuated by recombinant human EREG (Figure 4D). Altogether, these results suggest that in addition to fibroblasts, malignant cells are another source of EREG in the tumor microenvironment, and EREG production constitutes a negative feedback loop for cancer cell survival when MUC1 expression or activity is suppressed.
Inverse association of MUC1 and EREG expression in human lung cancer
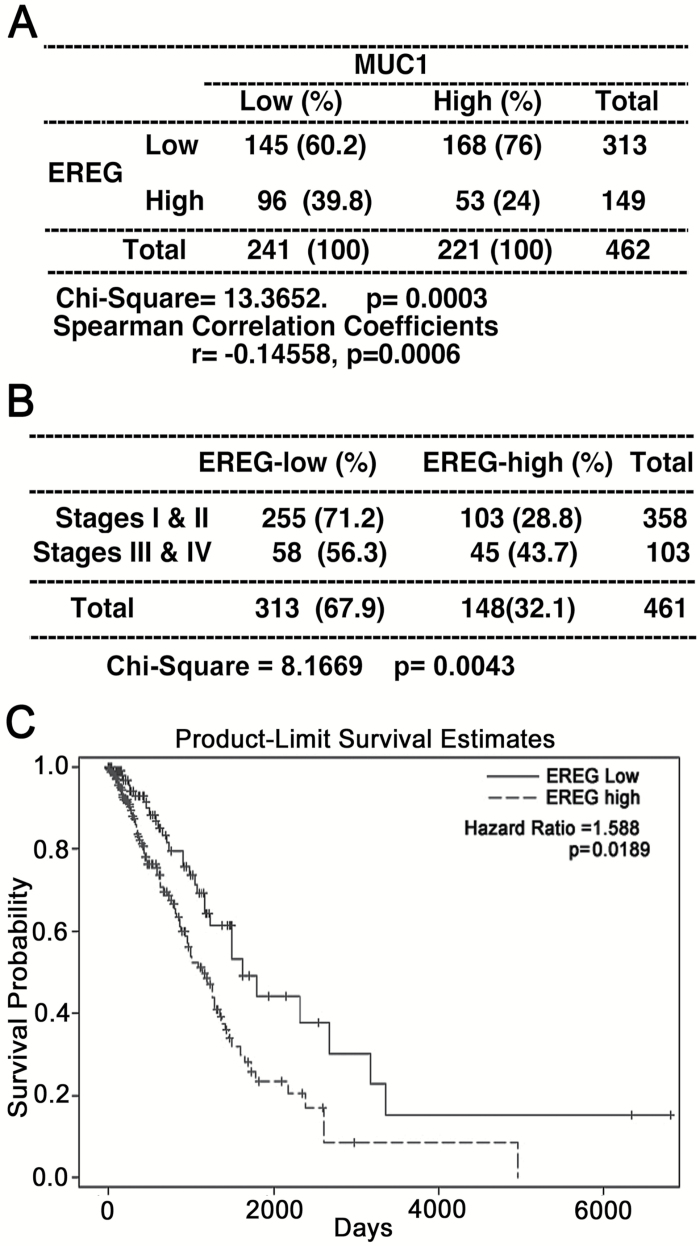
To further investigate the clinical relevance of the finding, we analyzed the correlation of steady-state mRNA expression between MUC1 and EREG in human lung cancer using RNA-seq data from The Cancer Genome Atlas (TCGA) database. The median expression of EREG (normalized read = 84) and MUC1 (normalized read = 14467) in 109 normal lung tissues was selected as the cutoff to define high versus low expression in tumor samples. Out of 462 lung adenocarcinomas, 149 and 221 cases had elevated expression of EREG or MUC1, respectively (Figure 5A). An inverse correlation between MUC1 and EREG was detected in 462 cases of adenocarcinoma (Spearman correlation r = −0.15, P = 0.0006, Figure 5A), which suggests that MUC1 may negatively regulate EREG expression and its mediated human lung carcinogenesis. Within the 461 cases having disease stage information, higher EREG expression (normalized read > 84) was associated with advanced tumor stage (stages 3 and 4 versus stages 1 and 2, P = 0.0043, Figure 5B). Kaplan–Meier curve suggested that patients with higher EREG expression had worse overall survival compared to those with low EREG expression (Figure 5C). Moreover, using Cox regression model with adjustment for age, sex, smoking status, and tumor stage, lung adenocarcinoma patients with high EREG expression (normalized read > 84) had 59% increased rate of death from lung cancer in up to 17.5 years of follow-up compared to those with low expression (Hazard Ratio = 1.59, P = 0.0189).
Figure 5.
Inverse association of MUC1 and EREG expression in human lung adenocarcinoma. (A) RNA-seq data was acquired for lung adenocarcinomas from The Cancer Genome Atlas (TCGA) database. EREG and MUC1 expressions were transformed to binary outcomes using the median expression levels in the 109 normal lungs and were compared using chi square test. Correlation between EREG and MUC1 expression as continuous variables was analyzed using Spearman correlation analysis. (B) Association of EREG expression with tumor stages. Higher EREG expression was associated with advanced tumor stages (stages 3 and 4). Note: one case was excluded due to missing stage data. (C) The Kaplan–Meier estimator was used to estimate the survival function between high versus low EREG expression. A further adjustment for important covariates using COX regression model confirmed that patients with greater EREG expression in tumor had a worse prognosis. HR and P value are shown.
Discussion
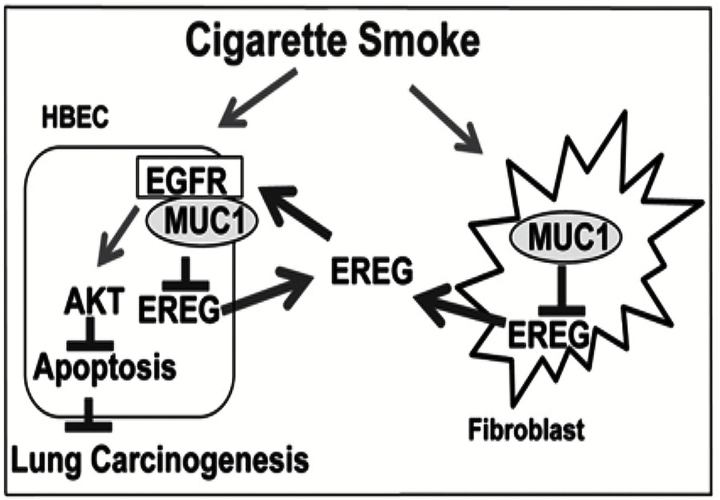
In this study, we surprisingly found that lung tumor multiplicity induced by NNK was significantly increased in whole-body Muc1 KO A/J mice, which was associated with activation of the EGFR/AKT pathway. These results suggest a tumor-suppressing effect of Muc1 on lung carcinogenesis, which is contradictory to the well-accepted oncogenic role of MUC1. In exploration of the underlying mechanism, we found the expression of the EGFR ligand EREG was increased in the Muc1 KO mouse lung, and EREG was able to stimulate proliferation and protected against CSE-induced cytotoxicity in cultured HBECs in vitro. Additionally, we determined that MUC1 was expressed in fibroblasts and that suppressing MUC1 genetically (knockdown) or pharmacologically (with GO-201) enhanced EREG production. Together with the oncogenic effects of MUC1 in epithelial and macrophage cells determined previously (10,12), this study highlights a promiscuous role of MUC1 in lung cancer development that may be related to cell-type specific functions of MUC1 in the tumor microenvironment. MUC1 deficiency in fibroblasts and malignant cells may increase EREG production that activates the EGFR pathway to compensate for MUC1 loss in epithelial cells, thereby promoting the EGFR/AKT pathway for lung carcinogenesis (Figure 6).
Figure 6.
A proposed model of MUC1 and EREG interaction in lung carcinogenesis. Cigarette smoke induces MUC1 expression that suppresses EREG in fibroblasts and transformed cells. Suppression of MUC1 facilitates EREG production from fibroblasts and transformed cells, which activates the EGFR/AKT pathway to compensate for MUC1 loss in epithelial cells, promoting lung carcinogenesis.
MUC1 is expressed in epithelial cells and hematopoietic cells including macrophages, T lymphocytes and dendritic cells (2,30–32). Here we determined that MUC1 is also expressed in pulmonary fibroblasts, which is consistent with the report of MUC1 expression in skin fibroblasts (33). Stromal cells including macrophages and fibroblasts in the tumor microenvironment play pivotal roles in regulating carcinogenesis. Growth factors and cytokines produced in stromal cells can be pro- or anti-oncogenic (34), and targeting the crosstalk between tumor and stromal cells has emerged as a new anticancer strategy (35). Our findings that MUC1 is involved in regulating TNF-α secretion from macrophages and EREG production in fibroblasts and malignant cells strongly suggest that MUC1 participates in modulation of the microenvironment during lung carcinogenesis.
Many in vitro studies consistently suggest an oncogenic role of MUC1 in different types of cancers (8,9), some of which was validated with Muc1 KO mice in vivo (36). Muc1 enhanced tumor progression in a mouse model of spontaneous pancreatic adenocarcinoma (37). Muc1 KO significantly delayed murine mammary tumor progression (18,38). In a KrasG12D-driven pancreatic ductal adenocarcinoma mouse model, Muc1 KO profoundly suppressed tumor growth and metastasis (39). Consistently, transgenic expression of human Muc1 in IL-10−/− mice accelerated progression of inflammatory bowel disease to colon cancer (40). Knowing that transgenic expression of human MUC1 in C57Bl/6 mice seemed to increase urethane-induced lung tumors (41,42), we were surprised to see a lung tumor-suppressing effect of Muc1 with the whole-body Muc1 KO mice in the NNK A/J mouse lung cancer model. This surprising but interesting observation implies the complexity of MUC1’s function in lung carcinogenesis, while its underlying mechanism is currently unknown. MUC1 suppresses EREG production in fibroblasts and malignant cells, which could be one of the pieces of this puzzle. Supporting this notion, an inverse association between MUC1 and EREG was detected and EREG expression was inversely associated with patient survival in lung adenocarcinoma. Our results suggest that MUC1 negatively regulates EREG-mediated human lung carcinogenesis.
EREG binds EGFR to activate the downstream pathways, including Akt, with higher potency than EGF in promoting cell proliferation and DNA synthesis (22,43). EREG derived from tumor-associated fibroblasts promoted growth of colitis-associated neoplasms (23), and was able to transform polarized Madin–Darby canine kidney epithelial cells (44). In non-small cell lung cancer patients, intratumoral EREG could be a marker of advanced disease (45). Here, we found a significant inverse association between EREG expression and lung cancer patient survival. Thus, increased EREG production may be responsible for increased lung carcinogenesis in the whole-body Muc1 KO mice. Although EREG production in normal epithelial cells was marginal (data not shown), it is produced in lung cancer cells. While losing MUC1 in epithelial cells results in suppression of EGFR signaling (12), the increased EREG production in fibroblasts and malignant cells may establish a negative feedback loop for compensating Muc1 loss for EGFR signaling in epithelial cells. EREG produced by fibroblasts and malignant cells may cooperatively stimulate EGFR in a higher magnitude for oncogenic signaling that shifts the balance from the expected tumor suppression to the surprising tumor promotion in Muc1 KO mice. Because GO-201, a MUC1 activity inhibitor that suppresses several cellular signaling pathways (28,29), enhanced EREG production in lung cancer cells, it is highly likely that MUC1 suppresses EREG through intracellular signaling. Further investigation on the precise mechanism of MUC1-mediated suppression on EREG production is warranted.
However, our model does not exclude other potential mechanisms. For example, MUC1 is expressed in immune cells, and thus, it is possible that an immune-related mechanism is involved. Indeed, Muc1 suppression increased tumor-promoting myeloid-derived suppressor cells (MDSCs) during experimental colitis, and depletion of MDSCs reduced colon tumor formation (30). Other mechanisms such as interaction of Ras and EGFR/Akt are also possible (46). Nevertheless, our results strongly suggest that the role of MUC1 in lung cancer development is complex, which may involve distinct functions of MUC1 in epithelial, stromal and cancer cells in the tumor microenvironment. Understanding the cell-type specific roles of MUC1 with cell-type specific Muc1 knockout mice may hold the key for elucidating the mechanism of lung carcinogenesis involving MUC1. Giving that directly targeting MUC1 for cancer therapy is under investigation, the complex roles of MUC1 in lung cancer suggested in this report warrant careful investigation to avoid unexpected deleterious effects from MUC1 suppression.
It is not uncommon that a factor has contradictory functions in cancer. For example, TGFβ has dual roles in carcinogenesis: tumor-suppressing at early stages and tumor-promoting at late stages (47). Similarly, while IL-6 and its downstream factor STAT3 are well-accepted oncoproteins, both of them were recently reported to have contradictory functions in lung carcinogenesis: prevents cancer initiation by maintaining pulmonary homeostasis under oncogenic stress, and facilitates cancer progression by promoting cancer cell growth (48,49). These reports suggest that some genes have distinct functions at different timing during carcinogenesis. Whether MUC1 similarly has temporally distinct roles in lung carcinogenesis deserves further study.
Supplementary Material
Supplementary data are available at Carcinogenesis online.
Funding
This study was supported by grants from NIEHS/NIH (R01ES017328) and NCI/NIH (1R21CA193633).
Supplementary Material
Acknowledgements
We thank Dr Sandra J Gendler from Mayo Clinic (Scottsdale, AZ) for providing the Muc1 knockout mouse strain (C57BL/6J background).
Conflict of Interest Statement: None declared.
Abbreviations
- CS
cigarette smoke
- CSE
cigarette smoke extract
- EREG
epiregulin
- KO
knockout
- HBEC
human bronchial epithelial cell
- MUC1
Mucin 1
- TNF-α
tumor necrosis factor-alpha
References
- 1. Kyo Y., et al. (2012) Antiinflammatory role of MUC1 mucin during infection with nontypeable Haemophilus influenzae. Am. J. Respir. Cell Mol. Biol., 46, 149–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baldus S.E., et al. (2004) MUC1 and the MUCs: a family of human mucins with impact in cancer biology. Crit. Rev. Clin. Lab. Sci., 41, 189–231. [DOI] [PubMed] [Google Scholar]
- 3. Woenckhaus M., et al. (2008) Prognostic value of FHIT, CTNNB1, and MUC1 expression in non-small cell lung cancer. Hum. Pathol., 39, 126–136. [DOI] [PubMed] [Google Scholar]
- 4. Schroeder J.A., et al. (2004) MUC1 overexpression results in mammary gland tumorigenesis and prolonged alveolar differentiation. Oncogene, 23, 5739–5747. [DOI] [PubMed] [Google Scholar]
- 5. Guddo F., et al. (1998) MUC1 (episialin) expression in non-small cell lung cancer is independent of EGFR and c-erbB-2 expression and correlates with poor survival in node positive patients. J. Clin. Pathol., 51, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Acres B., et al. (2015) Targeted immunotherapy designed to treat MUC1-expressing solid tumour. Curr Top Microbiol Immunol. [DOI] [PubMed] [Google Scholar]
- 7. Kimura T., et al. (2013) MUC1 immunotherapy is here to stay. Expert Opin. Biol. Ther., 13, 35–49. [DOI] [PubMed] [Google Scholar]
- 8. Kufe D.W. (2013) MUC1-C oncoprotein as a target in breast cancer: activation of signaling pathways and therapeutic approaches. Oncogene, 32, 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nath S., et al. (2014) MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med., 20, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu X., et al. (2014) MUC1 in macrophage: contributions to cigarette smoke-induced lung cancer. Cancer Res., 74, 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hattrup C.L., et al. (2008) Structure and function of the cell surface (tethered) mucins. Annu. Rev. Physiol., 70, 431–457. [DOI] [PubMed] [Google Scholar]
- 12. Xu X., et al. (2012) MUC1 contributes to BPDE-induced human bronchial epithelial cell transformation through facilitating EGFR activation. PLoS One, 7, e33846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Russo A., et al. (2015) A decade of EGFR inhibition in EGFR-mutated non small cell lung cancer (NSCLC): Old successes and future perspectives. Oncotarget, 6, 26814–26825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schneider M.R. (2014) The magnificent seven: epidermal growth factor receptor ligands. Semin. Cell Dev. Biol., 28, 1. [DOI] [PubMed] [Google Scholar]
- 15. Sunaga N., et al. (2013) Oncogenic KRAS-induced epiregulin overexpression contributes to aggressive phenotype and is a promising therapeutic target in non-small-cell lung cancer. Oncogene, 32, 4034–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bauer A.K., et al. (2016) Epiregulin is required for lung tumor promotion in a murine two-stage carcinogenesis model. Mol Carcinog., 56, 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wells A. (1999) EGF receptor. Int. J. Biochem. Cell Biol., 31, 637–643. [DOI] [PubMed] [Google Scholar]
- 18. Spicer A.P., et al. (1995) Delayed mammary tumor progression in Muc-1 null mice. J. Biol. Chem., 270, 30093–30101. [DOI] [PubMed] [Google Scholar]
- 19. Ramirez R.D., et al. (2004) Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res., 64, 9027–9034. [DOI] [PubMed] [Google Scholar]
- 20. Chen W., et al. (2012) Low-dose gamma-irradiation inhibits IL-6 sretion from human lung fibroblasts that promotes bronchial epithelial cell transformation by cigarette-smoke carcinogen. Carcinogenesis, 33, 1368–1374. [DOI] [PubMed] [Google Scholar]
- 21. Toyoda H., et al. (1995) Epiregulin. A novel epidermal growth factor with mitogenic activity for rat primary hepatocytes. J. Biol. Chem., 270, 7495–7500. [DOI] [PubMed] [Google Scholar]
- 22. Riese D.J., II, et al. (2014) Epiregulin: roles in normal physiology and cancer. Semin. Cell Dev. Biol., 28, 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neufert C., et al. (2013) Tumor fibroblast-derived epiregulin promotes growth of colitis-associated neoplasms through ERK. J. Clin. Invest., 123, 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cho W.C., et al. (2011) The role of inflammation in the pathogenesis of lung cancer. Expert Opin. Ther. Targets, 15, 1127–1137. [DOI] [PubMed] [Google Scholar]
- 25. Peebles K.A., et al. (2007) Inflammation and lung carcinogenesis: applying findings in prevention and treatment. Expert Rev. Anticancer Ther., 7, 1405–1421. [DOI] [PubMed] [Google Scholar]
- 26. Wang Q., et al. (2013) RIP1 potentiates BPDE-induced transformation in human bronchial epithelial cells through catalase-mediated suppression of excessive reactive oxygen species. Carcinogenesis, 34, 2119–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Raina D., et al. (2009) Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res., 69, 5133–5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kufe D.V. (2008) Targeting the human MUC1 oncoprotein: A tale of two proteins. Cancer Biol Ther, 7, 81–84. [DOI] [PubMed] [Google Scholar]
- 29. Kharbanda A., et al. (2014) Targeting the oncogenic MUC1-C protein inhibits mutant EGFR-mediated signaling and survival in non-small cell lung cancer cells. Clin. Cancer Res., 20, 5423–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Poh T.W., et al. (2013) Downregulation of hematopoietic MUC1 during experimental colitis increases tumor-promoting myeloid-derived suppressor cells. Clin. Cancer Res., 19, 5039–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Correa I., et al. (2003) Form and pattern of MUC1 expression on T cells activated in vivo or in vitro suggests a function in T-cell migration. Immunology, 108, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taki C., et al. (2002) MUC1 mucin expression in follicular dendritic cells and lymphoepithelial lesions of gastric mucosa-associated lymphoid tissue lymphoma. Pathol. Int., 52, 691–701. [DOI] [PubMed] [Google Scholar]
- 33. Kumar P., et al. (2014) MUC1 Is Expressed by Human Skin Fibroblasts and Plays a Role in Cell Adhesion and Migration. Biores. Open Access, 3, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hanahan D., et al. (2011) Hallmarks of cancer: the next generation. Cell, 144, 646–674. [DOI] [PubMed] [Google Scholar]
- 35. Rampias T., et al. (2016) Targeting tumor-stroma crosstalk: the example of the NT157 inhibitor. Oncogene, 35, 2562–2564. [DOI] [PubMed] [Google Scholar]
- 36. Joshi S., et al. (2015) Genetically engineered mucin mouse models for inflammation and cancer. Cancer Metastasis Rev., 34, 593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tinder T.L., et al. (2008) MUC1 enhances tumor progression and contributes toward immunosuppression in a mouse model of spontaneous pancreatic adenocarcinoma. J. Immunol., 181, 3116–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Al Masri A., et al. (2005) Muc1 affects c-Src signaling in PyV MT-induced mammary tumorigenesis. Oncogene, 24, 5799–5808. [DOI] [PubMed] [Google Scholar]
- 39. Besmer D.M., et al. (2011) Pancreatic ductal adenocarcinoma mice lacking mucin 1 have a profound defect in tumor growth and metastasis. Cancer Res., 71, 4432–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Beatty P.L., et al. (2007) Cutting edge: transgenic expression of human MUC1 in IL-10−/− mice accelerates inflammatory bowel disease and progression to colon cancer. J. Immunol., 179, 735–739. [DOI] [PubMed] [Google Scholar]
- 41. Wurz G.T., et al. (2013) Antitumor effects of L-BLP25 antigen-specific tumor immunotherapy in a novel human MUC1 transgenic lung cancer mouse model. J. Transl. Med., 11, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kao C.J., et al. (2014) Antitumor effects of cisplatin combined with tecemotide immunotherapy in a human MUC1 transgenic lung cancer mouse model. Cancer Immunol. Res., 2, 581–589. [DOI] [PubMed] [Google Scholar]
- 43. Wilson K.J., et al. (2012) EGFR ligands exhibit functional differences in models of paracrine and autocrine signaling. Growth Factors, 30, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Singh B., et al. (2013) Transformation of polarized epithelial cells by apical mistrafficking of epiregulin. Proc. Natl. Acad. Sci. USA, 110, 8960–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang J., et al. (2008) Intratumoral epiregulin is a marker of advanced disease in non-small cell lung cancer patients and confers invasive properties on EGFR-mutant cells. Cancer Prev. Res. (Phila)., 1, 201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Minjgee M., et al. (2011) K-RAS(V12) induces autocrine production of EGFR ligands and mediates radioresistance through EGFR-dependent Akt signaling and activation of DNA-PKcs. Int. J. Radiat. Oncol. Biol. Phys., 81, 1506–1514. [DOI] [PubMed] [Google Scholar]
- 47. Gold L.I. (1999) The role for transforming growth factor-beta (TGF-beta) in human cancer. Crit. Rev. Oncog., 10, 303–360. [PubMed] [Google Scholar]
- 48. Qu Z., et al. (2015) Interleukin-6 Prevents the Initiation but Enhances the Progression of Lung Cancer. Cancer Res., 75, 3209–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou J., et al. (2015) Differential roles of STAT3 in the initiation and growth of lung cancer. Oncogene, 34, 3804–3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.