Abstract
The computational prediction of drug responses based on the analysis of multiple types of genome-wide molecular data is vital for accomplishing the promise of precision medicine in oncology. This will benefit cancer patients by matching their tumor characteristics to the most effective therapy available. As larger and more diverse layers of patient-related data become available, further demands for new bioinformatics approaches and expertise will arise. This article reviews key strategies, resources and techniques for the prediction of drug sensitivity in cell lines and patient-derived samples. It discusses major advances and challenges associated with the different model development steps. This review highlights major trends in this area, and will assist researchers in the assessment of recent progress and in the selection of approaches to emerging applications in oncology.
Keywords: drug sensitivity, computational prediction models, cancer, translational bioinformatics, precision medicine
Introduction
The accurate computational prediction of the response of cancer patients to therapies based on the patient’s molecular and clinical profiles is vital in the era of precision medicine [1–5]. This is crucial to assist clinicians in making decisions on the most effective and least toxic therapeutic options available, and will enable a smarter selection and monitoring of patients in clinical trials [6–8]. This has been motivated in large part by an important transformation in cancer classification: Moving from solely analyzing histopathologic characteristics of tumors, into analyzing molecular features that are indicative of treatment response.
These molecular features comprise different types of genomic aberrations, ranging from point mutations, deletions, insertions and translocations of gene sequences. Such alterations may represent the direct targets of therapies. This comprises treatments already approved for clinical use and others undergoing further investigations [2, 5]. Examples of clinically actionable alterations are those involving EGFR and ALK genes, which may be targeted with kinase inhibitor drugs [9]. In lung cancers, point mutations and deletions in EGFR and EML4-ALK fusions may be found in a minority of patients [9]. Annotated catalogues of clinically relevant genomic alterations are available at the ClinVar (http://www.ncbi.nlm.nih.gov/clinvar) and COSMIC databases (http://cancer.sanger.ac.uk/cosmic).
Moreover, the computational prediction of drug responses can significantly contribute to preclinical research as in silico drug screening tools [10–12]. These tools can help biologists to prioritize candidate compounds in their research, and may represent useful strategies for efficiently planning experiments and reducing costs. These opportunities have been investigated in different preclinical and clinical application domains in oncology with diverse computational approaches and ‘omics’ data types.
The computational prediction of drug responses in cancer involves significant research challenges. It is a biological challenge because of the complexity of cancers as highly heterogeneous and multi-factorial diseases. It is a data challenge because of the complexity of potentially useful data sets available: in terms of volume, noise and heterogeneity. Furthermore, as the need for integrating data increases, new challenging technical questions arise, such as the harmonization and normalization of data originating from multiple sources. It is anticipated that these challenges, and opportunities, will be augmented as the cost of data generation is reduced and societal expectations on the promise of precision medicine grow.
Critical questions in the development of computational models for drug response prediction include: Which data sets should be selected for training and testing models? Are models specific to cancer types or generalizable as pan-cancer models? Which computational approaches are suitable for application? How such models are evaluated and validated? Furthermore, other user-centric issues, such as model interpretability and reporting, are crucial not only from the standpoint of bioinformaticians, but also from that of biologists and clinicians.
This article reviews the application of computational models for predicting drug responses in cancer research. It addresses the above-mentioned challenges and questions by reviewing key resources, approaches and examples with relevance to preclinical and clinical research. Although most applications reported to date are based on gene expression data, models based on other data types, e.g. DNA-level aberrations and protein expression, are also discussed here. This article dose not aim to cover all design aspects of computational modeling, and does not emphasize a particular technique or cancer area. Rather, it discusses central distinguishing features of data sources, modeling techniques and applications. It discusses challenges relating to the generation and validation of these models, as well as critical concerns about their reporting and interpretability. This review highlights the prediction of drug sensitivity using clinically relevant in vitro models and patient-derived data as inputs to supervised learning techniques. The latter represents the most used and representative prediction strategy to date. A detailed discussion of machine learning methods is out of the scope of our article. A review of the topic in the context of genomic research has been recently published elsewhere [13]. Other aspects related to drug response, such as side effects or cell type-specific drug effects, have been reviewed in other journals [14, 15]. Although the prediction of drug responses is relevant to drug repositioning, i.e. the use of ‘old’ drugs for new disease applications, the latter is based on specific design requirements and prediction goals that are not discussed here. In a typical drug repositioning setting, researchers are not a priori interested in a specific set of candidate drugs. In the application scenarios discussed here, the focus on specific candidate compounds usually drives both model design and evaluation. The models resulting from drug repositioning investigations may offer descriptions of drug mode of action. Conversely, while the models discussed here may provide the basis for further mechanistic understanding, their main objective is to accurately estimate the response of a given biological sample to a particular drug. A review on current trends in drug repositioning was recently published in this journal [16].
Overall computational strategy
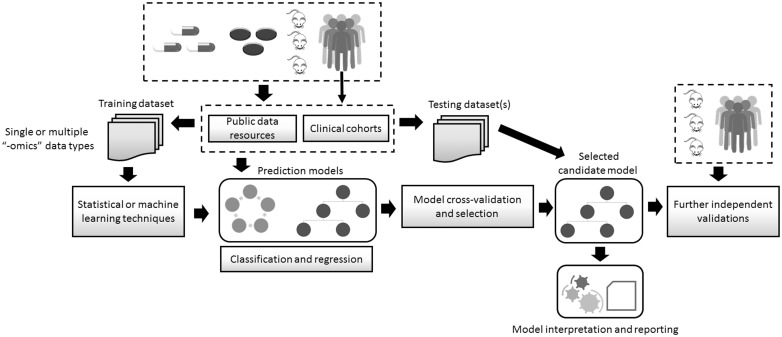
The development of computational models for predicting drug response requires four essential steps, which draw in part from the standard strategy for developing machine learning models (Figure 1) [13]. In the first step, data sets are selected and preprocessed. This involves the expert- or computer-driven selection of potentially relevant data sub-sets, their normalization and initial filtering of noisy or irrelevant data features. This comprises the detection of putative significant associations between molecular features and the drug response to be predicted. There is no universal solution to the problem of feature selection. The potential relevance of the selected features depends on the characteristics of the prediction problem investigated, including factors such as the amount of data available and the representation of the outcome variable. This may be done with different statistical methods, such as univariate correlations between gene expression levels and drug sensitivity measurements (e.g. IC50 values). Moreover, the selection of significant molecular features may also be part of, or embedded into, the model training phase. Reviews on feature selection and dimensionality reduction have been published in this journal.
Figure 1.
Key steps in the development of computational models for predicting drug response. Data obtained from cell lines, animals or humans are stored in different data repositories, including public databases. These resources also include drug response information. Data sets are obtained to be subsequently used as training data sets, and may contain one or more types of ‘omics’ data, e.g. transcriptomics and DNA sequence. Such data are used as inputs to statistical or machine learning techniques. The prediction problem may be defined as either a classification or a regression problem, and a variety of techniques may be applied. The predictive performance of the models is assessed with cross-validation sampling techniques. The most-promising models are selected and evaluated using testing data sets, which were not used during the training phase. The model and its predictions undergo human expert interpretation and their reporting to stakeholders follows. Further independent validations using clinically relevant data are required to continue bridging the gap between the laboratory and the clinic.
Drug sensitivity investigations often include samples exhibiting ‘extreme’ responses to treatments, and typically a non-standard hard thresholding for sensitivity versus resistance is defined. However, as discussed in more detail below, recent models are incorporating a wider range of cell lines or clinical samples, which more accurately represent tumor heterogeneity and treatment responses. Furthermore, prediction models that make numerical estimations of sensitivity without prior selection of response thresholds are also being investigated.
The second step comprises the training phase of the model chosen to address the prediction problem. To achieve this task, standard procedures for fitting models to data and for measuring the predictive quality of the resulting models are available. A wide variety of techniques derived from the statistical and machine learning research areas can be applied, and their selection is constrained by different factors, such as the type of input data and the characteristics of the drug response prediction problem, as discussed below.
After completing model training and selecting a model that appropriately fits the data, researchers implement multiple tests on independent data. This process, also referred to as independent evaluation, verifies that candidate models can accurately predict responses on unseen samples, and ideally is applied to multiple data sets obtained by different laboratories or measurement platforms. These training-testing phases may be based on the analysis of data obtained from clinical studies or from in vitro experiments using cancer cell lines, as discussed below. At the end of a training-testing cycle, new training-testing iterations may also be required for model refining or for incorporating prior biological knowledge into the model. Although thousands of cell lines necessary for training and testing prediction models have become publicly available, key limitations remain. One of them is that such data sets do not offer extensive cell line collections for all cancer types, which makes the implementation of accurate cancer type-specific models hard to achieve. Additionally, major data repositories emphasize cytotoxic drugs or targeted ones already in clinical use.
In a preclinical research context, models are typically trained on data coming from in vitro experiments using cell lines, and the most promising model is then tested on either new data generated by in vitro experiments or from animals. This testing phase may be followed by the application of the model to data that more closely resemble clinically relevant characteristics of a particular cancer type. For example, models trained on cell line-derived data can be tested on solid or liquid biopsies directly obtained from patients. In the case of models trained on patient-derived samples, the resulting model may be further applied to a different cancer sub-type or patient cohort. Other critical issues are the implementation of suitable methods for reporting predictions and for facilitating their interpretation by clinically oriented experts. The following sections will discuss these steps, summarized in Figure 1, in more detail.
Public data resources for building computational prediction models
Drug response prediction models are typically trained on data sets generated by different research consortia. When a consortium is driven by a specific clinical need in a particular oncology domain, this scenario enables applications that are tailored to specific biological hypotheses or patient characteristics. This setting is, however, potentially limited by problems of scale: number of patients analyzed, number of molecular readouts measured or the types of (omics and clinical) data that may be incorporated into model development. An alternative approach that is gaining significant support from the community is the training (and/or testing) of models based on publicly available data generated by large research consortia.
Two of the most significant resources of publicly available data for investigating drug responses are the Cancer Cell Line Encyclopedia (CCLE) [17] and the Genomics of Drug Sensitivity in Cancer (GDSC) project [18], which offer baseline data (i.e. coming from untreated samples) obtained from different ‘omic’ modalities and diverse measurements of drug sensitivity in cancer cell lines (Table 1). The CCLE contains mutation, gene copy numbers and gene expression data from >1K cell lines from 36 tumor sites. Moreover, drug sensitivity data from >11K experiments that tested 24 anticancer drugs on >500 cell lines are also publicly available. The GDSC offers around 75K experiments that tested 138 anticancer drugs on >1K cell lines from diverse cancer types. The baseline data include gene copy number and expression data, as well as somatic mutations in 75 genes relevant to cancer. Another key resource is the NCI-60 cancer cell line collection [19], which is a pioneer in the characterization of drugs in vitro and whose cell lines are also included in the CCLE and GDSC. The NCI-60 provides drug screening data for thousands of drugs with potential applications in cancer therapy and 60 cell lines representing nine cancers (Table 1). The cell lines are described by different types of data, including those included in the CCLE and GDSC, as well as protein expression data. These three resources and associated projects also offer Web sites or programmatic access for querying or downloading their data [17, 20–22], and new interactive resources that contain subsets of those data are becoming available [23, 24].
Table 1.
Summary of key public resources for enabling the development of computational models for predicting drug response
| Attribute | CCLE | GDSC | NCI-60 |
|---|---|---|---|
| # cell lines | >1000 | >1000 | 60 |
| # compounds | 24 | 138 | >15 K |
| # drug tests | >11 K | >75 K | >100 K |
| Main omics data sets | Mut, Gcn, Gexp | Mut, Gcn, Gexp | Mut, Gcn, Gexp, Prot |
| # cancers | 36 | >15 | 9 |
| Reference | [17] | [18] | [19] |
| Website | http://www.broadinstitute.org/ccle | http://www.cancerrxgene.org/ | discover.nci.nih.gov/cellminer/ |
Note. CCLE = Cancer Cell Line Encyclopedia; GDSC = genomics of drug sensitivity in cancer; NCI-60 = the US National Cancer Institute 60 human tumor cell line drug screen database; # = number of; Mut = mutations; Gcn = gene copy numbers; Gexp = gene expression; Pexp = protein expression.
In principle, all these data sets can be integrated to build drug sensitivity prediction models. In practice, researchers have typically developed models that focus, for example, on a particular database for model training and the resulting model is tested on privately owned data. Conversely, after training a model on public data, the resulting model is applied to a second public data set. Critical issues that need to be considered when combining data sets or using any of them to ‘validate’ the prediction capability of a model are: the statistical harmonization of data sets, processing of data present in one data set but missing in another one and data re-scaling procedures. Moreover, even if different databases use the same carefully characterized cell lines, it is important to note that different experimental and/or mathematical procedures may have been used to estimate drug responses, i.e. drug sensitivity measures. These factors are crucial to investigate the predictive performance and reproducibility of models. Although concerns have been previously reported about potential inconsistencies between the CCLE and the GDSC in terms of the drug response information stored in these data sets [25], other research has highlighted their concordance at different analysis levels. A recent joint study by the CCLE and the GDSC demonstrated a considerable concordance between these resources, not only in terms of drug response measures, but also regarding predictive features for those responses [26].
Input data types for prediction modeling
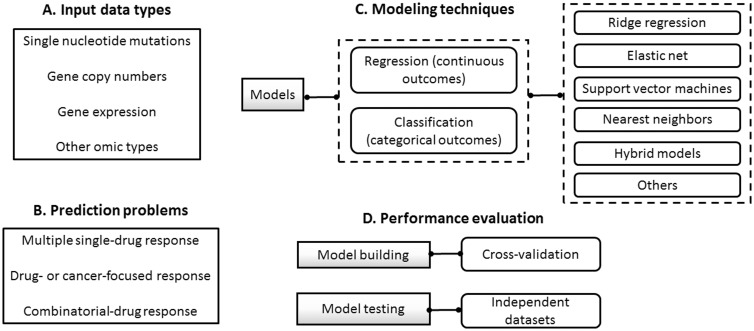
Models may also be characterized on the basis of the data types used as their inputs (Figure 2A). Several models have been developed with publicly and privately owned data sets consisting of single-nucleotide mutations, gene copy numbers and gene expression [27, 28]. Different examples of models based on single or multiple sources, e.g. gene expression data only or their combination with gene copy number data, have been reported with gene expression data as the most widely used source [29, 30]. Comparative analyses have usually shown that gene expression data encode the most powerful predictive features, and that integrated models may only marginally increase the accuracy of drug response predictions [27, 28]. However, such observations mainly refer to global tendencies across cell lines, and the level of predictive capability is constrained by the type of cancer investigated, training data size, algorithm choice and drugs selected for modeling [31]. In multiple myeloma, for instance, Amin et al. [32] reported that gene expression data alone are insufficient to predict responses for a small number of drugs and using a variety of models. Nevertheless, this study focused on the classification of samples in terms of two treatment response classes only: ‘complete’ versus ‘partial’ responses. A more recent study, by Cortes-Ciriano et al. [33], showed that the combination of compound structure, protein, gene and miRNA expression values can indeed offer added value in the prediction of drug sensitivity trained on the NCI-60 data set. Such a diversity of results underlines the need for further studies using different data and applications. In practice, the identification of the most predictive data sets is a context-specific problem, which depends on both cancer type and compounds investigated.
Figure 2.
A graphical synthesis of the diversity of computational models available for the prediction of drug responses. (A) List of data types most commonly used. (B) Categorization of models on the basis of the prediction problems addressed. (C) General hierarchy of statistical and machine learning techniques most commonly investigated. (D) Fundamental data sampling strategies for assessing model prediction capability.
In an effort to enable accurate and interpretable predictions, computational models based on the analysis of protein or compound structure, methylation, protein expression data and the activity of hallmark signaling pathways in cancer are increasingly becoming the focus of investigation [24, 33–37]. Fey et al. [35], for instance, recently reported a novel approach based on patient-specific simulations of signaling pathway activity, with a focus on the c-Jun N-terminal kinase pathway under stress. Network structure and logic were estimated with prior knowledge and protein expression experiments in a zebrafish model of neuroblastoma. Simulations of network perturbations, including those mimicking drug effects, were run on the resulting network. To validate this model in patients, the authors tackled an important requirement: Only gene expression data were available in the patient cohorts. The authors showed reasonable levels of correlation between the expression of the proteins included in the animal model and patient-derived transcript expression. This allowed independent tests on multiple cohorts consisting of hundreds of patients, and the demonstration of the prognostic utility of the model. This study exemplifies not only a novel computational modeling approach, but also the combination of different data types to meet study requirements and constraints.
Oncology application domains
Researchers have benefited from omics and drug response data available in public repositories across all cancer types for building (pan-cancer and cancer-specific) prediction models. Once these models have been trained, researchers can test them on their own data sets (or others) for the particular cancer types or tissue sites of interest. To assess their potential clinical relevance, drug response prediction models are evaluated with tumor data originating from patients [38, 39]. Moreover, this may be preceded by testing the model's predictive capacity on clinically relevant tumor xenografts implanted in animals, such as rodents [40] and zebrafish [35]. Although there is still a need to advance this practice, individual research groups and large consortia are increasingly reporting such patient-oriented investigations for enabling clinical decision-making support.
The following training-testing application scenarios have been the most commonly reported: (i) models are trained on public cell line-derived data resources and tested on publicly available patient data from clinical trials or other cohorts [30, 41]; (ii) models are trained on privately owned cell line-derived data and tested on patient data from clinical trials or other patient cohorts [42]; and (c) models are both trained and tested on patient-derived data from clinical trials or other patient cohorts [38, 39].
In these settings, different clinical outcomes are used as indicators of treatment response, such as survival and disease recurrence times. In applications in which models are trained on cell line data and tested on clinical cohorts, model predictions are interpreted as surrogates of patient response. This means that the assessment of prediction capacity, in the test mode, is based on the analysis of the predicted responses, e.g. drug sensitivity, and their relation to the observed patient responses. This is done by applying the model evaluation techniques introduced below, and includes analysis of correlations between predicted and observed responses.
Geeleher et al. [30] trained models on gene expression data and drug responses from the Cancer Genome Project, part of the GDSC, and performed independent tests on publicly available data from clinical trials in myeloma and non-small-cell lung cancers. Other application scenarios have focused on (either public or private) data that are relevant to a particular cancer for both training and testing purposes. For instance, Daemen et al. [29] generated drug sensitivity data for 138 drugs and molecular profiles from 70 cell lines in breast cancer, which were used to train models for classifying samples into two categories: good and poor treatment responders. Then, using independent patient-derived data, they validated their model on two drugs used for treating breast cancers (tamoxifen and valproic acid). Although further independent validations are warranted, Daemen et al. [29] and Geeleher et al. [30] showed that their cell line-based models can accurately predict therapeutic response in patients, including relapse-free survival.
Using lung cancer cell lines and patient data from the BATTLE study, Byers et al. [40] demonstrated how a 76-gene (expression) signature could distinguish between epithelial and mesenchymal tumor samples. Next, they correlated such groups with drug responses, which were defined as IC50 values in the cell lines and as disease control categories in the patients. Although the analysis focused on a relatively small number of patients with a specific phenotype (EGFR/KRAS wild-type patients), the authors made a solid case for the association between the epithelial-like signature with good clinical outcome. The study also enhanced its potential clinical applicability by showing that the signature is robust across different microarray platforms, and its selection was based on a combination of hypothesis- and data-driven approaches.
In colorectal cancer, Guinney et al. [41] implemented an elastic net model based on gene expression data, which accurately classified colorectal cancer samples according to their RAS phenotype. The model improves on the traditional clinical classification that relies on KRAS mutation status. They trained and tested models using multiple data sets from patients, cell lines and mouse xenografts. For instance, they tested the model’s capacity to predict resistance to cetuximab, an anti-EGFR antibody used to treat colorectal cancer, on mouse xenografts and three independent patient cohorts. This study exemplifies the potential clinical utility of modeling specific cancer phenotypes and molecular activity, and demonstrates the power of tapping into a diversity of publicly available data sets.
In gynecological cancers, Pereira et al. [43] reported evidence that posttreatment survival may be accurately estimated based on the amount of tumor DNA detected in the blood of patients. However, additional work is warranted owing to the limited number of samples (10 patients) and the lack of independent validation. In ovarian cancer, Chen et al. [38] showed the potential relevance of a 61-transcript expression signature for predicting patient’s response to platinum-taxane chemotherapy. More specifically, the model accurately assigned patients to two poor and one good survival groups. Moreover, when the expression signature was combined with BRCA1/2 mutation status, which is a traditional prognostic marker in this disease, a better patient stratification of clinical response was achieved. Another distinguishing feature of this study was the relatively large amounts of independent patient data used for model training (data from The Cancer Genome Atlas [44]) and testing (eight data sets from different studies).
Other disease- or drug-specific modeling approaches have been investigated for breast [37], colorectal [41, 45], lung [46], diffuse large B-cell lymphomas [42] and neuroblastoma [47] cancers.
Although the usefulness of cell lines in cancer research is indisputable and they offer opportunities for developing drug response prediction models, their translation into clinical applications poses major challenges [48]. It has been shown that prediction models based on these samples may not always have clinical or translational value [11, 49]. Among the reasons for the failure of previous research are that cell lines often tend to be more similar to each other than to the primary samples that they supposedly model, and that cell lines from specific anatomical sites may incorrectly match tumors originating from different locations [50, 51]. Moreover, in vitro-derived data do not take into account the influence of the tumor microenvironment on drug responses. In general, a crucial challenge is the selection of cell lines that accurately reflect the ‘omic’ profiles observed in tumors, a problem that is compounded by a substantial molecular diversity across and within cancers [48]. The latter means that drug response prediction models applicable to one cancer type may not always be successfully translated to another type. Another important factor that explains translational failure is the lack of sufficient numbers of cell lines that are representative of specific cancer types and sub-types [52], which significantly limits the feasibility of developing accurate predictions for specific tumors and drugs.
More recently, it has been shown that cell lines directly obtained from fresh patient material and kept under short-term culture can better capture tumor heterogeneity and maintain the genomic profile of the parental tumor [53, 54]. The similarity between cell lines and their tumors of origin, as well as their potential clinical relevance, have been carefully assessed for a few cancer types and is the topic of ongoing investigations [55, 56].
Despite these and other advances, model validations using clinically focused patient-derived data are still needed across cancer domains. These efforts will be hampered by restrictions to appropriately annotated data sets. Thus, there is a pressing need for sharing patient-derived data from clinical trials, including omics profiles and treatment response data, through public or user-restricted access.
Typical prediction problems and outcomes
Computational models typically estimate drug sensitivity for the single drugs included in a training data set. Hence, during testing, predictions may be made for each drug (or their combinations when possible) and for each sample included in a test data set (Figure 2B). To date, the application scenario most widely investigated involves building models that make sensitivity predictions for multiple single compounds. Here these models are referred to as pan-cancer single-drug response prediction models [27, 57]. Models trained and tested on the CCLE data are representative examples in this category [17, 27].
Other models focus on making predictions for one or a few treatments that undergo investigations in a specific patient cohort or clinical trial. This could involve, for example, models for predicting sensitivity (or resistance) to either a particular drug class or tailored to a specific cancer type [38, 46]. Here these models are referred to as drug- or cancer-focused response prediction models. Blumenschein et al.’s [39] model for predicting response to sorafenib in lung cancer patients from the BATTLE trial illustrates a novel and clinically relevant application in this category.
More recently, different efforts have been invested in the prediction of sensitivity for combinations of individual drugs [58, 59]. Here these models are referred to as combinatorial-drug response prediction models. As part of the DREAM Challenges initiative, for example, 31 models for predicting the response to drug combinations (pairs) were reported [59]. In this setting, models were trained on proprietary gene expression data from treated and untreated cell lines, including 14 compounds. During training, models were provided with drug response information derived from single compounds, and afterward the models made predictions for 91 compound pairs. The best performing method, DIGRE [58], predicted (sequentially applied) combinations based on the hypothesis that the transcriptional effects induced by the first drug also contribute to the effects of the second drug. Thus, DIGRE first calculated the similarity of the gene expression effects individually induced by two drugs, and then used this information together with dose-response curves as inputs to a mathematical model that estimated the residual effect. The last step involved the calculation of combinatorial effect scores for each of the two treatment sequences [59]. Alternative combinatorial-drug response prediction models and applications will increasingly be developed as larger data sets become accessible [60, 61].
Overview of techniques for building prediction models
The most widely applied computational approaches to building models for drug response prediction are based on supervised learning techniques (Figure 2C). Although here I focus on these techniques, it is important to note that unsupervised learning, including standard clustering techniques, can provide the basis for generating prediction models. They enable important tasks, such as data selection and visualization, before the implementation of supervised learning [24, 40, 62].
The wide spectrum of supervised learning approaches available can be divided into regression (also known as continuous) and classification (also known as categorical) models. The former are applied to generate numerical estimates of drug sensitivity, e.g. activity areas above drug–dose response curves or their IC50 values [42, 63]. Classification models make predictions of sensitivity according to predetermined response levels, such as high versus low sensitivity [27, 64]. Jang et al. [27] compared different regression and classification models, and showed that (i) there is no general solution, and (ii) one approach can outperform the other depending on the training data set (e.g. CCLE or CGP) and the measure of drug sensitivity used (e.g. IC50 or activity areas). A variety of applications based on methods originating from statistics and machine learning have been reported, ranging from multiple linear regression and support vector machines to random forests and k-nearest neighbors (KNN) models and others [32, 33, 38, 46, 65]. Comparative analyses using public and proprietary data have underscored the lack of all-purpose solutions [31]. Although no single approach can consistently outperform others on different data sets and across different drugs, it has been shown that regression models, such as elastic net and ridge regression [26], tend to offer good and robust performance in different settings [27]. In a recent study, an elastic net model was trained on the GDSC data and tested on the CCLE data, then vice versa, and a reasonable consistency of response predictors was reported [26].
Models can also be categorized according to their capacity to handle multiple types of inputs (numerical, nominal, missing values). Additionally, different model building strategies may be applied when dealing with multiple types of input data, e.g. gene expression and gene copy number. The most commonly reported approach involves training a single model that incorporates all the features available in a data set [29, 63]. Another promising approach that has received relatively less attention is the generation of model ensembles, i.e. models that achieve more accurate predictions based on the combination of predictions originating from multiple individual (data source specific) models [33, 57]. Other studies have also boosted prediction capacity through the implementation of ensembles of different techniques or multi-task learning frameworks [28, 57, 66].
Another integrative approach consists of combining the data sets as (or during) a preprocessing step, i.e. data sets are first harmonized and then combined into a single input (e.g. vector based) representation [26]. Other examples, such as those based on network-based data representations merit further investigations in this area [35, 36, 67]. An interesting example of the latter was reported by Zhang et al. [36], who generated similarity networks, between cell lines and between drugs independently, based on their gene expression and (1D and 2D) structural correlations, respectively. These two networks were integrated by linking the cell lines in the first network to their corresponding (previously tested) drugs in the second network. For a given cell line, sensitivity predictions were obtained in each network independently using the observed responses for the neighboring nodes (either cell lines or drugs) in the networks. The network-specific predictions were then combined in a weighted model to report the final response prediction.
Different adaptations of standard techniques are also being investigated. For instance, Neto et al. [63] proposed the STREAM algorithm that combines a Bayesian inference model with ridge-regression to develop prediction models trained and tested on public data (CCLE and CGP data separately). In another investigation, using expression data, Park et al. [68] improved prediction robustness by combining the elastic net and principal component analysis. Furthermore, they showed that the analysis of the data in the principal component space allows a more effective detection of sample outliers.
Readers who are interested in additional examples of the application of specific computational techniques may refer to [27, 28] and the DREAM challenges Web site (http://dreamchallenges.org).
Computational evaluation and selection of prediction models
In practice, different computational models are trained, compared and those showing the most promising predictive performance are selected for subsequent independent testing (Figure 2D). Different statistical indicators of predictive performance are available and chosen according to the aim of the prediction task investigated. Given training and testing data sets containing the sensitivity measure experiments, a general strategy is to use multiple indicators of error that compare the ‘goodness’ of the computational predictions versus the observed experimental values. In the regression modelling context, it is common to use correlations, root mean squared errors and the coefficient of determination as measures of error. For (categorical) classification models, standard indicators, such as accuracy, precision and recall are required, together with more detailed assessments including areas under the receiver operating characteristic curve and precision-recall curves [69–71].
An essential objective of model training is to generate a model that is generalizable beyond the data used for building the model. When a model does not achieve this objective, one may conclude that the model ‘over-fitted’ the data or that over-fitting occurred. Multiple factors, including the amount and diversity of the training data, determine the occurrence of overfitting, and are key research topics in the machine learning community [72]. Despite the complexity of the problem, a standard strategy has been established for estimating predictive performance: model cross-validation [73, 74]. This strategy basically consists of dividing the full data set available for training into two major disjoint data sub-sets: one for implementing the actual training and the other for ‘validating’ the resulting model. In the machine learning and bioinformatics literature, the latter data set is commonly referred to as either the validation, evaluation or test data sets, but should not be confused with the independent data set(s) used for testing the selected model (Figure 1D).
In drug response prediction modeling, the most widely applied cross-validation schemes are the K-fold (KF-CV) and the leave-one-out (LOO-CV) cross-validation methods. In the KF-CV, the full training data set is divided, by random sampling, into K partitions (folds), and (K-1) of the resulting partitions are used for training. After fitting the model to these data, the model is then tested on the left-out data partition. This process is repeated until all folds have been used as test data sets. Thus, prediction performance indicators are estimated for each test set independently, and an indicator of overall performance across the K-folds (typically the mean values) are reported. When insufficient amounts of data are available, the random partitioning procedure is implemented multiple times and overall performance indicators across such partitions are summarized. The LOO-CV may be seen as a version of KF-CV, in which the left-out data set contains a single sample from the data set, i.e. K = N, where N is the total number of samples in the data set. A more detailed discussion of cross-validation is provided in [73, 74].
Model interpretability and reporting
Another important challenge, which deserves more attention from the translational bioinformatics community, is to ensure that prediction models meet a minimum level of interpretability by end-users, especially clinicians. Model interpretability refers to the idea of allowing users to clearly visualize and understand the outcomes of the model. According to the prediction aims, this may also entail the implementation of models whose parameters are both statistically relevant and clinically meaningful. In other situations, bioinformaticians may also be required to provide models with some prediction explanation capability, i.e. descriptions of the evidence used to come up with a particular prediction or of the mechanism applied to make an inference. These issues are also relevant to the reporting of results in articles for the translational and clinical research communities.
Some of the most commonly investigated prediction models possess intrinsic properties that are amenable for supporting end-user interpretability and even explanatory capabilities. For example, multiple regression models and KNN learning approaches fall into this category. In the case of the former, a model may be represented in a relatively compact manner: a formula that contains inputs and outputs, with regression coefficients that may be interpreted as associations between specific input features, e.g. gene markers, and the prediction outcome, i.e. drug sensitivity measure. In the case of KNN models, predictions for a particular sample may be explained by visually representing the ‘previous cases’ used to make the prediction, i.e. the nearest neighbors that were retrieved from the training data set to estimate the drug response for the query sample. Both techniques are also relatively easily (or intuitively) understandable to researchers, and the KNN technique when presented as a ‘case-based reasoning process’ may especially appeal to clinicians.
The granularity and quality of information reported for model interpretation and reproducibility are critical issues that will continue necessitating careful consideration. Although the community is increasingly aware of the importance of data and software sharing for drug response prediction modeling [21, 22, 75], there is still room for improving transparency and accessibility. In this and other data-driven research areas, key questions deserve further attention: Is a software implementation of the model available for peer-reviewers and the community at large? Is the code for such implementation or at least a sufficiently detailed algorithmic description available? Are training and testing data sets, or at least versions of them, shared at the time of publication? Positive answers to these questions will need stronger incentives for researchers, while considering constraints and expectations from research managers, funders and publishers [76]. Better sharing and transparency practice will not only facilitate the reproducibility of results, but also the understandability of models. In the long-term, these factors will contribute to the acceptability of models by end-users in the clinical domain.
Conclusions
The development of computational models for predicting drug response remains a crucial challenge. Advances in this area are necessary for delivering on the promise of precision medicine. Moreover, as larger and more complex ‘omic’ data sets become available, the greater the expectations and opportunities for generating novel applications. Such systems are also likely to be based on new investigations about the interplay of multiple types of molecular data and their capacity for predicting drug responses. These models will progressively incorporate information encoded at various levels of biological control (e.g. tumor epigenetics), resolution analyses (e.g. multiple biopsies per tumor) and the tumor microenvironment. Also, as new advances in cancer immunotherapy materialize, there will be additional opportunities for developing prediction models tailored to this type of treatments or their combination with small-molecule drugs.
Although new research may build on existing statistical and machine learning techniques, it will also require the formulation of alternative approaches to representing, integrating and analyzing multiple data sets. For instance, unbiased systems-based approaches, which tap into the power of networks for representing data and discovering new knowledge, merit further investigations. Further research on the influence of cell types or specific therapies on drug response prediction is also needed, i.e. biological context-specific models [77, 78]. To date, prediction models have been primarily based on pan-cancer data analyses, which do not consider such context specificity owing to the relatively small data sets available for specific tissues or drugs.
There are strong reasons to continue investigating prediction models based on cell line-derived data. However, bioinformaticians should be well informed about the limitations of cell lines for clinically relevant research. The selection of cell lines that poorly reflect tumor biology, and the lack of sufficient numbers of cell lines for modeling response in specific cancers, are crucial failure factors to consider.
To bring these models closer to the clinic, it is important to require high levels of expertise and involvement from bioinformaticians and clinically oriented researchers during all key development phases. Similarly, as new opportunities and expectations arise, there will be further needs for powerful, interpretable and reproducible computational methods in different cancer domains, especially those with limited treatment options for patients. This will be accompanied by a higher demand for researchers who can design, adapt and evaluate these models, as well as for clinicians well trained to interpret results and to facilitate the creation of stronger bridges between the lab and the clinic.
Key Points
Large data sets, which contain pretreatment molecular profiles and drug response data, are publicly available and facilitate the development of computational models for predicting drug sensitivity.
Computational models built on cell line-derived data are applicable to the prediction of clinical responses in patient cohorts.
A diversity of techniques from statistics and machine learning have been investigated and will continue to be adapted to new applications in oncology.
There is still a need for model validations using clinically relevant animal models and patient data.
Funding
F.A. acknowledges support from LIH’s Connect2Predict project.
Francisco Azuaje works at the intersection of computational, biological and translational research. His team combines in vitro, in vivo and in silico approaches. A top priority is the development of novel computational models for cancer patient stratification and drug response prediction using multi-omics data sets.
References
- 1. Adams JU. Genetics: big hopes for big data. Nature 2015;527(7578):S108–9. [DOI] [PubMed] [Google Scholar]
- 2. Schmidt C. Cancer: reshaping the cancer clinic. Nature 2015;527(7576):S10–1. [DOI] [PubMed] [Google Scholar]
- 3. Rubin MA. Health: make precision medicine work for cancer care. Nature 2015;520(7547):290–1. [DOI] [PubMed] [Google Scholar]
- 4. Kohane IS. Health care policy. Ten things we have to do to achieve precision medicine. Science 2015;349(6243):37–8. [DOI] [PubMed] [Google Scholar]
- 5. Baselga J, Bhardwaj N, Cantley LC, et al. AACR cancer progress report 2015. Clin Cancer Res 2015;21(Suppl 19):S1–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simon R. Drug-diagnostics co-development in oncology. Front Oncol 2013;3:315.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Relling MV, Evans WE.. Pharmacogenomics in the clinic. Nature 2015;526(7573):343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aronson SJ, Rehm HL.. Building the foundation for genomics in precision medicine. Nature 2015;526(7573):336–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339(6127):1546–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boehm JS, Golub TR.. An ecosystem of cancer cell line factories to support a cancer dependency map. Nat Rev Genet 2015;16(7):373–4. [DOI] [PubMed] [Google Scholar]
- 11. Caponigro G, Sellers WR.. Advances in the preclinical testing of cancer therapeutic hypotheses. Nat Rev Drug Discov 2011;10(3):179–87. [DOI] [PubMed] [Google Scholar]
- 12. Klijn C, Durinck S, Stawiski EW, et al. A comprehensive transcriptional portrait of human cancer cell lines. Nat Biotechnol 2015;33(3):306–12. [DOI] [PubMed] [Google Scholar]
- 13. Libbrecht MW, Noble WS.. Machine learning applications in genetics and genomics. Nat Rev Genet 2015;16(6):321–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kidd BA, Wroblewska A, Boland MR, et al. Mapping the effects of drugs on the immune system. Nat Biotechnol 2016;34(1):47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boland MR, Jacunski A, Lorberbaum T, et al. Systems biology approaches for identifying adverse drug reactions and elucidating their underlying biological mechanisms. Wiley Interdiscip Rev Syst Biol Med, 2016;8(2):104–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li J, Zheng S, Chen B, et al. A survey of current trends in computational drug repositioning. Brief Bioinform 2016;17(1):2–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barretina J, Caponigro G, Stransky N, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483(7391):603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Garnett MJ, Edelman EJ, Heidorn SJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012;483(7391):570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6(10):813–23. [DOI] [PubMed] [Google Scholar]
- 20. Yang W, Soares J, Greninger P, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res 2013;41:D955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reinhold WC, Sunshine M, Liu H, et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res 2012;72(14):3499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luna A, Rajapakse VN, Sousa FG, et al. rcellminer: exploring molecular profiles and drug response of the NCI-60 cell lines in R. Bioinformatics, 2016;32(8):1272–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Basu A, Bodycombe NE, Cheah JH, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell 2013;154(5):1151–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moghaddas Gholami A, Hahne H, Wu Z, et al. Global proteome analysis of the NCI-60 cell line panel. Cell Rep 2013;4(3):609–20. [DOI] [PubMed] [Google Scholar]
- 25. Haibe-Kains B, El-Hachem N, Birkbak NJ, et al. Inconsistency in large pharmacogenomic studies. Nature 2013;504(7480):389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cancer Cell Line Encyclopedia Consortium; Genomics of Drug Sensitivity in Cancer Consortium. Pharmacogenomic agreement between two cancer cell line data sets. Nature 2015;528(7580):84–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jang IS, Neto EC, Guinney J, et al. Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data. Pac Symp Biocomput 2014;63–74. [PMC free article] [PubMed] [Google Scholar]
- 28. Costello JC, Heiser LM, Georgii E, et al. A community effort to assess and improve drug sensitivity prediction algorithms. Nat Biotechnol 2014;32(12):1202–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Daemen A, Griffith OL, Heiser LM, et al. Modeling precision treatment of breast cancer. Genome Biol 2013;14(10):R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geeleher P, Cox NJ, Huang SR.. Clinical drug response can be predicted using baseline gene expression levels and in vitro drug sensitivity in cell lines. Genome Biol 2014;15(3):R47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bayer I, Groth P, Schneckener S.. Prediction errors in learning drug response from gene expression data – influence of labeling, sample size, and machine learning algorithm. PLoS One 2013;8(7):e70294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amin SB, Yip WK, Minvielle S, et al. Gene expression profile alone is inadequate in predicting complete response in multiple myeloma. Leukemia 2014;28(11):2229–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cortés-Ciriano I, van Westen GJ, Bouvier G, et al. Improved large-scale prediction of growth inhibition patterns using the NCI60 cancer cell line panel. Bioinformatics 2016;32(1):85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menden MP, Iorio F, Garnett M, et al. Machine learning prediction of cancer cell sensitivity to drugs based on genomic and chemical properties. PLoS One 2013;8(4):e61318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fey D, Halasz M, Dreidax D, et al. Signaling pathway models as biomarkers: patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci Signal 2015;8(408):ra130. [DOI] [PubMed] [Google Scholar]
- 36. Zhang N, Wang H, Fang Y, et al. Predicting anticancer drug responses using a dual-layer integrated cell line-drug network model. PLoS Comput Biol 2015;11(9):e1004498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Niepel M, Hafner M, Pace EA, et al. Profiles of basal and stimulated receptor signaling networks predict drug response in breast cancer lines. Sci Signal 2013;6(294):ra84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen P, Huhtinen K, Kaipio K, et al. Identification of prognostic groups in high-grade serous ovarian cancer treated with platinum-taxane chemotherapy. Cancer Res 2015;75(15):2987–98. [DOI] [PubMed] [Google Scholar]
- 39. Blumenschein GR Jr., Saintigny P, Liu S, et al. Comprehensive biomarker analysis and final efficacy results of sorafenib in the BATTLE trial. Clin Cancer Res 2013;19(24):6967–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Byers LA, Diao L, Wang J, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res 2013;19(1):279–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guinney J, Ferté C, Dry J, et al. Modeling RAS phenotype in colorectal cancer uncovers novel molecular traits of RAS dependency and improves prediction of response to targeted agents in patients. Clin Cancer Res 2014;20(1):265–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Falgreen S, Dybkær K, Young KH, et al. Predicting response to multidrug regimens in cancer patients using cell line experiments and regularised regression models. BMC Cancer 2015;15(1):235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pereira E, Camacho-Vanegas O, Anand S, et al. Personalized circulating tumor DNA biomarkers dynamically predict treatment response and survival in gynecologic cancers. PLoS One 2015;10(12):e0145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474(7353):609–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zheng Y, Zhou J, Tong Y.. Gene signatures of drug resistance predict patient survival in colorectal cancer. Pharmacogenomics J 2015;15(2):135–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tran TP, Ong E, Hodges AP, et al. Prediction of kinase inhibitor response using activity profiling, in vitro screening, and elastic net regression. BMC Syst Biol 2014;8:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liang J, Tong P, Zhao W, et al. The REST gene signature predicts drug sensitivity in neuroblastoma cell lines and is significantly associated with neuroblastoma tumor stage. Int J Mol Sci 2014;15(7):11220–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goodspeed A, Heiser LM, Gray JW, et al. Tumor-derived cell lines as molecular models of cancer pharmacogenomics. Mol Cancer Res 2016;14(1):3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ross NT, Wilson CJ.. In vitro clinical trials: the future of cell-based profiling. Front Pharmacol 2014;5:121.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gillet JP, Calcagno AM, Varma S, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci USA 2011;108(46):18708–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gillet JP, Varma S, Gottesman MM.. The clinical relevance of cancer cell lines. J Natl Cancer Inst 2013;105(7):452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilding JL, Bodmer WF.. Cancer cell lines for drug discovery and development. Cancer Res 2014;74(9):2377–84. [DOI] [PubMed] [Google Scholar]
- 53. Tentler JJ, Tan AC, Weekes CD, et al. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol 2012;9(6):338–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Day CP, Merlino G, Van Dyke T.. Preclinical mouse cancer models: a maze of opportunities and challenges. Cell 2015;163(1):39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Domcke S, Sinha R, Levine DA, et al. Evaluating cell lines as tumour models by comparison of genomic profiles. Nat Commun 2013;4:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chen B, Sirota M, Fan-Minogue H, et al. Relating hepatocellular carcinoma tumor samples and cell lines using gene expression data in translational research. BMC Med Genomics 2015;8(Suppl 2):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wan Q, Pal R.. An ensemble based top performing approach for NCI-DREAM drug sensitivity prediction challenge. PLoS One 2014;9(6):e101183.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yang J, Tang H, Li Y, et al. DIGRE: drug-induced genomic residual effect model for successful prediction of multidrug effects. CPT Pharmacometrics Syst Pharmacol 2015;4(2):e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bansal M, Yang J, Karan C, et al. A community computational challenge to predict the activity of pairs of compounds. Nat Biotechnol 2014;32(12):1213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao B, Pritchard JR, Lauffenburger DA, et al. Addressing genetic tumor heterogeneity through computationally predictive combination therapy. Cancer Discov 2014;4(2):166–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao B, Hemann MT, Lauffenburger DA.. Intratumor heterogeneity alters most effective drugs in designed combinations. Proc Natl Acad Sci USA 2014;111(29):10773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nicolau M, Levine AJ, Carlsson G.. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci USA 2011;108(17):7265–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Neto EC, Jang IS, Friend SH, et al. The STREAM algorithm: computationally efficient ridge-regression via Bayesian model averaging, and applications to pharmacogenomic prediction of cancer cell line sensitivity. Pac Symp Biocomput 2014;27–38. [PMC free article] [PubMed] [Google Scholar]
- 64. Fersini E, Messina E, Archetti F.. A p-median approach for predicting drug response in tumour cells. BMC Bioinformatics 2014;15(1):353.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stetson LC, Pearl T, Chen Y, et al. Computational identification of multi-omic correlates of anticancer therapeutic response. BMC Genomics 2014;15(Suppl 7):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gonen M, Margolin AA.. Drug susceptibility prediction against a panel of drugs using kernelized Bayesian multitask learning. Bioinformatics 2014;30(17):i556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang B, Mezlini AM, Demir F, et al. Similarity network fusion for aggregating data types on a genomic scale. Nat Methods 2014;11(3):333–7. [DOI] [PubMed] [Google Scholar]
- 68. Park H, Shimamura T, Miyano S, et al. Robust prediction of anti-cancer drug sensitivity and sensitivity-specific biomarker. PLoS One 2014;9(10):e108990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Baldi P, Brunak S, Chauvin Y, et al. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics 2000;16(5):412–24. [DOI] [PubMed] [Google Scholar]
- 70. Berrar D, Flach P.. Caveats and pitfalls of ROC analysis in clinical microarray research (and how to avoid them). Brief Bioinform 2012;13(1):83–97. [DOI] [PubMed] [Google Scholar]
- 71. Saito T, Rehmsmeier M.. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One 2015;10(3):e0118432.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cawley GC, Talbot NLC.. On over-fitting in model selection and subsequent selection bias in performance evaluation. J Mach Learn Res 2010;11:2079–107. [Google Scholar]
- 73. Baek S, Tsai CA, Chen JJ.. Development of biomarker classifiers from high-dimensional data. Brief Bioinform 2009;10(5):537–46. [DOI] [PubMed] [Google Scholar]
- 74. Varma S, Simon R.. Bias in error estimation when using cross-validation for model selection. BMC Bioinformatics 2006;7:91.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Geeleher P, Cox N, Huang RS.. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 2014;9(9):e107468.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vihinen M. No more hidden solutions in bioinformatics. Nature 2015;521(7552):261.. [DOI] [PubMed] [Google Scholar]
- 77. Chen BJ, Litvin O, Ungar L, et al. Context sensitive modeling of cancer drug sensitivity. PLoS One 2015;10(8):e0133850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jaeger S, Duran-Frigola M, Aloy P.. Drug sensitivity in cancer cell lines is not tissue-specific. Mol Cancer 2015;14:40.. [DOI] [PMC free article] [PubMed] [Google Scholar]