Summary
Genome-wide DNA methylation analysis indicated that DNA methylation alterations are already present even at the precancerous NASH stage, clearly differing from such alterations in viral hepatitis or cirrhosis, and possibly continuing to participate in NASH-related multistage hepatocarcinogenesis.
Abstract
The aim of this study was to clarify the significance of DNA methylation alterations during non-alcoholic steatohepatitis (NASH)-related hepatocarcinogenesis. Single-CpG-resolution genome-wide DNA methylation analysis was performed on 264 liver tissue samples using the Illumina Infinium HumanMethylation450 BeadChip. After Bonferroni correction, 3331 probes showed significant DNA methylation alterations in 113 samples of non-cancerous liver tissue showing NASH (NASH-N) as compared with 55 samples of normal liver tissue (NLT). Principal component analysis using the 3331 probes revealed distinct DNA methylation profiles of NASH-N samples that were different from those of NLT samples and 37 samples of non-cancerous liver tissue showing chronic hepatitis or cirrhosis associated with hepatitis B virus (HBV) or hepatitis C virus (HCV) infection (viral-N). Receiver operating characteristic curve analysis identified 194 probes that were able to discriminate NASH-N samples from viral-N samples with area under the curve values of more than 0.95. Jonckheere-Terptsra trend test revealed that DNA methylation alterations in NASH-N samples from patients without hepatocellular carcinoma (HCC) were inherited by or strengthened in NASH-N samples from patients with HCC, and then inherited by or further strengthened in 22 samples of NASH-related HCC (NASH-T) themselves. NASH- and NASH-related HCC-specific DNA methylation alterations, which were not evident in viral-N samples and 37 samples of HCC associated with HBV or HCV infection, were observed in tumor-related genes, such as WHSC1, and were frequently associated with mRNA expression abnormalities. These data suggested that NASH-specific DNA methylation alterations may participate in NASH-related multistage hepatocarcinogenesis.
Introduction
It is well known that not only genomic but also epigenomic alterations participate in carcinogenesis in various human organs (1,2). In hepatocarcinogenesis, we have shown that DNA methylation alterations not present in normal liver can be detected even in liver tissue showing chronic hepatitis or cirrhosis associated with hepatitis B virus (HBV) or hepatitis C virus (HCV) infection (3), which are widely considered to be precancerous conditions for hepatocellular carcinoma (HCC). This report was one of the earliest to document alterations of DNA methylation at the precancerous stage in the liver. Subsequently, we showed that DNA methylation alterations evident at the precancerous stage are inherited by the cancers that develop in the same affected individuals, and determine tumor aggressiveness and patient outcome (4,5). Now, DNA methylation biomarkers for carcinogenetic risk estimation are being sought in patients with viral hepatitis and subsequent liver cirrhosis, and also for prognostication in patients with HCC (6–8).
Although HBV or HCV infection followed by chronic hepatitis and liver cirrhosis used to be the main cause of HCC, there is evidence that non-alcoholic steatohepatitis (NASH) is becoming another precancerous condition for HCC. NASH, a hepatic manifestation of metabolic syndrome (9) resulting in the development of liver cirrhosis, has shown an alarming increase in recent years. In addition, epigenome alterations have recently attracted a great deal of attention as the molecular basis of not only cancer but also metabolic disorders (10). In fact, studies using animal models of NASH have indicated that features of its pathophysiology, such as aberrant lipogenesis, may result in epigenome alterations (11). In the light of these facts, it would be informative to understand the significance of DNA methylation alterations in NASH-related hepatocarcinogenesis, especially at the precancerous stage.
Accordingly, in this study, we performed genome-wide DNA methylation analysis (methylome analysis) using the Infinium assay on 55 samples of normal liver tissue (NLT), 113 samples of non-cancerous liver tissue showing NASH (NASH-N), 22 samples of NASH-related HCC (NASH-T), 37 samples of non-cancerous liver tissue showing chronic hepatitis or cirrhosis associated with HBV or HCV infection (viral-N) and 37 samples of HCC associated with HBV or HCV infection (viral-T) (12). We then compared the DNA methylation alterations during NASH-related hepatocarcinogenesis to those during hepatitis virus-related hepatocarcinogenesis.
Materials and methods
Patients and tissue samples
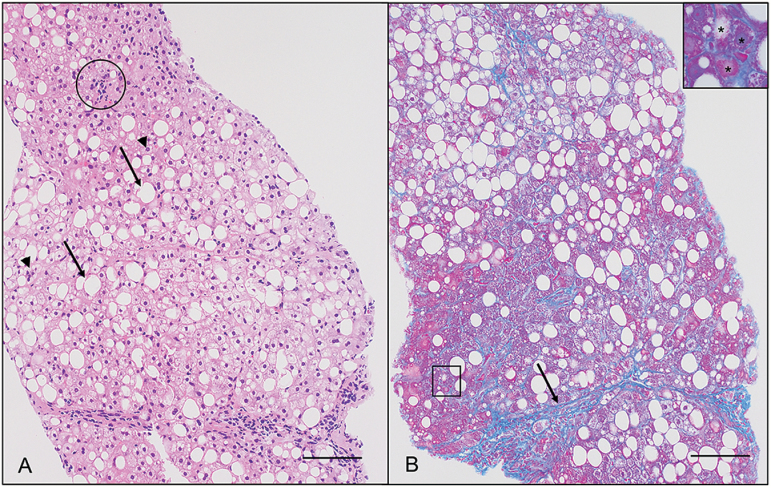
Liver biopsy specimens were obtained from 110 morbidly obese patients during bariatric surgery under general anesthesia at the Yotsuya Medical Cube. These patients had been informed of the risk of obesity-associated NASH (9), the therapeutic benefit of early diagnosis and the invasiveness of the liver biopsy procedure, and provided written informed consent. A proportion of these biopsy specimens were kept frozen and used for the present study. None of the 110 patients had HBV or HCV infection, or HCC. Microscopic evaluation revealed no pathologic findings in the liver biopsy specimens from 19 of the patients, and histological features compatible with NASH were seen in those from 91 of the patients, although none had shown symptoms of NASH before bariatric surgery (13). Thus, 19 samples of NLT and 91 samples of NASH-N were obtained from this cohort (Supplementary Figure 1, available at Carcinogenesis Online). NASH stage was evaluated microscopically on the basis of the histological scoring system for NASH (14) and the Brunt classification (15). Representative photos of the histological features of NASH are shown in Figure 1. The age, sex and clinicopathological backgrounds of these patients are summarized in Supplementary Table 1A, available at Carcinogenesis Online.
Figure 1.
Histopathological appearance of a liver biopsy specimen from a representative patient with non-alcoholic steatohepatitis. (A) Steatosis involves about 35% of hepatocytes in this case: Representative large lipid droplets (macrovesicular steatosis) and small lipid droplets (microvesicular steatosis) are indicated by arrows and arrowheads, respectively. A focus of lobular inflammation (circle) is also observed. Scale bar, 100 μm (hematoxylin and eosin staining). (B) Bridging fibrosis (arrow) and perivenular/pericellular fibrosis (square) are observed. High-power view of the area marked with the square (inset) indicates that hepatocytes (*) are surrounded and downsized by fibrosis. Scale bar, 100 μm (azan staining).
An additional 36 samples of NLT were obtained from partial hepatectomy specimens from 36 patients with liver metastases of primary colorectal cancers without HBV or HCV infection, chronic hepatitis, liver cirrhosis or HCC. Microscopy examination of non-cancerous liver tissue from 74 patients with HCC, but without HBV or HCV infection, revealed histological features compatible with NASH in 22 of them. Therefore, 22 paired samples of non-cancerous liver tissue and cancerous tissue from these 22 patients were considered to be representative of NASH-N and NASH-T, and subjected to further analysis (Supplementary Figure 1, available at Carcinogenesis Online). For comparison, 37 paired samples of viral-N and viral-T were obtained from partial hepatectomy specimens from 37 HCC patients with HBV or HCV infection (13 patients HBV surface antigen positive, 23 patients anti-HCV antibody positive, and one patient positive for both) (Supplementary Figure 1, available at Carcinogenesis Online). These patients did not receive any preoperative treatment, underwent partial hepatectomy at the National Cancer Center Hospital, and gave written informed consent for inclusion of their specimens in this study. Histological diagnosis of HCCs was made in accordance with the World Health Organization classification (16), and in accordance with the Tumor-Node-Metastasis classification (17). The age, sex and clinicopathological backgrounds of these patients are summarized in Supplementary Table 1, available at Carcinogenesis Online. Fresh frozen tissue samples from partial hepatectomy specimens were provided by the National Cancer Center Biobank.
This study was approved by the Ethics Committees of the National Cancer Center, the National Center for Global Health and Medicine, and Yotsuya Medical Cube.
Infinium assay
High-molecular weight DNA from fresh frozen tissue samples was extracted using either QIAamp DNA Micro kit (Qiagen, Valencia, CA), or phenol-chloroform, followed by dialysis. Five-hundred-nanogram aliquots of DNA were subjected to bisulfite conversion using an EZ DNA Methylation-GoldTM Kit (Zymo Research, Irvine, CA). DNA methylation status at 485 764 CpG loci was examined at single-CpG resolution using the Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA) (12). After hybridization, the specifically hybridized DNA was fluorescence-labeled by a single-base extension reaction and detected using an iScan reader (Illumina) in accordance with the manufacturer’s protocols. The data were then assembled using GenomeStudio methylation software (Illumina). At each CpG site, the ratio of the fluorescence signal was measured using a methylated probe relative to the sum of the methylated and unmethylated probes, i.e. the so-called β-value, which ranges from 0.00 to 1.00, reflecting the methylation level of an individual CpG site. The results of the Infinium assay have been deposited in the Integrative Disease Omics Database (http://gemdbj.ncc.go.jp/omics/), and some of the Infinium data have also been deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo/, Accession number GSE89852).
Gene ontology (GO) enrichment analysis and pathway analysis
In order to reveal the function of proteins encoded by genes showing DNA methylation alterations and the biological processes in which such proteins participate, GO enrichment analysis was conducted using the GeneGO MetaCore™ software package (version 6.7) (Thomson Reuters, New York, NY). Pathway analysis using genes showing NASH-N-specific DNA methylation alterations was also performed using the same package.
Real-time quantitative reverse transcription (RT)-PCR analysis
Using TRIzol reagent (Life Technologies, Carlsbad, CA), total RNA was isolated from 21 paired samples of NASH-N and NASH-T from 21 patients with NASH-related HCCs and 31 paired samples of viral-N and viral-T from 31 HCC patients with HBV or HCV infection, for which additional tissue specimens had been available after DNA extraction. cDNA was reverse-transcribed from total RNA using random primers and Superscript III RNase H-Reverse Transcriptase (Life Technologies). Levels of expression of mRNA for the WHSC1 gene were analyzed using custom TaqMan Expression Assays on the 7500 Fast Real-Time PCR System (Life Technologies) employing the relative standard curve method. The probes and PCR primer sets employed are summarized in Supplementary Table 2, available at Carcinogenesis Online. Experiments were performed in triplicate, and the mean value for the three experiments was used as the CT value. All CT values were normalized to that of GAPDH in the same sample.
Statistical analysis
In the Infinium assay, the call proportions (P < 0.01 for detection of signals above the background) for 822 probes (shown in Supplementary Table 3, available at Carcinogenesis Online) in all of the tissue samples examined were less than 90%. Since such a low proportion may have been attributable to polymorphism at the probe CpG sites, these 822 probes were excluded from the present assay. In addition, to avoid any gender-specific methylation bias, all 369 CpG sites on chromosomes X and Y, and 28 292 probes for which DNA methylation levels reportedly showed sexual dimorphism (18,19) were excluded, leaving a final total of 444 907 autosomal CpG sites.
Infinium probes showing significant differences in DNA methylation levels between the 55 NLT samples and 113 NASH-N samples, the 113 NASH-N samples and 37 viral-N samples, and 22 NASH-T samples and 37 viral-T samples, were defined by Welch’s t test and adjusted by Bonferroni correction. The DNA methylation profiles of the 55 NLT samples, 113 NASH-N samples and 37 viral-N samples were analyzed using principal component analysis (PCA) and unsupervised hierarchical clustering (maximum distance, complete linkage method). The receiver operating characteristic (ROC) curve was generated, and the Youden index (20) for each probe was used as a cutoff value for examining distinctions between the 113 NASH-N samples and 37 viral-N samples. Ordered differences from the 55 NLT samples to the 113 NASH-N samples (91 from patients without HCC to 22 from patients with HCC) and then to the 22 NASH-T samples were examined by Jonckheere-Terptsra (JT) trend test. All statistical analyses were performed using programming language R.
Results
Distinct DNA methylation profiles of NASH-N samples
Since the 55 NLT samples consisted of two groups, 19 specimens from patients with morbid obesity and 36 specimens from patients with liver metastases of primary colorectal cancers, we confirmed whether or not heterogeneity of DNA methylation profiles was evident among the 55 NLT samples using PCA and unsupervised hierarchical clustering based on the results of the Infinium assay. Using all 444 907 probes, PCA and unsupervised hierarchical clustering did not reveal any such heterogeneity among the NLT samples: the 19 with morbid obesity were not discriminated from the 36 with liver metastases of primary colorectal cancers (Supplementary Figure 2, available at Carcinogenesis Online).
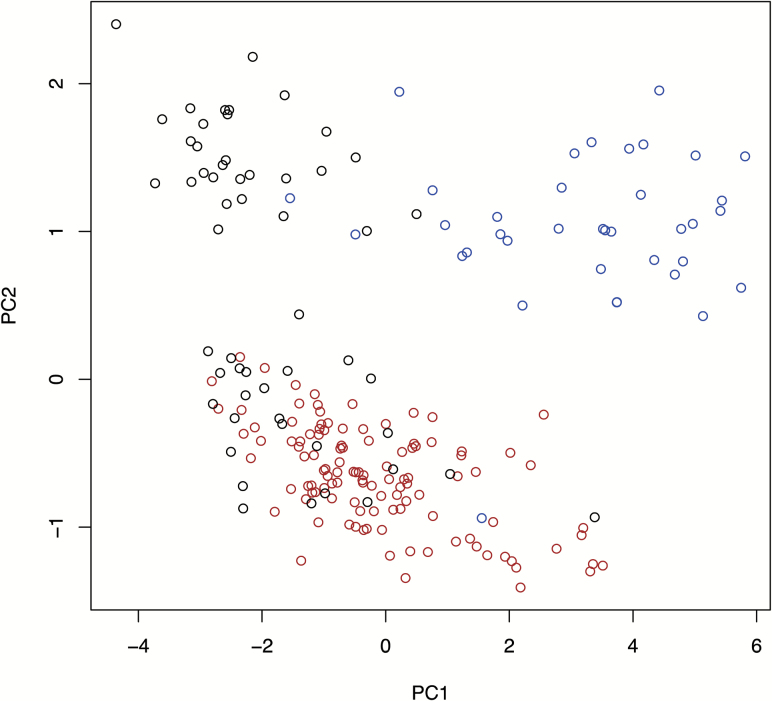
Then 3331 probes that were aberrantly methylated in the 113 NASH-N samples (91 from biopsy specimens and 22 from partial hepatectomy specimens) in comparison with the 55 NLT samples (Welch’s t test, adjusted Bonferroni correction [α = 1.12 × 10−7] and a ΔβNASH-N - NLT value of more than 0.05 or less than −0.05) were identified, indicating that DNA methylation alterations had occurred in NASH relative to normal liver. When we performed PCA using the DNA methylation levels of these 3331 probes, including 37 viral-N samples together with 55 NLT and 113 NASH-N samples, the NASH-N samples showed a DNA methylation profile that differed distinctly from the DNA methylation profiles of both NLT samples and viral-N samples (Figure 2).
Figure 2.
DNA methylation profiles of NLT, non-cancerous liver tissue showing non-alcoholic steatohepatitis (NASH-N), and non-cancerous liver tissue showing chronic hepatitis or cirrhosis associated with HBV or HCV infection (viral-N). PCA was performed using the 3331 probes showing significant DNA methylation alterations in NASH-N samples relative to NLT samples, in NLT samples (n = 55, black), NASH-N samples (n = 113, red) and viral-N samples (n = 37, blue).
When 19 NLTs with morbid obesity and 36 NLTs with liver metastases of primary colorectal cancers were separately plotted in the PCA using the same 3331 probes as those in Figure 2 (Supplementary Figure 3, available at Carcinogenesis Online), the DNA methylation profiles of the 19 NLTs with morbid obesity (gray circles) were more similar to the NASH-N samples than the 36 NLTs with liver metastases of primary colorectal cancers. Since we cannot rule out the possibility that morbid obesity and metastases of colorectal cancers affect the DNA methylation profiles of NLT samples, we employed 82 samples of adult NLT from two data sets deposited in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) (GSE61258 and GSE69852) (no history of primary or metastatic cancer, or morbid obesity, having been described in their metadata). We identified 9150 probes that were aberrantly methylated in the 113 NASH-N samples relative to control NLT data from the public database (Welch’s t test, adjusted Bonferroni correction [α = 1.28 × 10−7] and a ΔβNASH-N − control from the database value of more than 0.05 or less than −0.05). PCA using the DNA methylation levels of these 9150 probes again showed that the DNA methylation profiles of NASH samples differed distinctly from those of both control NLT samples from the public database and viral-N samples (Supplementary Figure 4, available at Carcinogenesis Online).
NASH-N sample-specific DNA methylation alterations
Next, we identified 45 214 probes that showed significant differences in DNA methylation levels between the 113 NASH-N samples and 37 viral-N samples (Welch’s t test, adjusted Bonferroni correction [α = 1.12 × 10−7] and a ΔβNASH-N – vial-N value of more than 0.05 or less than −0.05), indicating that the DNA methylation status of NASH differed from those of viral hepatitis or cirrhosis. Probes showing NASH-specific DNA methylation alterations were defined as those (a) included among the 3331 probes aberrantly methylated in the 113 NASH-N samples in comparison with the 55 NLT samples, (b) included among the 45 214 probes showing significant differences in DNA methylation levels between the 113 NASH-N samples and 37 viral-N samples, and (c) any of the following three cases: not showing significant differences in DNA methylation levels between the 55 NLT samples and 37 viral-N samples, showing DNA hypermethylation in the 113 NASH-N samples and DNA hypomethylation in the 37 viral-N samples in comparison with the 55 NLT samples, or showing DNA hypomethylation in the 113 NASH-N samples and DNA hypermethylation in the 37 viral-N samples (Supplementary Figure 5, available at Carcinogenesis Online). Accordingly, we identified 375 probes satisfying the above criteria (a), (b) and (c) (Supplementary Table 4, available at Carcinogenesis Online) as those showing NASH-specific DNA methylation alterations. Among these 375 probes, 116 showed DNA hypermethylation in NASH-N samples relative to NLT samples (βNASH-N > βNLT), and the remaining 259 showed DNA hypomethylation in NASH-N samples relative to NLT samples (βNASH-N < βNLT). One hundred and nineteen of the 375 probe CpG sites were annotated around the transcription start sites (TSSs) of any genes, such as the region from 200 bp upstream of the TSS to 1500 bp upstream of it (TSS1500), the region from TSS to 200 bp upstream of TSS (TSS200) or the first exon (Supplementary Table 4, available at Carcinogenesis Online). Multiple Infinium probes were designed for 264 of the 265 genes, for which the 375 probes had been designed, and such multiple probes for the AJUBA, ARHGAP22, HTRA1, RPS6KA2, SLC35C1, TSSC1 and ZNF311 genes simultaneously showed NASH-N-specific DNA methylation alterations.
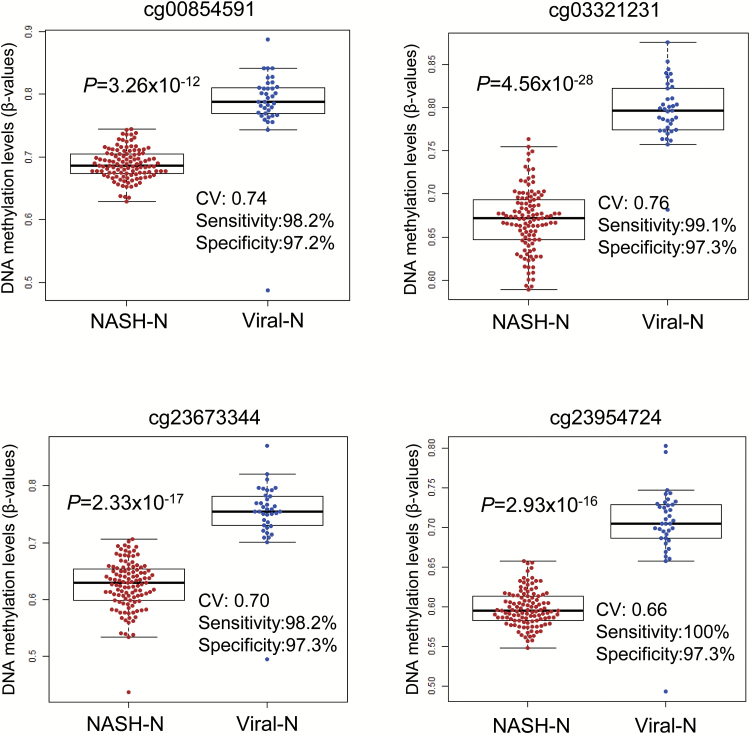
In order to further confirm the differences in DNA methylation profiles between NASH and viral hepatitis or cirrhosis, ROC curves were constructed for the 375 probes specifically affected in NASH-N using the 113 NASH-N samples and 37 viral-N samples, and the corresponding AUC values (21) were calculated. Among the probes, 194 (summarized in Supplementary Table 5, available at Carcinogenesis Online) showed AUC values larger than 0.95, and 72 of them showed DNA hypermethylation in NASH-N samples relative to viral-N samples (βNASH-N > βviral-N); the remaining 122 probes showed DNA hypomethylation in NASH-N samples relative to viral-N samples (βNASH-N < βviral-N). The Youden index was used as a cutoff value for each of the 194 probes in order to discriminate NASH-N samples from viral-N samples (Supplementary Table 5, available at Carcinogenesis Online). As shown by the representative scattergrams in Figure 3, the use of such cutoff values was able to discriminate NASH-N samples from viral-N samples with sufficient sensitivity and specificity (97.2–100% in Figure 3). The sensitivity and specificity of each cutoff value for the DNA methylation levels of all 194 probes are shown in Supplementary Table 5, available at Carcinogenesis Online.
Figure 3.
Scattergrams of DNA methylation levels for representative probes showing area under the curve values of more than 0.95 in ROC curve analysis for discrimination between non-cancerous liver tissue showing non-alcoholic steatohepatitis (NASH-N) and non-cancerous liver tissue showing chronic hepatitis or cirrhosis associated with HBV or HCV infection (viral-N). Using each probe and its cutoff value (CV) described in Supplementary Table 5, available at Carcinogenesis Online, NASH-N samples (n = 113, red) were discriminated from viral-N samples (n = 37, blue) with sufficient sensitivity and specificity (97.2-100%).
GO enrichment analysis using genes specifically affected in NASH-N
Two hundred and sixty-five genes, for which the 375 NASH-N-specific probes were designed, were evaluated for protein function by enrichment analysis using the MetaCore software, and compared with the protein function distribution of genes within the GeneGo databases (Table 1). Although, as seen in Table 1, the distribution of gene products that were specifically affected in NASH-N covered various protein classes, 14 out of the top 20 statistically significant GO biological processes were involved in protein-protein binding or binding of proteins to other molecules (Table 2). All 187 gene products specifically affected in NASH and also included in the top 20 GO biological processes (Table 2) are listed in Supplementary Table 6, available at Carcinogenesis Online. According to the MetaCore software, 31, 19 and 15 gene products included in Supplementary Table 6, available at Carcinogenesis Online, were DNA binding proteins, RNA binding proteins and chromatin regulators (indicated by the superscripts c, d and e, respectively, in Supplementary Table 6, available at Carcinogenesis Online).
Table 1.
Gene ontology enrichment analysis (GO protein function) of the 275 genes, for which the 375 probes specifically related to NASH-N (listed in Supplementary Table 4, available at Carcinogenesis Online) were designed, using MetaCore software
| Protein class | r | n | R | N | Expected | Ratio | P |
|---|---|---|---|---|---|---|---|
| Transcription factors | 21 | 258 | 1210 | 38 371 | 8.136 | 2.581 | 7.967 × 10−5 |
| Phosphatases | 4 | 258 | 238 | 38 371 | 1.600 | 2.5 | 7.760 × 10−2 |
| Kinases | 9 | 258 | 655 | 38 371 | 4.404 | 2.044 | 3.415 × 10−2 |
| Proteases | 6 | 258 | 582 | 38 371 | 3.913 | 1.533 | 1.996 × 10−1 |
| Enzymes | 28 | 258 | 2778 | 38 371 | 18.68 | 1.499 | 2.122 × 10−2 |
| Receptors | 13 | 258 | 1672 | 38 371 | 11.24 | 1.156 | 3.358 × 10−1 |
| Ligands | 3 | 258 | 518 | 38 371 | 3.483 | 0.8613 | 5.393 × 10−1 |
| Other | 175 | 258 | 30 772 | 38 371 | 206.9 | 0.8458 | 1.716 × 10−6 |
r, number of objects from the present data set for a given protein class; n, total number of objects from the present data set; R, number of background objects from the database for a given class; N, total number of background objects from the database; Expected, mean value for hypergeometric distribution (n × R/38 371); Ratio, the ratio of actual/expected. If the ratio is more than 1, P < 0.05 mean significant enrichment, and these are underlined.
Table 2.
Top 20 statistically significant GO biological processes revealed by MetaCore software analysis using the 275 genes, for which the 375 probes specifically related to NASH-N (listed in Supplementary Table 4, available at Carcinogenesis Online) were designed
| Biological process | P value |
|---|---|
| Protein bindinga | 3.777 × 10−7 |
| Molecular function regulator | 2.337 × 10−6 |
| Guanyl-nucleotide exchange factor activity | 4.976 × 10−5 |
| Actin bindinga | 9.468 × 10−5 |
| Structure-specific DNA bindinga | 1.027 × 10−4 |
| Macromolecular complex bindinga | 1.136 × 10−4 |
| Chromatin bindinga | 1.259 × 10−4 |
| Bindinga | 1.521 × 10−4 |
| Actin filament bindinga | 1.869 × 10−4 |
| Enzyme regulator activity | 2.290 × 10−4 |
| Ras guanyl-nucleotide exchange factor activity | 3.121 × 10−4 |
| Corticotropin hormone receptor bindinga | 3.479 × 10−4 |
| Type 5 melanocortin receptor bindinga | 3.479 × 10−4 |
| DNA bindinga | 4.297 × 10−4 |
| Chromatin DNA bindinga | 4.411 × 10−4 |
| Transcriptional repressor activity, RNA polymerase II activating transcription factor binding | 6.468 × 10−4 |
| Transcriptional activator activity, RNA polymerase II transcription factor bindinga | 6.468 × 10−4 |
| Core promotor activity | 8.838 × 10−4 |
| Enzyme bindinga | 1.341 × 10−3 |
| Cytoskeletal protein bindinga | 1.540 × 10−3 |
aGO biological processes involved in protein–protein binding or binding of proteins to other molecules.
In addition, MetaCore pathway analysis revealed that the 265 genes designed for the 375 NASH-N-specific probes were significantly (P < 0.05) accumulated in molecular pathways frequently disturbed during carcinogenesis, such as ‘Development_NOTCH-induced EMT (P = 1.408 × 10−7)’ and ‘Development_EGFR signaling pathway (P = 1.198 × 10−4)’ (Table 3). All 163 significant pathways have been summarized in Supplementary Table 7, available at Carcinogenesis Online.
Table 3.
Top 20 statistically significant pathway maps revealed by MetaCore software analysis using the 275 genes, for which the 375 probes specifically related to NASH-N (listed in Supplementary Table 4, available at Carcinogenesis Online) were designed
| Pathway maps | P value |
|---|---|
| Development_NOTCH-induced EMT | 1.408 × 10−7 |
| Development_Notch Signaling Pathway | 1.027 × 10−5 |
| Development_NOTCH1-mediated pathway for NF-KB activity modulation | 2.699 × 10−5 |
| Development_Gastrin in cell growth and proliferation | 6.243 × 10−5 |
| Oxidative stress_ROS-mediated activation of MAPK via inhibition of phosphatases | 8.034 × 10−4 |
| Development_EGFR signaling pathway | 1.198 × 10−4 |
| Development_Thromboxane A2 signaling pathway | 3.674 × 10−4 |
| Development_Oligodendrocyte differentiation from adult stem cells | 3.967 × 10−4 |
| Immune response_T regulatory cell-mediated modulation of effector T cell and NK cell functions | 4.600 × 10−4 |
| Development_Mu-type opioid receptor signaling via Beta-arrestin | 5.635 × 10−4 |
| G-protein signaling_M-RAS regulation pathway | 5.635 × 10−4 |
| Development_Keratinocyte differentiation | 5.682 × 10−4 |
| G-protein signaling_TC21 regulation pathway | 6.372 × 10−4 |
| G-protein signaling_R-RAS regulation pathway | 6.372 × 10−4 |
| Neurogenesis_NGF/ TrkA MAPK-mediated signaling | 7.426 × 10−4 |
| Development_Mu-type opioid receptor regulation of proliferation | 8.944 × 10−4 |
| Immune response_Sublytic effects of membrane attack complex | 1.185 × 10−3 |
| Development_Positive regulation of STK3/4 (Hippo) pathway and negative regulation of YAP/TAZ function | 1.320 × 10−3 |
| Signal transduction_ERK1/2 signaling pathway | 1.328 × 10−3 |
| Reproduction_Gonadotropin-releasing hormone (GnRH) signaling | 1.466 × 10−3 |
DNA methylation alterations during NASH-related hepatocarcinogenesis
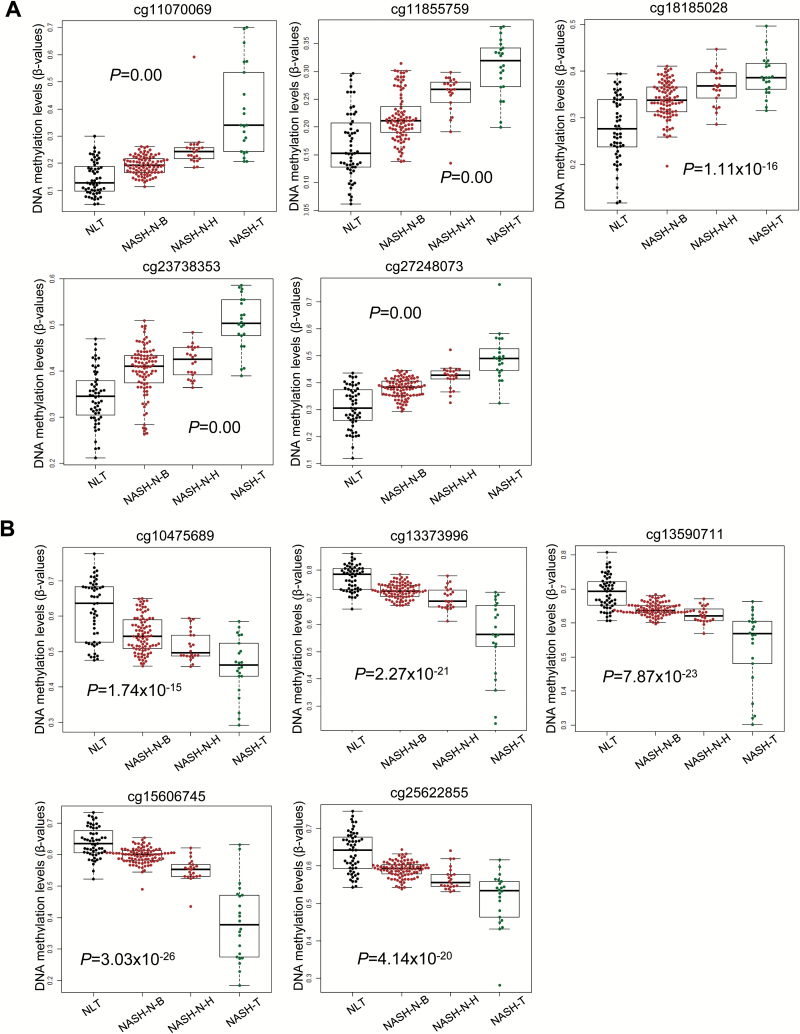
In order to clarify the DNA methylation status of the 375 probes during NASH-related carcinogenesis, JT trend test (P < 0.05) was performed. This revealed the ordered differences in DNA methylation status from NLT samples to NASH-N from biopsy specimens of patients with morbid obesity but without HCC, NASH-N from partial hepatectomy specimens of patients with HCC and then to NASH-T samples for 364 of the 375 probes, indicating that DNA methylation alterations in NASH-N from patients without HCC were inherited by or strengthened in NASH-N from patients with HCC, and then inherited by or further strengthened in NASH-T samples. Representative ordered differences are shown in Figure 4. DNA hypermethylation in NASH-N samples relative to NLT samples was inherited by or strengthened in NASH-T samples for 115 probes (Figure 4A), whereas DNA hypomethylation in NASH-N samples relative to NLT samples was inherited by or strengthened in NASH-T samples for the remaining 249 probes (Figure 4B).
Figure 4.
Ordered differences in DNA methylation levels for representative probes listed in Supplementary Table 4, available at Carcinogenesis Online, which were aberrantly methylated in non-cancerous liver tissue showing non-alcoholic steatohepatitis (NASH-N) relative to NLT and showed significant differences in DNA methylation levels between NASH-N and non-cancerous liver tissues showing chronic hepatitis or cirrhosis associated with HBV or HCV infection (viral-N). NASH-specific alterations of DNA methylation relative to NLT samples (n = 55) were observed in NASH-N samples from biopsy specimens from patients with morbid obesity but without hepatocellular carcinoma (HCC) (NASH-N-B, n = 91), and inherited by or strengthened in NASH-N samples from partial hepatectomy specimens from patients with HCC (NASH-N-H, n = 22), and then inherited by or further strengthened in NASH-related HCC (NASH-T, n = 22) itself. (A) Representative probes showing DNA hypermethylation by Jonckheere-Terpstra (JT) trend test. (B) Representative probes showing DNA hypomethylation by JT trend test.
Identification of genes affected by DNA methylation alterations in NASH-T samples
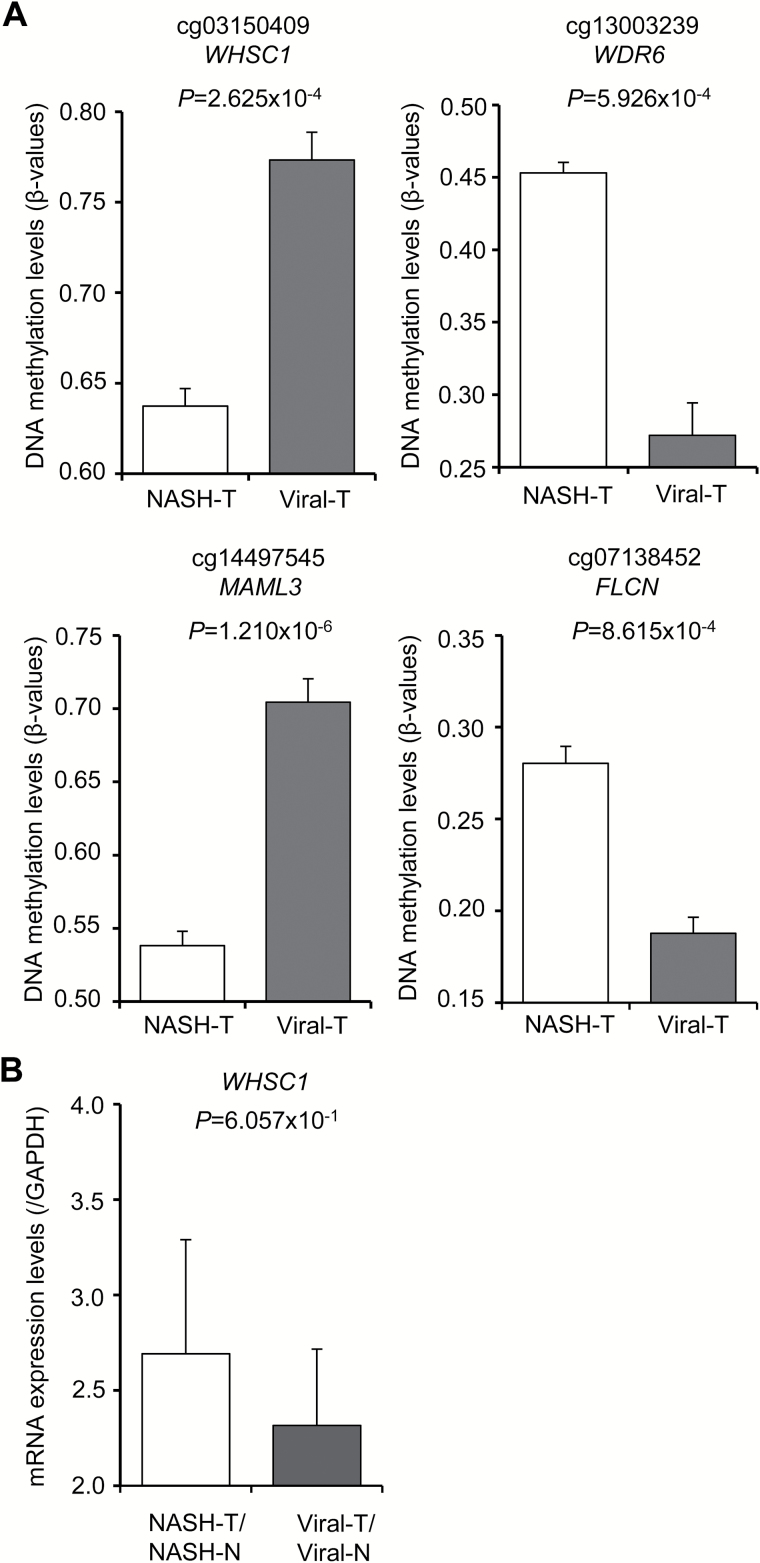
Furthermore, we identified 80 probes that were included among the 364 NASH-N-specifically affected probes whose DNA methylation alterations were inherited by or strengthened in NASH-T samples (JT trend test) and that showed significant differences in DNA methylation levels even between NASH-T samples and viral-T samples (Welch’s t test, adjusted Bonferroni correction [α = 1.12 × 10−7] and a ΔβNASH-T – viral-T value of more than 0.05 or less than −0.05) (Supplementary Table 8, available at Carcinogenesis Online). The DNA methylation levels for the representative probes are shown in Figure 5A: cg03150409 and cg14497545 showed significantly lower DNA methylation levels in NASH-T samples than in viral-T samples, whereas cg13003239 and cg07138452 showed significantly higher DNA methylation levels in NASH-T samples than in viral-T samples.
Figure 5.
DNA methylation levels and mRNA expression levels for representative probes that were included in the 80 NASH-specific probes (Supplementary Table 8, available at Carcinogenesis Online) for which DNA methylation alterations were inherited by NASH-related hepatocellular carcinoma (HCC) (NASH-T) and showed significant differences in DNA methylation levels between NASH-T samples and HCC associated with HBV or HCV infection (viral-T). (A) DNA methylation levels in NASH-T samples (n = 22) and viral-T samples (n = 37). Each gene symbol is shown under the Infinium probe ID. (B) Levels of mRNA expression for the WHSC1 gene in NASH-T samples (n = 21) and viral-T samples (n = 31).
When we examined 70 of the genes for which the above 80 probes were designed, using the public database (https://tcga-data.nci.nih.gov/tcga/) of The Cancer Genome Atlas (TCGA), inverse correlations (r < −0.2) between the DNA methylation levels and mRNA expression levels of 22 genes, ACOT7, ARF4, DHX36, FLCN, FOXN3, FYTTD1, MAML3, MICAL3, MRPS24, NQO2, PCNT, PKP4, PLAG1, PMPCA, PROSER3 RASA2, SPG7, TRAPPC10, WDR6, WHSC1, ZC3H14 and ZZEF1, were observed in human tissue samples, as shown in Supplementary Figure 6, available at Carcinogenesis Online. Such inverse correlation between DNA methylation levels and mRNA expression levels of the WHSC1 (P = 6.057 × 10−1) gene was then confirmed in our own NASH-T and viral-T samples using real-time quantitative RT-PCR analysis (Figure 5B), although the difference did not reach statistically significant levels, possibly due to the insufficient number of samples.
Discussion
Using a total of 264 samples of liver tissue, we have performed the Infinium assay to clarify the DNA methylation status of NASH-N and NASH-T. NASH has recently become a serious medical issue, rivaling viral hepatitis and cirrhosis as a precancerous stage for HCC. We and other groups have reported that aberrant DNA methylation participates even in the early and precancerous stages of carcinogenesis in different organs (22–27). Therefore, in the present study, we focused on the significance of DNA methylation alterations at the precancerous stage of NASH-related hepatocarcinogenesis.
First, the Infinium assay identified 3331 probes that were aberrantly methylated in NASH-N samples relative to NLT samples, indicating that DNA methylation alteration was already present even at the precancerous NASH stage. PCA analysis (Figure 2) indicated that the DNA methylation profile of NASH was clearly distinct from the well-studied DNA methylation profiles of other precancerous conditions for HCC, i.e. viral hepatitis and cirrhosis.
For the 375 probes (summarized in Supplementary Table 4, available at Carcinogenesis Online), DNA methylation levels were significantly different between both NASH-N samples and NLT samples, and NASH-N samples and viral-N samples. Therefore it was evident that the 375 probes showed NASH-specific DNA methylation abnormalities that were different from those of viral hepatitis and cirrhosis. In addition, the presence of 194 probes showing AUC values of more than 0.95 in ROC analysis again indicated that the DNA methylation status of NASH was clearly different from that of viral hepatitis and cirrhosis.
Even though NASH is usually diagnosed on the basis of clinicopathological parameters, and the present study did not aim to establish diagnostic criteria for NASH, we were able to discriminate NASH-N samples from viral-N samples using each cutoff value with a sensitivity and specificity of almost 100% (Figure 3). Although it may be possible to diagnose NASH-related HCC based on DNA methylation profiles even in patients with established cirrhosis and in whom non-cancerous liver tissue from hepatectomy specimens has already lost any histological features specific for NASH, such diagnostic criteria should be established and validated independently from the present study.
GO enrichment analysis indicated that many genes specifically affected in NASH-N encode DNA-binding proteins, RNA-binding proteins and chromatin regulators (Supplementary Table 6, available at Carcinogenesis Online). Such gene products could potentially affect the transcriptional regulation of various downstream genes including tumor-related genes. In addition, genes specifically affected in NASH-N were significantly accumulated in molecular pathways frequently disturbed during carcinogenesis (Table 3), though the significance of each pathway will need to be clarified by further examinations. These findings prompted us to examine whether DNA methylation status is simply altered at the precancerous NASH stage, or whether DNA methylation alterations at the precancerous stage are actually inherited by NASH-T.
NASH-N in biopsy specimens can be considered to be an early precancerous stage of multistage carcinogenesis at which patients are asymptomatic and HCC has not yet developed. On the other hand, NASH-N in partial hepatectomy specimens can be considered to be a more advanced precancerous stage at which HCC has already developed. JT trend test revealed that NASH-specific DNA methylation profiles were inherited from NASH-N in biopsy specimens (an early precancerous stage), were evident at a more advanced precancerous stage in partial hepatectomy specimens, and were then inherited by or further strengthened in NASH-T itself, on almost all (364/375) of the probes, suggesting that DNA methylation status is not simply altered as a precancer marker, but that DNA methylation alterations of the genes listed in Supplementary Table 6, available at Carcinogenesis Online, continuously participate in NASH-related multistage hepatocarcinogenesis.
On the other hand, we identified 24 632 probes designed for 5491 genes whose DNA methylation levels differed significantly between the 113 NASH-N samples and 22 NASH-T samples (data not shown). Although DNA methylation alterations of such genes may potentially be associated with progression from NASH to NASH-related HCC, further verification and validation are needed before the establishment of criteria for carcinogenetic risk estimation in patients with NASH.
The probe CpG sites showing NASH-N-specific DNA methylation alterations were frequently annotated to the regions potentially participating in gene expression regulation, such as TSS1500, TSS200 or the first exon (Supplementary Table 4, available at Carcinogenesis Online). We then examined 80 probes (70 genes) included in Supplementary Table 8, available at Carcinogenesis Online, for which NASH-N-specific DNA methylation alterations were inherited by NASH-T, and for which DNA methylation status differed significantly between not only NASH-N and viral-N but also between NASH-T and viral-T, using the TCGA database. This indicated that DNA methylation alterations in 22 of the 70 genes actually resulted in alterations of expression in human tissue specimens deposited in the database (Supplementary Figure 6, available at Carcinogenesis Online): DNA hypomethylation was potentially associated with overexpression of the ACOT7, FOXN3, MAML3, PCNT, PMPCA RASA2, SPG7 and WHSC1 genes, whereas hypermethylation was potentially associated with reduced expression of the ARF4, DHX36, FLCN, FYTTD1, MICAL3, MRPS24, NQO2, PKP4, PLAG1, PROSER3, TRAPPC10, WDR6, ZC3H14 and ZZEF1 genes.
Among the genes showing DNA hypomethylation during NASH-related carcinogenesis in comparison with both NLT samples and viral-N and viral-T samples, MAML3 is a critical transcriptional co-activator in the Notch signaling pathway (28) included in Table 3, and is also a co-activator for β-catenin-mediated transcription (29). MAML3 is recruited by β-catenin to Wnt target gene promoters and increases the transcriptional activity of β-catenin (29). Overexpression of MAML3 has been reported in endometrial (30) and pancreatic (31) cancers.
Moreover, the WHSC1 gene encodes a histone H3 lysine 36 (K36) methyltransferase (32). Generation of chimeric transcripts including WHSC1 and overexpression of WHSC1 were first reported in multiple myelomas with t(4;14) translocation (32). Since then, overexpression of WHSC1 has been reported in various human hematological malignancies (33) and epithelial cancers, such as those of the bladder, lung and head and neck (34,35). Overexpression of WHSC1 reportedly participates in carcinogenesis via activation of oncogenic pathways, such as the Wnt pathway, and through activation of NIMA-related kinase-7 (NEK7) due to increased H3K36 methylation (34,35). Moreover, a tendency for overexpression of WHSC1 was confirmed in our own NASH-T samples, suggesting that DNA hypomethylation of WHSC1 may participate in NASH-related carcinogenesis via overexpression of the gene.
Among the genes showing DNA hypermethylation during NASH-related carcinogenesis, the significance of the FLCN gene, a tumor suppressor for Birt-Hogg-Dubé syndrome-associated renal cell carcinoma (36), during hepatocarcinogenesis has been unclear to date. On the other hand, the WDR6 gene encodes a WD repeat family protein that interacts with the serine/threonine kinase STK11 and is implicated in cell growth arrest (37). During piperonyl butoxide-induced hepatocarcinogenesis in mice, especially in non-neoplastic liver tissue surrounding proliferative lesions, CpG island microarray and immunohistochemistry have revealed DNA hypermethylation and reduced expression of WDR6, respectively (38). Therefore, it is feasible that DNA hypermethylation of the WDR6 gene, even that present in NASH-N samples, participates in NASH-related hepatocarcinogenesis. Although the impacts of each of the above genes need to be investigated further, NASH-specific DNA methylation alterations observed at the precancerous stage were shown to be inherited by NASH-T and may continuously participate in NASH-related multistage hepatocarcingenesis, at least partly via alterations in the expression of the affected genes.
Funding
This study was supported by the Program for Promotion of Fundamental Studies in Health Sciences (10-42) of the National Institute of Biomedical Innovation (NiBio), the Research Program on Hepatitis (15fk0210034h0001) from the Japan Agency for Medical Research and Development (AMED), KAKENHI (JP16744879) from the Japan Society for the Promotion of Science, and a Grant (25D102) from the National Center for Global Health and Medicine. The National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund (26-A-1).
Conflict of Interest Statement: None declared.
Supplementary material
Supplementary Table 1–8 and Figures 1–6 can be found at Carcinogenesis Online.
Supplementary Material
Abbreviations
- GO
gene ontology
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- NASH
non-alcoholic steatohepatitis
- NASH-N
non-cancerous liver tissue showing NASH
- NASH-T
NASH-related HCC
- NLT
normal liver tissue
- PCA
principal component analysis
- ROC
receiver operating characteristic
- RT
reverse transcription
- TSS
transcription start site
- viral-N
non-cancerous liver tissue showing chronic hepatitis or cirrhosis associated with HBV or HCV infection
- viral-T
HCC associated with HBV or HCV infection
References
- 1. You J.S., et al. (2012) Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell, 22, 9–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baylin S.B., et al. (2011) A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer, 11, 726–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kanai Y., et al. (1996) Aberrant DNA methylation on chromosome 16 is an early event in hepatocarcinogenesis. Jpn. J. Cancer Res., 87, 1210–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Arai E., et al. (2010) DNA methylation profiles in precancerous tissue and cancers: carcinogenetic risk estimation and prognostication based on DNA methylation status. Epigenomics, 2, 467–481. [DOI] [PubMed] [Google Scholar]
- 5. Kanai Y. (2010) Genome-wide DNA methylation profiles in precancerous conditions and cancers. Cancer Sci., 101, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Utsunomiya T., et al. (2014) Specific molecular signatures of non-tumor liver tissue may predict a risk of hepatocarcinogenesis. Cancer Sci., 105, 749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagashio R., et al. (2011) Carcinogenetic risk estimation based on quantification of DNA methylation levels in liver tissue at the precancerous stage. Int. J. Cancer, 129, 1170–1179. [DOI] [PubMed] [Google Scholar]
- 8. Arai E., et al. (2009) Genome-wide DNA methylation profiles in liver tissue at the precancerous stage and in hepatocellular carcinoma. Int. J. Cancer, 125, 2854–2862. [DOI] [PubMed] [Google Scholar]
- 9. Asrih M., et al. (2015) Metabolic syndrome and nonalcoholic fatty liver disease: Is insulin resistance the link? Mol. Cell. Endocrinol., 418 Pt 1, 55–65. [DOI] [PubMed] [Google Scholar]
- 10. Keating S.T., et al. (2015) Epigenetics and metabolism. Circ. Res., 116, 715–736. [DOI] [PubMed] [Google Scholar]
- 11. Tian Y., et al. (2015) Histone deacetylase HDAC8 promotes insulin resistance and β-catenin activation in NAFLD-associated hepatocellular carcinoma. Cancer Res., 75, 4803–4816. [DOI] [PubMed] [Google Scholar]
- 12. Bibikova M., et al. (2009) Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics, 1, 177–200. [DOI] [PubMed] [Google Scholar]
- 13. Seki Y., et al. (2016) Prevalence of nonalcoholic steatohepatitis in Japanese patients with morbid obesity undergoing bariatric surgery. J. Gastroenterol., 51, 281–289. [DOI] [PubMed] [Google Scholar]
- 14. Kleiner D.E., et al. (2005) Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology, 41, 1313–1321. [DOI] [PubMed] [Google Scholar]
- 15. Brunt E.M., et al. (1999) Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. Am. J. Gastroenterol., 94, 2467–2474. [DOI] [PubMed] [Google Scholar]
- 16. Hirohashi S., et al. (2009) Hepatocellular carcinoma. World Health Organization classification of tumours. Pathology and genetics of tumours of the digestive system. IARC Press, Lyon. [Google Scholar]
- 17. Sobin L.H., et al. (2010) Liver-Hepatocellular Carcinoma. International Union Against Cancer (UICC) TNM classification of malignant tumours, 7 ed Wiley-Liss, New York. [Google Scholar]
- 18. Price M.E., et al. (2013) Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin, 6, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Y.A., et al. (2013) Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics, 8, 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fan J., et al. (2006) Understanding receiver operating characteristic (ROC) curves. CJEM, 8, 19–20. [DOI] [PubMed] [Google Scholar]
- 21. Akobeng A.K. (2007) Understanding diagnostic tests 3: Receiver operating characteristic curves. Acta Paediatr., 96, 644–647. [DOI] [PubMed] [Google Scholar]
- 22. Klebaner D., et al. (2016) X chromosome-wide analysis identifies DNA methylation sites influenced by cigarette smoking. Clin. Epigenetics, 8, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Planello A.C., et al. (2016) Pre-neoplastic epigenetic disruption of transcriptional enhancers in chronic inflammation. Oncotarget, 7, 15772–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamanoi K., et al. (2015) Epigenetic clustering of gastric carcinomas based on DNA methylation profiles at the precancerous stage: its correlation with tumor aggressiveness and patient outcome. Carcinogenesis, 36, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sato T., et al. (2014) Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: Its correlation with smoking history and chronic obstructive pulmonary disease. Int. J. Cancer, 135, 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sato T., et al. (2013) DNA methylation profiles at precancerous stages associated with recurrence of lung adenocarcinoma. PLoS One, 8, e59444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nishiyama N., et al. (2010) Genome-wide DNA methylation profiles in urothelial carcinomas and urothelia at the precancerous stage. Cancer Sci., 101, 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McElhinny A.S., et al. (2008) Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene, 27, 5138–5147. [DOI] [PubMed] [Google Scholar]
- 29. lves-Guerra M.C., et al. (2007) Mastermind-like 1 is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res., 67, 8690–8698. [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., et al. (2016) SOX17 is a tumor suppressor in endometrial cancer. Oncotarget. doi:10.18632/oncotarget.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Onishi H., et al. (2016) Hypoxia but not normoxia promotes smoothened transcription through upregulation of RBPJ and Mastermind-like 3 in pancreatic cancer. Cancer Lett., 371, 143–150. [DOI] [PubMed] [Google Scholar]
- 32. Chesi M., et al. (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood, 92, 3025–3034. [PubMed] [Google Scholar]
- 33. Haladyna J.N., et al. (2015) Epigenetic modifiers in normal and malignant hematopoiesis. Epigenomics, 7, 301–320. [DOI] [PubMed] [Google Scholar]
- 34. Toyokawa G., et al. (2011) Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate 1 is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia, 13, 887–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saloura V., et al. (2015) WHSC1 promotes oncogenesis through regulation of NIMA-related kinase-7 in squamous cell carcinoma of the head and neck. Mol. Cancer Res., 13, 293–304. [DOI] [PubMed] [Google Scholar]
- 36. Schmidt L.S., et al. (2015) Molecular genetics and clinical features of Birt-Hogg-Dubé syndrome. Nat. Rev. Urol., 12, 558–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie X., et al. (2007) Association of LKB1 with a WD-repeat protein WDR6 is implicated in cell growth arrest and p27(Kip1) induction. Mol. Cell. Biochem., 301, 115–122. [DOI] [PubMed] [Google Scholar]
- 38. Yafune A., et al. (2013) Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model. Toxicol. Lett., 222, 295–302. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.