Summary
We demonstrate that in colon, HFD stimulates transcripts associated with inflammation and tumorigenesis similarly seen in human colon cancer with inflammatory microsatellite instability. These findings allow a better understanding of colonic transcriptional changes mediated by obesity that lead to tumorigenesis.
Abstract
Obesity, an immense epidemic affecting approximately half a billion adults, has doubled in prevalence in the last several decades. Epidemiological data support that obesity, due to intake of a high-fat, western diet, increases the risk of colon cancer; however, the mechanisms underlying this risk remain unclear. Here, utilizing next generation RNA sequencing, we aimed to determine the high-fat diet (HFD) mediated expression profile in mouse colon and the azoxymethane/dextran sulfate sodium model of colon cancer. Mice on HFD had significantly higher colonic inflammation, tumor burden, and a number of differentially expressed transcripts compared to mice on regular diet (RD). We identified 721 transcripts differentially expressed in mouse HFD colon that were in a shared pattern with colonic tumors (RD and HFD). Importantly, in mouse colon, HFD stimulated an expression signature strikingly similar to human colon cancer, especially those with inflammatory microsatellite instability. Furthermore, pathway analysis of these transcripts demonstrated their association with active inflammation and colon cancer signaling, with leptin and Wnt as the top two transcripts elevated in mouse HFD colon shared with tumors. Moreover, in mouse colon, HFD-stimulated tumorigenic Wnt pathway activation was further validated by upregulation of β-catenin transcriptional targets. Finally, in human colon cancer, upregulation of leptin pathway members was shown with a large network of dysregulated transcripts being linked with worse overall survival.
Introduction
Obesity and overweight are immense epidemics that have doubled in prevalence in the last several decades and affect approximately half a billion adults in both the industrialized and developing world (Global Status Report on non-communicable diseases 2014. World Health Org: http://apps.who.int/iris/bitstream/10665/148114/1/9789241564854_eng.pdf). Emerging findings have revealed that obesity is associated with increased systemic inflammation and risk of cancer in different tissues, including colorectal cancer, the second leading cause of cancer-related death in the United States (1,2). While a complex network of factors including genetics and diet supports obesity-mediated colon cancer development, the mechanisms underlying this pathobiology remain unclear.
Obesity is often accompanied by the development of metabolic syndrome, a combination of health conditions including hypertriglyceridemia, low high-density lipoprotein, high blood pressure, high fasting blood glucose, a large waist circumference and insulin resistance (3,4). The health impacts associated with obesity fall into two main groups: those associated with carrying increased weight, including osteoarthritis and obstructive sleep apnea, and those linked with increased adiposity, as manifested by type 2 diabetes, cardiovascular disease, non-alcoholic fatty liver and certain types of cancer (3,5). Recently, obesity has been linked to an abnormal gut microbiota composition. Aberrant microbiota have also been found in genetic (ob/ob mice deficient in leptin production) or diet-induced (Zucker fa/fa rat) animal models of obesity (6–8), further highlighting the role of the intestine in obesity-related pathobiology. While knowledge concerning the role and function of microbiota in obesity-mediated pathology has been evolving, less is known about the changes affecting the intestinal epithelium.
Emerging studies have indicated that obesity increases the risk of a number of cancers including those of the endometrium, kidney, breast, esophagus and colon (1,2). Moreover, in obese patients affected with colon cancer elevated rates of recurrence, disease related death and a less effective response to chemotherapy have been recognized (2,9). While the underlying links between obesity and cancer are unclear, evidence suggests that insulin-like growth factor, adipose tissue-mediated systemic inflammation and production of adipose-related adipokines could, in part, be contributing to disease (1,10). It is plausible that a combination of these factors may support colon cancer progression, yet the changes in colonic gene expression induced by obesity remain unexplored.
Here, using next generation RNA sequencing and pathway analysis, we demonstrate that in the colon of mice, HFD stimulates an expression profile that promotes inflammation and tumorigenesis in a similar pattern with colonic tumors, and we confirmed shared, elevated levels of inflammatory leptin and tumorigenic Wnt. This HFD signature also shares a similar expression pattern with human colon cancer, especially those associated with an inflammatory microsatellite instability signature. These findings identify new transcripts and pathways stimulated by HFD in the colon, and, consequently could advance the understanding of obesity-mediated colon tumorigenesis.
Materials and methods
Animals
Mice, C57BL/6 strain obtained from Jackson laboratory (6 weeks old), were housed at Tulane University School of Medicine. All lean experimental mice were kept on a standard chow diet (regular diet, RD), whereas, all obese experimental animals were maintained on a high-fat chow diet (D12492, high-fat diet – HFD) consisting of 60% kcal/fat. For the HFD tumor group, mice were maintained on HFD for 8 weeks before azoxymethane (AOM)/DSS treatment. All experimental procedures were performed in accordance with the principles and guidelines adopted by NIH and approved by the Tulane IACUC.
AOM and DSS treatment
Colonic tumors were induced in experimental mice by a single AOM (Sigma) intraperitoneal injection of 10 mg/kg, followed by three separate 5-day cycles of 2.5% DSS (MP Biomedicals) added to drinking water (11,12), with 2-week intervals between treatments. Mice were monitored for their weight for 24 weeks and were fasted overnight prior to blood glucose measurements (Accu-Chek, Roche). At the end of the experiment, colonic tissues were collected.
RNA isolation
Total RNA from colonic tissues and tumors were isolated using the miRNeasy kit (Qiagen) according to manufacturer’s instructions. An Agilent Bioanalyzer (Agilent Technologies) was used to determine RNA quality, where RNA integrity numbers (RIN) obtained ranged from 7 to 8 for individual samples before RNA sequencing.
Next generation RNA sequencing
Next generation RNA sequencing (RNA-seq) and library preparation were performed at the University of Wisconsin Biotechnology Center DNA Sequencing Facility (http://www.biotech.wisc.edu/services/dnaseq). Poly-A selection was employed to enrich high quality mRNAs from total RNA samples (1 μg input per sample) and library preparation was accomplished using the Illumina Truseq Stranded mRNA preparation kit (Illumina Inc.). Briefly, following poly-A purification, mRNA underwent fragmentation and priming for cDNA synthesis followed by first and second strand cDNA synthesis, 3′ adenylation, and subsequent ligation of indexing adapters to the ends of cDNA. After PCR enrichment of DNA fragments with adapter molecules, an Agilent DNA 1000 Chip (Agilent Technologies) assay was performed to verify the library’s profile and size. Samples underwent single-end 100 bp strand-specific sequencing using an Illumina HiSeq 2500 instrument (Illumina Inc.).
Differential expression analysis of RNA-seq data
Data analysis was carried out in the Tulane Cancer Center Next Generation Sequence Analysis Core using core computational resources (www.tulane.edu/som/cancer/research/core-facilities/cancer-crusaders). RNA-seq reads were mapped to an index containing the mouse reference haploid genome sequence (Genome Reference Consortium murine genome build 38, GRCm38). The software package, RSEM (v1.2.25) (13), was used to quantify transcript expression from RNA-seq data. EBseq (14) was employed to identify differentially expressed transcripts at the whole gene level across two or more biological samples. Lists of differentially expressed transcripts were obtained with a false discovery rate (FDR) controlled at 0.05. These data along with design parameters have been submitted in NCBI’s Sequence Read Archive and are available through the study accession SRP093363. Comparison of the obtained HFD expression signature against available expression data from colon of AOM/DSS treated mice (GSE31106) and human colon cancer samples (GSE4183) was achieved using NCBI’s GEO2R (15), and only genes significantly expressed (P < 0.05) employing the Benjamini & Hochberg testing adjustment for false discovery rate were used in analysis (16). For comparison of the obtained HFD expression signature against expression data from human microsatellite instable (MSI) colon cancers cell lines, quantified RNA-seq data was downloaded from the Cancer Cell Line Encyclopedia (CCLE) (17) using OCG/CTD2 (https://ctd2.nci.nih.gov/dataPortal). Likewise, for evaluation of the HFD signature against human MSI colon cancers, quantified RNA-seq data was downloaded from The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/) (18) using Firebrowse (firebrowse.org).
Ingenuity pathway analysis
All network, function and heatmap analyses were generated through the use of QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN, www.qiagen.com/ingenuity). Transcripts input into IPA were expressed at a minimum threshold of >|1.5|-fold change relative to mice on RD diet with a FDR < 0.05 using n = 3 mice per condition.
cBioPortal for cancer genomics
Investigation of leptin pathway dysregulation in human colon cancer samples was performed by query of colon cancer TCGA data using cBioPortal for human cancer genomics (www.cbioportal.org) with a z-score threshold of ±2 for all RNA-seq analyses (19,20).
qPCR
RNA obtained from select colonic tissue and tumors were converted to cDNA (Life Technologies) according to manufacturer’s instructions and used for qPCR analysis. DNase treated mRNA was reverse transcribed with Oligo-dT12-18 primers employing the SuperScript® First-Strand Synthesis System (Invitrogen). For quantification of resulting cDNA, the C1000 Thermal Cycler system (Bio-Rad) and iQ SYBR Green DNA double-strand binding dye (iQ SYBR Green Supermix, Bio-Rad) were used. The following primers specifically amplifying mouse Leptin (mLep-FOR 5′-CCAGGATGACACCAAAACCCT-3′ mLep-REV 5′-GGATACCGA CTGCGTGTGTG-3′), Wnt10a (mWnt10a-FOR 5′-CCCATCTTCAGCCG AGGTTTT-3′ mWnt10a-REV 5′-AGCCTTCAGTTTACCCAGAGC-3′) and HPRT-1 (mHPRT-1-FOR 5′-GACCAGTCAACAGGGGACAT-3′ mHPRT-1-REV 5′-AACACTTCGTGGGGTCCTTTTC-3′) were utilized. To determine the relative levels of mRNA, the comparative Ct method was employed using HPRT-1 as a housekeeping control.
Histological analysis
Colonic tissue was fixed in 4% formaldehyde, paraffin embedded and sectioned for H&E staining. Inflammatory scores of mouse colonic tissue were determined by blinded pathology according to the following criteria: Lymphocyte: (i) infiltration (minimal – 0, mild – 1, moderate – 2, severe – 3); (ii) depth of infiltration (mucosal only – 1, mucosal and submucosal – 2, mucosal, submucosal and muscle – 3). Neutrophils: (a) infiltration (minimal – 0, mild – 1, moderate – 2, severe – 3); (b) depth of infiltration (mucosal only – 1, mucosal and submucosal – 2, mucosal, submucosal and muscle – 3); (c) extent of neutrophil infiltration (none – 0, ≤10% – 1, 10–50% – 2, ≥50% – 3). Goblet cell depletion (none – 0, mild – 1, moderate – 2, severe – 3). Vascular density (low – 1, high – 2) (21).
Tissue sections were immunohistostained after antigen retrieval with antibodies against leptin (1:500, 1 h; Abcam), Wnt10a (1:100, 1 h; Abcam), β-catenin (1:400, 1 h; Cell Signaling), c-Myc (1:400, 1 h, Abcam) and Ki67 (1:100, 45 min, BioCare Medical). Antibody labeled with HRP was added, followed by incubation with DAB+ chromogen and counterstaining with hematoxylin. Tissues were dehydrated in graded alcohol and xylene and cover-slipped using Permount (Biomeda). Images were obtained using the slide scanner Aperio CS2 (Leica) and prepared by Image Scope software.
Cells
Human colon cancer HT29, HCT116 and SW480 cells were obtained from the American Type Culture Collection (ATCC). HT29 and HCT116 cells were propagated in complete McCoy’s 5A media (Sigma-Aldrich) containing 10% fetal bovine serum (Gibco), while SW480 cells (ATCC) were grown in Leibovitz’s L-15 Medium containing 10% fetal bovine serum (Gibco). Human non-transformed colonic NCM460 cells were obtained from INCELL Corporation (INCELL) and were grown in M3Base medium (INCELL) containing 10% fetal bovine serum. Cells used in this study were authenticated by the vendor sources noted using short tandem repeat profiling and were passaged up to 10 times within four months following receipt or resuscitation. Cells were serum starved overnight prior to the experimental procedures.
Protein extraction and immunoblotting
Proteins were extracted from experimental cells and immunoblotted as previously described (22). Specific antibodies against leptin (Abcam), Wnt10a (Sigma) and β-actin (Sigma) were used. Proteins were visualized with anti-rabbit or anti-mouse IR Dye-conjugated secondary antibodies (LI-COR Biosciences) using an Odyssey infrared imaging system (LI-COR Biosciences).
Statistical analysis
All data are means ± SE for a series of experiments. Statistical analysis was performed by Student’s unpaired t-test or analysis of variance (ANOVA) and Student Newman-Keuls post-test using Graph Pad Instat 3 software (Graph Pad Software). A P < 0.05 was considered significant.
Results
HFD promotes colonic inflammation and tumor burden in AOM/DSS mice
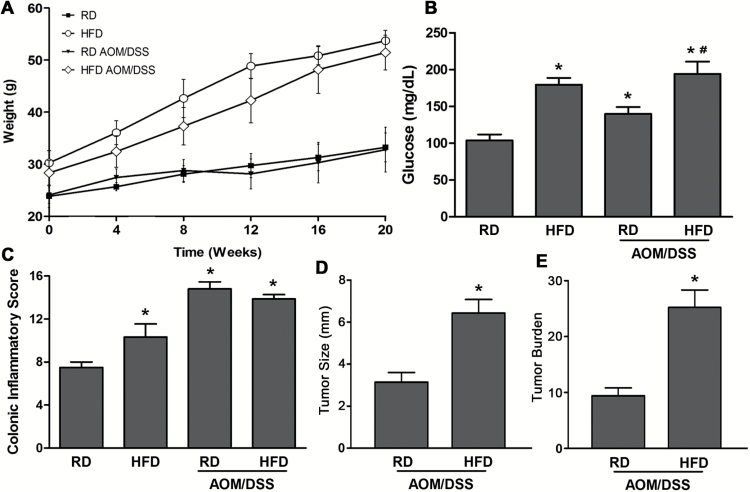
We utilized the AOM/DSS mouse model of colon cancer to investigate the contribution of HFD in promoting changes to colonic tissue and tumors (12). Consistent with an obese phenotype, mice on HFD gained weight (RD: 33 ± 3 g, HFD: 54 ± 2* g, n = 16, *P < 0.05) and had significantly elevated blood glucose levels (RD: 103 ± 8 mg/dl, HFD: 179 ± 9* mg/dl, n = 16, *P < 0.05) compared to their littermates on RD (Figure 1A and B). Additionally, in AOM/DSS-treated mice on either HFD or RD, elevated blood glucose was found, suggesting that dysregulation of glucose metabolism is not only associated with obesity, but also with colonic tumor growth. Total inflammatory scores revealed that mice on HFD exhibited significantly increased inflammation in the colon, which was higher in both AOM/DSS groups (RD and HFD) when compared to RD mice (Figure 1C). Moreover, we analyzed colonic tumor incidence and size in AOM/DSS mice on both diets. While tumor incidence between these two groups was not significantly different, tumor size was increased in mouse HFD colon (>5 mm) relative to RD (<5 mm), leading to a considerably elevated tumor burden (~2.5-fold) (Figure 1D and E). These data demonstrate that HFD promotes colonic inflammation and increases tumor burden in the AOM/DSS model. Next, we aimed to understand the changes in gene expression induced by HFD in mouse colon and its role in tumor promotion.
Figure 1.
HFD promotes colonic inflammation and tumor burden. (A, B) Mice on HFD untreated or subjected to AOM/DSS gained weight and had significantly elevated blood glucose levels relative to mice on RD (n = 16 per group, *#P < 0.05, *Compared to RD, #compared to RD Tumor). (C) Inflammatory scoring of mouse colon, control and AOM/DSS on HFD and RD (n = 12 per group, *P < 0.05, *compared to RD colon). (D, E) Tumor size and tumor burden per colon were compared between AOM/DSS mice on either HFD or RD (*P < 0.05, *compared to RD Tumor).
HFD promotes differential gene expression in colonic tissue and tumors
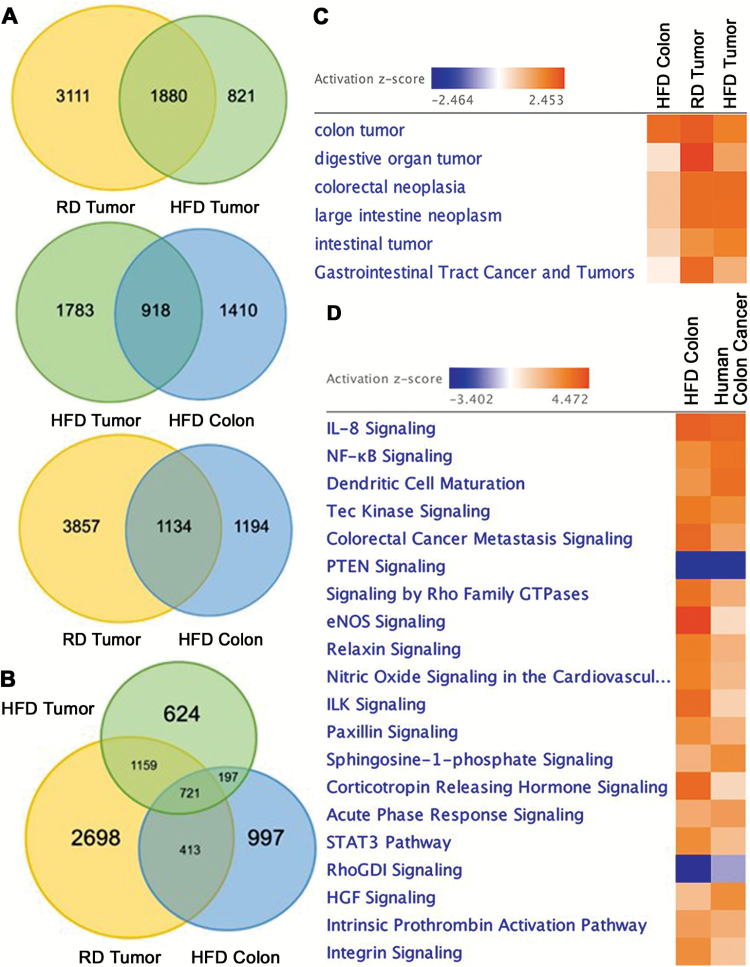
To understand the effects of HFD on gene expression in the colon and AOM/DSS induced tumors, next generation RNA sequencing was performed. As expected, we found a significant number of differentially expressed transcripts in tumors (HFD and RD) compared to RD colon (FDR < 0.05, EBseq) (Figure 2A). Compared to RD colon, 1880 transcripts were differentially expressed in colonic tumors of mice on both HFD and RD; however, differences in expression among tumors from each diet group were also detected (3111 in RD tumors and 821 in HFD tumors compared to RD colon). The variation in the number of differentially expressed transcripts amongst tumors could likely be due to tumor heterogeneity as previously reported by Zhang et al. (23). Moreover, close to half of the total transcripts differentially expressed in HFD colon shared an expression pattern with tumors (HFD tumors: 918 transcripts, RD tumors: 1134 transcripts) (Figure 2A). Further assessment revealed that in HFD colon, 721 transcripts were shared with tumors from both HFD and RD mice (Figure 2B) and IPA determined that these transcripts were associated with multiple forms of gastrointestinal malignancy (Figure 2C). Since we compared the expression signature of mouse HFD colon with colonic tumors induced by AOM/DSS, next we determined if the HFD-mediated transcriptional signature was similar to that of AOM/DSS-mediated inflammation or dysplasia prior to tumor development (GSE31106) (24). We found that in mouse colon, HFD stimulated an expression profile largely similar to acute inflammation and low-grade dysplasia associated with AOM/DSS treatment (FDR < 0.05, IPA) (Supplementary Material 1). However, as mouse colonic tumors were obtained 20 weeks after AOM/DSS initiation, the effect of the treatment is diminished (11); hence, the high expressional similarity found between HFD colon and tumors is most likely a consequence of transcriptional changes associated with tumorigenesis. Moreover, the expression similarity between mouse colon on HFD and tumors was further confirmed in human colon cancer. When comparing pathway activation in mouse HFD colon with human colon cancer (GSE4183), we found a considerable amount of transcriptional similarity (FDR < 0.05, IPA) (Figure 2D). Together, these data indicate that in mouse colon, HFD facilitates transcriptional changes similar to tumors that also resemble those found in human colon cancer.
Figure 2.
HFD promotes differential gene expression in colonic tissue that resembles colonic tumors. (A, B) Venn Diagrams depict the number of transcripts commonly or uniquely expressed in colon and tumors of mice on HFD and RD (n = 3 mice per condition, FDR < 0.05, EBseq). (C) Ingenuity Pathway Analysis (IPA) revealed the transcriptional signature of HFD colon is associated with numerous gastrointestinal malignancies. (D) Pathway analysis revealed a high degree of transcriptional similarity between mouse HFD colon and human colon cancer (GSE4183) (FDR < 0.05, IPA).
HFD stimulates in mouse colon pathways involved in inflammation and tumorigenesis shared with human colon cancer microsatellite instability
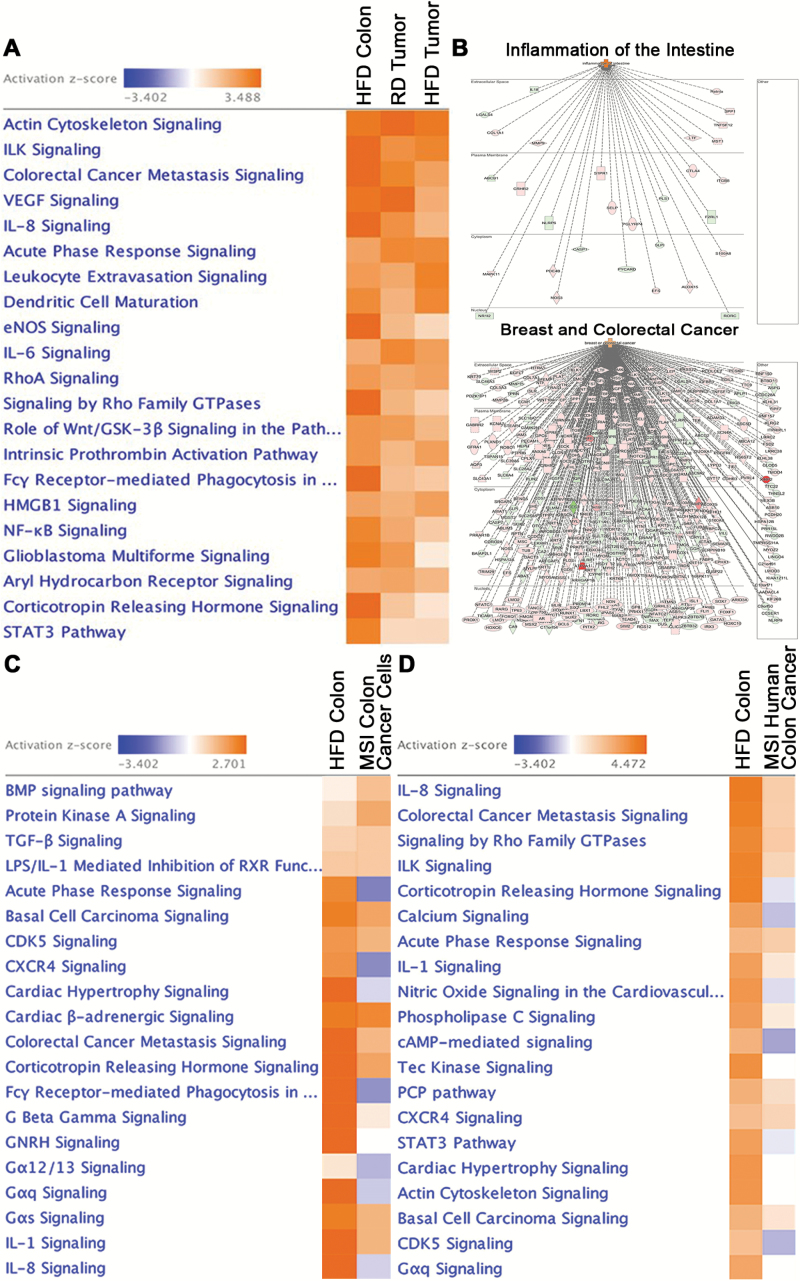
Next, we determined the functional role of the transcripts shared by HFD colon and tumors using pathway analysis. We found multiple pathways involved in diverse cellular functions (Figure 3A) (FDR < 0.05, IPA), including: elevated metabolic activity, cellular movement, inflammation, and cancer (Table 1). A list of 356 transcripts associated with these respective pathways has been incorporated in Supplementary Material 2, including those associated with dysregulation in intestinal barrier function. Of the activated inflammatory pathways in HFD colon shared with tumors, we found NF-kB, IL-6, IL-8 and dendritic cell and leukocyte signaling, while amongst tumorigenic pathways, colorectal cancer metastasis and VEGF signaling were both stimulated. Disease and function analysis demonstrated that the differentially expressed and shared transcripts were associated with intestinal inflammation (30 transcripts) and with breast and colorectal cancer (245 transcripts) (FDR < 0.05, IPA) (Figure 3B). It is plausible that the association with breast cancer may be due to adipose tissue playing an active role in breast cancer pathobiology (25), and thus, further supports a role for accumulated adipose tissue in obesity in promoting tumorigenesis in the colon. Moreover, as the mouse colonic HFD signature revealed activation of inflammatory signaling, we assessed a potential transcriptional overlap with human inflammatory colon cancer associated with microsatellite instability (MSI), characterized by ineffective mismatch repair leading to genetic hypermutation (26). Indeed, we found that 13 of the top 20 activated canonical pathways in mouse HFD colon were shared with inflammatory MSI human colon cancer cell lines (CCLE) and human colon cancer tissue (TCGA) (FDR < 0.05, IPA) (Figure 3C and D). These findings reveal that in colonic tissue, HFD induces activation of inflammatory and tumorigenic pathways similar to those found in human colon cancer with inflammatory microsatellite instability.
Figure 3.
HFD activates pathways involved in inflammation and carcinogenesis similar to those associated with microsatellite instability. (A) Top canonical pathways activated in colon of mice on HFD and tumors (RD and HFD) relative to colon on RD (FDR < 0.05, IPA). (B) Disease and function analysis of the differentially expressed transcripts revealed their association with intestinal inflammation and breast and colorectal cancer (activation color score: red: ↑ expression, green: ↓ expression) (FDR < 0.05, IPA). (C) Pathway analysis revealed that the transcriptional signature obtained from mouse HFD colon shared a high degree of similarity with MSI human colorectal cancer cell lines. The MSI colon cancer cell line expression signature was generated by comparing RNA-seq from 5 inflammatory MSI cell lines (HCT15, LoVo, LS411N, RKO, SW48) against 5 non-inflammatory MSS cell lines (SW480, SNUC1, HT55, HT29, COLO-201) (27) using EBseq (FDR < 0.05, IPA). (D) Pathway analysis revealed that the transcriptional signature obtained from mouse HFD colon shared a high degree of similarity with human MSI colon cancer. The MSI colon cancer expression signature was generated by comparing RNA-seq from human MSI against MSS colon cancers (TCGA) using EBseq (FDR < 0.05, IPA).
Table 1.
Signaling pathways upregulated in HFD colon of mice shared with tumors
| Metabolic activity | Cellular movement | Cancer | Inflammation |
|---|---|---|---|
| eNOS signaling | Actin cytoskeleton signaling | CRC metastasis | IL-8 signaling |
| Corticotropin hormone signaling | ILK signaling | VEGF signaling | Acute phase response |
| Intrinsic prothrombin activation | RhoA signaling | Role of Wnt/GSK-3β signaling | Leukocyte extravasation |
| Signaling by Rho family GTPases | HMGB1 signaling | Dendritic cell maturation | |
| NF-κB signaling | IL-6 signaling | ||
| Glioblastoma multiforme signaling | Fcγ receptor phagocytosis signaling | ||
| Aryl hydrocarbon signaling | HMGB1 signaling | ||
| NF-κB signaling |
HFD stimulates genes involved in inflammation and tumorigenesis
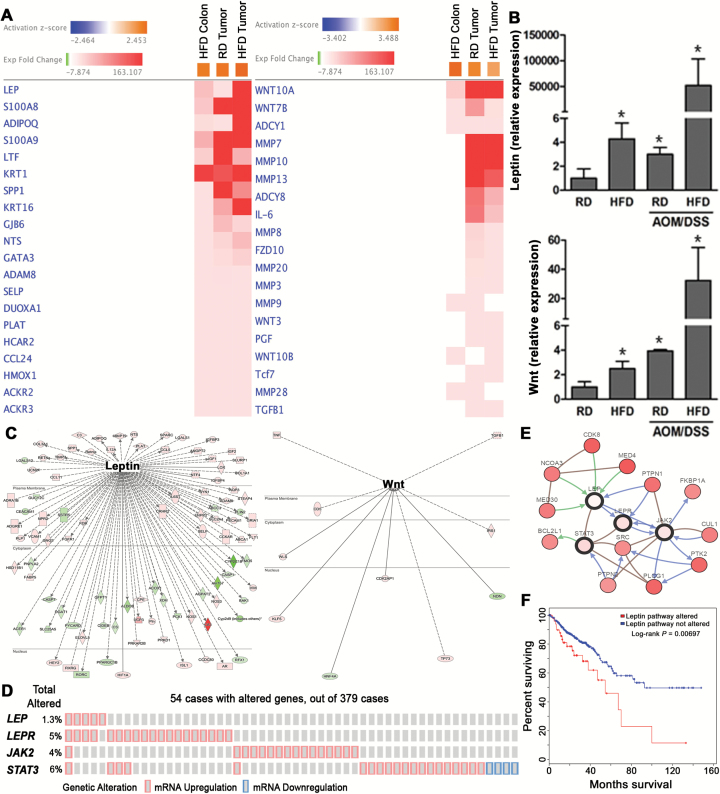
We further investigated expression of individual transcripts from select pathways shared by HFD colon and tumors. Heatmap gene analysis indicated that inflammatory leptin and tumorigenic Wnt were the top elevated transcripts in HFD colon shared with colonic tumors (RD and HFD) (Figure 4A), which was further confirmed by qPCR (Figure 4B). Moreover, pathway analysis indicated that leptin was connected with an additional 94 differentially expressed transcripts, while Wnt was associated with 10 (Figure 4C). Among the inflammatory transcripts, we found upregulated calprotectin (S100A8/A9) and chemokine ligand 24 (ccl24), shown to be elevated with inflammatory bowel disease (IBD) (28,29). Among elevated tumorigenic transcripts, we found matrix metalloproteases MMP 9 and 28, as well as Wnt 9 and 10B ligands, which are involved in colon cancer progression (30,31).
Figure 4.
HFD activates genes involved in inflammation and tumorigenesis. (A) Heatmap gene analysis of the transcripts significantly elevated in HFD colon and tumors (RD and HFD) indicated leptin and Wnt as the top expressed genes (FDR < 0.05, IPA). (B) qPCR confirmed upregulation of leptin and Wnt expression from colon and tumors of mice on RD and HFD (n = 3, *P < 0.05 relative to RD colon). (C) Pathway analysis of differentially expressed transcripts from HFD colon or tumors indicated that additional dysregulated transcripts were connected with leptin and with Wnt (activation color score: red: ↑ expression, green: ↓ expression) (FDR < 0.05, IPA). (D) Alterations in expression of leptin pathway members: leptin (LEP), leptin receptor (LEPR), janus kinase 2 (JAK2) and signal transducer and activator of transcription 3 (STAT3) in human colorectal cancer. Individual genes are represented as rows, whereas individual colon cancer cases are shown as columns (cBioPortal.org). (E) Network analysis indicated additional transcripts associated with leptin signaling as consistently upregulated in human colon cancer patients (cbioportal.org). (F) Survival analysis of colon cancer patients with leptin pathway alterations (cbioportal.org).
Since activation of the tumorigenic Wnt pathway in colon cancer progression has been established (31), we investigated changes associated with leptin expression in human colon cancer using TCGA and the cBioPortal for human cancer genomics. Our assessment of differentially expressed transcripts from human colon cancer compared to normal tissue (TCGA) indicated insignificant upregulation in leptin expression (data not shown). However, it has been demonstrated that leptin protein levels are increased in more than half of human colorectal cancer cases compared to matched control tissue (32). Thus, we further assessed human colon cancer tissue (TCGA) itself using cBioPortal for expression of members of the leptin pathway including: leptin, leptin receptor, and JAK-STAT (33). In 14% of 379 human colon cancer cases, we found upregulation of members of the leptin pathway, as well as leptin and leptin receptor expression tending toward co-occurrence (P < 0.05, cBioPortal.org) (Figure 4D). The significance of leptin pathway activation was reinforced by network analysis revealing that additional factors associated with leptin signaling were consistently upregulated in human colon cancer (Figure 4E). Importantly, leptin pathway alterations revealed a worse overall survival in human colon cancer patients (Figure 4F). Together, these data reveal that in mouse colon, HFD stimulates expression of a large network of transcripts associated with inflammation and tumorigenesis, with leptin and Wnt as the top expressed genes. Furthermore, in human colon cancer, alterations in leptin pathway expression and associated signaling factors are associated with worse overall survival.
Upregulation of inflammatory leptin and tumorigenic Wnt in mouse colon and human colonic cells by HFD
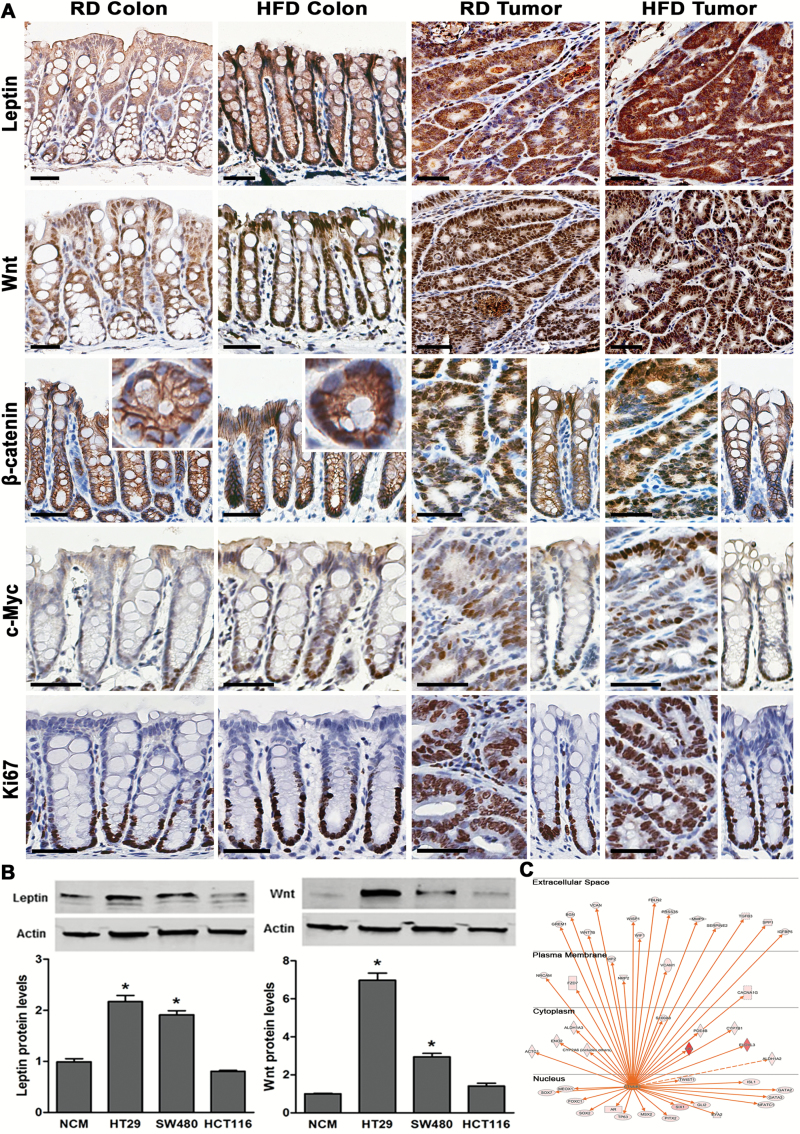
As the above analysis demonstrated increased transcription of leptin and Wnt in HFD colon and tumors (Figure 4A and B), we confirmed elevation in their protein levels through immunohistostaining (Figure 5A). The increase in leptin and Wnt protein was also observed in human colonic cancer HT29 and SW480 cells relative to non-transformed colonic NCM460 cells (Figure 5B). Furthermore, as a marker of Wnt activation we observed nuclear localization of β-catenin (34) and upregulation of its transcriptional target c-Myc (Figure 5A), as well as augmentation of additional transcripts dependent on β-catenin activation in mouse HFD colon (FDR < 0.05, IPA) (Figure 5C). Consequently, as a result of activated Wnt/β-catenin signaling, increased rates of proliferation were seen in HFD colonic crypts as determined by elevated Ki67 positive cells (Figure 5A) (proliferative index – RD colon: 26.7 ± 1.7%, HFD colon: 40.7 ± 3*%; 10 crypts per mouse, n = 3 mice per condition, *P < 0.05). These data further support that in mouse colon HFD stimulates increased expression of leptin and Wnt, and indicate activation of tumorigenic Wnt/β-catenin pathway.
Figure. 5.
HFD leads to increased expression of leptin and Wnt, as well as β-catenin activation in colon and colonic tumors. (A) Immunohistostainings of leptin, Wnt, β-catenin, c-Myc and Ki67 from colon and tumors of mice on RD and HFD (scale bar 40 µm). (B) Immunoblots of leptin and Wnt from human colonic non-transformed NCM460 cells and transformed HT29, SW480 and HCT116 cells (n = 3, *P < 0.05 relative to NCM460 cells). (C) Upstream regulator analysis indicated β-catenin targets as being expressed in HFD mice (activation color score: red: ↑ expression) (FDR < 0.05, IPA).
Discussion
Obesity is associated with increased inflammation and risk of cancer of different organs, including the colon (1,2). While multiple factors could contribute to this risk, colonic expressional changes induced by obesity are still not thoroughly explored. Here, using next generation RNA sequencing and pathway analysis we have identified a significant number of transcripts induced by HFD in colon of mice that are shared with colonic tumors. These transcripts are predominantly involved in inflammation and cancer progression, with leptin and Wnt as the top expressed transcripts from these categories, respectively. Notably, in mouse colon, HFD stimulates an expression signature with striking similarity to human colon cancer, especially those with inflammatory microsatellite instability. Thus, HFD induces expressional changes in the colon associated with inflammation and tumorigenesis that could be contributing to cancer progression.
Obesity is associated with increased chronic systemic inflammation, in part, due to release of inflammatory mediators by accumulated adipocytes. This systemic inflammation, due to the production of cytokines and adipokines like leptin, is believed to be a major driver of obesity-mediated pathology (35,36). In addition to systemic inflammation, however, in the colon, obesity facilitates local production of cytokines and adipokines, further contributing to tissue-specific pathobiology. For example, leptin is found to be elevated in individuals affected by chronic inflammatory bowel disease not only systemically, but also in affected colonic tissue (37,38). This elevation may further exacerbate intestinal inflammation as activation of the leptin receptor, which is normally expressed by colonic cells, leads to stimulation of inflammatory NF-kB signaling (37). Additionally, several studies have shown that obesity promotes local intestinal inflammation due to disturbances in barrier function, enhancing the overall burden of aberrant microbiota (39,40). Moreover, we found that these HFD induced transcriptional changes are similar to those detected in human colon cancer with inflammatory microsatellite instability. Microsatellite instability contributes to tumorigenesis associated with chronic inflammation seen in IBD (41) as a result of genetic instability mediated by an inflammatory microenvironment, leading to accumulation of random mutations (26). Though we showed that HFD stimulates tumor growth but not tumor incidence in mice, we speculate that in obese individuals exposed to multiple years of HFD, an inflammatory microenvironment would not only facilitate proliferation and survival of tumor cells, but also promote genetic instability leading to increased colonic cancer incidence.
Here, we demonstrate that transcriptional changes in the colon of mice on HFD are similar to those seen in colonic tumors. Evidence suggests that obesity-mediated cancer progression transpires, in part, through signaling occurring in tumor cells (1). For example, breast cancer cell proliferation is stimulated by the insulin pathway activation of ERK and PI3K signaling (42). Moreover, in breast cancer, leptin signaling activates the MAPK, PI3K and STAT pathways, which are critical for transformed cell survival, proliferation and differentiation (43). Leptin, acting as a growth factor for colon cancer cells, promotes TGF-β expression as well as VEGF-driven angiogenesis and vascular development (44). Additionally, diet-induced obesity in colonic tissue mediates activation of Wnt signaling which has been shown to directly promote proliferation of colonic stem cells through activation of PPAR-δ signaling, leading to tumorigenesis (45,46). However, one study suggested that obesity and physical inactivity are less likely to promote colon cancer through means of Wnt activation (47). Collectively, it is more likely that obesity-mediated increases in colon cancer depend on multiple pathways that favor tumorigenesis or facilitate the growth of tumors once they have been initiated. Moreover, while it is still unclear whether obesity-mediated colon tumorigenesis is caused indirectly via inflammation or through direct effects on colonic (stem) cells, our findings support that HFD stimulates transcriptional changes in the colon that are similar to those found in colonic tumors.
Obesity and overweight, public health crises affecting almost half a billion adults worldwide, are influenced by a complex network of factors including HFD, genetics and microbiota (3,5,6). Epidemiological data have associated obesity with increased risk of colon cancer, which is thought to be caused by aberrant microbiota, elevated insulin signaling, adipokine production, inflammation and oxidative stress (1,48); yet, the obesity-mediated changes within colonic cells that support tumor growth are not completely understood. Here, we provide RNA-seq and pathway analysis that indicate the colonic transcriptional changes brought on by HFD resemble those changes seen in mouse colonic tumors as well as human colon cancer, especially those associated with inflammatory microsatellite instability. Among the top pathways activated by HFD, inflammatory and colon cancer signaling and their associated transcripts, leptin and Wnt, were observed. These findings could contribute to a better understanding of the mechanisms driving obesity-associated colorectal tumorigenesis.
Supplementary material
Supplementary data are available at Carcinogenesis Online.
Funding
This work was supported by a NIH RO1 award (CA160809).
Supplementary Material
Acknowledgements
The authors thank Dr. Rania Makboul for providing histological analyses of tissue. Additionally, the authors thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for providing next generation sequencing facilities and services. H.P. and S.D.S. conception and design of research; H.P., S.H., and C.C. performed experiments; H.P., S.H., H.N., M.B., and S.D.S. analyzed data; H.P., S.E.C., E.F., and S.D.S. interpreted results of experiments; H.P. prepared figures; H.P., S.H., C.C., H.N., M.B., E.F., S.E.C., and S.D.S. approved final version of manuscript; H.P., and S.D.S. edited and revised manuscript; H.P. and S.D.S. drafted manuscript.
Conflict of Interest Statement: Suzana D. Savkovic wishes to disclose ownership in Pegasus Biosolution, LLC. No other conflicts of interest, financial or otherwise, are declared by the authors.
Abbreviations
- AOM
azoxymethane
- DSS
dextran sulfate sodium
- HFD
high-fat diet
- RD
regular diet
References
- 1. Pietrzyk L., et al. (2015) Obesity and obese-related chronic low-grade inflammation in promotion of colorectal cancer development. Asian Pac. J. Cancer Prev., 16, 4161–4168. [DOI] [PubMed] [Google Scholar]
- 2. Bardou M., et al. (2013) Obesity and colorectal cancer. Gut, 62, 933–947. [DOI] [PubMed] [Google Scholar]
- 3. Barnett R. (2005) Obesity. Lancet, 365, 1843. [DOI] [PubMed] [Google Scholar]
- 4. Haslam D.W., et al. (2005) Obesity. Lancet, 366, 1197–1209. [DOI] [PubMed] [Google Scholar]
- 5. Bray G.A. (2004) Medical consequences of obesity. J. Clin. Endocrinol. Metab., 89, 2583–2589. [DOI] [PubMed] [Google Scholar]
- 6. Ley R.E., et al. (2005) Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA, 102, 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turnbaugh P.J., et al. (2006) An obesity-associated gut microbiome with increased capacity for energy harvest. Nature, 444, 1027–1031. [DOI] [PubMed] [Google Scholar]
- 8. Waldram A., et al. (2009) Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J. Proteome Res., 8, 2361–2375. [DOI] [PubMed] [Google Scholar]
- 9. Scarpa M., et al. (2014) Obesity is a risk factor for multifocal disease and recurrence after colorectal cancer surgery: a case-control study. Anticancer Res., 34, 5735–5741. [PubMed] [Google Scholar]
- 10. Barb D., et al. (2007) Adiponectin in relation to malignancies: a review of existing basic research and clinical evidence. Am. J. Clin. Nutr., 86, s858–s866. [DOI] [PubMed] [Google Scholar]
- 11. Okayasu I., et al. (1996) Promotion of colorectal neoplasia in experimental murine ulcerative colitis. Gut, 39, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Greten F.R., et al. (2004) IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell, 118, 285–296. [DOI] [PubMed] [Google Scholar]
- 13. Li B., et al. (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leng N., et al. (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics, 29, 1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barrett T., et al. (2013) NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res., 41, D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoav B., et al. (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol., 57, 289–300. [Google Scholar]
- 17. Barretina J., et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature, 483, 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cancer Genome Atlas, N. (2012) Comprehensive molecular characterization of human colon and rectal cancer. Nature, 487, 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gao J., et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal., 6, pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cerami E., et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov., 2, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erben U., et al. (2014) A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol., 7, 4557–4576. [PMC free article] [PubMed] [Google Scholar]
- 22. Snoeks L., et al. (2009) Tumor suppressor FOXO3 participates in the regulation of intestinal inflammation. Lab. Invest., 89, 1053–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang L., et al. (1997) Gene expression profiles in normal and cancer cells. Science, 276, 1268–1272. [DOI] [PubMed] [Google Scholar]
- 24. Tang A., et al. (2012) Dynamic activation of the key pathways: linking colitis to colorectal cancer in a mouse model. Carcinogenesis, 33, 1375–1383. [DOI] [PubMed] [Google Scholar]
- 25. Simpson E. R., et al. (2013) Minireview: obesity and breast cancer: a tale of inflammation and dysregulated metabolism. Mol. Endocrinol. (Baltimore, Md, 27, 715–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Colotta F., et al. (2009) Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis, 30, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 27. Sadanandam A., et al. (2013) A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat. Med., 19, 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lin J.F., et al. (2014) Meta-analysis: fecal calprotectin for assessment of inflammatory bowel disease activity. Inflamm. Bowel Dis., 20, 1407–1415. [DOI] [PubMed] [Google Scholar]
- 29. Manousou P., et al. (2010) Increased expression of chemokine receptor CCR3 and its ligands in ulcerative colitis: the role of colonic epithelial cells in in vitro studies. Clin. Exp. Immunol., 162, 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zucker S., et al. (2004) Role of matrix metalloproteinases (MMPs) in colorectal cancer. Cancer Metastasis Rev., 23, 101–117. [DOI] [PubMed] [Google Scholar]
- 31. Burgess A.W., et al. (2011) Wnt signaling and colon tumorigenesis–a view from the periphery. Exp. Cell Res., 317, 2748–2758. [DOI] [PubMed] [Google Scholar]
- 32. Koda M., et al. (2007) Overexpression of the obesity hormone leptin in human colorectal cancer. J. Clin. Pathol., 60, 902–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fruhbeck G. (2006) Intracellular signalling pathways activated by leptin. Biochem. J., 393, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reguart N., et al. (2005) The role of Wnt signaling in cancer and stem cells. Future Oncol., 1, 787–797. [DOI] [PubMed] [Google Scholar]
- 35. Lyon C.J., et al. (2003) Minireview: adiposity, inflammation, and atherogenesis. Endocrinology, 144, 2195–2200. [DOI] [PubMed] [Google Scholar]
- 36. Comstock S.S., et al. (2014) Adipokines and obesity are associated with colorectal polyps in adult males: a cross-sectional study. PLoS One, 9, e85939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sitaraman S., et al. (2004) Colonic leptin: source of a novel proinflammatory cytokine involved in IBD. FASEB J., 18, 696–698. [DOI] [PubMed] [Google Scholar]
- 38. Biesiada G., et al. (2012) Expression and release of leptin and proinflammatory cytokines in patients with ulcerative colitis and infectious diarrhea. J. Physiol. Pharmacol., 63, 471–481. [PubMed] [Google Scholar]
- 39. Brun P., et al. (2007) Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am. J. Physiol. Gastrointest. Liver Physiol., 292, G518–G525. [DOI] [PubMed] [Google Scholar]
- 40. Teixeira T.F., et al. (2012) Potential mechanisms for the emerging link between obesity and increased intestinal permeability. Nutr. Res., 32, 637–647. [DOI] [PubMed] [Google Scholar]
- 41. Tahara T., et al. (2005) Clinical significance of microsatellite instability in the inflamed mucosa for the prediction of colonic neoplasms in patients with ulcerative colitis. J. Gastroenterol. Hepatol., 20, 710–715. [DOI] [PubMed] [Google Scholar]
- 42. Frasca F., et al. (2008) The role of insulin receptors and IGF-I receptors in cancer and other diseases. Arch. Physiol. Biochem., 114, 23–37. [DOI] [PubMed] [Google Scholar]
- 43. Dieudonne M.N., et al. (2002) Leptin mediates a proliferative response in human MCF7 breast cancer cells. Biochem. Biophys. Res. Commun., 293, 622–628. [DOI] [PubMed] [Google Scholar]
- 44. Endo H., et al. (2011) Leptin acts as a growth factor for colorectal tumours at stages subsequent to tumour initiation in murine colon carcinogenesis. Gut, 60, 1363–1371. [DOI] [PubMed] [Google Scholar]
- 45. Liu Z., et al. (2012) Diet-induced obesity elevates colonic TNF-α in mice and is accompanied by an activation of Wnt signaling: a mechanism for obesity-associated colorectal cancer. J. Nutr. Biochem., 23, 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beyaz S., et al. (2016) High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature, 531, 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morikawa T., et al. (2013) Prospective analysis of body mass index, physical activity, and colorectal cancer risk associated with β-catenin (CTNNB1) status. Cancer Res., 73, 1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deng T., et al. (2016) Obesity, inflammation, and cancer. Annu. Rev. Pathol., 11, 421–449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.