Summary
Mucins are multi-domain high-molecular-weight glycoproteins encoded by large multi-exon genes that are aberrantly overexpressed in inflammation and cancer. Alternative splicing, high mutational rates, and polymorphisms in mucin genes in cancer and inflammation can potentially generate a large repertoire of functionally diverse, disease-specific mucin gene products that can serve as potential biomarkers and therapeutic targets. This review summarizes the current understanding of the genetic variants of mucins in cancer and inflammation, identifies the gaps in our knowledge and discusses the future course of research to discern their functional role and exploit their diagnostic and therapeutic potential.
Abstract
Alternative gene splicing, occurring ubiquitously in multicellular organisms can produce several protein isoforms with putatively different functions. The enormously extended genomic structure of mucin genes characterized by the presence of multiple exons encoding various domains may result in functionally diverse repertoire of mucin proteins due to alternative splicing. Splice variants (Svs) and mutations in mucin genes have been observed in various cancers and shown to participate in cancer progression and metastasis. Although several mucin Svs have been identified, their potential functions remain largely unexplored with the exception of the Svs of MUC1 and MUC4. A few studies have examined the expression of MUC1 and MUC4 Svs in cancer and indicated their potential involvement in promoting cancer cell proliferation, invasion, migration, angiogenesis and inflammation. Herein we review the current understanding of mucin Svs in cancer and inflammation and discuss the potential impact of splicing in generating a functionally diverse repertoire of mucin gene products. We also performed mutational analysis of mucin genes across five major cancer types in International Cancer Genome Consortium database and found unequal mutational rates across the panel of cancer-associated mucins. Although the functional role of mucins in the pathobiology of various malignancies and their utility as diagnostic and therapeutic targets remain undisputed, these attributes need to be reevaluated in light of the potentially unique functions of disease-specific genetic variants of mucins. Thus, the expressional and functional characterization of the genetic variants of mucins may provide avenues to fully exploit their potential as novel biomarkers and therapeutic targets.
Introduction
Mucins are expressed at the apical surfaces of polarized epithelial cells either as transmembrane or as secretory glycoproteins, characterized by high molecular weight and extensive O-linked glycans (1,2). The presence of polymorphic Variable Number of Tandem Repeats (VNTR) rich in Proline, Threonine and Serine residues (PTS domain) is a hallmark of all mucins (3). A total of 21 mucins have been identified in humans to date, of which 12 are membrane tethered (MUC1, 3A, 3B, 4, 12, 13, 14, 15, 16, 17, 21 and 22), 7 are secreted (MUC2, 5, 6,7,19, and 20), due to the lack of a transmembrane domain and domain architecture is unknown for MUC8 and MUC9. Although the expression pattern of various mucins in the respiratory, gastrointestinal and genitourinary tracts has been characterized and their involvement in multiple pathologies extensively studied, the biology of mucins remains poorly understood.
Mucins not only protect the epithelial lining from the external environment but they are also an important class of molecules aberrantly overexpressed in various pathologies and have been mechanistically implicated in inflammatory disorders and epithelial cancers. By the same token, they serve as useful prognostic and diagnostic markers, as well as potential therapeutic targets (1,4–6). Mucins are transcribed from large genes containing multiple exons that encode various functionally unique domains, including sperm protein enterokinase agrin (SEA), epidermal growth factor-like (EGF-like), Nidogen-like (NIDO), the von Willebrand factor D like (vWD) and the cytoplasmic tail (CT) and are hence structurally and functionally heterogeneous. These domains facilitate interactions with cell surface proteins like integrins and receptor tyrosine kinases, and the components of extracellular matrix (ECM) that allow mucins to mediate diverse roles under physiological and pathological conditions including protection and lubrication of epithelial surfaces, regulation of cell-to-cell interactions, environmental sensing, and immune modulation (7,8).
Like other eukaryotic genes, mucins undergo extensive splicing. Due to their large genomic size and presence of multiple exons encoding various domains, alternative splicing of mucins genes can potentially create a large repertoire of structurally and functionally diverse splice variants (Svs). In fact, discovery and evaluation of multiple transcripts have revealed that mucins Svs do play a critical role under various pathological conditions. The functional evaluation of various MUC1 Svs, namely MUC1/Y, MUC1/A and MUC1/B in different cancers and inflammatory disorders, discussed in detail in subsequent sections, best exemplifies the critical role of mucin Svs. In addition to splicing, mucins genes carry multiple mutations in various functional domains. Further, polymorphisms in their VNTR region can facilitate various mechanisms that may contribute to different pathologies. The implications of structural and functional heterogeneity of mucins resulting from a combination of alternative splicing, mutations and polymorphisms in physiology and disease have been poorly understood and understudied. This review summarizes the studies on mucin splicing, discusses its contribution to various pathological conditions in the context of the current understanding of the functions of mucin domains and emphasizes the need to comprehensively evaluate their biological significance.
Mucin domains
Mucins contain multiple domains arranged in a specific order to facilitate their putative functions. Many of these domains like SEA, NIDO, vWD, and Adhesion-associated domain in MUC4 and Other Protein (AMOP) are evolutionarily conserved in mucin like proteins, suggesting an important role played by these domains (9–11). Here, we briefly describe the structural attributes of domains commonly present in the majority of mucins and their known and putative functions.
SEA domain
The SEA domain is present in the majority of membrane-bound mucins, including MUC1, MUC3, MUC12, MUC13, MUC14, MUC16 and MUC17. Most of these mucins have a single SEA domain, except MUC16, which has 33 SEA domains arranged in tandem (12). The precise function of SEA domain is unknown, but based on its presence in different proteins along with its subcellular localization, it is suggested to be involved in cell–ECM interactions. Therefore, the SEA domain potentially contributes to the critical role played by mucins during cell migration and cancer metastasis.
vWD domain
The vWD domain is present in MUC2, MUC4, MUC5AC, MUC5B, MUC6 and MUC19. It was first identified in the von Willebrand factor in blood and later in multiple proteins involved in extracellular adhesion functions. In a pathological context, this domain has been most commonly associated with bleeding disorders (13). However, in the context of malignant disease, the upregulation of von Willebrand factor may confer a pathological advantage to circulating tumor cells.
NIDO domain
This extracellular domain consists of ~180 amino acids and is present in proteins that interact with matrix components (14,15). The NIDO domain is only present in MUC4 and is believed to participate in the metastatic dissemination of pancreatic cancer cells by compromising the integrity of the basement membrane (16). Deletion of NIDO from MUC4 resulted in significant reduction in extravasation of pancreatic cancer cells in vitro as well as a significant decrease in liver metastasis in vivo (16). The MUC4–NIDO domain interacts with fibulin-2 to facilitate metastasis (16).
AMOP domain
The AMOP extracellular domain is present only in MUC4 and contains ~100 residues with eight conserved cysteine residues that are suggested to be involved in cross-linking through disulphide bridges (10). This domain was identified in MUC4 by a bioinformatics PSI-BLAST search analysis for the sequence between the NIDO and vWD domains (1,10). The presence of AMOP domain is restricted to proteins that contain other cell adhesion domains like NIDO and vWD domains.
EGF-like domain
The EGF-like domain is a small, evolutionary conserved domain of 30–40 residues that is present extracellularly in membrane-bound and secreted proteins (17). The EGF-like domain is present either singularly or in tandem, which folds together into a single solenoid functional unit (17–19). MUC3 contains two EGF-like domains and MUC4 has three, arranged in tandem at the 3′ end of the molecule (1,20). MUC13 contains three EGF-like domains, which flank the SEA domain (21). The importance of EGF-like domains in mucins is not clear; however, with the loss of polarity and redistribution of surface molecules under pathological conditions, these domains can act as ligands for EGF receptor family members and may potentiate proliferative signaling.
Cytoplasmic tail
All membrane-tethered mucins have a cytoplasmic domain consisting of residues that can undergo posttranslational modifications and interact with adaptor proteins to participate in cell signaling pathways. The MUC1 CT is the most studied one, which interacts with beta-catenin, ZAP70 and heat shock protein-70, and participates in Wingless, the Drosophila melanogaster segment-polarity gene, and integrase-1 (Wnt), and nuclear factor kappa light-chain enhancer of activated B cells signaling pathways (2,5). Recently, MUC16 CT has also been shown to increase the metastatic potential of pancreatic cancer cells by mediating JAK2 translocation to the nucleus and upregulation of LMO2 and NANOG (22).
Mucin splice variants
Due to the highly regulated processing of messenger RNA (mRNA), even limited genetic information can give rise to an enormous repertoire of functional proteins (23). Alternative splicing of precursor mRNA is a process whereby exons and introns are differentially arranged through mechanisms that involve cassette exons, alternative 5′ and 3′ splice sites, exon skipping, intron retention and mutually exclusive exons (24,25). The resulting Svs may acquire new functions, novel interacting partners and different subcellular localization, leading to a pathological phenotype. The splicing process, under both physiological and pathological conditions is regulated by cis-regulatory sequences and RNA binding proteins. Modulations in the expression, activity, and mutations in RNA binding proteins are predominant mechanisms contributing to perturbed splicing during cancer progression that give rise to cancer-specific Svs. Among various RNA binding proteins, the overexpression of huRNApA1, huRNPA2, SRSF1, SRSF3 and PTB has been linked with the multiple cancers including lung, breast, stomach, colon and liver. Similarly, mutations in the components of spliceosome like U2AF1, SF3B1, ZRSR2 and SRSF2 lead to the selection of defective splice sites that may predispose to multiple cancers (26). The underlying mechanisms of alternative splicing in cancer have been extensively reviewed in several articles (24–26) and hence not discussed in detail here.
Mucin1
MUC1 is located on chromosome 1 in the q22 region, spanning 4–7 kb, and encoded by seven exons (E). This protein consists of a VNTR region with 20–125 repeats of 20 amino acids and other important domains like SEA and the cytoplasmic tail (27). MUC1 is synthesized as a single polypeptide chain but undergoes cleavage in the SEA domain due to molecular stress; the resulting two fragments are held together through non-covalent interactions (27). MUC1 has extensively been investigated and shown to play a role in various physiological and pathological conditions such as infection, inflammation and cancer (3,28).
Alternative Svs of MUC1
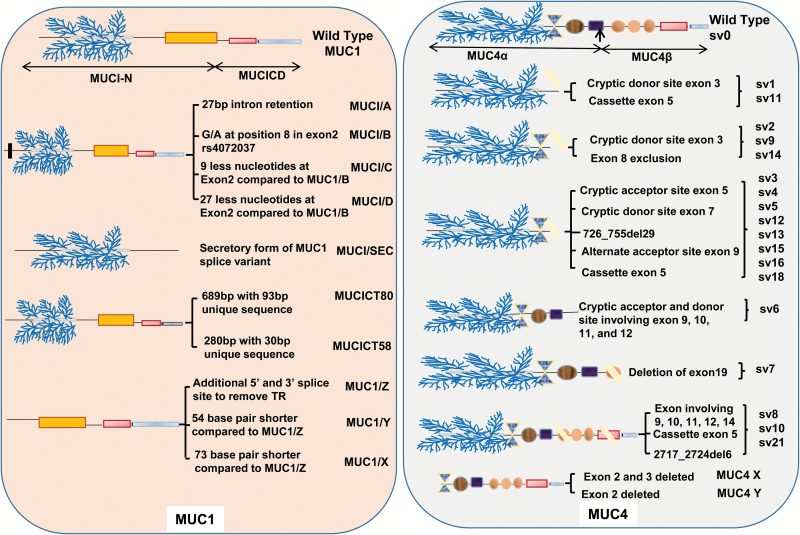
MUC1 mRNA is differentially spliced under various pathological conditions and in a tissue-specific manner (Figure 1). Of the seven exons E1–E7, E1 codes for signal sequence and is retained in all known Svs, whereas E2 codes for the VNTR region and E4–E7 code for the MUC-1β region, which consists of a transmembrane domain and a cytoplasmic tail. There are a total of 78 known Svs for MUC1 generated due to exon skipping, alternative use of 5′ and 3′ splice sites and the complete or partial retention of introns (29). Among all the exons, E4 has the highest skipping rate (29). Multiple conventional and cryptic splice sites are used during the processing of MUC1 transcript. The most commonly used cryptic splice sites in intron 1 are at positions 3096 and 3222 at the 5′ and 3252, 3261, 3471, 3507 and 3534 at the 3′ ends, respectively. The use of a cryptic 5′ splice site at 3600 and 3′ splice sites at 3934, 4120, 4192, 4205, 4235, 4259 and 4278 in E2 result in VNTR exclusion, which leads to the generation of significantly shorter transcripts called MUC1/Y, X, Z and ZD (29). Similarly, the 3′ cryptic splice site at position 4945 in E5 is involved in E3 and E4 skipping and the 5′ and 3′ cryptic sites in intron 6 results in E2 retention in the mature transcript.
Figure 1.
Splice variants of MUC1 and MUC4. Mucins are large-molecular-weight glycoproteins expressed at the apical surface of epithelial cells and consist of multiple functional domains [Tandem repeat ( ), NIDO (
), NIDO ( ), EGF (
), EGF ( ), AMOP (
), AMOP ( ), SEA (
), SEA ( ) and cytoplasmic tail (
) and cytoplasmic tail ( )] capable of interacting with multiple proteins, including cell surface receptors, ECM proteins, signaling molecules and nuclear proteins. Transmembrane MUC1 and MUC4 have multiple Svs, which are generated due to exclusion and inclusion of exon/introns, exon skipping and the complete or partial retention of introns, and the use of alternative 5′ and 3′ splice sites. The expression of predominant MUC1 splice variants are demonstrated along with the loss and gain of major functional domains. The MUC1 Svs have been implicated in the cancer pathology, inflammatory disorders and immune modulation. Similarly, the 24 splice variants identified for MUC4 are expressed in a variety of tissues, some of which are specifically expressed under pathological conditions (sv10 is only detected in the pancreatic cancer). MUC4 Svs are depicted along with the mechanisms leading to their generation and the loss and gain of major functional domains. Recent studies have demonstrated the critical role of MUC4/X and MUC4/Y Svs in pancreatic cancer progression and metastasis.
)] capable of interacting with multiple proteins, including cell surface receptors, ECM proteins, signaling molecules and nuclear proteins. Transmembrane MUC1 and MUC4 have multiple Svs, which are generated due to exclusion and inclusion of exon/introns, exon skipping and the complete or partial retention of introns, and the use of alternative 5′ and 3′ splice sites. The expression of predominant MUC1 splice variants are demonstrated along with the loss and gain of major functional domains. The MUC1 Svs have been implicated in the cancer pathology, inflammatory disorders and immune modulation. Similarly, the 24 splice variants identified for MUC4 are expressed in a variety of tissues, some of which are specifically expressed under pathological conditions (sv10 is only detected in the pancreatic cancer). MUC4 Svs are depicted along with the mechanisms leading to their generation and the loss and gain of major functional domains. Recent studies have demonstrated the critical role of MUC4/X and MUC4/Y Svs in pancreatic cancer progression and metastasis.
The VNTR in the wild-type MUC1 (MUC1/B or MUC1/REP) is spliced by an in-frame removal, leading to the generation of MUC1/Y. The E2 containing VNTR region is spliced preferentially due to the presence of the cryptic 5′ and 3′ splicing sites, branch points, cis-regulatory intron splice enhancer elements, multiple splicing factors binding sites (YCAY) and the A/G single nucleotide polymorphism (SNP; i.e. rs4072037) (29,30). These elements, collectively, give the characteristic feature of an intron to the E2, which results in VNTR exclusion during pre-mRNA processing or even from the mature transcript as a secondary event. A/G silent SNP (i.e. no change in the amino acid coded) at position 3506 is also involved in the selection of 3′ splice site. The presence of A or G directs the selection of the 3498 and 3471 3′ splice sites, respectively, resulting in different splice forms with a nine amino acid insertion in the signal peptide. Other splice forms lacking the VNTR region, such as MUC1/X, Z and ZD, also contain a deletion of 19 base pairs as well as retention of an additional 54 base pairs (31).
In addition to VNTR exclusion, mature transcripts are generated by comparatively shorter addition and deletion of exons and introns in wild-type MUC1. For example, the MUC1 transcript A is generated due to retention of 27 base pairs of intron 1 at the 3′ end; however, variant C and D have 9 and 27 fewer base pairs, respectively, than wild-type MUC1 at the start of the E2. As with shorter MUC1 splice forms, MUC1/A, C and D variants are also influenced by the A/G SNP (i.e. rs4072037) (31). The presence of G and A allele is associated with the presence of MUC1/A and MUC1/B transcripts, respectively. In AA homozygous alleles, there is no MUC1/A transcript. Additionally, the GG homozygous condition is associated with the absence of MUC1/B. Similarly, minor transcripts MUC1/C and D are also associated with A/G SNP (31). Baruch et al. reported another Sv, MUC1/SEC (secreted form). The cleaved products of MUC1/REP and MUC1/SEC act as the ligands for MUC-1/Y and promote phosphorylation of tyrosine residues in its cytoplasmic tail. Both MUC1/SEC and cleaved MUC1/REP share a binding site for MUC1/Y isoform (31,32).
Mucin 4
MUC4 is another membrane-bound mucin that is normally expressed in colon, salivary glands and respiratory and reproductive tracts. MUC4 is mapped to the chromosome 3q29 where it spans 59 kb region. MUC4 gene is encoded by 26 exons, which vary from 65 bp to 22 kb in length and its molecular weight ranges from 550 to 930 kDa due to the polymorphic VNTR region. MUC4 apoprotein contains four functionally unique domains termed NIDO, AMOP, vWD and three EGF-like domains downstream of the central VNTR region. MUC4 structure and biology has seen comprehensively reviewed by us previously (1).
Alternative Svs of MUC4
Similar to MUC1, exon skipping is the most common splicing event, followed by the use of a cryptic donor and acceptor splice sites to generate a wide array of MUC4 Svs (Figure 1). Here, sv0 is designated as the wild-type MUC4. Comparison of the sequence of different Svs with sv0 highlights mechanisms for generating splice forms that involve multiple events. The most common splicing events include 309_386del76, 474_475ins209 and 474_631del156 [Sv nomenclature is described in reference (33)] involving E4–E7 (34,35). The most prevalent splicing event, 309_386del76, observed in MUC4 Svs, sv3, sv4, sv5, sv12, sv13, sv15 and sv16, include the use of a cryptic acceptor site of E5, which results in the deletion of 14 and 76 bp at positions 3284 and 3420, respectively, causing a change in the reading frame.
The other two common events resulting in MUC4 sv1, sv10, sv11, sv15, sv16 and sv18 involve 474_475ins209 and 474_631del156. The event 474_475ins209 involves insertion of an E5, cassette exon, resulting in an additional 211 bp and a consequent change in the reading frame. However, the splicing event 474_631del156 involves an in-frame exclusion of 156 bp of E6, which results due to the use of an alternative splice acceptor site at 3584 position in E7. The least common splicing events include 966_1396del429, 1020_1699del1678 and 2218_2587del368 involving E9–E12, E9–E14 and E17–E20 that result in sv6, sv8 and sv14, respectively (34–36). Analogous to MUC1/X and MUC1/Y Svs, we identified two shorter forms of the MUC4, which are devoid of VNTR region. Using primers targeted to the ATG region of MUC4 and EGF-like domains (34,35,37), two amplicons of 2849 and 2696 bp were generated that were found to contain an in-frame deletion of E2 (MUC4/X) and E2 and E3 (MUC4/Y), respectively. A total of 17 secreted Svs have been identified, of which variants like sv4, sv9 and sv11–sv17 have modified reading frames with 94 to 201 unique amino acids at the C-terminal end. Two additional variants, sv18 and sv19, have 202 and 243 amino acids less at C-terminal ends with no changes in their reading frames.
Svs of other mucins
Crawley et al. identified a 524 bp long full-length transcript along with three splice forms of MUC3, which were generated due to an exclusion of E3 (408 bases), E4 (459 bases) and E5 (375 bases) in human fetal and adult small intestine and colon (20). Similarly, MUC2 Svs lacking tandem repeat (MUC2.1) have been reported both in normal and in colon cancer cell lines (38).
Mucin Svs in cancer and inflammation
Aberrant mucin expression is a characteristic feature of multiple epithelial cancers. However, the precise mechanistic contribution of mucin Svs to the overall pathology of cancer is poorly understood. Due to its higher incidence, most studies have evaluated the role of various MUC1 Svs in cancer and inflammatory responses.
Mucin Svs in cancer
Zhang et al. investigated the expression of MUC1 Svs in multiple cell lines including HeLa, MCF7, Jurkat and activated T cells and identified 78 Svs, including MUC1/Y and MUC1/Z, and 1 new Sv, MUC1/Y-LSP (29). Of these 78 Svs, 31 Svs were in-frame, 21 forms maintained their transmembrane domain and 10 encoded for the secreted forms. The study demonstrated tissue-specific expression of MUC1 variants; 10 were exclusively expressed in T cells, 12 in HeLa, 14 in MCF7 and 27 variants in the Jurkat cells (29). Interestingly, different MUC1 Svs can potentially interact together to form a ligand–receptor complex. For example, when HBL100 cells, a normal breast cell line expressing MUC1/Y, were incubated with conditioned media from DA3 cells expressing MUC1/SEC, there was formation of receptor–ligand complexes (MUC1/Y:MUC1/SEC) that modulated the cell morphology and growth pattern in HBL100 cells (32). Notably, the 6E6/2 antibody specifically mapped to the MUC1/Y splice site region (junctional sequence) does not react with MUC1/REP (39). Using this antibody, the expression of MUC1/Y was confirmed in breast and ovarian cancer cells lines. The binding affinity of 6E6/2 was also validated under in vivo conditions by injecting a radiolabeled 6E6/2 antibody into mice bearing DA3 murine mammary cells expressing MUC1/Y in their mammary fat pad, thus demonstrating the clinical potential for this antibody (39). The presence of MUC1/Y in breast cancer patients was further verified by western blot analysis. This study also verified that MUC1/Y, which is devoid of the tandem repeat, is not cleaved proteolytically like wild-type MUC1/REP (39).
Another study by Zrihan-Licht et al. demonstrated that breast tumors express very low levels of MUC1 protein and that the expression of MUC1/Y varies among patients. However, MUC1/Y expression was specific to the tumors and was absent in the tumor-adjacent normal tissues (40). The expression of MUC1/SEC Sv in breast cancer is enhanced by estrogen treatment due to utilization of different region of the MUC1 promoter for transcription (41). MUC1/SEC is also expressed in endometrial cancer cell lines (42), but absent in ovarian cancer tissues. The expression of other MUC1 Svs has been detected in ovarian cancer. MUC1/X expression in ovarian cancer was associated with better response to chemotherapy (43).
In high-grade serous ovarian carcinoma, MUC1 splicing has also been shown to produce a chimeric RNA, involving TRIM46 and KRTCAP2 genes. Sequencing analysis of 59 high-grade serous ovarian carcinoma patients revealed six different Svs of this chimeric RNA due to the utilization of different splice sites. The expression of the chimeric proteins lead to the synthesis of unglycosylated and cytoplasmically localized forms of MUC1 (44). Further, the inclusion of MUC1 Svs Y and Z to a panel consisting of cytology, CEA and Ep-CAM improved the sensitivity and specificity for detecting cancerous cells in malignant serous effusion to 94.5% and 93%, respectively (45).
Weiss et al. reported that 7 of the 8 fine needle aspirate samples of papillary thyroid cancer were positive for an alternative MUC1 mRNA product that was absent in all 13 benign samples. This Sv termed MUC1/A was found to differ from the expected sequence by a 27 bp due to the use of an alternative splice acceptor site in E2 and E1 and is associated with SNP (46). The MUC1 Svs A, D, X, Y and Z were also reported in ovarian cancer patients with 98% frequency. However, the presence of MUC1/A was not associated with the response to chemotherapy or overall survival in the ovarian cancer patients (43).
In cervical cancer, MUC1 Svs A, B, C and D were reported with varying intensities in multiple cell lines, including C-4 II, C33A, DoTc24510, C-4I, SiHa, Hat3, Hs 636 and Hela. This study also identified two additional Svs, C and D (47). The expression of MUC1/C, D and Z splice forms was shown to correlate with tumor progression in esophageal cancer, whereas wild-type MUC1 demonstrated protective effects and a better prognosis (48). Hinojosa-Kurtzberg et al. described two important Svs of MUC1 in cystic fibrosis transmembrane conductance regulator-deficient mice with varying lengths of the cytoplasmic tail. MUC1 CT58 and MUC1 CT80 Svs were detected in jejunum, ileum and colon of the cystic fibrosis mice. The expression profile of MUC1CT80 at the protein level was further confirmed using a mouse polyclonal antibody generated against the unique sequence present in CT80 Sv. The sequence analysis revealed that the first 48 amino acids in CT of MUC1, CT58 and CT80 are same, however, due to change in reading frame both CT58 and CT80 contain 10 and 32 unique amino sequences at C-terminal, respectively, and lack β-catenin and Grb2 binding sites (49).
We have evaluated the expression of MUC4 Svs in pancreatic cancer cell lines, testis, lung and trachea. The majority of pancreatic cancer cell lines (79%) showed higher expression of wild-type MUC4 along with the expression of 12 variants (i.e. sv4, sv9, sv10–sv18 and sv21). Normal testis, lung and trachea also expressed sv4, sv19 and sv20-MUC4 Svs. However, the sv10 Sv was absent in normal tissue but was specifically present in the pancreatic cancer cell lines (50). In 2014, Xie et al. characterized the role of MUC4/Y in the pancreatic cancer cell line, MIAPaCa-2 using a lentivirus-based overexpression system. In this study, overexpression of MUC4/Y Sv significantly accelerated the tumor progression, suppressed apoptosis, altered cell morphology and interfered with cell cycle regulation under both in vitro and in vivo conditions. Further MUC4/Y activated JNK and AKT pathways but had an insignificant effect on the phosphorylation of Her-2 (51). Another study has demonstrated that the deletion of AMOP domain in MUC4/Y Sv reduces the angiogenic and metastatic potential of MUC4/Y in pancreatic cancer (52). On similar lines, MUC4/X is also expected to play a significant role in cancer pathology.
Wild-type MUC3 is present in the human tissues as well as colon cell lines, including Caco-2, LoVo, LIM1899, LIM 1215 LS 174T, HCT116, SW116 and KM12SM. Svs lacking E3 showed distribution pattern similar to wild-type in the intestines and colon; however, E4 and E5 lacking transcripts showed more restricted distribution. E5 lacking transcripts were absent in fetal small intestine and fetal and adult colon (20). A genome-wide association study analyzed the expression of rare and low-frequency single-nucleotide variants in 350 inflammatory bowel disease samples and demonstrated that a rare variant of MUC19 was associated with higher risk for the disease (53).
SNP, in addition to facilitating splicing events, also predisposes to cancer progression. Patients with SNP at rs2070803 and rs4072037 in MUC1 gene have higher risk of developing diffuse-type gastric cancer (54). The presence of G/G phenotype at rs4072037 SNP is more frequently present in the colorectal patients compared with the controls (55). The frequency of C allele at rs1104760, and rs2688513, A allele at rs2258447 and G allele at rs2246901 in MUC4 is associated with the advance stage of the endometriosis (56). Similarly, homozygous SNP rs12984471 in MUC16 showed poor survival of ovarian cancer patients and higher circulating levels of CA125 (57). Further, homozygous CC at position rs2547065 is associated with higher risk of ovarian cancers (58). Three SNPs at rs11564245, rs4768261 and rs2933353 in MUC19 are significantly associated with the development of the Crohn’s disease (59).
Mucin Svs in immune modulation
Expression of MUC1/A is associated with the protection and lubrication of the ocular surfaces. Low levels of MUC1/A have been implicated in dry eye syndrome. In COS-7 cells, overexpression of MUC1/A increased the expression of interleukin 8 and interleukin 1β after 24 h of treatment with tumor necrosis factor alpha, contrary to MUC1/B, which has no impact on the expression of these cytokines. MUC1/A further increased the basal expression of transforming growth factor beta, thereby modulating the inflammatory response to dry eye syndrome (60–62). The MUC1 VNTR region has also been shown to form complexes with the nuclear factor kappa light-chain enhancer of activated B cells p65 subunit and upregulate the expression of the pro-inflammatory cytokines, including interleukin 6 and tumor necrosis factor alpha. Further, expression of nuclear factor kappa light-chain enhancer of activated B cells correlated with the number of tandem repeats present in the MUC1 VNTR region (63). The anti-tumor response of MUC1 Svs has been investigated in breast cancer. DA3 mammary tumor cells expressing the MUC1/SEC Sv failed to grow in immune-competent mice but developed tumors in T-cell deficient animals. Further evaluation demonstrated that MUC1/SEC upregulated the expression of chemokine (C–C motif) ligand 2 that is responsible for T-cell recruitment (64,65). Moreover, the DA3 cells expressing MUC1/SEC recruit significantly less myeloid-derived suppressor cells compared with DA3-derived tumor expressing wild-type MUC1. MUC1/SEC expression downregulated the levels of urokinase Plasminogen Activator (uPA) in tumor cells via STAT1-dependent pathway, resulting in decreased myeloid-derived suppressor cells recruitment. Further, MUC1/SEC repressed the expression of arginase1 and generation of reactive oxygen species required for the suppression of the anti-tumor response by myeloid-derived suppressor cells (66).
Mutational profile of cancer-associated mucins
In addition to alternative splicing, site-specific variations due to mutations may further contribute to the transcriptional heterogeneity observed in the expression of cancer-associated mucins. Indeed, in colorectal cancer, microsatellite instability resulting from deficiencies in mismatch repair mechanisms may lead to an increase in variation within cancer-associated mucins (67).
The increased frequency of genetic changes in cancer-associated mucins may play an important role in cancer pathogenesis. To identify the source of variability among mucins, mucin-specific somatic mutations were profiled among breast, ovarian, pancreatic, colorectal and lung cancers. This in silico investigation was performed using International Cancer Genome Consortium database. Sample information corresponding to 1333 breasts, 677 ovarian, 999 pancreatic, 675 colorectal and 1062 lung cancers was obtained from the database. Specifically, MUC1, MUC4, MUC16 and MUC5AC were queried for the presence of somatic mutations and their corresponding incidence rates were measured. Mapping of mutations to the affected regions was performed in comparison with reference sequence grch37.p13. Multigroup comparisons were done by the use of Kruskal–Wallis test (one-way analysis of variance) with a P value <0.05 being considered statistically significant. Subsequently, Conover method was used for post hoc nonparametric analysis of the subsets (68).
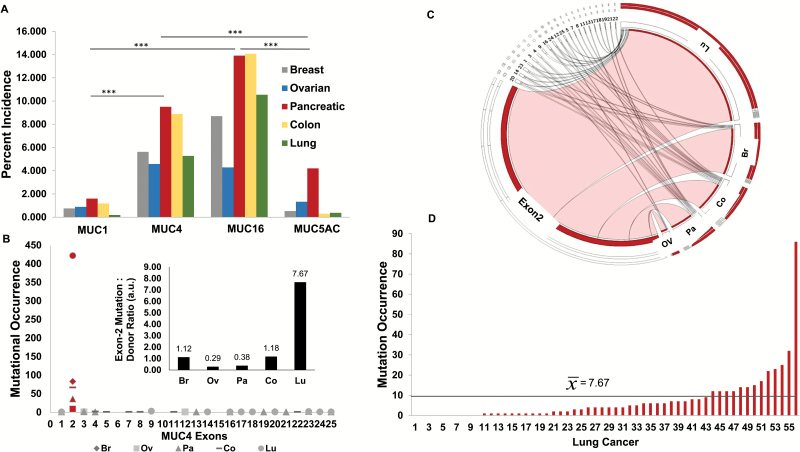
Analysis across the five cohorts revealed an unequal incidence of mutations throughout the panel of cancer-associated mucins. Most notably, a statistically significant increase in the number of samples harboring MUC4 and MUC16 mutations was observed compared with samples exhibiting MUC1 or MUC5AC mutations (Figure 2A). Due to aberrant overexpression of MUC4 in multiple cancers, a more in-depth assessment of the MUC4 mutation bearing samples was performed, and the mutational incidence was subsequently resolved to the exonic regions affected. Consequently, mapping the precise locus affected to the corresponding exons revealed a sparse distribution of mutational incidences throughout the MUC4 transcriptional sequence. Interestingly, across all tissue sites examined, an island of mutational accumulation indicative of microsatellite instability was identified within E2 corresponding to the tandem repeat domain (Figure 2B and 2C).
Figure 2.
Mucin signatures and mutational profiles in various cancers. (A) Percent incidence of mutations in cancer-associated mucins within breast (Br), ovarian (Ov), pancreatic (Pa), colon (Co) and lung (Lu) cancer cohorts (***P < 0.01). The mutational analysis on 1333 Br, 677 Ov, 999 Pa, 675 Co and 1062 Lu cancers was performed using International Cancer Genome Consortium database. Multigroup comparisons were performed by Kruskal–Wallis test (one-way analysis of variance). (B) Exon mapping of MUC4-specific mutations in Br, Ov, Pa, Co and Lu cancer cohorts. There is higher frequency of the mutations in the E2 of MUC4 compared with other exons (red). (inset: depiction of the ratio of E2 mutation per donor stratified by the site of the cancer). (C) Circos plot illustrating the distribution of MUC4-specific mutations in Br, Ov, Pa, Co and Lu cancer cohorts with respect to exon. Exons from right to left (starting from exon 2) are 20, 14, 23, 1, 3, 4, 9, 16, 24, 12, 25, 5, 7, 8, 11, 13, 17, 18, 19, 21 and 22. (D) MUC4, E2 mutational occurrence in Lu cancer in a 55 sample cohort. There is an increase in accumulation of genetic insult in E2 region in a subset of lung cancer patients and distribution of mutational occurrence among various cancer samples.
Furthermore, although a general increase in somatic mutations was observed within E2, lung cancer specifically showed a significantly higher abundance (P < 0.05) of mutations in E2 (Table 1). Indeed, within the 55 lung cancer subset, there were 422 mutations within E2. Moreover, although breast, ovarian, pancreatic and colorectal cancer sample subsets exhibited between 0.29 to 1.18 E2 mutations per sample, lung cancer samples exhibited an average of 7.67 E2 mutations per sample (Figure 2B inset). Examination of the frequency of mutational occurrence per patient showed a subpopulation that possess an aberrantly increased accumulation of genetic insults to this region (Figure 2D).
Table 1.
A representative cohort of samples from various tropisms was analyzed for the MUC4 exon mutational distribution and incidence
| Tissue site | n | Average rank | Exon 2 (P < 0.05) |
|---|---|---|---|
| Breast (1) | 74 | 178.44 | (3)(4)(5) |
| Colorectal (2) | 57 | 185.31 | (3)(4)(5) |
| Lung (3) | 56 | 231 | (1)(2)(4)(5) |
| Ovarian (4) | 31 | 91.24 | (1)(2)(3) |
| Pancreatic (5) | 95 | 101.15 | (1)(2)(3) |
Multigroup comparisons were done by the use of Kruskal–Wallis test with a P value <0.05 being considered statistically significant. Subsequently, post hoc nonparametric analysis of subsets was performed revealing significant accumulation of mutations in exon 2 of MUC4 with lung cancer samples exhibiting the greatest increase. n indicates patients with the mutations in MUC4 gene.
Although E2 mutations appear to be pathognomonic in MUC4, there are tissue-specific sample subsets that exhibit exceptionally robust and cumulative changes at this genetic locus. Furthermore, although changes in the magnitude of gene transcription and alternative splicing may significantly contribute to the phenotypic alterations, the compounding effects of accumulating mutations within a particular region or domain may be of comparable functional significance. The exome sequence analysis from ~3000 tissues samples revealed that MUC16 is among 12 most mutated genes regardless of tumor type. However, the functional significance or contribution of these mutations to tumor pathogenesis has not yet been determined (69).
Conclusion and perspective
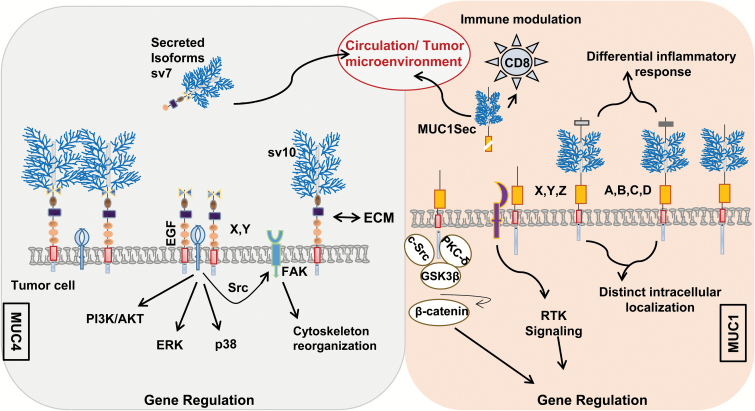
Mucins project up to 200 nm into the extracellular space due to their exceptionally large size and are enriched in ECM-interacting domains like NIDO, SEA, AMOP and vWD. Hence, mucins are uniquely endowed to interact with the extracellular environment as compared with other cell surface proteins. Alternative splicing involving exons encoding ECM-interacting domains can confer unique combinations of functional domains that can potentially facilitate novel interactions with the ECM proteins like collagen, fibulin-2 and galectins to promote tumor progression and metastasis. Moreover, alternative splicing involving transmembrane domains can generate truncated secretory forms of otherwise transmembrane mucins, which could be exploited as potential biomarkers in liquid biopsies for risk stratification, prognosis and evaluating response to therapies (Figure 3). Functionally, such soluble and truncated variants may also compete with the co-expressed wild-type forms for interacting partners and can thus modulate cell–cell or cell–matrix interactions and regulate mucin-based signaling. The tandem repeat domains, which are the hallmark of mucins, are decorated with diverse glycosylated structure that mediate both homotypic and heterotypic cell–cell interactions and can thus participate in critical processes like metastasis, immune evasion and angiogenesis. Genetic variants of mucins that lack tandem repeat domains or harbor mutations in this region, as observed in MUC4, can result in altered glycosylation patterns and modulate cellular interactions and signaling mediated by mucin glycans and thus impart novel functions to the mucins.
Figure 3.
Putative contributions of mucin Svs under pathological conditions. The aberrant splicing events can potentially create a novel combinations of functional domains [Tandem repeat ( ), NIDO (
), NIDO ( ), EGF (
), EGF ( ), AMOP (
), AMOP ( ), SEA (
), SEA ( ) and cytoplasmic tail (
) and cytoplasmic tail ( )], which can change the mucin interactome as well as tissue- and cell-specific functions under normal and various pathological conditions. Absence of the heavily glycosylated tandem repeat region in mucins will reduce the stearic hindrance and facilitate improved interaction of mucins with surface receptors/proteins through various functional domains. These interactions can initiate oncogenic signaling leading to cancer cell proliferation, migration and resistance to apoptosis. Deletion of the transmembrane domain (
)], which can change the mucin interactome as well as tissue- and cell-specific functions under normal and various pathological conditions. Absence of the heavily glycosylated tandem repeat region in mucins will reduce the stearic hindrance and facilitate improved interaction of mucins with surface receptors/proteins through various functional domains. These interactions can initiate oncogenic signaling leading to cancer cell proliferation, migration and resistance to apoptosis. Deletion of the transmembrane domain ( ) due to alternative splicing will lead to the secretion of membrane-tethered mucins in the body fluids that can serve as biomarkers for various cancer and inflammatory disorders. These alternatively spliced cancer-associated mucins can be used for the risk stratification, tumor grading, predicting response and resistance to therapy and prognosis. The cartoon is the pictorial demonstration of Svs specific to MUC4 and MUC1 with their designated and putative functions.
) due to alternative splicing will lead to the secretion of membrane-tethered mucins in the body fluids that can serve as biomarkers for various cancer and inflammatory disorders. These alternatively spliced cancer-associated mucins can be used for the risk stratification, tumor grading, predicting response and resistance to therapy and prognosis. The cartoon is the pictorial demonstration of Svs specific to MUC4 and MUC1 with their designated and putative functions.
Cancer-specific changes in the reading frame or mutations generate unique amino acid sequences that can be targeted to specifically differentiate malignant phenotype from benign pathologies and inflammatory disorders. Concurrently, unique alternative splice junctional sequences in the mucin ectodomain can easily be targeted using radionuclide and toxin-tagged junction-specific antibodies for both imaging and therapeutic purposes. This has been demonstrated by targeting alpha/beta junctions of MUC1 by Pseudomonas toxin (PE38)-tagged DMC209 antibody in breast cancer (70). The unique sequences resulting from alternative spicing or mutations in mucin genes can potentially elicit an immune response in cancer patients. If the genetic variants occur early during tumorigenesis, it would be interest to test whether specific auto-antibodies directed against these unique sequences exist in patients and have potential as diagnostic or prognostic biomarkers. The unique sequences in the genetic variants of mucins can also be used to design vaccines to effectively break tolerance and develop Sv-specific immunotherapy approaches. Understanding of the expression pattern, mechanistic contribution and mechanism of mucins splicing will provide additional tools to develop novel therapeutic interventions for cancer and other pathologies. It is important to emphasize that our current understanding regarding the expression patterns of mucin Svs is based on transcript analysis and their existence and relative abundance at protein level remains to be determined in biological samples due to lack of specific antibodies. Similarly, the knowledge regarding the functional role of a handful of mucin Svs is based on ectopic overexpression studies. Therefore, extensive efforts are needed to develop antibodies and small interfering RNAs directed against unique sequences in the Svs and capable of distinguishing them from wild-type forms. Subsequently, it will also be of interest to develop genetically engineered mouse models of disease-relevant mucin Svs to determine their functional impact on pathobiology. Such reagents and models will not only provide deeper insight into the abundance and functional relevance mucin Sv proteins but also serve as a critical resource for the developing and testing diagnostic and therapeutic modalities targeting mucin Svs. Further, the absolute changes in the expression of cancer-associated mucins and increased Svs production may be under the diagnostic thresholds of detection. Thus, combined use of tissue-specific signatures of cancer-associated mucin Svs and profiling of the microsatellite instability may confer an additional diagnostic and prognostic benefit. Finally, long untranslated mucin Svs mRNA can function similar to long noncoding RNA and act as a scaffold for assembly of the multimeric protein complexes involved in the regulation of multiple cellular processes. Together, the unexplored world of mucin Svs provides a unique opportunity to comprehend their biological significance, utility as biomarkers and pathology-specific targeting.
Funding
National Institute of Health (R01 CA195586, R01 CA78590, EDRN U01 CA200466, EDRN U01 CA111294, R01 CA133774, R01 CA131944, SPORE P50 CA127297, T32 CA009476 and U54 TMEN CA163120).
Conflict of Interest Statement: None declared.
Abbreviations
- AMOP
adhesion-associated domain in MUC4 and other protein
- CT
cytoplasmic tail
- ECM
extracellular matrix
- EGF-like
epidermal growth factor-like
- mRNA
messenger RNA
- NIDO
Nidogen-like
- SEA
sperm protein enterokinase agrin
- SNP
single nucleotide polymorphism
- Svs
splice variants
- VNTR
variable number of tandem repeats
- vWD
von Willebrand Factor D
References
- 1. Chaturvedi P., et al. (2008) Structure, evolution, and biology of the MUC4 mucin. FASEB J., 22, 966–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaur S., et al. (2013) Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol. Hepatol., 10, 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joshi S., et al. (2015) Genetically engineered mucin mouse models for inflammation and cancer. Cancer Metastasis Rev., 34, 593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baruch A., et al. (1997) Preferential expression of novel MUC1 tumor antigen isoforms in human epithelial tumors and their tumor-potentiating function. Int. J. Cancer, 71, 741–749. [DOI] [PubMed] [Google Scholar]
- 5. Kufe D.W. (2009) Mucins in cancer: function, prognosis and therapy. Nat. Rev. Cancer, 9, 874–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rachagani S., et al. (2009) Current status of mucins in the diagnosis and therapy of cancer. Biofactors, 35, 509–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Joshi S., et al. (2014) Altered mucins (MUC) trafficking in benign and malignant conditions. Oncotarget, 5, 7272–7284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Paszek M.J., et al. (2014) The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature, 511, 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bork P., et al. (1995) The SEA module: a new extracellular domain associated with O-glycosylation. Protein Sci., 4, 1421–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ciccarelli F.D., et al. (2002) AMOP, a protein module alternatively spliced in cancer cells. Trends Biochem. Sci., 27, 113–115. [DOI] [PubMed] [Google Scholar]
- 11. Duraisamy S., et al. (2006) Distinct evolution of the human carcinoma-associated transmembrane mucins, MUC1, MUC4 AND MUC16. Gene, 373, 28–34. [DOI] [PubMed] [Google Scholar]
- 12. Maeda T., et al. (2004) Solution structure of the SEA domain from the murine homologue of ovarian cancer antigen CA125 (MUC16). J. Biol. Chem., 279, 13174–13182. [DOI] [PubMed] [Google Scholar]
- 13. James P.D., et al. (2011) von Willebrand disease. Genet. Med., 13, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kimura N., et al. (1998) Entactin-2: a new member of basement membrane protein with high homology to entactin/nidogen. Exp. Cell Res., 241, 36–45. [DOI] [PubMed] [Google Scholar]
- 15. Sasaki T., et al. (1998) Inhibition of glycosaminoglycan modification of perlecan domain I by site-directed mutagenesis changes protease sensitivity and laminin-1 binding activity. FEBS Lett., 435, 169–172. [DOI] [PubMed] [Google Scholar]
- 16. Senapati S., et al. (2012) Role of MUC4-NIDO domain in the MUC4-mediated metastasis of pancreatic cancer cells. Oncogene, 31, 3346–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Appella E., et al. (1988) Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett., 231, 1–4. [DOI] [PubMed] [Google Scholar]
- 18. Davis C.G. (1990) The many faces of epidermal growth factor repeats. New Biol., 2, 410–419. [PubMed] [Google Scholar]
- 19. Hommel U., et al. (1992) Human epidermal growth factor. High resolution solution structure and comparison with human transforming growth factor alpha. J. Mol. Biol., 227, 271–282. [DOI] [PubMed] [Google Scholar]
- 20. Crawley S.C., et al. (1999) Genomic organization and structure of the 3’ region of human MUC3: alternative splicing predicts membrane-bound and soluble forms of the mucin. Biochem. Biophys. Res. Commun., 263, 728–736. [DOI] [PubMed] [Google Scholar]
- 21. Maher D.M., et al. (2011) Mucin 13: structure, function, and potential roles in cancer pathogenesis. Mol. Cancer Res., 9, 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Das S., et al. (2015) Carboxyl-terminal domain of MUC16 imparts tumorigenic and metastatic functions through nuclear translocation of JAK2 to pancreatic cancer cells. Oncotarget, 6, 5772–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Coelho M.B., et al. (2014) Regulation of alternative pre-mRNA splicing. Methods Mol.Biol., 1126, 55–82. [DOI] [PubMed] [Google Scholar]
- 24. Chen M., et al. (2009) Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol., 10, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Germann S., et al. (2012) Splicing programs and cancer. J. Nucleic Acids, 2012, Article ID 269570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang J., et al. (2013) Misregulation of pre-mRNA alternative splicing in cancer. Cancer Discov., 3, 1228–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brayman M., et al. (2004) MUC1: a multifunctional cell surface component of reproductive tissue epithelia. Reprod. Biol. Endocrinol., 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nath S., et al. (2014) MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med., 20, 332–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang L., et al. (2013) Human mucin MUC1 RNA undergoes different types of alternative splicing resulting in multiple isoforms. Cancer Immunol. Immunother., 62, 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng L., et al. (2013) Functional polymorphism rs4072037 in MUC1 gene contributes to the susceptibility to gastric cancer: evidence from pooled 6,580 cases and 10,324 controls. Mol. Biol. Rep., 40, 5791–5796. [DOI] [PubMed] [Google Scholar]
- 31. Ng W., et al. (2008) Genetic regulation of MUC1 alternative splicing in human tissues. Br. J. Cancer, 99, 978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baruch A., et al. (1999) The breast cancer-associated MUC1 gene generates both a receptor and its cognate binding protein. Cancer Res., 59, 1552–1561. [PubMed] [Google Scholar]
- 33. den Dunnen J.T., et al. (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum. Mutat., 15, 7–12. [DOI] [PubMed] [Google Scholar]
- 34. Choudhury A., et al. (2001) Alternate splicing at the 3’-end of the human pancreatic tumor-associated mucin MUC4 cDNA. Teratog. Carcinog. Mutagen., 21, 83–96. [DOI] [PubMed] [Google Scholar]
- 35. Moniaux N., et al. (2000) Alternative splicing generates a family of putative secreted and membrane-associated MUC4 mucins. Eur. J. Biochem., 267, 4536–4544. [DOI] [PubMed] [Google Scholar]
- 36. Gamazon E.R., et al. (2014) Genomics of alternative splicing: evolution, development and pathophysiology. Hum.Genet., 133, 679–687. [DOI] [PubMed] [Google Scholar]
- 37. Escande F., et al. (2002) Genomic organization of MUC4 mucin gene. Towards the characterization of splice variants. Eur. J. Biochem., 269, 3637–3644. [DOI] [PubMed] [Google Scholar]
- 38. Sternberg L.R., et al. (2004) Alternative splicing of the human MUC2 gene. Arch. Biochem. Biophys., 421, 21–33. [DOI] [PubMed] [Google Scholar]
- 39. Hartman M., et al. (1999) MUC1 isoform specific monoclonal antibody 6E6/2 detects preferential expression of the novel MUC1/Y protein in breast and ovarian cancer. Int. J. Cancer, 82, 256–267. [DOI] [PubMed] [Google Scholar]
- 40. Zrihan-Licht S., et al. (1994) Characterization and molecular cloning of a novel MUC1 protein, devoid of tandem repeats, expressed in human breast cancer tissue. Eur. J. Biochem., 224, 787–795. [DOI] [PubMed] [Google Scholar]
- 41. Zaretsky J.Z., et al. (2006) MUC1 gene overexpressed in breast cancer: structure and transcriptional activity of the MUC1 promoter and role of estrogen receptor alpha (ERalpha) in regulation of the MUC1 gene expression. Mol. Cancer, 5, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hey N.A., et al. (2003) Transmembrane and truncated (SEC) isoforms of MUC1 in the human endometrium and Fallopian tube. Reprod. Biol. Endocrinol., 1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Obermair A., et al. (2002) Expression of MUC1 splice variants in benign and malignant ovarian tumours. Int. J. Cancer, 100, 166–171. [DOI] [PubMed] [Google Scholar]
- 44. Kannan K., et al. (2015) Aberrant MUC1-TRIM46-KRTCAP2 Chimeric RNAs in High-Grade Serous Ovarian Carcinoma. Cancers (Basel)., 7, 2083–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Passebosc-Faure K., et al. (2005) Evaluation of a panel of molecular markers for the diagnosis of malignant serous effusions. Clin. Cancer Res., 11, 6862–6867. [DOI] [PubMed] [Google Scholar]
- 46. Weiss M., et al. (1996) Preoperative diagnosis of thyroid papillary carcinoma by reverse transcriptase polymerase chain reaction of the MUC1 gene. Int. J. Cancer, 66, 55–59. [DOI] [PubMed] [Google Scholar]
- 47. Obermair A., et al. (2001) Novel MUC1 splice variants are expressed in cervical carcinoma. Gynecol. Oncol., 83, 343–347. [DOI] [PubMed] [Google Scholar]
- 48. Kahkhaie K.R., et al. (2014) Specific MUC1 splice variants are correlated with tumor progression in esophageal cancer. World J. Surg., 38, 2052–2057. [DOI] [PubMed] [Google Scholar]
- 49. Hinojosa-Kurtzberg A.M., et al. (2003) Novel MUC1 splice variants contribute to mucin overexpression in CFTR-deficient mice. Am. J. Physiol. Gastrointest. Liver Physiol., 284, G853–G862. [DOI] [PubMed] [Google Scholar]
- 50. Choudhury A., et al. (2000) Human MUC4 mucin cDNA and its variants in pancreatic carcinoma. J. Biochem., 128, 233–243. [DOI] [PubMed] [Google Scholar]
- 51. Xie K., et al. (2014) Upregulation of the splice variant MUC4/Y in the pancreatic cancer cell line MIA PaCa-2 potentiates proliferation and suppresses apoptosis: new insight into the presence of the transcript variant of MUC4. Oncol. Rep., 31, 2187–2194. [DOI] [PubMed] [Google Scholar]
- 52. Tang J., et al. (2016) The role of the AMOP domain in MUC4/Y-promoted tumour angiogenesis and metastasis in pancreatic cancer. J. Exp. Clin. Cancer Res., 35, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rivas M.A., et al. (2011) Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat.Genet., 43, 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Saeki N., et al. (2011) A functional single nucleotide polymorphism in mucin 1, at chromosome 1q22, determines susceptibility to diffuse-type gastric cancer. Gastroenterology, 140, 892–902. [DOI] [PubMed] [Google Scholar]
- 55. Kupcinskas J., et al. (2015) Common Genetic Variants of PSCA, MUC1 and PLCE1 Genes are not Associated with Colorectal Cancer. Asian Pac. J. Cancer Prev., 16, 6027–6032. [DOI] [PubMed] [Google Scholar]
- 56. Chang C.Y., et al. (2011) MUC4 gene polymorphisms associate with endometriosis development and endometriosis-related infertility. BMC Med., 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Williams K.A., et al. (2014) Polymorphisms of MUC16 (CA125) and MUC1 (CA15.3) in relation to ovarian cancer risk and survival. PLoS One, 9, e88334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bouanene H., et al. (2011) Polymorphisms in the MUC16 gene: potential implication in epithelial ovarian cancer. Pathol. Oncol. Res., 17, 295–299. [DOI] [PubMed] [Google Scholar]
- 59. Kumar V., et al. (2013) Genome-wide association study signal at the 12q12 locus for Crohn’s disease may represent associations with the MUC19 gene. Inflamm. Bowel Dis., 19, 1254–1259. [DOI] [PubMed] [Google Scholar]
- 60. Imbert-Fernandez Y., et al. (2011) MUC1/A and MUC1/B splice variants differentially regulate inflammatory cytokine expression. Exp. Eye Res., 93, 649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Imbert Y., et al. (2006) MUC1 splice variants in human ocular surface tissues: possible differences between dry eye patients and normal controls. Exp. Eye Res., 83, 493–501. [DOI] [PubMed] [Google Scholar]
- 62. Imbert Y., et al. (2009) MUC1 and estrogen receptor alpha gene polymorphisms in dry eye patients. Exp. Eye Res., 88, 334–338. [DOI] [PubMed] [Google Scholar]
- 63. Cascio S., et al. (2011) MUC1 protein expression in tumor cells regulates transcription of proinflammatory cytokines by forming a complex with nuclear factor-κB p65 and binding to cytokine promoters: importance of extracellular domain. J. Biol. Chem., 286, 42248–42256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Grosso J.F., et al. (2004) MUC1/sec-expressing tumors are rejected in vivo by a T cell-dependent mechanism and secrete high levels of CCL2. J. Immunol., 173, 1721–1730. [DOI] [PubMed] [Google Scholar]
- 65. Ilkovitch D., et al. (2013) Mechanisms of antitumor and immune-enhancing activities of MUC1/sec, a secreted form of mucin-1. Immunol. Res., 57, 70–80. [DOI] [PubMed] [Google Scholar]
- 66. Ilkovitch D., et al. (2009) Urokinase-mediated recruitment of myeloid-derived suppressor cells and their suppressive mechanisms are blocked by MUC1/sec. Blood, 113, 4729–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pastrello C., et al. (2005) MUC gene abnormalities in sporadic and hereditary mucinous colon cancers with microsatellite instability. Dis. Markers, 21, 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Conover W.L. (1999) Practical Nonparametric Statistics. 3rd edn. John Wiley and Sons, New York, NY, p. 584. [Google Scholar]
- 69. Kim N., et al. (2013) Somatic mutaome profile in human cancer tissues. Genomics Inform., 11, 239–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rubinstein D.B., et al. (2009) The MUC1 oncoprotein as a functional target: immunotoxin binding to alpha/beta junction mediates cell killing. Int. J. Cancer, 124, 46–54. [DOI] [PubMed] [Google Scholar]