Abstract
The digitization of biocollections is a critical task with direct implications for the global community who use the data for research and education. Recent innovations to involve citizen scientists in digitization increase awareness of the value of biodiversity specimens; advance science, technology, engineering, and math literacy; and build sustainability for digitization. In support of these activities, we launched the first global citizen-science event focused on the digitization of biodiversity specimens: Worldwide Engagement for Digitizing Biocollections (WeDigBio). During the inaugural 2015 event, 21 sites hosted events where citizen scientists transcribed specimen labels via online platforms (DigiVol, Les Herbonautes, Notes from Nature, the Smithsonian Institution's Transcription Center, and Symbiota). Many citizen scientists also contributed off-site. In total, thousands of citizen scientists around the world completed over 50,000 transcription tasks. Here, we present the process of organizing an international citizen-science event, an analysis of the event's effectiveness, and future directions—content now foundational to the growing WeDigBio event.
Keywords: biodiversity informatics, biodiversity research collections, citizen science, crowdsourcing, natural history collections
Biodiversity collections (“biocollections”) are invaluable to society. They provide the data crucial to investigating climate and other environmental changes (e.g., Labay et al. 2011, Robbirt et al. 2011, Lavoie 2013), conservation biology (e.g., Gaubert et al. 2006, Swenson et al. 2012, Scheper et al. 2014), population genetics and genomics (e.g., Wandeler et al. 2007, Bi et al. 2013, Holmes et al. 2016), and even public health and safety (Suarez and Tsutsui 2004, Pinto et al. 2010). However, the majority of biocollection specimen data remain difficult to access, locked in the cabinets of museum and university collections in analog format, presenting the biocollections community with many years of digitization work (Page et al. 2015). Digitization typically involves curation, imaging, image processing, the electronic capture of label and ledger data, and georeferencing (Nelson et al. 2012), all of which require people power and other resources. Recent funding at local, national, and international scales has provided institutions the ability to hire digitization technicians (AIBS 2013), but the workload is greater than what can be readily accomplished with current funding and technologies. Public participation has the potential to advance digitization and has the additional benefits of improving science literacy among contributors, community support for biocollections, and the sustainability of digitization activities (Ellwood et al. 2015). In October 2015, we piloted the Worldwide Engagement for Digitizing Biocollections event (WeDigBio 2015) to mobilize citizen scientists for biocollection digitization and provide the biocollections community with a large-scale education and outreach opportunity. In October of the following years, we organized WeDigBio 2016 and 2017. Here, we document the process of organizing a citizen-science event of this scale, assess the event's success, convey the lessons learned, and discuss the future directions of WeDigBio as an annual event.
WeDigBio emerged from the December 2014 CITStitch Hackathon at iDigBio, the National Science Foundation's National Resource for Advancing the Digitization of Biodiversity Collections. The goal of the 24-person CITStitch Hackathon was to build the interoperability among projects that enables public participation in the digitization of biodiversity research specimens in useful and exciting ways (Mast et al. 2014). We decided to combine our efforts to produce a global event with a big outreach push in the biocollections and citizen-science communities as a crucial step for the community, leading toward greater efficiencies and effectiveness. Biodiversity citizen-science projects with somewhat similar goals, such as National Geographic's Great Nature Project, the National Audubon Society's Christmas Bird Count (Dunn et al. 2005), BioBlitzes (Lundmark 2003) and Personal BioBlitzes (Pollock et al. 2015), and eBird's Global Big Day (www.ebird.org/globalbigday), had already demonstrated past success with comparable models that integrate on-site (e.g., in the field) and online (e.g., at a computer or with a mobile app) activities (table 1). However, ours is the first project of global scale to create data about historically collected biodiversity specimens rather than create observations. This distinction is often made when assessing fitness for use in downstream research, because specimen data are more rigorously verifiable.
Table 1.
Comparison of international biodiversity-related citizen-science projects that have at least some on-site component.
| Event | Short description | Frequency and year established | Geographic range | Number of participants and/or completed tasks | Online or on-site | References |
|---|---|---|---|---|---|---|
| BioBlitzes | Survey all living species within an area over a set amount of time (usually 24 hours). Unlike the other projects featured in this table, BioBlitzes are not managed by a parent organization and any group can organize a “BioBlitz.” | Varied, depending on host; 1996 | Global | Unknown. Numbers vary with each BioBlitz and without a governing body there are no summative data. | On-site | www.pwrc.usgs.gov/blitz.html, www.wikipedia.org/wiki/BioBlitz |
| eBird's Global Big Day | Record the number of bird species seen on 1 day and upload checklists to Cornell Lab of Ornithology's eBird. | Annual; 2015 | Global | 16,679 participants, 6307 species, 145 countries during 2016 event. | Main activities on-site with results shared online | www.ebird.org/ebird/globalbigday |
| National Audubon Society's Christmas Bird Count | Census of birds within a 24-km-diameter area completed by groups of at least 10 volunteers | Annual, on 1 day between 14 December and 5 January; 1900 | Western Hemisphere | 72,653 observers, 2106 species from 24 countries during 2015 event | On-site | www.audubon.org/conservation/science/christmas-bird-count |
| National Geographic's Great Nature Project | Document biodiversity around you by uploading photos of plants and animals to the project website. | Two events (2013 and 2015); 2013 (no longer active). | Global. 102 countries during 2015 event. | 40,396 observations of 8000 species, from over 3000 users, from 102 countries during the second and final 2015 event | Main activities on-site with results shared online at iNaturalist | www.greatnatureproject.org; www.voices.nationalgeographic.com/2015/06/04/40000-observations-of-biodiversity-in-11-days |
| WeDigBio | Transcribe biocollections information using online platforms | Annual; 2015 | Global | 51,822 transcription tasks from users in over 100 countries during 2015 event | Main activities online with activities available at on-site locations | www.wedigbio.org; present study |
Note: For the purposes of this research, we have defined “on-site” as “at the physical location of the data source” and generally include field sites outdoors, as well as indoor venues such as museums and universities. Values provided are most current available data.
Because of the relative maturity of online transcription platforms, the inaugural WeDigBio event focused on the transcription step of the digitization process. Transcription platforms include DigiVol (supported by the Australian Museum and the Atlas of Living Australia; https://volunteer.ala.org.au), Les Herbonautes (supported by the Muséum National d’Histoire Naturelle, Paris, France; http://lesherbonautes.mnhn.fr), the Smithsonian Institution's Transcription Center (SITC; https://transcription.si.edu), Notes from Nature (part of the Zooniverse suite of projects; www.notesfromnature.org), and the Symbiota crowdsourcing module (www.symbiota.org). The goal of each of these platforms is the same: to facilitate volunteer transcription of biodiversity specimen information from digital images that include specimen labels. However, their approaches vary (reviewed by Ellwood et al. 2015) in ways that became relevant to interpreting WeDigBio event statistics. For example, DigiVol, Les Herbonautes, SITC, and Symbiota employ an approach in which one volunteer transcribes and then a second validates the transcription (Rouhan et al. 2014). In contrast, Notes from Nature asks three different volunteers to transcribe a specimen, and then the transcriptions are reconciled using Notes from Nature tools to derive a final output (Matsunaga et al. 2016; see also www.github.com/juliema/label_reconciliations).
Online citizen-science projects, such as volunteer transcription platforms, have several known strengths and weaknesses (Lukyanenko et al. 2011, Newman et al. 2012). A tremendous strength of online projects is the ability to reach an Internet-scale audience. Anyone with a computer and an Internet connection can participate. This is invaluable to researchers who require assistance in collecting or processing large amounts of data. It is also beneficial to volunteers, who can learn about and make contributions to scientific research without spending money and time for transportation (Simpson et al. 2012, Price and Lee 2013, Hennon et al. 2015). Research projects from a diversity of scientific and humanities disciplines, including human biology (e.g., EyeWire, www.eyewire.org; Foldit, www.fold.it), astronomy (e.g., Globe at Night, www.globeatnight.org; Agent Exoplanet, www.lcogt.net/agentexoplanet), archaeology (e.g., Micropasts, www.micropasts.org), and history (e.g., National Libraries of Israel, www.nlics.org), have successfully engaged citizen scientists in online research activities.
A potential weakness of online citizen-science projects, compared with on-site projects at an institution or field site, is a lack of connection between researchers and volunteers or between volunteers and the local biocollection or natural ecosystem, which may lessen volunteer motivation. Furthermore, researchers are unable to provide personalized training to volunteers, and volunteers may feel isolated by working alone. Designers of online projects have attempted to address these limitations by integrating social features (Jackson et al. 2015). For example, some projects have forums in which participants and researchers can learn about the platform, communicate, ask questions, and share noteworthy items from their online work (e.g., groups.google.com/d/forum/inaturalist and https://forum.ispotnature.org). Three of the WeDigBio transcription platforms maintain forums as well: www.zooniverse.org/projects/zooniverse/notes-from-nature/talk, www.volunteer.ala.org.au/forum, and www.lesherbonautes.mnhn.fr/discussions/all. These tools function to help build and maintain communities of scientists, developers, and volunteers.
In creating the WeDigBio event, we viewed bringing citizen scientists together to participate in online activities as a logical step in building volunteer communities. In this scenario, the participants could either directly participate in or remotely interact with on-site activities at museums, universities, classrooms, etc. On-site events provide richer engagement opportunities, such as interactions with scientists and specimens, and may build stronger links with local communities and biocollections. Hybrid events that merged online specimen digitization with on-site activities had already been piloted by a few of the planners of the WeDigBio event at Florida State University, the Smithsonian Institution, Valdosta State University, and the Australian Museum. From these events, the importance of media attention for recruiting participants became clear, as did the opportunity to speak to scientists and the value of games that gave participants a reason to think more deeply about specimen information and the aggregate picture that was forming as they created digital biodiversity data.
The inaugural WeDigBio 2015 event occurred Thursday, 22 October–Sunday, 25 October, and subsequent events in 2016 and 2017 occurred over similar 4-day periods in October. These dates made it possible for activities to take place on weekdays in formal education settings and on the weekend in informal education settings or at home. In this paper, we ask the following questions about WeDigBio 2015: (a) Did WeDigBio 2015 result in increased transcription activity over its 4 days? Were there noticeable spikes in transcription activity during on-site events? Did transcription activity “stick” by increasing the rate of transcription 1 month after the event as compared with 1 month before the event? (b) Did the event broaden online engagement at the transcription platforms, as was measured, for example, by numbers of new registrations and countries to which IP addresses can be mapped? (c) Did on-site events increase public understanding of the value of biocollections? Which elements did the event participants consider to be most important to their overall on-site experience? (d) What resources were expended to host on-site events, and are these opportunity costs offset by the transcription activity and increased understanding by citizen scientists? Answers to these questions will help shape WeDigBio as an annual event, establishing a place for biocollection digitization on the global citizen-science and biocollections calendars. Because we would also like to see it serve as a model for other communities that are interested in building hybrid online and on-site citizen-science events, we document here the process of planning WeDigBio, the lessons learned, and our thoughts on future directions for the event.
Preparation
The WeDigBio Event was organized over the course of several meetings, especially the CITStitch Hackathon in December 2014 (Mast et al. 2014) and the WeDigBio planning workshop at the Smithsonian Institution in March 2015 (Mast et al. 2015). We worked with a graphic designer, Jeremy Spinks at Jelly Bean Communications Design, to create a logo (figure 1). WeDigBio organizers participated in one or more of four working groups that persisted for the duration of event planning: (1) website, (2) materials and resources, (3) evaluation and statistics, and (4) recruitment and advertisement.
Figure 1.
The WeDigBio logo. This image was used on official WeDigBio documents, as well as for stickers and temporary tattoos that were distributed at on-site events. In 2016, augmented reality features were added such that a praying mantis popped up from the logo when viewed through the Libraries of Life app (www.libraries-of-life.org).
Website working group
This working group produced the website www.wedigbio.org to communicate the goals of WeDigBio, direct citizen scientists to online projects and on-site events in which they could participate, make logistical and educational resources available for event hosts, and provide a dynamic dashboard displaying digitization activity during the event (figure 2). A content management system, Drupal 7.x, deployed on a Linux/Apache/MySQL/PHP host, served this content to site visitors. A custom theme and dashboard elements were developed using different visualization components, including the D3 JavaScript plotting library and the web-mapping tools from Carto (www.carto.com).
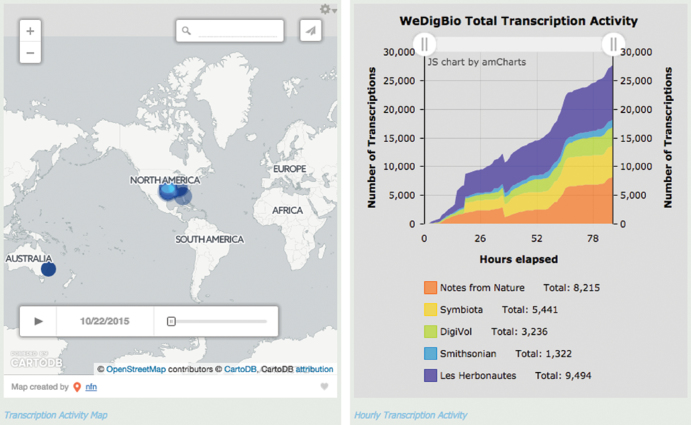
Figure 2.
A screenshot of the www.wedigbio.org dashboard during WeDigBio 2015. The image on the left shows the approximate location of the transcriber, as was determined by IP address. The image on the right shows the tally of transcriptions, by platform, as time elapsed during the event. This screenshot was taken before the end of the event and as such does not reflect the final transcription tallies. Furthermore, the approximate counts and errors in the display of these preliminary results were addressed in later aggregation of the data for analysis in the present research. For example, the number of transcriptions shown for the Smithsonian is completed transcriptions (i.e., those that have been transcribed by one or more of the participants and also reviewed). The comparable SITC data in figure 3 include transcriptions that were still in process.
Materials and resources working group
This working group produced content for the on-site event hosts. The group created planning documents, example games, a lesson plan for undergraduate classes, and a press kit that on-site public event organizers could use to recruit and advertise in newsletters, organizational communications, and local media. The first three items relied heavily on the group's experience with single-institution on-site events held at Florida State University, the Smithsonian Institution, Valdosta State University, and the Australian Museum.
Evaluation working group
The working group produced a 24-question survey for the on-site participants (supplemental appendix S1) and an 8-question survey for the on-site event hosts (supplemental appendix S2). The surveys for the on-site participants assessed their motivations and enjoyment and sought feedback for improvement. The host surveys focused on determining what resources were required for the event, the hosts’ impressions on which aspects of events were most successful, and what lessons they learned through hosting an event.
Recruitment and advertisement working group
This working group recruited on-site event hosts and citizen scientists for the WeDigBio event. The on-site event hosts were recruited through posts on domain-relevant listservs, talks at professional conferences, and phone calls to colleagues. Social media was the primary tool to recruit participants because it has proven to be especially useful for organizing targeted campaigns focused on a particular goal, such as completing a set of transcriptions tailored to specific interests or related to activities outside of the platform itself (Parilla and Ferriter 2016).
Evaluation
In total, 21 on-site events were held in 2015. Three of these occurred in formal education settings on Thursday or Friday (Cornerstone Learning Community's middle-school science classes, Florida State University's Field Botany class, and University of Florida's Plant Taxonomy class), and 18 of these were held on Saturday and Sunday in informal education settings at museums (e.g., The Field Museum, the Smithsonian's National Museum of Natural History, and the Australian Museum) and universities (e.g., Yale University and the University of California, Berkeley; supplemental appendix S3).
Transcription activity
We tested whether transcription activity increased during WeDigBio 2015 as compared with before it and whether activity persisted above pre-event baselines. Given that the participants could contribute from any of the world's time zones, we considered transcriptions that occurred during the event to include time periods when it was 22–25 October anywhere in the world (i.e., transcriptions occurring between 12:00 p.m. UTC on 21 October through 11:59 a.m. UTC on 26 October). We will refer to this time period as during or during the event. We considered the before time period to be an identical length of time 4 weeks prior to the event (i.e., 12:00 p.m. UTC on 23 September through 11:59 a.m. UTC on 28 September) and the after time period to be 4 weeks following the event (i.e., 12:00 p.m. UTC on 18 November through 11:59 a.m .UTC on 23 November).
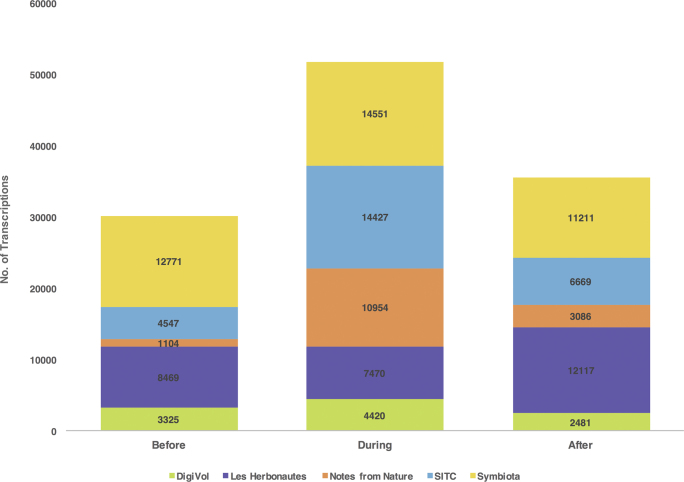
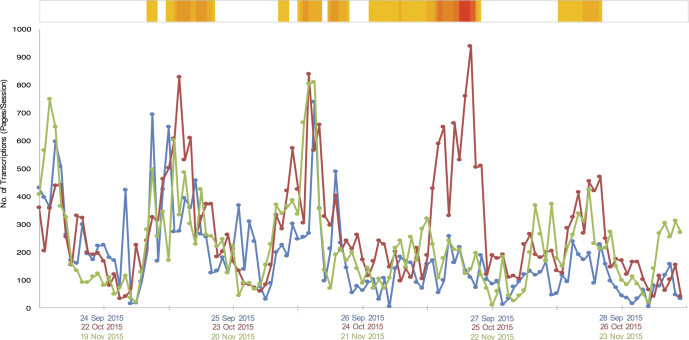
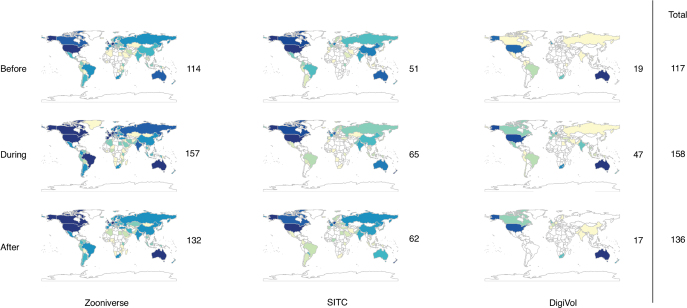
The total number of transcriptions completed across the five platforms increased by 72% during WeDigBio 2015, from 30,216 (the before time period) to 51,822 (figure 3). Much of this increase of 21,606 transcription tasks occurred on Saturday, when most on-site events occurred (figure 4). Specifically, between 12:00 p.m. and 11:00 p.m. UTC on Saturday, transcription activity increased more than 3.5 times (362%), highlighting the value of the on-site events to the overall success of the WeDigBio event. We suspect that these not only led to contributions by those in attendance on-site but also created an exciting social media environment for those participating individually off-site. We observed increases in transcription rates for each of the platforms individually as well, with the exception of Les Herbonautes (figure 3). The greatest hourly transcription rates during WeDigBio occurred on Saturday (8:00 p.m.–8:59 p.m. UTC, 937 transcriptions), Friday (2:00 p.m.–2:59 p.m. UTC, 836 transcriptions), and Thursday (2:00 p.m.–2:59 p.m. UTC, 827 transcriptions). Peak activity for the before and after time periods proved to be similar to the during time period, with the striking exception of the Saturday spike, which was conspicuously absent outside of the WeDigBio event (figure 4). The number of countries from which participants engaged in the activity increased with the WeDigBio event (figure 5) but also far exceeded the small number of countries (4) in which on-site events were available. Many of the IP addresses mapped to Europe and North America, as did nearly all on-site events (figure 5 and appendix S3).
Figure 3.
The total number of transcription tasks completed on each online platform in the before, during, and after time periods of the event. In the case of SITC, the values include transcriptions that were still in process, such as those that have been transcribed but not yet reviewed.
Figure 4.
The summed total of hourly transcription tasks for all platforms before (blue), during (red), and after (green) the event. The transcription counts are based on the respective way each platform calculates tasks (as we described in “Methods”), with the exception of SITC. The transcription rates were not available at the hourly scale through SITC; therefore, the Google Analytics statistic of pages per session was used as a proxy. The pages per session reflects how active a participant was on the website and is therefore the closest approximation available in the absence of hourly transcription data. The heat bar at the top reflects the number of on-site events that were taking place during the event, ranging from one event (yellow) to a maximum of eight events (red). All the submission times have been normalized to UTC. Dates on the x-axis are at midnight on the given date for each period of time: before (the top date on the x-axis label), during (the middle date on the label), and after (the bottom date on the label). The tick marks in between each date therefore represent noon on the given date.
Figure 5.
Transcription activity by country for Zooniverse projects (left panel), SITC projects (center panel), and DigiVol (right panel) for before (top row), during (middle row), and after (bottom row). Lower activity is shown in lighter shades and higher activity in darker shades. The numbers to the right of each map indicate the number of countries with participants during each time period for each transcription platform. The numbers in the far right column are the total number of unique countries for each time period.
We expected that the higher rates of productivity and engagement reached during the WeDigBio event would not be sustainable after the event, but we were pleased to discover some “stickiness” to the transcription platforms. When we compared the values of each of our metrics 4 weeks after the WeDigBio event to 4 weeks prior to it, all of them had increased. For example, the total number of transcription tasks rose 18% from the before time period to the after time period (35,564 transcriptions). There were no significant rollouts of other projects at those platforms just before or during our after time period to complicate our interpretation of the data. Further evidence of “stickiness” continues to emerge, such as the successes of The Field Museum's new 70-member Collections Club and the WeDigFLPlants collaboration between Florida herbaria and local amateur naturalist groups, both direct spin-offs of WeDigBio activities.
WeDigBio 2016 saw a doubling in the number of on-site events, and results point to modest gains in transcriptions. Because of the changes described below in how transcriptions are standardized and counted, the transcription counts in 2016 will represent the new baseline of activity for subsequent years.
Online engagement
We tallied the number of new-user registrations for each time period for all transcription platforms except Les Herbonautes and Symbiota, which could not provide these data. Not all of the platforms require registration to participate. For example, Notes from Nature and Les Herbonautes do not require registration but provide incentives (e.g., virtual badges or personal tallies of completed tasks) to encourage it. Volunteers with SITC may transcribe without registering, but they must register in order to be reviewers of transcriptions. All DigiVol and Symbiota volunteers are required to register.
We used Google Analytics to assess whether the event broadened the engagement of the online participants. The platforms using Google Analytics were DigiVol, SITC (which includes all SITC projects, not just those focused on biocollections), and Zooniverse (which includes all projects, not just Notes from Nature). We examined the total number of sessions (i.e., visits to a website) and the number of countries to which IP addresses could be mapped to determine whether the WeDigBio event led to more extensive online engagement than the before and after time periods. Google Analytics tallies the number of sessions and site visitors using slightly different metrics. For this reason, the total numbers of sessions and visitors are different.
The total new-user registrations for all platforms combined increased from 1479 new registrations in the before time period to 2629 during WeDigBio 2015, an increase of 82%. The number of new registrations in the after time period (1996) remained higher than the before time period. For those transcription platforms that could provide the data, the total number of sessions during the WeDigBio event (11,310) was almost four times greater than that of the before period (2990; table 2). After the event, the total number of sessions was double that of the before period (6101). In total, visitors from 158 countries visited DigiVol, SITC, and/or Zooniverse during the event (figure 5). The majority of visitors were from the United States (32,095), the United Kingdom (6986), Germany (4342), Canada (3473), and Australia (3149). Across platforms, there was an increase in the number of countries represented during the event compared with those of the before and after time periods (figure 5). Before the event, www.wedigbio.org had 66 sessions from 15 countries. During the event, there were 1843 sessions from 61 countries, and after the event, there were 212 sessions from 12 countries.
Table 2.
The number of sessions and average session duration (from Google Analytics) and new registrations for DigiVol, Notes from Nature (*inclusive of all Zooniverse projects), SITC (*inclusive of all SITC projects), and Symbiota.
| Transcription platform | Number of sessions | Average session duration | New registrations | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Before | During | After | Before | During | After | Before | During | After | |
| DigiVol | 806 | 1476 | 534 | 0:21:30 | 0:16:46 | 0:23:16 | 15 | 70 | 11 |
| Notes from Nature* | 199 | 6780 | 2983 | 0:05:55 | 0:05:47 | 0:06:28 | 1435 | 2449 | 1944 |
| SITC* | 796 | 1600 | 1364 | 0:06:58 | 0:14:50 | 0:15:53 | 22 | 61 | 33 |
| Symbiota | 1189 | 1454 | 1220 | n/a | n/a | n/a | 17 | 49 | 8 |
| Total | 2990 | 11,310 | 6101 | 1479 | 2629 | 1996 |
Note: Full descriptions of the metrics used can be found in the “Evaluation” section.
Surveys
At the end of on-site events, the volunteers were provided an anonymous link to a survey. To preserve anonymity, the volunteers were not asked to provide names, and no other identifiable information (e.g., email address) was collected. Qualtrics Survey Software was used for the surveys prepared by the evaluation working group.
One hundred thirty-nine participants at 12 on-site events completed the survey (appendix S1). A majority of the respondents reported a “higher” or “much higher” level of awareness about the number (67%), kinds (62%), and value (70%) of biodiversity specimens held in the collections. Three-quarters (74%) reported a “higher” or “much higher” awareness of the process of transcribing specimen labels. The motivations for participating were varied and included enjoyment or a personal interest in biodiversity (30%), a desire to help the scientific community and/or the host institution (32%), fulfilling a class requirement (32%), and an interest in volunteering (6%). The participants viewed lectures and collection tours as more important to their overall experience, on average, than games and take-home gifts (table 3).
Table 3.
The results of an event participant survey showing their ratings of how important various activities were to their enjoyment of the event.
| Very unimportant (1) | Unimportant (2) | Neither important nor unimportant (3) | Important (4) | Very important (5) | Total number of participants offered this activity | Average response | |
|---|---|---|---|---|---|---|---|
| Lecture | 2 (1.9%) | 0 (0%) | 10 (9.4%) | 42 (39.6%) | 52 (49.1%) | 106 | 4.35 |
| Collection tour | 7 (7.9%) | 0 (0%) | 8 (9.0%) | 22 (24.7%) | 52 (58.4%) | 89 | 4.26 |
| GeoLocator or timeline games | 4 (5.8%) | 0 (0%) | 18 (26.1%) | 32 (46.4%) | 15 (21.75%) | 69 | 3.78 |
| Bingo game | 6 (6.5%) | 6 (6.5%) | 35 (38%) | 30 (32.6%) | 15 (16.3%) | 92 | 3.46 |
| Take-home item | 8 (7.4%) | 9 (8.3%) | 35 (32.4%) | 32 (29.6%) | 24 (22.2%) | 108 | 3.51 |
The vast majority of the participants responded with “agree” or “strongly agree” to the statement, “The blitz was worth my time” (92%; table 4). They responded similarly when asked whether biodiversity research collections merit public funding (91%) and whether they would participate in another transcription blitz (87%).
Table 4.
The results of the event participant survey showing the degree to which participants valued the on-site event (the “blitz”), as well as volunteering and biodiversity collections more broadly.
| Strongly disagree (1) | Disagree (2) | Neither agree nor disagree (3) | Agree (4) | Strongly agree (5) | Total number of respondents | Average response | |
|---|---|---|---|---|---|---|---|
| The blitz was worth my time. | 2 (1%) | 2 (1%) | 6 (4%) | 56 (41%) | 70 (51%) | 136 | 4.40 |
| Time was appropriately distributed among different blitz activities. | 2 (1%) | 2 (1%) | 26 (19%) | 70 (51%) | 37 (27%) | 137 | 4.01 |
| Biodiversity research collections merit public funding. | 2 (1%) | 0 (0%) | 11 (8%) | 45 (33%) | 80 (58%) | 138 | 4.46 |
| How likely is it that you would participate in a transcription blitz in the future if given the chance? | 2 (1%) | 1 (1%) | 14 (10%) | 50 (36%) | 71 (51%) | 138 | 4.36 |
| How likely is it that you would volunteer to transcribe specimen labels on a regular basis? | 5 (4%) | 12 (9%) | 34 (25%) | 51 (37%) | 35 (26%) | 137 | 3.72 |
| How likely is it that you would volunteer to work in biodiversity collections to perform other tasks if given the opportunity? | 3 (4%) | 6 (4%) | 21 (15%) | 51 (38%) | 55 (40%) | 136 | 4.10 |
Note: The values shown represent the number (and percentage) of people who rated each statement.
Although the off-site participants were not offered activities per se and did not complete surveys, they were encouraged to follow the event online via social media and video feeds, such as those offered by the Smithsonian Institution. In planning WeDigBio 2016, we conferenced with an off-site power contributor who felt that her ability to watch a video lecture from a researcher, in this case Seán Brady discussing bee research at the Smithsonian Institution (https://youtu.be/odM3UDtOl8Q), improved her connection to the SITC project and to the WeDigBio event.
Ten of the 18 on-site event hosts completed a post-event survey (appendix S2). Most of the hosts spent the greatest amount of time on scientific tasks, such as curating specimens, barcoding, and updating labels (an average of 5.7 hours) and marketing or publicity (an average of 5.2 hours). Training and volunteer management was a close third (an average of 4.2 hours), with logistics (e.g., staffing a front desk or helping with food), IT support, and security falling in at fourth, fifth, and sixth for time consumption. An average of 20.2 hours were spent by the hosts for each event. The greatest number of individuals was involved in training and volunteer management, with events averaging three people involved in this task. Scientific tasks were completed by, on average, 2.9 people per event. The remaining tasks each involved approximately one individual.
Opportunity cost assessment
As we seek to grow the number of WeDigBio on-site events and as scientists seek to justify their participation to administrators and funding agencies, the topic of opportunity costs becomes quite important. Engaging the public in the process of science generally has opportunity costs for both citizen and professional scientists (Tulloch et al. 2013, Thornhill et al. 2016). For citizens, there are time commitments and often monetary costs for transportation and appropriate materials (e.g., binoculars, computer, and field clothing). For professional scientists, the time and monetary commitments can be significant in planning, conducting, and sustaining citizen-science activities (Tulloch et al. 2013). Like the citizen scientists participating in WeDigBio 2015, the on-site event hosts (teachers, museum education and outreach staff, and biocollections curators) were motivated by a range of factors. Because the support of on-site event hosts is crucial to the future of WeDigBio, we focus here on their opportunity costs. This opportunity cost is mainly other work that could have been completed had they opted not to participate in the WeDigBio event, because the costs of incentive gifts and refreshments were quite modest (or free in the case of those providing WeDigBio stickers and tattoos as incentives).
We view the most straightforward comparison to be between the number of specimens event organizers could have transcribed themselves in the time taken to prepare for the on-site event and the number transcribed by citizen scientists during the event. If the average preparation time for an on-site event is 20.2 hours and we assume that rates of transcription over a long period average 2 minutes per transcription for biocollections staff, then the on-site host will break even, on average, at 606 transcriptions. We did not track this individually for each event host, but the numbers suggest that this would have typically been reached. If the 21,606 extra transcriptions generated during the WeDigBio event were evenly distributed among the 17 institutions involved in hosting an on-site event with images from their own biocollection (appendix S3), there would have been, on average, 1271 additional transcriptions per biocollection. Although these citizen-scientist transcriptions would need validation either by other citizen scientists or staff, the contribution, as we further elucidate below, is substantial.
This calculation argues for the participation of biocollections in WeDigBio, but we should also point out some of the complexities that it ignores. The actual number of biocollections with specimens on an online transcription platform (17) was lower than the total participating, with the remaining biocollections being motivated to participate by factors related to education and outreach goals. Those goals, including the creation of strong local support and a volunteer base for biocollections, insight into career paths for students, general publicity, and building capacity for maintaining digitization activities outside of sponsored research grants, could be at least as compelling to biocollections curators as the actual number of additional transcriptions completed during the short WeDigBio event. The math also does not consider the fact that many of the on-site event coordinators serve as supervisors of digitizing technicians, with research, education, and outreach activities more often in their assignments of responsibilities than digitization itself. That is, those organizing the on-site events often would not themselves be digitizing specimens with the time that they would otherwise have available if not organizing an event. Further, the math completely ignores the lingering boost in digitization rates that appear to have followed the WeDigBio event. Finally, in our experience, preparation time for on-site events drops significantly—by one-third to one-half—with the second on-site event, making the break-even point lower for returning hosts at future WeDigBio events.
Challenges and future directions
We recognize that annual preparation for a WeDigBio event could catalyze significant innovations in the areas of standardization and engagement.
Standardization
Standardization in reporting progress across transcription platforms emerged as an important topic and has implications for, for example, assessing alternative engagement strategies. In 2015, the lack of a common understanding for a unit of progress (transcription of a single field versus all fields for a specimen label versus all fields plus validation) had significant implications for the interpretation of progress toward eventwide goals and comparisons across platforms. This is unlikely to be an issue for the projects in table 1 that employ a single eventwide data-entry form. In our instance, it was more of a consensus-building challenge after WeDigBio 2015 than a technical challenge, and the platforms have since agreed that a single pass at transcribing or proofreading all targeted fields from a specimen's label(s) will equal one unit of activity for future WeDigBio counting purposes. For example, in Notes from Nature, which requires three transcriptions of a specimen's label content before a specimen is considered complete, each transcription now counts as one unit, so a completed specimen counts as three units. Similarly, a transcription and a validation in SITC count as two units. During WeDigBio 2016, we piloted this method of counting standardization with most of the transcription platforms and continue to use it. Because counting standardizations across platforms were not applied to the WeDigBio 2015 data reported here, it is best to focus on each platform's before, during, and after periods in turn rather than comparing activity among platforms.
A second area where standardization is expected to benefit WeDigBio in the long term involves sharing information about transcription projects (e.g., WeDigFLPlants) among www.wedigbio.org and the go-to sites for learning about available citizen-science projects (e.g., https://scistarter.org). We are building the capacity to consume information about relevant digitization projects in the current version of the PPSR CORE standard of the Citizen Science Association (www.citizenscience.org/2015/10/09/ppsr_core-metadata-standard), after which we can ask transcription-project creators to manage their project descriptions at a site such as https://scistarter.org. Then, www.wedigbio.org would make application programming interface calls to https://scistarter.org for current information rather than asking project managers to maintain their information as up to date at both sites. This should lead to greater scalability in terms of transcription project numbers and, potentially, online sites on which one might advertise the projects.
Engagement
Video conferencing can enable on-site event hosts to broadcast to off-site participants the lectures and collection tours that the on-site participants found most important (appendix S1). This was piloted during WeDigBio 2015, when two middle-school science classrooms at Cornerstone Learning Community (in Tallahassee, Florida) received virtual tours of the Smithsonian's National Museum of Natural History (in Washington, DC) using Adobe Connect. In 2016, we expanded this concept using Sococo online software (www.sococo.com), which allowed us to simultaneously host numerous video-conference rooms in the same virtual space. A quarter of the on-site events experimented with this in 2016, and we are eager to provide more research talks, improved scheduling of video participation, and intersite transcription games for a richer online experience in future years.
The individual transcription projects at each platform were somewhat heterogeneous: One could have transcribed specimen labels of insects, spiders, plants, and marine invertebrates. We welcome this heterogeneity, because it caters to the diversity of hosts and citizen-scientist interests and broadens the impact of the event. However, we plan to experiment in the future with rotating research themes that might highlight the relatedness of multiple digitization projects for citizen scientists and (especially) the media. Themes could focus on a grand scientific challenge (e.g., climate change or invasive species) or provide researchers with a focused data set to complete analyses, such as risk assessments for vector-borne diseases. Focusing on a particular research topic has the potential to mobilize large amounts of research-ready data in a short amount of time. A novel application of these data would be for event organizers to draft a skeletal manuscript ahead of WeDigBio and then use the data to complete an analysis and write-up of research findings immediately following the event, with rapid publication and recognition of citizen-science contributors. Researchers have made considerable use of data produced by other international citizen-science projects, such as the National Audubon Society's Christmas Count (Dunn et al. 2005 and references therein) and eBird (Sullivan et al. 2009). A specimen-to-publication model made possible through WeDigBio (similar to the rapid publication that resulted from a BioBlitz and work of international taxonomic experts; Telfer et al. 2015) would go far to demonstrate the immediate need for biocollections data and the whole of the scientific process.
We would also like to help the biocollections community build its capacity to sustain high rates of digitization beyond the WeDigBio event by supporting the establishment of interest (dubbed WeDigInterest) groups. These would represent partnerships with potentially large organizations whose memberships are motivated to digitize subsets of biocollection specimens. The new Biospex platform (https://biospex.org) is partnering with WeDigBio to provide tools for this type of sustained campaign of transcription projects and events (e.g., https://biospex.org/project/wedigflplants). This strategy is not unlike that of eBird, which promotes Global Big Day as an annual event, and other smaller-scale monthly challenges, project highlights, and timely research projects (ebird.org).
Outreach to new countries, biocollections, and classrooms is underway. We welcome participation from online digitization projects around the world and are prioritizing international partnerships in our outreach and recruitment efforts. Resources such as multilingual website pages and materials, a greater variety of lesson plans, and year-round support for interest groups are being discussed with the goal that hosts and participants around the world have the necessary tools to successfully participate in WeDigBio.
Conclusions
We consider the inaugural WeDigBio 2015 to have been a success by each of our measures of productivity, engagement, satisfaction, and opportunity costs. In particular, we were impressed by the “stickiness” of the event—the extent to which online digitization was enhanced after the event ended, both by increased online participation and elevated on-site interest. WeDigBio appears promising as both an annual event for the global citizen-science and biocollections calendars and as a framework for greater collaboration across the citizen-science platforms for the biocollections digitization domain. Likewise, we are enthusiastic to contribute to established citizen-science events, such as Citizen Science Day (www.citizenscience.org/events/citizen-science-day). We hope that WeDigBio will serve as a model for large-scale, hybrid online or on-site citizen-science events.
We invite you to join our efforts to digitize biocollections information and involve the public in scientific activities as a WeDigBio event organizer or participant.
Supplementary Material
Acknowledgments
We sincerely thank all of the event hosts and citizen scientists at on-site events and online around the world who participated in the inaugural WeDigBio event and subsequent events; the success of WeDigBio is driven by their efforts and brilliant enthusiasm. We are grateful to the supporters, including the participants in the CITStitch Hackathon and the first WeDigBio planning meetings, and to members of the Society for the Preservation of Natural History Collections International Collaboration Working Group for early discussions about collaborative transcription parties. Special thanks to Siobhan Leachman for her incredible dedication to biocollections transcription and her insightful discussions about the event. Chris Dell was instrumental in developing a working WeDigBio dashboard.
This research was funded by the National Science Foundation via Cooperative Agreement no. EF-1115210 (for iDigBio from the Advancing Digitization of Biodiversity Collections Program) and grant nos. DBI 1458527 (for Notes from Nature), DBI 1458550 (for Biospex), DBI 1410069 (for the SERNEC Thematic Collections Network), DBI 1209149 (for the New England Vascular Plant TCN at Yale University), DBI 1115002 (for Lichens and Bryophytes TCN at The Field Museum), DBI 1458082 (for the North Carolina Museum of Natural Sciences), DBI 1502735 (for the Microfungi Collections Consortium), and DBI 1054366 (for Valdosta State University). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. The Smithsonian Transcription Center team would like to thank the National Museum of Natural History Departments of Botany and Entomology for opening their collaboration and the Office of the Chief Information Officer and the Libraries and Archives Support Services Branch team for their ongoing support. A special thank you and acknowledgment to the digital volunteers who share their time, energy, and discoveries with us all as we work together to unlock and connect biodiversity specimen data #BeeByBee and beyond. The Les Herbonautes program is part of the French infrastructure e-RECOLNAT (no. ANR-11-INBS-004) coordinated by the Museum National d’Histoire Naturelle (France) and cofunded by the Fondation de la Maison de la Chimie. DigiVol would like to thank the Australian Museum and Atlas of Living Australia for their continuing support of DigiVol, in particular the Australian Museum Foundation for its assistance with funding. DigiVol would also like to thank all our amazing volunteers around the world who contribute their time and skills to support biodiversity science.
Supplemental material
Supplementary data are available at BIOSCI online.
References cited
- [AIBS] American Institute of Biological Sciences 2013. Implementation Plan for the Network Integrated Biocollections Alliance. AIBS. [Google Scholar]
- Bi K, Linderoth T, Vanderpool D, Good JM, Nielsen R, Moritz C. 2013. Unlocking the vault: Next-generation museum population genomics. Molecular Ecology 22: 6018–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn EH, Francis CM, Blancher PJ, Drennan SR, Howe MA, Lepage D, Robbins CS, Rosenberg KV, Sauer JR, Smith KG. 2005. Enhancing the scientific value of the Christmas bird count. Auk 122: 338–346. [Google Scholar]
- Ellwood ER, et al. 2015. Accelerating the digitization of biodiversity research specimens through online public participation. BioScience 65: 383–396. [Google Scholar]
- Gaubert P, Papes M, Peterson AT. 2006. Natural history collections and the conservation of poorly known taxa: Ecological niche modeling in central African rainforest genets (Genetta spp.). Biological Conservation 130: 106–117. [Google Scholar]
- Hennon CC, et al. 2015. Cyclone Center: Can citizen scientists improve tropical cyclone intensity records? Bulletin of the American Meteorological Society 96: 591–607. [Google Scholar]
- Holmes MW, et al. 2016. Natural history collections as windows on evolutionary processes. Molecular Ecology 25: 864–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CB, Osterlund C, Mugar G, Hassman KD, Crowston K. 2015. Motivations for sustained participation in crowdsourcing: Case studies of citizen science on the role of talk. 1624–1634 in Bui TX, Sprague RH Jr, eds. Proceedings of the 48th Annual Hawaii International Conference on System Sciences. Institute of Electrical and Electronics Engineers. [Google Scholar]
- Labay B, Cohen AE, Sissel B, Hendrickson DA, Martin FD, Sarkar S. 2011. Assessing historical fish community composition using surveys, historical collection data, and species distribution models. PLOS ONE 6 (art. e25145). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie C. 2013. Biological collections in an ever changing world: Herbaria as tools for biogeographical and environmental studies. Perspectives in Plant Ecology Evolution and Systematics 15: 68–76. [Google Scholar]
- Lukyanenko R, Parsons J, Wiersma Y. 2011. Citizen science 2.0: Data management principles to harness the power of the crowd. 465–473 in Jain H, Sinha AP, Vitharana P, eds. Service-Oriented Perspectives in Design Science Research. Springer. [Google Scholar]
- Lundmark C. 2003. BioBlitz: Getting into backyard biodiversity. BioScience 53: 329. [Google Scholar]
- Mast AR, Ellwood ER, Guralnick R. 2014. The sitch with the Stitch—The CITStitch Hackathon. iDigBio: Integrated Digitized Biocollections. (22 November 2017; www.idigbio.org/content/sitch-stitch%E2%80%94-citstitch-hackathon)
- Mast AR, Ellwood ER, Kimberly P. 2015. Planning the Worldwide Engagement for Digitizing Biocollections (WeDigBio) Event. iDigBio: Integrated Digitized Biocollections. (22 November 2017; www.idigbio.org/content/planning-worldwide-engagement-digitizing-biocollections-wedigbio-event) [DOI] [PMC free article] [PubMed]
- Matsunaga A, Mast A, Fortes JAB. 2016. Workforce-efficient consensus in crowdsourced transcription of biocollections information. Future Generation Computer Systems 56: 526–536. [Google Scholar]
- Nelson G, Paul D, Riccardi G, Mast A. 2012. Five task clusters that enable efficient and effective digitization of biological collections. ZooKeys 209: 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman G, Wiggins A, Crall A, Graham E, Newman S, Crowston K. 2012. The future of citizen science: Emerging technologies and shifting paradigms. Frontiers in Ecology and the Environment 10: 298–304. [Google Scholar]
- Page LM, MacFadden BJ, Fortes JA, Soltis PS, Riccardi G. 2015. Digitization of biodiversity collections reveals biggest data on biodiversity. BioScience 65: 841–842. [Google Scholar]
- Parilla L, Ferriter M. 2016. The impact of coordinated social media campaigns on online citizen science engagement. Notes and News from the BHL Staff. Biodiversity Heritage Library. (22 November 2017; http://blog.biodiversitylibrary.org/2016/02/the-impact-of-coordinated-social-media.html)
- Pinto CM, Dnate’Baxter B, Hanson JD, Méndez-Harclerode FM, Suchecki JR, Grijalva MJ, Fulhorst CF, Bradley RD. 2010. Using museum collections to detect pathogens. Emerging Infectious Diseases 16: 356–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock NB, Howe N, Irizarry I, Lorusso N, Kruger A, Himmler K, Struwe L. 2015. Personal BioBlitz: A new way to encourage biodiversity discovery and knowledge in K–99 education and outreach. BioScience 65: 1154–1164. [Google Scholar]
- Price CA, Lee H-S. 2013. Changes in participants’ scientific attitudes and epistemological beliefs during an astronomical citizen science project. Journal of Research in Science Teaching 50: 773–801. [Google Scholar]
- Robbirt KM, Davy AJ, Hutchings MJ, Roberts DL. 2011. Validation of biological collections as a source of phenological data for use in climate change studies: A case study with the orchid Ophrys sphegodes. Journal of Ecology 99: 235–241. [Google Scholar]
- Rouhan G, Chagnoux S, Dennetière B, Shchäfer V, Pignal M. 2016. The herbonauts website: Recruiting the general public to acquire the data from herbarium labels. 143–148 in Rakotoarisoa NR, Blackmore S, Riera B, eds. Botanists of the Twenty-First Century: Roles, Challenges and Opportunities. United Nations Educational, Scientific and Cultural Organisation. [Google Scholar]
- Scheper J, Reemer M, van Kats R, Ozinga WA, van der Linden GTJ, Schaminee JHJ, Siepel H, Kleijn D. 2014. Museum specimens reveal loss of pollen host plants as key factor driving wild bee decline in The Netherlands. Proceedings of the National Academy of Sciences 111: 17552–17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson RJ, et al. 2012. The Milky Way Project First Data Release: A bubblier Galactic disc. Monthly Notices of the Royal Astronomical Society 424: 2442–2460. [Google Scholar]
- Suarez AV, Tsutsui ND. 2004. The value of museum collections for research and society. BioScience 54: 66–74. [Google Scholar]
- Sullivan BL, Wood CL, Iliff MJ, Bonney RE, Fink D, Kelling S. 2009. eBird: A citizen-based bird observation network in the biological sciences. Biological Conservation 142: 2282–2292. [Google Scholar]
- Swenson JJ, et al. 2012. Plant and animal endemism in the eastern Andean slope: Challenges to conservation. BMC Ecology 12 (art. 18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, et al. 2015. Biodiversity inventories in high gear: DNA barcoding facilitates a rapid biotic survey of a temperate nature reserve. Biodiversity Data Journal 3 (art. e6313). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill I, Loiselle S, Lind K, Ophof D. 2016. The citizen science opportunity for researchers and agencies. BioScience 66: 720–721. [Google Scholar]
- Tulloch AIT, Possingham HP, Joseph LN, Szabo J, Martin TG. 2013. Realising the full potential of citizen science monitoring programs. Biological Conservation 165: 128–138. [Google Scholar]
- Wandeler P, Hoeck PEA, Keller LF. 2007. Back to the future: Museum specimens in population genetics. Trends in Ecology and Evolution 22: 634–642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.