Abstract
Staphylococcal enterotoxin A is well known as a superantigen and able to be used for cancer immunotherapy. In this study, recombinant Staphylococcal enterotoxin A was genetically conjugated to epidermal growth factor to produce a chimeric protein recombinant Staphylococcal enterotoxin A–epidermal growth factor expressed in Escherichia coli. The recombinant Staphylococcal enterotoxin A–epidermal growth factor protein was purified using Strep-Tactin affinity chromatography and Endotoxin Removal Resin and identified by sodium dodecyl sulfate-polyacrylamide gel electropheresis and liquid chromatography-tandem mass spectrometry analysis. Furthermore, in vitro experiments showed purified recombinant Staphylococcal enterotoxin A–epidermal growth factor could successfully bind to the human nasopharyngeal carcinoma cell line CNE2, significantly promote the proliferation of human peripheral blood mononuclear cells, and enhance the secretion of several cytokines that have broad antitumor activities, such as interferon-γ, tumor necrosis factor-α, and interleukin-2 . Importantly, recombinant Staphylococcal enterotoxin A–epidermal growth factor significantly inhibited proliferation of CNE2 cells and promoted apoptosis in CNE2 cells when cocultured with peripheral blood mononuclear cells. Finally, both the binding of recombinant Staphylococcal enterotoxin A–epidermal growth factor and the toxicity of recombinant Staphylococcal enterotoxin A–epidermal growth factor-activated peripheral blood mononuclear cells were demonstrated as specific and only effective on high epidermal growth factor receptor-expressing cell lines. In all, our work suggests that recombinant Staphylococcal enterotoxin A–epidermal growth factor serves as a promising novel immunotherapeutic agent. More in vivo and in vitro studies are needed to verify its antitumor potency, as well as investigate the underlying mechanisms in cancer immunotherapy.
Keywords: superantigen fusion protein, staphylococcal enterotoxin A, epidermal growth factor, cancer immunotherapy, ligand-targeted therapeutics, nasopharyngeal cancer.
Introduction
Over the past half century, the clinical management of cancer has relied on 3 conventional anticancer therapies—surgery, radiation therapy, and chemotherapy—but, unfortunately, current clinicians and scientists are still struggling to cope with the multiple challenges such as lack of cancer epigenetic profiling and drug specificity, metastasis, and drug resistance.1–3 Over the past 2 decades, immunotherapy, also called as biologic therapy, has been considered as a new and exciting field of cancer treatment from basic scientific research to clinical applications. Immunotherapies with a variety of approaches, including oncolytic viruses, cancer vaccines, checkpoint inhibitors, chimeric antigen receptor (CAR) T cells, natural killer cells, and genetically modified dendritic cells, and so on, either stimulate the responses of the innate immune system (“active” therapies) or counteract signals produced by cancer cells that suppress or avoid immune activities (“passive” therapies).2–4 Currently, there are quite a few immune checkpoint inhibitors that have been approved by the Food and Drug Administration.2–6 In all, at least 2 immunotherapeutic approaches, checkpoint inhibition and cellular therapy with CAR T cells, have already shown promising antitumor effects either when used alone or when combined with existing conventional therapies.2–6
Cancer vaccines have been used to both treat and prevent cancer by targeting the immune system.2,3,7 Similar to classic vaccines, most therapeutic cancer vaccines are also made using protein or peptide antigens. Staphylococcal enterotoxins (SEs) are powerful peptide antigens produced by Staphylococcus aureus and there are more than 20 distinct SEs which share a number of genetic, biochemical, and functional characteristics.8 Staphylococcal enterotoxin A and SEB are 2 SEs that have been well characterized. They nonspecifically cross-link the major histocompatibility complex class II molecules on antigen-presenting cells and specific V regions of T-cell receptors, thus resulting in hyperactivation of both T lymphocytes and monocytes/macrophages.8 These activated host cells produce excessive amounts of cytokines and chemokines, that is, tumor necrosis factor α (TNF-α), interleukin 1 (IL-1), IL-2, interferon γ (IFN-γ), and macrophage chemoattractant protein 1 (or CCL2), and so on, that are involved in the pathogenesis of a number of inflammatory and/or autoimmune disorders, and even cancer.8–10
In the past, both SEA and SEB were fused to either Fab fragments of tumor reactive monoclonal antibodies (mAbs) or to ligands that bind to receptors expressed or overexpressed on the target cells and were named “ligand-targeted therapeutics (LTTs).”2,3 These LTTs have been used in animal studies and effectively inhibited breast cancer,11–13 bladder cancer,14 melanoma,12,13,15 and so on. Recent studies have defended that the use of SEs in immunotherapy is not “outdated,” but actually a great treatment option for cancer therapy as long as we learn more about efficient modes of SE engineering and administration and the dynamics of the SE-induced host antitumor immune response.7–9 Nasopharyngeal cancer is believed to be Epstein-Barr virus associated and rare in the west but endemic in the east (ie, in Southern China, the annual incidence is over 30 per 100 000 persons).16,17 Although nasopharyngeal cancer is highly sensitive to radiotherapy and chemotherapy in its early stage, outcomes are poor in its advanced stage and better treatments are urgently needed.16–18 Up to now, there hasn’t been any report regarding introducing SEs or SE-induced LTTs in treating nasopharyngeal cancer.
Epidermal growth factor (EGF) is the natural ligand that binds to the EGF receptor (EGFR) and activates more than 200 downstream signal molecules to initiate or modulate various intracellular processes.18–21 Most nasopharyngeal cancer cell lines and patients overexpress EGFR, which was thus proposed as a new target for cancer treatment.18–21 Therefore, in this study, we produced a fusion protein from SEA genetically conjugated to EGF forming a chimeric construct (referred to as recombinant SEA-EGF). The fusion protein was expressed in Escherichia coli. The purified fusion protein sequence was then identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, its ability to bind to the cancer cells was determined, and its in vitro antitumor effect was examined. To our knowledge, this is the first report of an antitumor effect on nasopharyngeal cancer by a superantigen fusion protein.
Materials and Methods
Materials and Reagents
The E. coli strain JM-109 (Promega) was used for plasmid proliferation and cloning. The E. coli strain Rosetta-gami 2 (Novagen) was used as a host for the production of recombinant proteins. Human nasopharyngeal carcinoma cell line CNE2 and human embryonic kidney cell line (HEK 293T) were purchased from the American Type Culture Collection (ATCC). Plasmid pET-44a was purchased from Promega. All primers used in this study were synthesized by Biosune Biology and Technology Company (Shanghai, China). NdeI and XhoI restriction endonucleases, T4 DNA ligase, and PrimeSTAR HS DNA polymerase were purchased from Takara Biotechnology (Dalian, China). Human IL-2, IFN-γ, and TNF-α enzyme linked immunosorbent assay (ELISA) kits were purchased from R&D Systems (Minneapolis, Minnesota, USA). β-tubulin rabbit mAb and EGFR rabbit mAb were purchased from Cell Signaling Technology (Danvers, Massachusetts, USA). Strep-Tag II mAb was purchased from Merck (Billerica, Massachusetts, USA). (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt) (MTS) reagent for cell proliferation assay was purchased from Promega (Madison, Wisconsin, USA). Annexin V-conjugated Alexa Fluor 488 (Alexa488) Apoptosis Detection Kit was purchased from BD Pharmingen (San Diego, California, USA).
Staphylococcal Enterotoxin A–EGF Gene Amplification and Plasmid Construction
The chimeric gene was constructed according to our previous work.22 The SEA gene was amplified from genomic DNA of S aureus strain ATCC-13565 using primers F1 and R1 (F1: 5′-AGCGAGAAAAGCGAAGAAAT-3′; R1: 5′-ACTTGTATATAAATATATAT-3′). Epidermal growth factor gene was amplified from complementary DNA (cDNA) of human parotid gland using primers F2 and R2 (F2: 5′-AATAGTGACTCTGAATGTCCCCTGTC-3′; R2: 5′-GCGCAGTTCCCACCACTTCA -3′). To fuse EGF to the C terminal of SEA, primers R3 and F3 were annealed and extended by splice overlap extension polymerase chain reaction (PCR) using R4 and F4 (R3: GGACATTCAGAGTCACTATTACTTGTATATAAATATATAT; F3: ATATATATTTATATACAAGTAATAGTGACTCTGAATGTCC). The resultant products were SEA-EGF. Polymerase chain reaction was carried out to add the NdeI restriction site, XhoI restriction site, and Strep-tag II for purification using the F4 and R4 (F4: 5′- GGAATTCCATATGAGCGAGAAAAGCGAAGAAAT-3′, with NdeI site underlined; R4: 5′-CCGCTCGAGTTATTATTTTTCGAACTGCGGGTGGCTCCAGCGCAGTTCCCACCACTTCA-3′, with XhoI site underlined and Strep-Tag II sequence in bold italic). The digested amplified products were inserted into the NdeI and XhoI sites of the pET44a vector and transformed into E coli strain JM109. Clones were proliferated and harvested for plasmid extraction. The positive clones were identified by double digestion, colony PCR, and sequencing analysis. Subsequently, the pET44a-SEA-EGF plasmid was transformed into E. coli strain Rosetta-gami 2 for protein expression.
Expression of Recombinant SEA-EGF (rSEA-EGF) in E. coli
The cultures were grown at 37°C until the OD600 (OD absorbance-600nm) reached 0.6 to 0.8, followed by the induction of protein expression with 0.5 mmol/L Isopropyl β-D-1-thiogalactopyranoside (IPTG) for 4 to 5 hours at 37°C. The inducible expression of rSEA-EGF was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and verified by Western blotting. Afterward, the cell pellets were resuspended in lysis buffer (25 mmol/L phosphate buffer, 150 mmol/L NaCl, 2 mmol/L EDTA, and 10% glycerol; pH 7.5) and lysed by sonication. Inclusion bodies were recovered by centrifugation (6000 rpm, 30 minutes, 4°C), washed twice in the wash buffer (2% Triton X-100, 50 mmol/L Tris, and 1 mmol/L EDTA; pH 7.5), and dissolved in a denaturation solution (6 mol/L guanidine hydrochloride, 10 mmol/L dithiothreitol, 100 mmol/L Tris, and 2 mmol/L EDTA; pH 8.0).
Refolding and Purification of rSEA-EGF
To obtain the renatured rSEA protein, the denatured inclusion bodies (final concentration 0.5 mg/mL) were refolded by dialysis using the renaturation solution (150 mmol/L NaCl, 50 mmol/L Tris, 1 mmol/L EDTA, 1 mmol/L cysteine, 3 mmol/L Glutathione disulfide (GSSG), 3 mmol/L Glutathione (GSH), 0.2% l-arginine and 10% glycerol; pH 8.0). The renatured rSEA-EGF protein was purified using Strep-Tactin affinity chromatography (GE Healthcare, Pittsburgh, Pennsylvania, USA) following dialyzed against binding buffer (150 mmol/L NaCl, 50 mmol/L Tris, 1 mmol/L EDTA; pH 8.0). The purity of rSEA-EGF was analyzed by SDS-PAGE. To remove any contaminating LPS (Lipopolysaccharide), rSEA-EGF protein was purified by high-capacity endotoxin removal resin (Thermo Fisher Scientific, Waltham, Massachusetts, USA). The LPS content was analyzed by the tachypleus amebocyte lysate (TAL) method. Samples that contained less than 0.5 EU/ mg LPS were deemed to be suitable for use in the following assays.
Liquid Chromatography-tandem Mass Spectrometry Analysis
Liquid Chromatography-tandem Mass Spectrometry analysis was conducted according to the reference by Guan et al23 with some modification. Recombinant SEA-EGF was digested by trypsin at a work ratio of 1:10 (enzyme to protein) at 37°C for 4 hours. Next, digested peptides were separated on an Easy-spray column (15 cm × 75 μm ID, 3-μm C18 particles) by using the EASY-nLC system (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and were then eluted with 0.1% formic acid solution in a linear gradient (0%-90%) of methanol at the flow rate of 300 nL/min for 20 minutes. Finally, a Q Exactive mass spectrometer (Thermo Fisher Scientific, Waltham, Massachusetts, USA) with a resolution of 60 000 at m/z 350 to 1800 was used to acquire high-resolution spectra. All the measured MS/MS (tandem mass spectrometry) results in an mascot generic format (MGF) file were studied with the Analyst software 1.3.2 Mascot script. Using the Mascot MS/MS ion search engine (Matrix Science, Boston, Massachusetts), the unknown peptides were identified.
Proliferation of Human Peripheral Blood Mononuclear Cells Stimulated by rSEA-EGF
This procedure was performed similar to our previous work.10 Briefly, heparinized blood was obtained from healthy donors, and PBMCs were separated by Ficoll-Paque PLUS (GE Healthcare, Pittsburgh, Pennsylvania, USA) density gradient centrifugation. Freshly isolated PBMCs were suspended in Roswell Park Memorial Institute (RPMI) 1640 culture medium and then distributed into 96-well plates (105 cells/well) with the addition of rSEA-EGF at various concentrations from 1 pg/mL to 1 μg/mL. After 7 days of coculture, cell viability was detected by MTS assay. A reference of absorbance at 630 nm served as the blank and absorbance at 490 nm was recorded. The effect of rSEA-EGF on cell viability was assessed as percentage of cell viability compared with control cells, which were arbitrarily assigned 100%. The mean value of 5 wells was calculated, and each experiment was repeated 3 times.
Cytokine Analysis by ELISA
Freshly isolated PBMCs in 24-well plates (5 × 105 cells/well) were stimulated by rSEA-EGF at 100 ng/mL. After 24 hours cell culture, supernatants were harvested and cytokine levels were measured by ELISA. The concentrations of IFN-γ, TNF-α, and IL-2 in cell culture supernatants were measured using ELISA assay (Quantikine; R&D Systems, Minneapolis, Minnesota, USA), following the manufacturer’s instructions.
Real-Time Quantitative PCR Assay
To determine the presence of endogenous EGFR messenger RNA (mRNA) in CNE2 or HEK 293 T cells, total RNA was extracted and purified using an RNeasy Kit (Qiagen, Venlo, Netherlands).Total RNA was reverse transcribed to cDNA using PrimeScript RT Master Mix (Takara, Dalian, China). Real-time PCR was carried out with SYBR Green Master Mix (Applied Biosystems, Redwood, California,USA). All samples were assayed in duplicate. Polymerase chain reaction (heating at 95°C for 10 seconds and 60°C for 30 seconds) was performed. The expression of target mRNA was normalized to the expression of β-actin. Specific intron-spanning primers were designed with the NCBI Primer-BLAST. The primers used are: EGFR (F: 5′- GCAAAGTGCCTATCAAGTG-3′; R: 5′- CCAAACGGTCACCCCGTA -3′) and β-actin (F: 5′- ATTGCCGACAGGATGCAGA-3′; R: 5′- GAGTACTTGCGCTCAGGAGGA -3′).
RT-PCR Assay
Reverse transcription polymerase chain reaction (RT-PCR) with 3% agarose gel electrophoresis was performed using Taq DNA Polymerase (Takara, Dalian, China) with 1 μL cDNA as a template per reaction. The optimized PCR conditions were as follows: 95°C 5 minutes, 95°C 30 seconds, 60°C 30 seconds, 72°C 30 seconds for 25 cycles, 72°C 10 minutes.
Western Blot Analysis
To determine the presence of endogenous EGFR, CNE2 or HEK 293 T cells were solubilized in cell lysis solution containing protease inhibitor cocktail (Roche, Branford, Connecticut, USA). The protein concentration of the soluble extracts was determined by BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Separation of 50 mg of total protein was done on 12% SDS-polyacrylamide gels and transferred to a polyvinylidene difluoride membrane before immunoblotting with EGFR rabbit antibodies and β-tubulin rabbit antibodies as indicated. The specific proteins were detected by the enhanced chemiluminescence detection system (Thermo Fisher Scientific, Waltham, Massachusetts, USA).
Tumor Cell Binding Assay
Binding of rSEA-EGF or rSEA to EGFR was detected by ELISA as described.24 Briefly, 1 × 104 CNE2 or HEK 293 T cells per well were seeded into 96-well flat-bottomed plates overnight, then incubated with serial concentrations of rSEA-EGF or rSEA (25, 12.5, 6.25, 3.12, 1.56, 0.78 μg/mL) at 37°C for 1 hour and then fixed with 4% neutral paraformaldehyde (10 mmol/L Phosphate-buffered saline (PBS), 4% paraformaldehyde, pH 7.4) at room temperature for 1 hour. The cells were blocked with 5 mg/mL bovine serum albumin for 2 hours. After being washed 5 times with PBST (10 mmol/L PBS pH 7.4, 0.1% Tween-20), the cells were incubated with Strep-Tag II mAb (1:1000) overnight, followed by horseradish peroxidase-conjugated rabbit anti-mouse Immunoglobulin G (1:5000) and washed as previously. Finally, the color was developed using tetramethylbenzidine solution, and the absorbance value at 450 nm was obtained by ELISA reader.
CNE2 or HEK 293 T Cell Growth Inhibition Assay
Peripheral blood mononuclear cells (PBMCs) were incubated with rSEA-EGF at a series of concentrations (from 10 pg/mL to 1 μg/mL) at 37°C in 5% CO2 for 7 days. After 7 days of stimulation, the PBMC and rSEA-EGF mixtures (unwashed PBMC including supernatant, 2.5 × 104 cells/well) were added to CNE2 or HEK 293 T cells (5 × 103 cells/well in 96-well flat-bottomed plates) to a total volume of 200 μL. Target cells (CNE2 or HEK 293 T cells) and effector cells (SEA pretreated PBMCs) were cocultured at 37°C in 5% CO2 for 72 hours after which the cell supernatant and suspended cells were removed, and fresh medium with MTS reagent was added to the remaining viable cells for a 1-hour incubation. Using a microplate reader (490 nm) to detect absorbance, the remaining viable tumor cells were determined using the MTS assay according to manufacturer’s instruction.
Quantification of Apoptotic and Dead Cells
Recombinant SEA-EGF-induced cell death in CNE2 cells cocultured with PBMCs was determined by flow cytometry using the Annexin V-conjugated Alexa Fluor 488 (Alexa488) Apoptosis Detection Kit (BD Pharmingen, San Diego, California, USA) using a procedure similar to our previous work.10 Briefly, PBMCs were pretreated with rSEA-EGF (100 ng/mL) for 7 days and then the rSEA-EGF-treated PBMCs were cocultured with CNE2 cells for 48 hours. The cells were harvested after the addition of 0.25% trypsin-EDTA (Life Technologies, Gibco, Frederick, Maryland, USA) immediately followed by the addition of fetal bovine serum to terminate the reaction and then centrifuged. The cell pellets were washed 2 times with 1× PBS, washed once with 1× binding buffer, then resuspended in 1× binding buffer, and incubated with annexin V and propidium iodide (PI) for cellular staining at room temperature for 15 minutes in the dark. Cells were analyzed by flow cytometry on a FACSCalibur instrument (BD Biosciences, San Diego, California, USA) within 1 hour.
Statistical Analysis
GraphPad Prism software was used for statistical comparisons with unpaired t test or 1-way analysis of variance followed by post hoc analysis. All quantitative data were presented as mean ± standard error of the mean. Statistical significance was concluded if P value <.05.
Results
Construction of rSEA-EGF
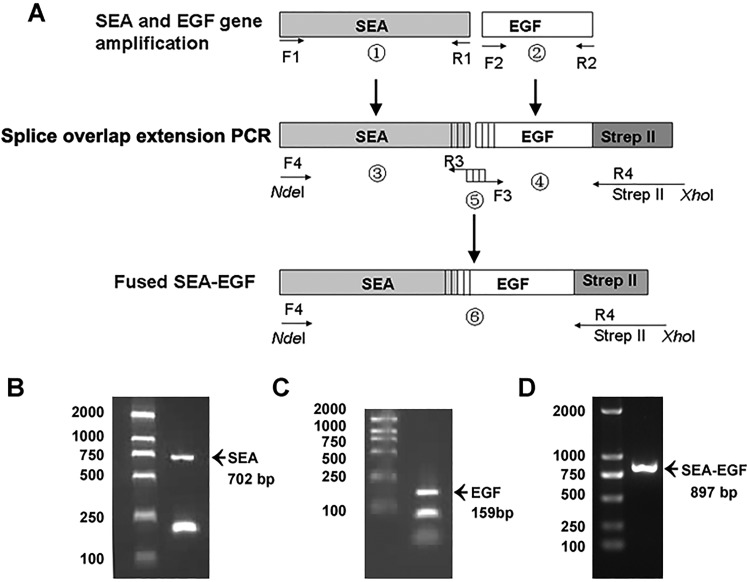
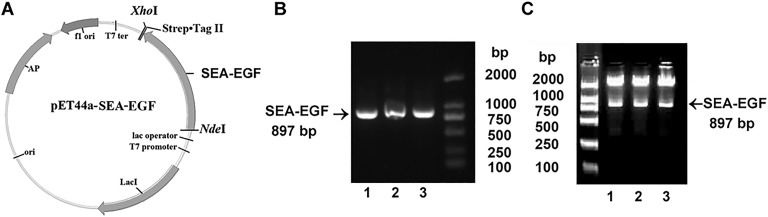
The SEA gene of S aureus ATCC-13565 was amplified by PCR and resulted in a 702 bp amplicon (Figure 1B). The EGF gene was amplified by PCR and resulted in a 159 bp amplicon (Figure 1C). The SEA-EGF fragment was amplified by splice overlap extension PCR (Figure 1A) and resulted in an 897 bp amplicon (Figure 1D). The entire coding region of SEA-EGF was then cloned into a pET44a vector (Figure 2A), clones were screened by PCR assay and NdeI and XhoI restriction enzyme digestion, and the verifications were in accordance with results as expected (Figure 2B and C). The candidate positive clones were then sequenced, which showed consistency with the sequences identified using the Basic Local Alignment Search Tool program.
Figure 1.
Cloning of Staphylococcal enterotoxin A-epidermal growth factor (SEA-EGF) gene. A, Schematic presentation of splice overlap extension polymerase chain reaction (PCR) for fused SEA-EGF. Epidermal growth factor was fused to the C-terminal of Staphylococcal enterotoxin A (SEA) and tagged with Strep-tag II at the C-terminal. B-D, Polymerase chain reaction amplified products of SEA gene (B), EGF gene (C), and SEA-EGF gene (D), respectively. DNA ladder is indicated on the left side, arrows indicating the PCR products, and their expected sizes are labeled below as well.
Figure 2.
Identification of the pET44a Staphylococcal enterotoxin A–epidermal growth factor (SEA-EGF) plasmid. A, Plasmid map of the recombinant construct of pET44a-SEA-EGF. B, Identification of the pET44a-SEA-EGF by PCR assay. Lanes 1-3 are agarose gel electrophoresis of PCR products of different clones after using primers for PCR detection. C, Identification of the pET44a-SEA-EGF by restrictive enzyme digestion assay with NdeI and XhoI. Lanes 1-3 are agarose gel electrophoresis of products of different clones after restrictive enzyme digestion.
Expression and Identification of rSEA-EGF Protein
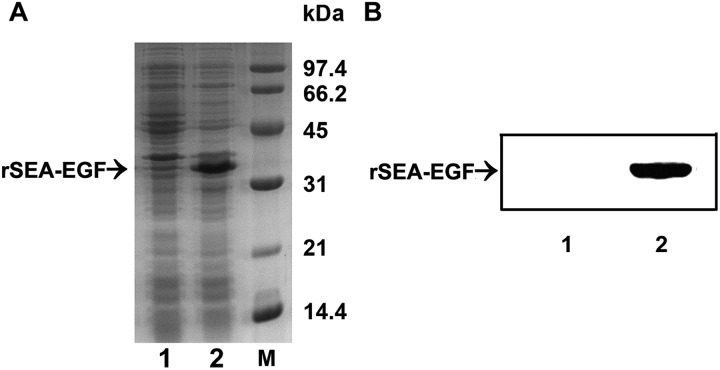
After sequencing confirmation of the correct reading frame of the target protein gene in the expression vector, the recovered plasmids were then successfully transformed into E coli strain Rosetta-gami 2 cells. Following induction and further incubation at 37°C for 6 hours, the recombinant protein was analyzed by SDS-PAGE and found to be identical to the reported protein with a molecular weight of ∼34.5 kDa (Figure 3A). The SDS-PAGE results indicated that the level of rSEA-EGF in E coli was approximately 65% of the total bacterial protein (Figure 3A, lane 2). The expression of rSEA-EGF was confirmed using anti-Strep-Tag II mouse mAb as the primary antibody. A specific band with an apparent molecular mass of ∼34.5 kDa showed that the protein induced was of rSEA-EGF (Figure 3B).
Figure 3.
SDS-PAGE and immunoblot analysis of the recombinant Staphylococcal enterotoxin A–epidermal growth factor (rSEA-EGF). A, SDS-PAGE analysis of inducible expression of rSEA-EGF. Lanes 1 and 2: lysates of E coli cells harboring pET44a-SEA before (lane 1) and after (lane 2) IPTG induction. Lane M: Low-molecular-weight standard. B, Western blotting analysis of rSEA-EGF protein with anti-Strep-Tag II antibody. Lanes 1 and 2: Lysates of Escherichia coli cells harboring pET44a SEA-EGF before (lane 1) and after (lane 2) IPTG induction.
Refolding and Purification of rSEA-EGF Protein
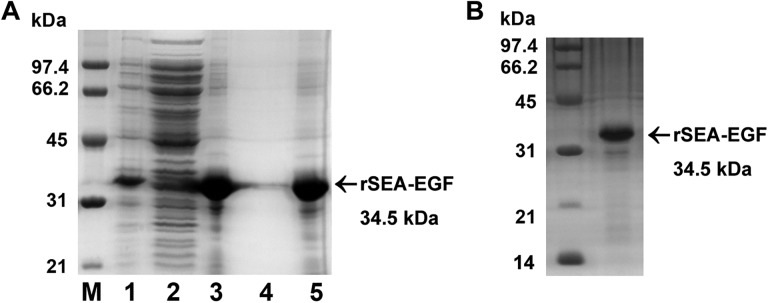
After the bacterial cells were lysed, rSEA-EGF remained in the precipitate in the form of inclusion bodies. After washing 2 times, the purity of the inclusion bodies was found to be greater than 85% (Figure 4A). The rSEA-EGF protein was refolded by dialysis with the renaturation solution and purified by a Strep-Tactin affinity chromatography. The target protein bound on the gel was collected by elution and verified by SDS-PAGE analysis. The purity of the final rSEA-EGF protein was found to be greater than 95% (Figure 4B). After purification using high-capacity endotoxin removal resin, the concentration of LPS was 0.037 EU/mg as detected by the TAL method.
Figure 4.
Identification of refolded and purified recombinant Staphylococcal enterotoxin A-epidermal growth factor (rSEA-EGF) protein. A, SDS-PAGE analysis of inclusion bodies after 2 washes with the wash buffer. Lane M: Low-molecular-weight standard; lane 1: lysates of Escherichia coli cells harboring pET44a Staphylococcal enterotoxin A-epidermal growth factor (SEA-EGF) after IPTG induction. Lane 2, supernatant after cell lysis; Lane 3, pellet after cell lysis (inclusion bodies); Lane 4, supernatant after washing of inclusion bodies; Lane 5, inclusion bodies after washing step. B, SDS-PAGE analysis of the purified rSEA-EGF protein by Strep-Tactin affinity chromatography.
Liquid Chromatography-tandem Mass Spectrometry Analysis
For successful protein identification, rSEA-EGF was processed with tryptic digestion and subsequent LC-MS/MS analysis. The overlap analysis showed that the coverage of digested rSEA-EGF peptides matched with the known SEA amino acid sequences is 88% (Figure 5A) and the coverage of digested rSEA-EGF peptides matched with the known EGF sequences is 47% (Figure 5B). Such high protein sequence coverage guarantees higher affinity and specificity for rSEA-EGF binding to the human EGFR-expressing carcinoma cells and subsequently can enhance the therapeutic effects while reducing potential damage to normal cells and tissues. The details of the identified peptide sequences from rSEA-EGF matched with known SEA and EGF sequences are shown in Table 1.
Figure 5.
Sequence identification of recombinant Staphylococcal enterotoxin A–epidermal growth factor (rSEA-EGF) using liquid chromatography with mass spectrometry (LC-MS/MS) analysis. Amino acid sequence coverage of peptides digested from rSEA-EGF matched with the known SEA (A) and EGF (B) sequences. Matching identified peptides are indicated in bold red.
Table 1.
Identified Peptide Sequences From Recombinant SEA-EGF Protein.
| Peptide Sequence | Matched protein name | Accession No | Start-End |
|---|---|---|---|
| SEKSEEINEKDLR | SEA | AAA26681.1 | 1-13 |
| KKSELQGTALGNLK | SEA | AAA26681.1 | 14-27 |
| QIYYYNEKAK | SEA | AAA26681.1 | 28-37 |
| TENKESHDQFLQHTILFK | SEA | AAA26681.1 | 38-55 |
| GFFTDHSWYNDLLVDFDSK | SEA | AAA26681.1 | 56-74 |
| VDLYGAYYGYQCAGGTPNK | SEA | AAA26681.1 | 85-103 |
| TACMYGGVTLHDNNR | SEA | AAA26681.1 | 104-118 |
| LTEEKKVPINLWLDGK | SEA | AAA26681.1 | 119-134 |
| KVPINLWLDGKQNTVPLETVK | SEA | AAA26681.1 | 124-144 |
| QNTVPLETVKTNKK | SEA | AAA26681.1 | 135-148 |
| KNVTVQELDLQAR | SEA | AAA26681.1 | 148-160 |
| RYLQEKYNLYNSDVFDGK | SEA | AAA26681.1 | 161-178 |
| VQRGLIVFHTSTEPSVNYDLF GAQGQYSNTLLR | SEA | AAA26681.1 | 179-211 |
| IYRDNK | SEA | AAA26681.1 | 212-217 |
| YACNCVVGYIGERCQYR | EGF | AFA26280.1 | 29-45 |
| CQYRDLK | EGF | AFA26280.1 | 42-48 |
| WWELR | EGF | AFA26280.1 | 49-53 |
Abbreviations: EGF, epidermal growth factor; SEA, Staphylococcal enterotoxin A.
Recombinant SEA-EGF Induces Proliferation of Human PBMCs
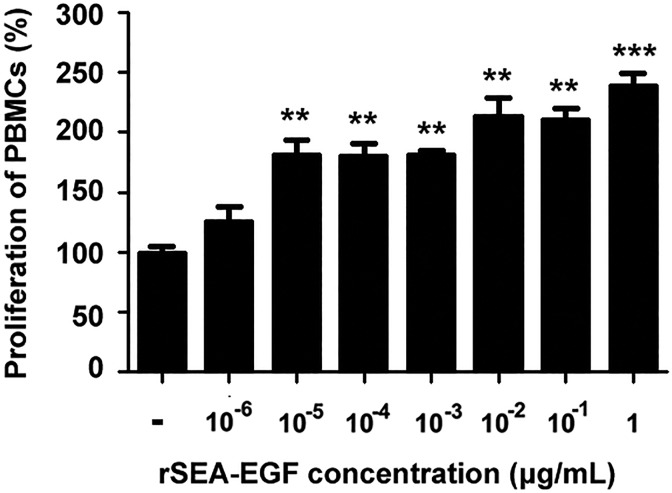
To examine the stimulating potency of rSEA-EGF on the proliferation of lymphocytes, human PBMCs were incubated with rSEA-EGF (1 pg/mL~∼1μg/mL) at 37°C for 7 days. The data indicated that rSEA-EGF significantly promoted human PBMC proliferation at almost all concentrations (1 pg/mL~∼1 μg/mL) tested, with the most effective dose at 100 ng/mL. Recombinant SEA-EGF (1 μg/mL) increased human PBMCs proliferation from 100% to 238% when compared with vector stimulated PBMCs (Figure 6).
Figure 6.
Proliferation of peripheral blood mononuclear cells (PBMCs) stimulated by different concentrations of recombinant Staphylococcal enterotoxin A–epidermal growth factor (rSEA-EGF). Cells were incubated in the presence of different concentrations of rSEA-EGF (1 pg/mL-1 μg/mL) for 7 days at 37°C in 5% CO2. MTS reagent was added for the last 4 hours of the 7-day incubation. **P < .01, ***P < .001, the rSEA-EGF-stimulated human PBMCs compared with the negative control (PBS).
Recombinant SEA-EGF Induces Cytokines Secretion by Human PBMCs
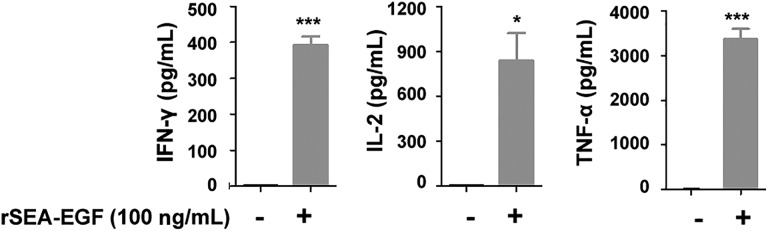
Secretion of several key cytokines essential for antitumor or cancer-related inflammation8–10 by rSEA-EGF-stimulated human PBMC was also evaluated using ELISA assay. The results indicated that IFN-γ, TNF-α, and IL-2 levels were significantly increased in the cell culture supernatant (Figure 7) when PBMCs were stimulated by rSEA-EGF (100 ng/mL).
Figure 7.
Cytokine secretion of peripheral blood mononuclear cells (PBMCs) stimulated by different concentrations of recombinant Staphylococcal enterotoxin A–epidermal growth factor (rSEA-EGF). After 24 hours in culture, the supernatants were harvested and IFN-γ, TNFα, and IL-2 concentration was measured by ELISA assay. The rSEA-EGF-stimulated human PBMCs were compared with the negative control (PBS, *P < .05, ***P < .001) PBMCs. The mean ± standard error of the mean of triplicate determinations is shown.
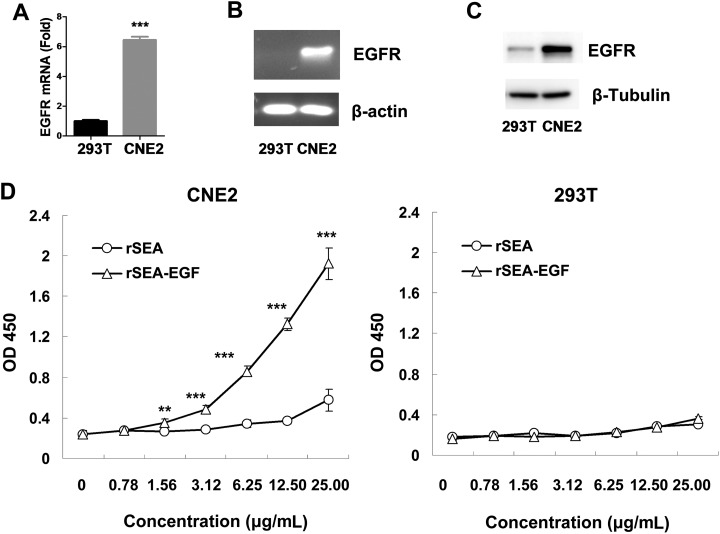
Recombinant SEA-EGF Binding Affinity to the EGFR in CNE2 and 293 T cells
To investigate whether rSEA-EGF binds to EGFR specifically, EGFR expression levels in CNE2 and HEK 293 T cells were detected by quantitative PCR, RT-PCR, and Western blot analysis. Recombinant SEA-EGF binding to the CNE2 and HEK 293 T cells was analyzed using cell ELISA assay. Our real-time quantitative PCR (Figure 8A), RT-PCR (Figure 8B), and Western blot (Figure 8C) data indicated that there were high levels of EGFR mRNA and protein expression in CNE2 cells but much lower levels in HEK 293 T cells (Figure 8A-C). The cell ELISA assay data showed that rSEA did not bind or only weakly bound to both CNE2 and HEK 293 T cells (Figure 8D) and that rSEA-EGF also showed very poor binding ability to HEK 293 T cells at all doses tested (Figure 8D, right). However, rSEA-EGF showed expected binding ability to CNE2 cells in a dose-dependent manner (Figure 8D, left).
Figure 8.
Recombinant Staphylococcal enterotoxin A–epidermal growth factor (rSEA-EGF) binds to epidermal growth factor receptor (EGFR) in CNE2 and HEK 293 T cells. A-C, Expression of EGFR in CNE2 and HEK 293 T cell lines was detected by quantitative PCR (A), RT-PCR assay (B), and Western blot (C). D, The binding of rSEA or rSEA-EGF fusion protein to EGFR. The binding of various concentrations of rSEA or rSEA-EGF fusion proteins to CNE2 cancer cells or HEK 293 T cells was detected by cell ELISA assay. Values represented the mean (standard deviation) of results from 4 samples. **P < .01, ***P < .001, compared with rSEA.
Recombinant SEA-EGF Inhibited Tumor Cell Growth In Vitro
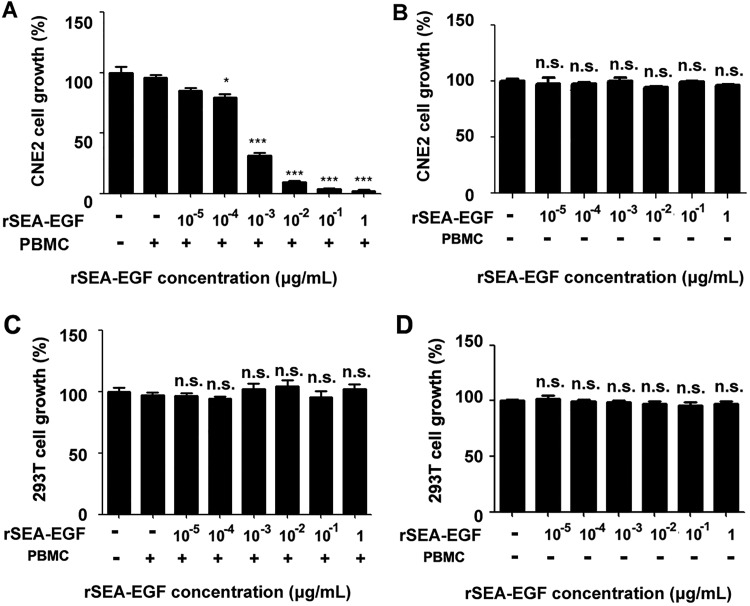
The antitumor effect of rSEA-EGF-treated PBMCs on human nasopharyngeal carcinoma CNE2 cells or HEK 293 T cells was examined by MTS cell proliferation assay (Figure 9). At an effector/target (E/T) ratio of 5:1, we found that rSEA-EGF significantly inhibited the growth of CNE2 cells in a dose-dependent manner (100 pg/mL-1 μg/mL), with the most effective inhibitory dose being from 100 ng/mL to 1 μg/mL. The percentage inhibition of 1 μg/mL rSEA-EGF on CNE2 cells was as high as 97.8% (Figure 9A). There was no effect of rSEA-EGF on the proliferation of CNE2 cells or HEK 293 T cells at any of the doses tested in the absence of PBMCs (Figure 9B and D). Recombinant SEA-EGF also had no effect on the proliferation of HEK 293 T cells at any dose tested when cocultured with PBMCs (Figure 9C).
Figure 9.
Effect of recombinant Staphylococcal enterotoxin A–epidermal growth factor (rSEA-EGF) on proliferation of CNE2 and HEK 293 T cells. A, Peripheral blood mononuclear cells (PBMCs) were treated with rSEA-EGF at different concentrations (10 pg/mL-1 μg/mL) for 7 days. Pretreated PBMCs were then cocultured with CNE2 cells (at a density of 5 × 103 cells per well) in a total volume of 200 μL in 96-well flat-bottomed plates, at 37°C in 5% CO2 for 72 hours. MTS reagent was added for the last 4 hours of incubation. The mean value of 5 wells was calculated, and each experiment was from at least 1 donor and repeated 3 times. *P< .05, *** P< .001 compared with PBMCs treated with 0 μg/mL rSEA-EGF. B, The same procedure performed as that in Figure 9A but in the absence of PBMCs. C-D, The same procedures performed as that in Figure 9A and 9B on HEK 293 T cells with or without the addition of PBMCs. n.s. indicates nonsignificant.
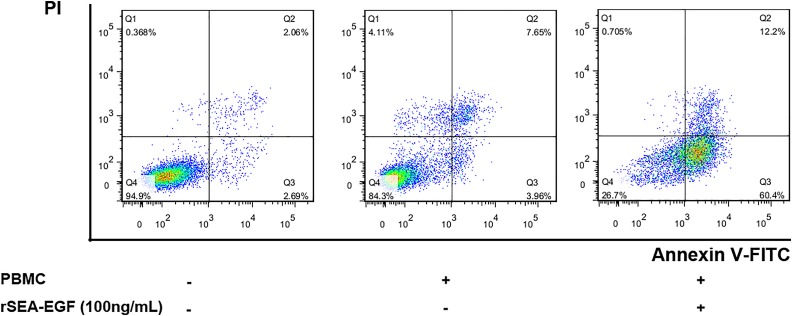
Recombinant SEA-EGF-Induced Apoptosis in CNE2 Cells
Annexin V and PI staining has been widely used to identify cell viability and to distinguish between apoptotic and necrotic cell death. In this study, we performed annexin V/PI double staining and measured apoptosis and necrotic cell death of CNE2 cells by flow cytometry after 48 hours coculturing with rSEA-EGF-treated (100 ng/mL) PBMCs. We found that when CNE2 cells were cocultured with rSEA-EGF-treated PBMCs, only 0.71% of the total cells were detected in the necrotic cell region (Q1, annexin V−, PI+), but 72.6% of the total cells were apoptotic cells (Q2 + Q3, annexin V+), with 60.4% of them undergoing early (Q3, annexin V+, PI−) and 12.2% of them undergoing late (Q2, annexin V+, PI+) apoptosis (Figure 10). Nonetheless, rSEA-EGF-treated PBMCs markedly increased the percentage of early and late apoptotic cells. Coculture of CNE2 cells with untreated PBMCs also increased the percentage of apoptotic CNE2 cells but with much less potency than if PBMCs were pretreated with rSEA-EGF (Figure 10).
Figure 10.
Effect of recombinant Staphylococcal enterotoxin A–epidermal growth factor (rSEA-EGF) on apoptosis in CNE2 cells. Peripheral blood mononuclear cells (PBMCs) were pretreated with rSEA-EGF for 7 days, then added into CNE2 cells and cocultured for 48 hours, followed by annexin V and PI staining and flow cytometry assay. Necrotic cells (Q1, annexin V–, PI+), viable cells (Q4, annexin V– PI–), early (Q3, annexin V+ PI–), and late (Q3, annexin V+ PI+) apoptotic cells are defined following the instructions from the Apoptosis Detection Kit (BD Pharmingen), and their percentages are given in the respective regions.
Discussion
In humans, the number of natural tumor-reactive T cells is often low. Preclinical and clinical studies have shown that stimulation of T cells by immunogenic proteins has an ability to interfere with established growing cancer.25 In the tumor-targeted superantigen concept, bacterial superantigens, the most potent known activators of human T cells, are used to activate and recruit large numbers of T cells to the targeted tumors.8–10 Cancer immunotherapy through a tumor cell–specific small-molecule ligand, or LTTs, has been arousing enthusiasm for many reasons, that is, LTTs were believed to be easily accessible, less expensive, less antigenic than mAbs, and, more importantly, easily enhance the ability of drugs getting into solid tumors but decreasing drug delivery to normal cells.26–28 In fact, LTTs could not only be used as a specific delivery system for cancer immunotherapy but also represent a novel concept for the delivery of immune agents for many other diseases, such as food poisoning,8,28,29 skin inflammation,30 toxic shock syndrome, autoimmune diseases,28, and so on. Therefore, in this study, we constructed, expressed, and characterized a novel recombinant SEA-EGF and demonstrated that purified rSEA-EGF could stimulate the proliferation of human PBMCs, thereby releasing cytokines such as IFN-γ, TNF-α, IL-2 and significantly inhibiting the proliferation of CNE2 cells, at least via promoting their apoptosis. To our knowledge, this is the first study to report using a superantigen chimeric protein against nasopharyngeal cancer.
Challenges in LTTs require high delivery specificity and minimum potential damage to healthy cells,31 which we carefully considered in our work. By splice overlap extension PCR22, we successfully combined SEA with EGF while retaining EGF binding ability. In our pilot assay, the purified chimeric protein rSEA-EGF had excellent binding affinity when incubated with the CNE2 tumor cell line, had high stimulatory ability on PBMCs, and had ideal antitumor effects. Similar work has been accomplished by Yousefi et al,28,32 using a mature form of SEA and different lead sequence. As seen in our present work, the purified rSEA-EGF significantly elicited the secretion of tumoricidal or cancer-related inflammatory cytokines (IFN-γ, TNF-α, and IL-2) by PBMCs, which is an ideal indicators for the chimeric protein. Thus, this work is a good foundation for our future chimeric protein engineering research and also a possible direction for all targeted immunotoxin research.
Multiple cytokines and chemokines can be generated in different tissues as a response of the body to cancer, however, whether the cytokines and chemokines induced by rSEA-EGF can interact together through a form of cross-talk and affect immunotherapy responses has not been well characterized.8–10 Recently, we found that when cocultured with PBMCs, SEA induced crosstalk-dependent activation of extracellular regulated protein kinases and signal transducers and activators of transcription and the production of tumor-suppressive cytokines.10 We plan to use our rSEA-EGF fusion protein and coculture with PBMCs to investigate chemokine and chemokine receptor network-related therapeutic interventions with cancer cells, which may yield important information for understanding the immune-related mechanisms of cancer development that could subsequently lead to applications in cancer immunotherapy.
Conclusion
Our work introduces a new superantigen, rSEA-EGF fusion protein, for targeted treatment of EGFR-expressing cancer (ie, nasopharyngeal cancer) cells. It could be of great benefit to further explore with intensive studies its actual antitumor effects on animals and the molecular mechanisms involved.
Acknowledgments
The authors thank Lucinda Beck and Joseph Jeffry for their editing and critical reading of the paper and important advice on flow cytometry experiments.
Abbreviations
- ATCC
American Type Culture Collection
- cDNA
complementary DNA
- CAR
chimeric antigen receptor
- EGF
epidermal growth factor
- EGFR
EGF receptor
- EGFR
epidermal growth factor receptor
- IL-1
interleukin-1
- IFN-γ
interferon-γ
- LC-MS/MS
liquid chromatography with mass spectrometry
- LTTs
ligand-targeted therapeutics
- mAbs
monoclonal antibodies
- mRNA
messenger RNA
- MTS
(3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxy-methoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium inner salt)
- PBMCs
peripheral blood mononuclear cells
- PCR
polymerase chain reaction
- PI
propidium iodide
- rSEA-EGF
recombinant SEA-EGF
- rSEA
recombinant SEA
- SEA
Staphylococcal enterotoxin A
- SEs
Staphylococcal enterotoxins
- TAL
tachypleus amebocyte lysate
- TNF-α
tumor necrosis factor-α.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Natural Science Foundation of China (Nos. 81301948, 30640033, 81500017) and Guangdong Special Fund for Public Welfare Research and Capacity Building (2014A020212646).
References
- 1. Chakraborty S, Rahman T. The difficulties in cancer treatment. Ecancermedicalscience. 2012;6:ed16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Trapani JA, Darcy PK. Immunotherapy of cancer. Aust Fam Physician. 2017;46(4):194–199. [PubMed] [Google Scholar]
- 3. Kamta J, Chaar M, Ande A, Altomare DA, Ait-Oudhia S. Advancing cancer therapy with present and emerging immuno-oncology approaches. Front Oncol. 2017;7:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gotwals P, Cameron S, Cipolletta D, et al. Prospects for combining targeted and conventional cancer therapy with immunotherapy. Nat Rev Cancer. 2017;17(5):286–301. [DOI] [PubMed] [Google Scholar]
- 5. Ramagopal UA, Liu W, Garrett-Thomson SC, et al. Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab. Proc Natl Acad Sci U S A. 2017;114(21): E4223–E4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tartari F, Santoni M, Burattini L, Mazzanti P, Onofri A, Berardi R. Economic sustainability of anti-PD-1 agents nivolumab and pembrolizumab in cancer patients: recent insights and future challenges. Cancer Treat Rev. 2016;48:20–24. [DOI] [PubMed] [Google Scholar]
- 7. Kumai T, Kobayashi H, Harabuchi Y, Celis E. Peptide vaccines in cancer-old concept revisited. Curr Opin Immunol. 2017;45:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pinchuk IV, Beswick EJ, Reyes VE. Staphylococcal enterotoxins. Toxins(Basel). 2010;2(8):2177–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krakauer T. Update on staphylococcal superantigen-induced signaling pathways and therapeutic interventions. Toxins(Basel). 2013;5(9):1629–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu X, Zeng L, Zhao Z, et al. PBMC activation via the ERK and STAT signaling pathways enhances the anti-tumor activity of Staphylococcal enterotoxin A. Mol Cell Biochem. 2017;434(1-2):75–87. [DOI] [PubMed] [Google Scholar]
- 11. Yousefi F, Siadat SD, Saraji AA, et al. Tagging staphylococcal enterotoxin B (SEB) with TGFaL3 for breast cancer therapy. Tumour Biol. 2016;37(4):5305–5316. [DOI] [PubMed] [Google Scholar]
- 12. Yu J, Tian R, Xiu B, et al. Antitumor activity of T cells generated from lymph nodes draining the SEA-expressing murine B16 melanoma and secondarily activated with dendritic cells. Int J Bio Sci. 2009;5(2):135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sundstedt A, Celander M, Hedlund G. Combining tumor-targeted superantigens with interferon-alpha results in synergistic anti-tumor effects. Int Immunopharmacol. 2008;8(3):442–452. [DOI] [PubMed] [Google Scholar]
- 14. Han C, Hao L, Chen M, et al. Target expression of Staphylococcus enterotoxin A from an oncolytic adenovirus suppresses mouse bladder tumor growth and recruits CD3+ T cell. Tumour Bio. 2013;34(5):2863–2869. [DOI] [PubMed] [Google Scholar]
- 15. Jeudy G, Salvadori F, Chauffert B, Solary E, Vabres P, Chluba J. Polyethylenimine-mediated in vivo gene transfer of a transmembrane superantigen fusion construct inhibits B16 murine melanoma growth. Cancer Gene Ther. 2008;15(11):742–749. [DOI] [PubMed] [Google Scholar]
- 16. Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387(10022):1012–1024. [DOI] [PubMed] [Google Scholar]
- 17. Lee AW, Ma BB, Ng WT, Chan AT. Management of nasopharyngeal carcinoma: current practice and future perspective. J Clin Oncol. 2015;33(29):3356–3364. [DOI] [PubMed] [Google Scholar]
- 18. Chen W, Hu GH. Biomarkers for enhancing the radiosensitivity of nasopharyngeal carcinoma. Cancer Biol Med. 2015;12(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan Y, Zhou X, Song J, et al. Expression and clinical significance of epidermal growth factor receptor and type 1 insulin-like growth factor receptor in nasopharyngeal carcinoma. Ann Otol Rhinol Laryngol. 2008;117(3):192–200. [DOI] [PubMed] [Google Scholar]
- 20. Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19(13):3159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clinical Oncol. 2015;12(1):11–26. [DOI] [PubMed] [Google Scholar]
- 22. Tao AL, He SH. Bridging PCR and partially overlapping primers for novel allergen gene cloning and expression insert decoration. World J Gastroenterol. 2004;10(14):2103–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guan F, Uboh CE, Soma LR, et al. LC-MS/MS method for confirmation of recombinant human erythropoietin and darbepoetin alpha in equine plasma. Anal Chem. 2007;79(12):4627–4635. [DOI] [PubMed] [Google Scholar]
- 24. Xu Q, Zhang X, Yue J, et al. Human TGFalpha-derived peptide TGFalphaL3 fused with superantigen for immunotherapy of EGFR-expressing tumours. BMC Biotechnol. 2010;10:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Disis ML, Bernhard H, Jaffee EM. Use of tumour-responsive T cells as cancer treatment. Lancet. 2009;373(9664):673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sapra P, Allen TM. Ligand-targeted liposomal anticancer drugs. Prog Lipid Res. 2003;42(5):439–462. [DOI] [PubMed] [Google Scholar]
- 27. Srinivasarao M, Galliford CV, Low PS. Principles in the design of ligand-targeted cancer therapeutics and imaging agents. Nat Rev Drug Discov. 2015;14(3):203–219. [DOI] [PubMed] [Google Scholar]
- 28. Yousefi F, Mousavi SF, Siadat SD, et al. Preparation and in vitro evaluation of antitumor activity of TGFalphaL3-SEB as a ligand-targeted superantigen. Technol Cancer Res Treat. 2016;15(2):215–226. [DOI] [PubMed] [Google Scholar]
- 29. Gerlach K, Tomuschat C, Finke R, et al. Experimental arthritis in the rat induced by the superantigen staphylococcal enterotoxin A. Scand J Immunol. 2017;85(3):191–196. [DOI] [PubMed] [Google Scholar]
- 30. Kim BS, Choi JK, Jung HJ, et al. Effects of topical application of a recombinant staphylococcal enterotoxin A on DNCB and dust mite extract-induced atopic dermatitis-like lesions in a murine model. Eur J Dermatol. 2014;24(2):186–193. [DOI] [PubMed] [Google Scholar]
- 31. Muro S. Challenges in design and characterization of ligand-targeted drug delivery systems. J Control Release. 2012;164(2):125–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Agheli R, Emkanian B, Halabian R, Fallah Mehrabadi J, Imani Fooladi AA. Recombinant staphylococcal enterotoxin type a stimulate antitumoral cytokines. Technol Cancer Res Treat. 2017;16(1):125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]