Abstract
Alzheimer's disease is characterized by amyloid β peptide (Aβ)-loaded plaques in the brain. Aβ is a cleavage fragment of amyloid-β protein precursor (APP) and over production of APP may lead to amyloidogenesis. The regulatory region of APP gene contains consensus sites recognized by transcription factor SP1, which has been shown to be required for the regulation of APP and Aβ. To understand the role of SP1 in APP biogenesis, herein we have characterized the relative distribution and localization of SP1, APP, and Aβ in various brain regions of rodent and primate models using immunohistochemistry. We observed that overall distribution and cellular localization of SP1, APP, and Aβ are similar and neuronal in origin. Their distribution is abundant in various layers of neocortex, but restricted to Purkinje cell layer of cerebellum, and pyramidal cell layer of hippocampus. These findings suggest that overproduction of Aβ in vivo may be associated with transcriptional pathways involving SP1 and APP gene.
Keywords: Abeta, Amyloidogenesis, APP, Brain, Immunohistochemistry, Monkey, SP1, Transcription
Introduction
The presence of abundant deposition of amyloid-beta (Aβ) in the brain is a hallmark pathological feature of Alzheimer's disease (AD), the most common neurodegenerative disorder found in elderly populations. Amyloidogenesis, the process of Aβ formation, is crucial in the development of AD pathology. Aβ is generated by the proteolysis of a transmembrane protein called amyloid-β protein precursor (APP) [11, 18, 22, 30, 41]. Anatomical characterization of AD brain shows atrophy of specific regions of the brain due to the loss of neurons, neuronal processes and synapses in the regions associated with higher intellectual functioning, including cerebral cortex and hippocampus [6, 52].
To understand the regulation of the APP gene, the 5'-untranslated region (5-UTR) as well as the 5'-flanking regulatory region, including the promoter, of the APP gene have been characterized [21, 26, 29, 38]. The 5'-flanking regulatory region of the APP gene lacks a characteristic TATA box, is rich in GC box elements, and contains consensus sites that are recognized by several transcription factors, including specificity protein 1 (SP1) [21, 26, 38]. SP1 binds to the human [15], rhesus [42, 43] and cynomolgus [32] monkey, and rat [4] APP promoters (Fig. 1) and accelerates the production of APP mRNA, which can be further spliced to generate several cell-specific species [16, 34]. SP1 belongs to a family of zinc finger protein (ZFP) transcription factors that includes other members such as SP2, SP3 and SP4 [13, 14, 17, 20, 37]. Several studies have demonstrated the presence and importance of SP1 in the expression of APP as well as β-APP cleaving enzyme 1 (BACE1), an enzyme that participates in the first step of APP proteolysis and is crucial for the formation of Aβ [1, 5, 10, 23, 25, 39]. Notably, the profiles of SP1 DNA-binding activity over the lifespan of rodents are commensurate with the patterns of APP mRNA expression, and depletion of SP1 levels in vitro greatly diminishes APP promoter activity [1]. In addition, elevated levels of SP3 and SP4 have recently been discovered in AD brain samples [2].
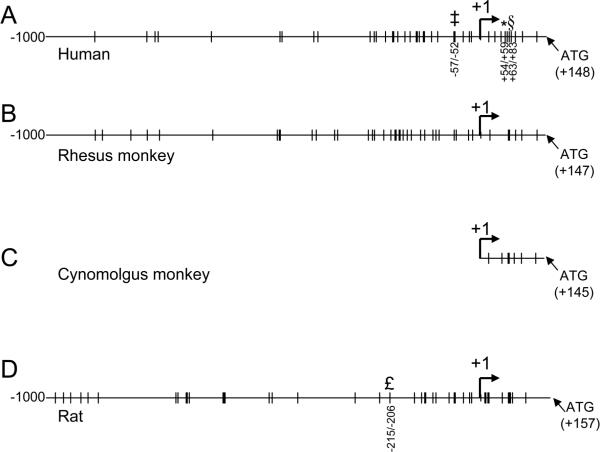
Fig. 1. SP1 consensus sites on human, rhesus and cynomolgus monkey, and rat APP promoter sequences.
Available sequences for human (GenBank #D87675), rhesus (GenBank #AF067971), cynomolgus (GenBank #M58727) and rat (RGSC v3.4) APP promoter sequences from -1000 bases upstream of the +1 transcription start site to the translation start “ATG” were scanned with the “TESS” utility [40] for the presence of SP1 consensus sequences at 95% minimum homology. Sites are indicated on the respective sequences. (A) Human sequence, experimentally-validated sites are indicated at positions -57/-52 [35], +54/+59 [8], and +63/+83 [50]. (B) Rhesus monkey sequence, (C) Available cynomolgus monkey sequence, (D) Rat sequence. Experimentally-validated SP1 site at -215/-206 [16] is indicated.
A series of studies have mapped SP1, APP and Aβ in rodent, primate, and human brains; however, these experiments limited analysis to one or two of these proteins at a time and to specific regions of the brain [3, 19, 27, 44, 48, 49]. Previous studies have also localized APP and Aβ on the transmembrane, within organelles and in the cytoplasmic compartment of cortical neurons [3, 16, 31, 48, 49]. In addition to an APP, Aβ and SP1 connection, the co-localization of Aβ and apolipoprotein E (apoE) has recently been demonstrated in the perivascular drainage channels of APP-transgenic mice suggesting an association of apoE in the perivascular clearance of Aβ [47].
In the current study, we examined the regional distribution and cellular location of SP1, APP, and Aβ in both rat and non-human primate brains. We further investigated the hypothesis that cells rich in SP1 contain high levels of APP and its amyloidogenic cleavage product, Aβ, thereby suggesting the role of transcriptional regulation in amyloidogenesis.
Materials and Methods
Animals & Tissue preparation, Rats
Timed-pregnant Long-Evans hooded rats were obtained from Charles River Laboratories (Wilmington, MA). Day of birth was considered postnatal day 0 (PND0). To randomize prenatal and genetic factors, pups from all litters were pooled and new litters were reconstituted by the random selection of 9-10 male pups per litter on PND1. Litters were maintained at a constant litter size over the course of lactation with the addition of female filler pups if one of the original male pups died. The animals were housed at ambient temperature (21 ± 2°C) and relative humidity (50 ± 10 %) with a 12-hour light/dark cycle (0700 - 1900 hours). Food and water were freely available throughout the study. Pups were weaned on PND21 and placed in group housing (3 animals/cage) for three months and then housed individually. All animal procedures were conducted according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Rhode Island.
On PND 600 (20 months), five animals (one from each litter) were used for the study. These animals were deeply anesthetized with an intraperitoneal injection of sodium pentobarbital (40 mg/kg body weight) and transcardially perfused with 100 cc of perfusion wash (0.8% sodium chloride, 0.4% dextrose, 0.8% sucrose, 0.023% calcium chloride and 0.034% anhydrous-sodium cacodylate) followed by 100 cc perfusion fix (4% sucrose, 4% paraformaldehyde and 1.07% anhydrous sodium cacodylate, pH 7.2). The brains were removed and post-fixed in the same fixative (perfusion fix) overnight. The collected brains were subjected to sectioning. Coronal sections (40 μm) were collected using MultiBrain Technology (NeuroScience Associates, Knoxville, TN). Sections spanning the cerebellum and hippocampus were maintained in a preservative fixative at -20°C. These sections were subjected to immunohistochemical analysis as described in subsequent sections.
Animals & Tissue preparation, Monkeys
Twenty-three year-old cynomolgus (Macaca fascicularis) female monkeys, housed at the NIH Poolesville, MD, primate facility over the last decade of life, were euthanized via overdose of pentobarbital, with a veterinary confirmation of death. The brains were excised, cut along the mid-sagittal plane, and each hemisphere was cut into 10 mm sections and rapidly immersion-fixed in 10% neutral buffered formalin. The brain sections containing the frontal association cortex were rinsed and taken through a graded series of ethanol concentrations, processed, and embedded in paraffin. Eight micron serial sections through the region were collected on charged slides.
Immunohistochemistry
To determine the cellular distribution of APP, SP1, and Aβ, immunohistochemistry was conducted using free-floating brain tissue of 20 month-old rodents and the paraffin embedded brain tissue of 23 year-old cynomolgus monkeys. Free-floating sections of brain tissues obtained from rodents were taken from the preservative fixative and briefly washed in distilled water to rinse away the fixative prior to PBS rinses. Both free-floating rodent and paraffin embedded monkey brain tissues were subjected to brief washes in 1X Phosphate Buffer Saline (PBS) and 3% Hydrogen peroxide (H2O2). After rinsing, the sections were incubated in PBS with 2% Bovine Serum Albumin (BSA) and 1% Triton X-100 blocking solution for 30 min., and incubated in the presence of either rabbit IgG specific to the N terminus (Fig. 2A) of APP (1:200; Sigma-Aldrich, St. Louis, MO), monoclonal antibody 6E10 for APP/Aβ (Fig. 2A) (1:50; Sigma-Aldrich), or rabbit antiserum to an internal epitope within SP1 (Fig. 2B) (1:100; Santa Cruz Biotechnology, Santa Cruz, CA), overnight at 4° C. Sections were washed with PBS and incubated along with the species-specific biotinylated secondary antibody mouse/rabbit (1:200;Vector Labs, Burlingame, CA) for 30 min. The sections were incubated with Streptavidin (Vector Labs) for 30 min, rinsed briefly with PBS, and immunoreactivity detected with the substrate, 3-3′ Diaminobenzidine tetra hydrochloride (DAB) (Vector Labs). The free-floating rodent sections were mounted on microscope slides using procedures provided by Neuroscience Associates, Knoxville, TN. Coverslips were mounted with Permanent Mounting Medium (Vector Labs) on the rodent and monkey microslides.
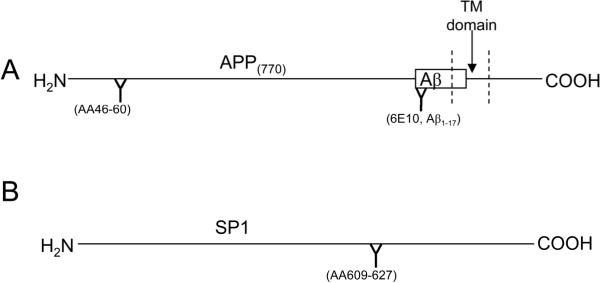
Fig. 2. Epitopes of antisera/antibodies used in this study.
Schematic diagrams of (A) APP (indicating location of Aβ peptide) and (B) SP1 proteins with binding sites of antisera/antibodies shown.
Immunofluorescent staining
Free-floating sections (40 microns) of rat hippocampus, cortex and cerebellum were removed from storage solution, washed with distilled water, 1X PBS and 3% H2O2 and incubated in 2% BSA/1% Triton X-100/PBS blocking solution for 30 min. Sections were incubated overnight with the primary antibody for either APP (1:200), SP1 (1:100), or Aβ (1:50), and washed with PBS. The sections were incubated with the species-specific biotinylated secondary antibody against mouse/rabbit (1:200; Vector Labs) for 30 min., and rinsed with buffer. Sections were incubated with Texas Red Streptavidin (1:100; Vector Labs) for 30 min to facilitate the attachment of the fluorescent tag to target proteins. After a brief rinse in PBS, sections were mounted on microscope slides according to standard procedures of Neuroscience Associates. The slides were secured with cover slips using the Fluorescence Mounting Medium (Vector Labs) to preserve the quality of fluorescence immunoreactivity.
Double fluorescence staining was conducted for SP1 and APP in the monkey brain sections. These sections were first immunostained for SP1 and then labeled with Texas Red using the immunofluorescent procedure as described above. After a brief rinse in PBS, the previous steps were repeated using the primary APP antibody labeled with Fluorescein (green) streptavidin (1:200; Vector Labs) to distinguish the presence of APP from SP1. After a brief rinse in PBS, sections were dried and cover-slipped with Fluorescence Mounting Medium (Vector Labs) to preserve the quality of fluorescence immunoreactivity. This double labeling of SP1 (Texas Red) and APP (Fluorescein) was performed in the same section to show co-localization in the neurons.
In all cases, negative controls were run on slides from each animal by omitting the respective primary antibody incubation step. No signal was evident after incubation of tissues in secondary antibody alone.
Microscopy
All sections were examined using a Nikon Eclipse E600 microscope with an attached Diagnostic Instruments digital camera, using SPOT Diagnostic Instruments system and software (Nikon, Melville, NY). Fluorescent images were viewed using FITC (green) and Rhodamine (red) filters in the Nikon Epi-fluorescence filter set to visualize the immunoreactivity of target proteins. Further studies are needed to quantify the protein expression, cell loss, and damage.
Results
The premise of the present work is that SP1 consensus sites are present on human, rhesus and cynomolgus monkey, and rat APP promoter sequences (Fig. 1). The data presented in the following subsequent studies were based on the observations made with at least 3 sections/slides per each staining process in the presence of different primary antibodies raised against epitopes of APP, Aβ, and SP1 proteins as shown in Fig. 2.
Rodent Studies of SP1, APP, and Aβ immunoreactivity in different regions of the brain
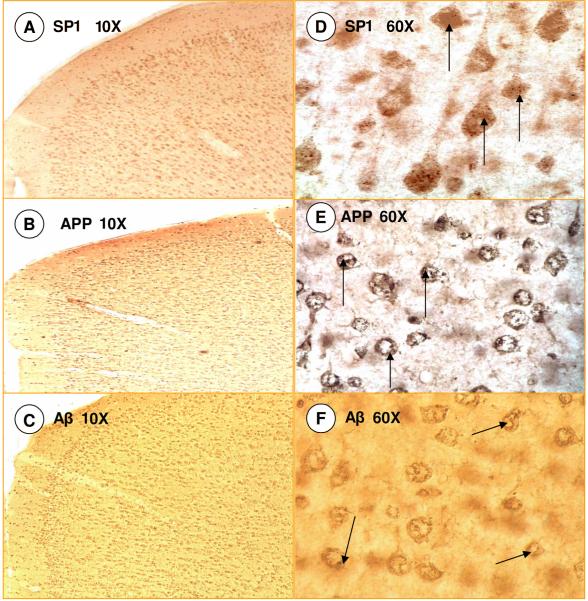
The staining patterns of SP1, APP, and Aβ were visualized in various brain regions of the rat including cerebral cortex, hippocampus, and cerebellum. The general pattern of immunoreactivity, as visualized by density of staining, in the cerebral cortex was similar for SP1, APP, and Aβ with a distribution throughout the cortical layers (Fig. 3) and sparse staining in the molecular layer (Fig. 3 A, B, C). That is to say, the cells that show intense SP1 staining also show a similar greater intensity of APP staining, whereas areas where SP1 staining is absent are also deficient in APP staining. Higher magnification (60X objective) revealed that SP1 was localized in the soma of pyramidal and globular neurons of multiple cortical layers (Fig. 3D). APP and Aβ stainings were detected at the cell membrane and within the organelles and cytoplasmic compartment of cortical neurons (Fig. 3E & F) as previously reported [3, 16, 31, 48, 49].
Fig. 3. SP1, APP and Aβ Immunoreactivity in Rat Cortex.
(A-C) Similarity of staining pattern of SP1, APP and Aβ; (D-F). Cellular characteristics of SP1, APP and Aβ at higher magnification.
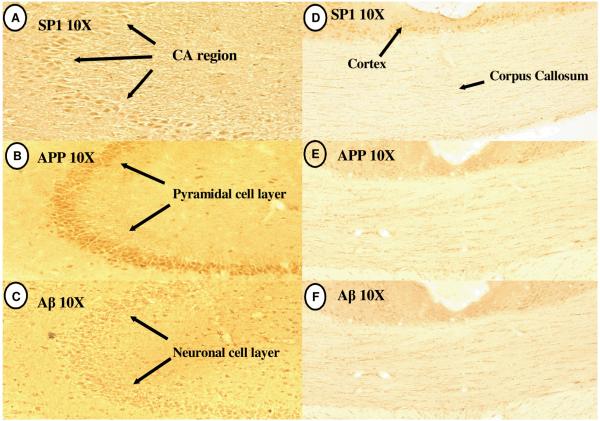
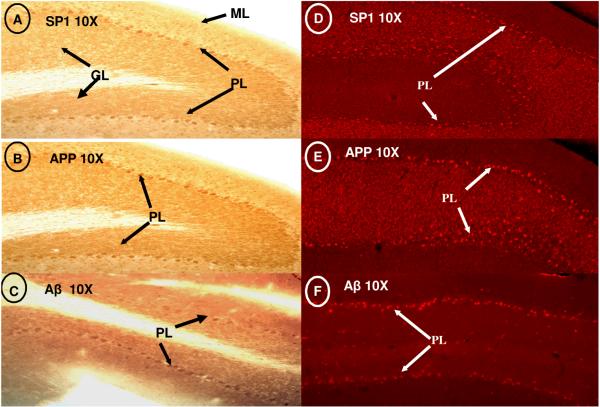
In the hippocampus, immunoreactivity for SP1, APP, and Aβ was restricted to the pyramidal cell layer of CA regions 1-3 (Fig. 4A-C); however distinct staining was not found in the corpus callosum (Figure 2D-F). Distinct localization of SP1, APP, and Aβ expression was found in the globular Purkinje Cells (PC) of the cerebellar lobule; some scattered staining was also noted in the molecular layer (Fig. 5A-C). Fluorescence labeling in the cerebellum further confirmed the defined neuronal expression of these components in the Purkinje cell layers (Fig. 5D-F). It is noteworthy that the regions of the cerebrum, hippocampus, and cerebellum in which the cells over-expressed SP1 also contained higher levels of APP and an overabundance of Aβ.
Fig. 4. SP1, APP and Aβ immunoreactivity in hippocampus and corpus callosum of rat.
(A-C) Similarity of staining in the CA region of the Hippocampus; (D-F) Absence of staining for SP1, APP and Aβ in Corpus Callosum.
Fig. 5. SP1, APP and Aβ Immunoreactivity in rat cerebellum.
(A-C) Similarity of SP1, APP, and Aß staining in the Purkinje cell layer of the cerebellum; (D-F) Fluorescent micrographs of the Purkinje cell layer. ML= molecular layer. PL= Purkinje cell layer. GL= granular cell layer.
Primate Studies of SP1, APP, and Aβ immunoreactivity in different regions of the brain
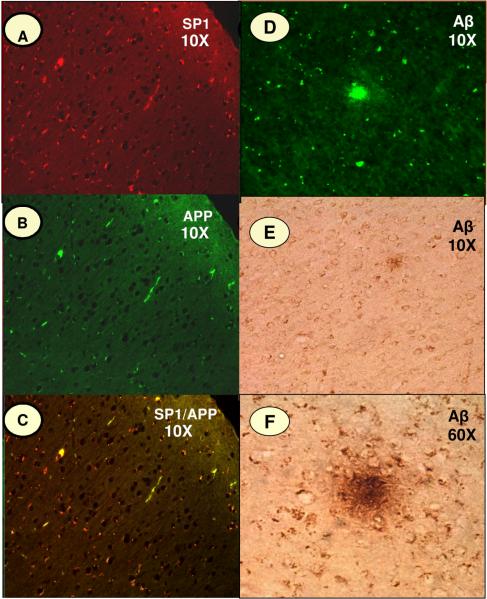
To investigate whether the aforementioned results with a rodent species are species-specific, immunostaining pattern for SP1, APP, and Aβ was further analyzed by fluorescence tagging in paraffin-fixed slides prepared from the frontal association cortex of cynomolgus monkeys. The fluorescence tagging confirmed that the expression of these proteins was localized primarily in neuronal cells (Figure-4). Texas red (SP1) and APP fluorescein (APP) were used to distinguish the presence of each protein, with the merged image (yellow) indicating overlapping presence for SP1 and APP in the same cells (Fig. 6A-C). 6E10 staining (APP/Aβ) highlighted areas of plaque formation in the cortical sections (Fig. 6E and F), indicating that a significant portion of the 6E10-generated signal was extracellular and distinct from N-terminal APP signal.
Fig. 6. SP1, APP and Aβ Immunoreactivity in Monkey Cortex.
Intra-neuronal staining of SP1, APP and Aβ in the frontal association cortex of cynomolgus monkey (Macaca fascicularis). The fluorescence immunoreactivity of (A) SP1, (B) APP, and (D) Aβ; (C) shows the co-localization of SP1 and APP; (E & F) reveals the staining patterns of Aβ in senile plaques.
Discussion
A series of studies have provided evidence that SP1 is involved in the regulation of the APP gene, with SP1 proposed as an essential transcription factor in the expression of APP [1, 8, 28, 33, 36, 50]. Our previous study suggested that, with a concurrent activation of APP (Fig. 7) and BACE1 expression, SP1 could also be involved in amyloidogenesis. Notably, siRNA knockdown of SP1 abolished about 70% of the activity of the human APP promoter in transfected PC12 cells and SP1 activity across lifespan mirrors that of APP gene expression [1]. Furthermore, the ability of SP1 to regulate the expression of BACE1, the crucial β-secretase associated with the onset of AD, has also been demonstrated [5, 45]. Recent studies also suggested that intracellular accumulation of Aβ in neuronal cells may promote AD pathology [6, 7, 9, 19, 46, 51]. However, these experiments limited analysis to one or two of these proteins at a time and to specific regions of the brain of one species. To bridge this gap of knowledge and extend the study from rodent to primate models, the current investigation was designed to examine the localization of SP1, Aβ, and APP in distinct brain regions of both rodent and primate. Our immunohistological characterizations ought not be taken as sole evidence for the relationship between these proteins but are to provide corroboration of the relationship between SP1 and APP established in previous studies. The staining pattern displayed in the rodent brain was suggestive of a primary neuronal localization, with a notable lack of SP1 and APP signals in glia. This neuronal localization was present in all regions examined. In addition, merged images for both APP and SP1 demonstrated localization within a subpopulation of neurons.
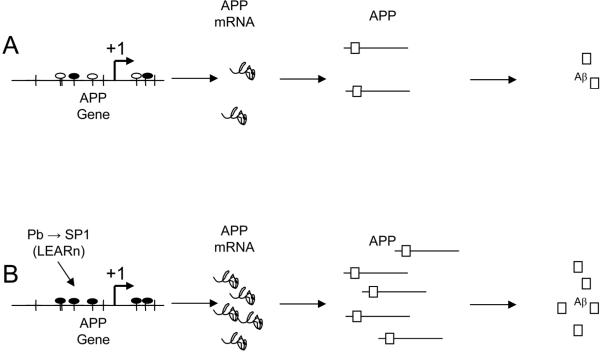
Fig. 7. Model of SP1 action upon the APP gene, requiring co-localization.
(A) Non-pathogenic model of SP1 effects on APP expression. SP1 transcription factor (black oval) interacts with the APP gene promoter to stimulate transcription at levels leaving potential SP1 consensus sites (empty oval) unstimulated. APP mRNA is translated to APP protein, which is processed by secretases to produce (among other products) Aβ peptide. (B) In the pathogenic pathway, SP1 promoter activity is increased either acutely or in a latent (LEARn) fashion by early-life exposure to environmental insult [1, 24]. Increased SP1 expression leads to greater SP1 TF availability, which stimulates higher levels of APP transcription. Increased APP levels lead to more available APP, which is processed by secretases to Aβ.
This study showed that SP1, APP, and Aβ immunoreactivity in the rat cerebral cortex, hippocampus, and cerebellum are similar and appear to be localized exclusively to neurons (Fig. 3-6). In addition, within the hippocampus, the highly dense granule cells do not exhibit any staining beyond background. Similarly, in the cerebellum, both SP1 and APP proteins are expressed only in the Purkinje cell layer, with no staining in the cerebellar granule cell layer. Our studies also suggested that the relationship between SP1 and APP is not species-specific and is also present in non-human primates. The co-localization of SP1 and APP in primates is highly relevant because unlike rodents, aging primates are known to express Aβ plaques. SP1 has a developmental pattern of expression and its activation is dependent on certain stimuli. [54]. Furthermore, were SP1 a general transcription factor, one would expect widespread initiation in all its target genes, however, we have previously found that SP1 target genes are selectively activated under certain conditions [1, 54]. Thus, these data strongly suggest that amyloidogenesis and plaque formation may be driven in part by a transcriptional pathway which involves interactions between SP1 and the APP gene.
Overall, these in vivo immunohistochemical findings validate the existence of a relationship between APP expression and SP1, and suggest that Aβ production occurs in cells where SP1 is strongly present. The predominant localization of these three proteins in the same neurons and in areas primarily affected by AD pathology suggests that SP1 may render some neurons more susceptible to amyloidogenic damage. Furthermore, SP1 is also thought to have a role in the regulation of the tau gene (MAPT) and thus could also contribute to the formation of neurofibrillary tangles. Tangles have been shown to co-exist with Aβ [12]. Thus, environmental models of AD should consider SP1 as a potential target that could influence the process of both amyloidogenesis and tau deposition. Taken together these results, along with the recently proposed role of SP1 in altering latent early-life associated regulation (LEARn) that affects the expression of genes associated with a later-manifest condition, could explain the progression of amyloidogenesis seen in AD [24]
Based on these results, we propose the following model of SP1 action upon the APP gene, requiring co-localization as shown in Fig. 7. Under a non-pathogenic model of SP1 effects on APP expression, SP1 transcription factor interacts with the APP gene promoter to stimulate transcription at levels, leaving some potential SP1 consensus sites unstimulated. APP mRNA is translated to APP protein, which is processed by secretases to produce (among other products) Aβ peptide. Under the pathogenic pathway, SP1 promoter activity is increased either acutely or in a latent (LEARn) fashion by early-life exposure to environmental insult, such as lead (Pb) [23, 24] as shown by our group in mice [1] and more recently in monkeys [53]. Increased SP1 expression leads to greater SP1 TF availability, which stimulates higher levels of APP transcription. Increased APP levels lead to more available APP, which is processed by secretases to Aβ.
Acknowledgements
This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (NIEHS) and by grants (ES013022 and AG027246) from the National Institutes of Health (NIH) awarded to NHZ. The research core facility was funded (P20RR016457) by the National Center for Research Resources (NCRR), a component of NIH. Work at DKL's laboratory was funded by the Alzheimer's association and NIH (AG18379 and AG18884). The authors thank Dr. Gordon Flake and Dr. Dixie-Ann Sawin for their helpful comments on an earlier version of this manuscript.
References
- [1].Basha MR, Wei W, Bakheet SA, Benitez N, Siddiqi HK, Ge YW, Lahiri DK, Zawia NH. The fetal basis of amyloidogenesis: exposure to lead and latent overexpression of amyloid precursor protein and beta-amyloid in the aging brain. J. Neurosci. 2005;25:823–9. doi: 10.1523/JNEUROSCI.4335-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Boutillier S, Lannes B, Buée L, Delacourte A, Rouaux C, Mohr M, Bellocq J-P, Sellal F, Larmet Y, Boutillier A-L, Loeffler J-P. Sp3 and Sp4 Transcription Factor Levels Are Increased in Brains of Patients with Alzheimer's Disease. Neurodegenerative Disease. 2007;4:413–423. doi: 10.1159/000107701. [DOI] [PubMed] [Google Scholar]
- [3].Cataldo AM, Petanceska S, Terio NB, Peterhoff CM, Durham R, Mercken M, Mehta PD, Buxbaum J, Haroutunian V, Nixon RA. Abeta localization in abnormal endosomes: association with earliest Abeta elevations in AD and Down syndrome. Neurobiol Aging. 2004;25:1263–72. doi: 10.1016/j.neurobiolaging.2004.02.027. [DOI] [PubMed] [Google Scholar]
- [4].Chernak JM. Structural features of the 5' upstream regulatory region of the gene encoding rat amyloid precursor protein. Gene. 1993;133:255–260. doi: 10.1016/0378-1119(93)90648-m. [DOI] [PubMed] [Google Scholar]
- [5].Christensen MA, Zhou W, Qing H, Lehman A, Philipsen S, Song W. Transcriptional regulation of BACE1, the beta-amyloid precursor protein beta-secretase, by Sp1. Mol Cell Biol. 2004;24:865–74. doi: 10.1128/MCB.24.2.865-874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].D'Andrea MR, Nagele RG, Wang HY, Lee DH. Consistent immunohistochemical detection of intracellular beta-amyloid42 in pyramidal neurons of Alzheimer's disease entorhinal cortex. Neurosci Lett. 2002;333:163–6. doi: 10.1016/s0304-3940(02)00875-3. [DOI] [PubMed] [Google Scholar]
- [7].D'Andrea MR, Nagele RG, Wang HY, Peterson PA, Lee DH. Evidence that neurones accumulating amyloid can undergo lysis to form amyloid plaques in Alzheimer's disease. Histopathology. 2001;38:120–34. doi: 10.1046/j.1365-2559.2001.01082.x. [DOI] [PubMed] [Google Scholar]
- [8].Docagne F, Gabriel C, Lebeurrier N, Lesne S, Hommet Y, Plawinski L, Mackenzie ET, Vivien D. Sp1 and Smad transcription factors co-operate to mediate TGF-beta-dependent activation of amyloid-beta precursor protein gene transcription. Biochem J. 2004;383:393–399. doi: 10.1042/BJ20040682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Echeverria V, Cuello AC. Intracellular A-beta amyloid, a sign for worse things to come? Mol Neurobiol. 2002;26:299–316. doi: 10.1385/MN:26:2-3:299. [DOI] [PubMed] [Google Scholar]
- [10].Ge Y-W, Maloney B, Sambamurti K, Lahiri DK. Functional characterization of the 5' flanking region of the BACE gene: identification of a 91 bp fragment involved in basal level of BACE promoter expression. FASEB J. 2004;18:1037–1039. doi: 10.1096/fj.03-1379fje. [DOI] [PubMed] [Google Scholar]
- [11].Glenner GG, Wong CW. Alzheimer's disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984:885–90. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- [12].Grundke-Iqbal I, Iqbal K. Neuronal cytoskeleton in the biology of Alzheimer disease. Prog Clin Biol Res. 1989;317:745–53. [PubMed] [Google Scholar]
- [13].Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem. 1995;270:24989–94. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- [14].Hagen G, Muller S, Beato M, Suske G. Cloning by recognition site screening of two novel GT box binding proteins: a family of Sp1 related genes. Nucleic Acids Res. 1992;20:5519–25. doi: 10.1093/nar/20.21.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Hattori M, Tsukahara F, Furuhata Y, Tanahashi H, Hirose M, Saito M, Tsukuni S, Sakaki Y. A novel method for making nested deletions and its application for sequencing of a 300 kb region of human APP locus. Nucleic Acids Res. 1997;25:1802–1808. doi: 10.1093/nar/25.9.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hoffman PW, Chernak JM. DNA binding and regulatory effects of transcription factors SP1 and USF at the rat amyloid precursor protein gene promoter. Nucleic Acids Res. 1995;23:2229–35. doi: 10.1093/nar/23.12.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51:1079–90. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- [18].Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Muller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–6. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- [19].Kimura N, Yanagisawa K, Terao K, Ono F, Sakakibara I, Ishii Y, Kyuwa S, Yoshikawa Y. Age-related changes of intracellular Abeta in cynomolgus monkey brains. Neuropathol Appl Neurobiol. 2005;31:170–80. doi: 10.1111/j.1365-2990.2004.00624.x. [DOI] [PubMed] [Google Scholar]
- [20].Kingsley C, Winoto A. Cloning of GT box-binding proteins: a novel Sp1 multigene family regulating T-cell receptor gene expression. Mol Cell Biol. 1992;12:4251–61. doi: 10.1128/mcb.12.10.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].La Fauci G, Lahiri DK, Salton SR, Robakis NK. Characterization of the 5'-end region and the first two exons of the beta-protein precursor gene. Biochem Biophys Res Commun. 1989;159:297–304. doi: 10.1016/0006-291x(89)92437-6. [DOI] [PubMed] [Google Scholar]
- [22].Lahiri DK, Farlow MR, Sambamurti K, Greig NH, Giacobini E, Schneider LS. A critical analysis of new molecular targets and strategies for drug developments in Alzheimer's disease. Curr. Drug Targets. 2003;4:97–112. doi: 10.2174/1389450033346957. [DOI] [PubMed] [Google Scholar]
- [23].Lahiri DK, Ge Y-W, Rogers JT, Sambamurti K, Greig NH. Taking down the unindicted co-conspirators of amyloid beta-peptide-mediated neuronal death: Shared gene regulation of BACE1 and APP genes interacting with CREB, Fe65 and YY1 transcription factors. Curr. Alzheimer Res. 2006;3:475–484. doi: 10.2174/156720506779025224. [DOI] [PubMed] [Google Scholar]
- [24].Lahiri DK, Maloney B, Basha MR, Ge YW, Zawia NH. How and when environmental agents and dietary factors affect the course of Alzheimer's disease: the “LEARn” model (Latent Early Associated Regulation) may explain the triggering of AD. Curr. Alzheimer Res. 2007;4:219–228. doi: 10.2174/156720507780362164. [DOI] [PubMed] [Google Scholar]
- [25].Lahiri DK, Maloney B, Ge YW. Functional domains of the BACE1 and BACE2 promoters and mechanisms of transcriptional suppression of the BACE2 promoter in normal neuronal cells. J Mol Neurosci. 2006;29:65–80. doi: 10.1385/JMN:29:1:65. [DOI] [PubMed] [Google Scholar]
- [26].Lahiri DK, Robakis NK. The promoter activity of the gene encoding Alzheimer beta-amyloid precursor protein (APP) is regulated by two blocks of upstream sequences. Brain Res. Mol. Brain Res. 1991;9:253–257. doi: 10.1016/0169-328x(91)90009-m. [DOI] [PubMed] [Google Scholar]
- [27].Langui D, Girardot N, El Hachimi KH, Allinquant B, Blanchard V, Pradier L, Duyckaerts C. Subcellular topography of neuronal Abeta peptide in APPxPS1 transgenic mice. Am J Pathol. 2004;165:1465–77. doi: 10.1016/s0002-9440(10)63405-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lukiw WJ, Rogaev EI, Wong L, Vaula G, McLachlan DR, George Hyslop P. Protein-DNA interactions in the promoter region of the amyloid precursor protein (APP) gene in human neocortex. Brain Res Mol Brain Res. 1994;22:121–31. doi: 10.1016/0169-328x(94)90039-6. [DOI] [PubMed] [Google Scholar]
- [29].Maloney B, Ge YW, Greig N, Lahiri DK. Presence of a "CAGA box" in the APP gene unique to amyloid plaque-forming species and absent in all APLP-1/2 genes: implications in Alzheimer's disease. FASEB J. 2004;18:1288–1290. doi: 10.1096/fj.03-1703fje. [DOI] [PubMed] [Google Scholar]
- [30].Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci U S A. 1985;82:4245–9. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Muresan Z, Muresan V. A phosphorylated, carboxy-terminal fragment of beta-amyloid precursor protein localizes to the splicing factor compartment. Hum Mol Genet. 2004;13:475–88. doi: 10.1093/hmg/ddh054. [DOI] [PubMed] [Google Scholar]
- [32].Podlisny MB, Tolan DR, Selkoe DJ. Homology of the amyloid beta protein precursor in monkey and human supports a primate model for beta amyloidosis in Alzheimer's disease. Am. J. Pathol. 1991;138:1423–1435. [PMC free article] [PubMed] [Google Scholar]
- [33].Pollwein P. Overlapping binding sites of two different transcription factors in the promoter of the human gene for the Alzheimer amyloid precursor protein. Biochem Biophys Res Commun. 1993;190:637–47. doi: 10.1006/bbrc.1993.1096. [DOI] [PubMed] [Google Scholar]
- [34].Pollwein P, Masters CL, Beyreuther K. The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res. 1992;20:63–8. doi: 10.1093/nar/20.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Querfurth HW, Jiang J, Xia W, Selkoe DJ. Enhancer function and novel DNA binding protein activity in the near upstream [beta]APP gene promoter. Gene. 1999;232:125–141. doi: 10.1016/s0378-1119(99)00091-8. [DOI] [PubMed] [Google Scholar]
- [36].Querfurth HW, Jiang J, Xia W, Selkoe DJ. Enhancer function and novel DNA binding protein activity in the near upstream betaAPP gene promoter. Gene. 1999;232:125–41. doi: 10.1016/s0378-1119(99)00091-8. [DOI] [PubMed] [Google Scholar]
- [37].Saffer JD, Jackson SP, Annarella MB. Developmental expression of Sp1 in the mouse. Mol Cell Biol. 1991;11:2189–99. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Salbaum JM, Weidemann A, Lemaire HG, Masters CL, Beyreuther K. The promoter of Alzheimer's disease amyloid A4 precursor gene. Embo J. 1988;7:2807–13. doi: 10.1002/j.1460-2075.1988.tb03136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sambamurti K, Kinsey R, Maloney B, Ge YW, Lahiri DK. Gene structure and organization of the human beta-secretase (BACE) promoter. FASEB J. 2004;18:1034–1036. doi: 10.1096/fj.03-1378fje. [DOI] [PubMed] [Google Scholar]
- [40].Schug J, Overton GC. Technical Reports of the Computational Biology and Informatics Laboratory. School of Medicine, University of Pennsylvania; 1998. TESS: Transcription Element Search Software on the WWW. CBIL-TR-1997-1001. [Google Scholar]
- [41].Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–66. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- [42].Song W, Lahiri DK. Functional identification of the promoter of the gene encoding the Rhesus monkey beta-amyloid precursor protein. Gene. 1998;217:165–176. doi: 10.1016/s0378-1119(98)00340-0. [DOI] [PubMed] [Google Scholar]
- [43].Song W, Lahiri DK. Molecular cloning of the promoter of the gene encoding the Rhesus monkey beta-amyloid precursor protein: structural characterization and a comparative study with other species. Gene. 1998;217:151–164. doi: 10.1016/s0378-1119(98)00337-0. [DOI] [PubMed] [Google Scholar]
- [44].Sparks DL. Intraneuronal beta-amyloid immunoreactivity in the CNS. Neurobiol Aging. 1996;17:291–9. doi: 10.1016/0197-4580(95)02067-5. [DOI] [PubMed] [Google Scholar]
- [45].Sun X, Wang Y, Qing H, Christensen MA, Liu Y, Zhou W, Tong Y, Xiao C, Huang Y, Zhang S, Liu X, Song W. Distinct transcriptional regulation and function of the human BACE2 and BACE1 genes. Faseb J. 2005;19:739–49. doi: 10.1096/fj.04-3426com. [DOI] [PubMed] [Google Scholar]
- [46].Tabira T, Chui DH, Nakayama H, Kuroda S, Shibuya M. Alzheimer's disease with spastic paresis and cotton wool type plaques. J Neurosci Res. 2002;70:367–72. doi: 10.1002/jnr.10392. [DOI] [PubMed] [Google Scholar]
- [47].Thal DR, Larionov S, Abramowski D, Wiederhold KH, Van Dooren T, Yamaguchi H, Haass C, Van Leuven F, Staufenbiel M, Capetillo-Zarate E. Occurrence and co-localization of amyloid beta-protein and apolipoprotein E in perivascular drainage channels of wild-type and APP-transgenic mice. Neurobiol Aging. 2006 doi: 10.1016/j.neurobiolaging.2006.05.029. [DOI] [PubMed] [Google Scholar]
- [48].Toledano A, Alvarez MI, Rivas L, Lacruz C, Martinez-Rodriguez R. Amyloid precursor proteins in the cerebellar cortex of Alzheimer's disease patients devoid of cerebellar beta-amyloid deposits: immunocytochemical study of five cases. J Neural Transm. 1999;106:1151–69. doi: 10.1007/s007020050231. [DOI] [PubMed] [Google Scholar]
- [49].Verbeek MM, Otte-Holler I, Fransen JA, de Waal RM. Accumulation of the amyloid-beta precursor protein in multivesicular body-like organelles. J Histochem Cytochem. 2002;50:681–90. doi: 10.1177/002215540205000509. [DOI] [PubMed] [Google Scholar]
- [50].Villa A, Santiago J, Belandia B, Pascual A. A response unit in the first exon of the beta-amyloid precursor protein gene containing thyroid hormone receptor and Sp1 binding sites mediates negative regulation by 3,5,3'-triiodothyronine. Mol Endocrinol. 2004;18:863–73. doi: 10.1210/me.2003-0260. [DOI] [PubMed] [Google Scholar]
- [51].Wang HY, D'Andrea MR, Nagele RG. Cerebellar diffuse amyloid plaques are derived from dendritic Abeta42 accumulations in Purkinje cells. Neurobiol Aging. 2002;23:21–23. doi: 10.1016/s0197-4580(01)00279-2. [DOI] [PubMed] [Google Scholar]
- [52].Wenk GL. Neuropathologic changes in Alzheimer's disease. J Clin Psychiatry. 2003;64(Suppl 9):7–10. [PubMed] [Google Scholar]
- [53].Wu J, Basha MR, Brock B, Maloney B, Cox D, Harry J, Cardozo-Paleaz F, Rice DC, Lahiri DK, Zawia NH. Alteration of Alzheimer-like pathology in aged monkeys by developmental exposure to lead (Pb) J. Neurosci. 2007 doi: 10.1523/JNEUROSCI.4405-07.2008. IN PRESS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zawia NH, Sharan R, Brydie M, Oyama T, Crumpton T. Sp1 as a target site for metal-induced perturbations of transcriptional regulation of developmental brain gene expression. Brain Res Dev Brain Res. 1998;107:291–8. doi: 10.1016/s0165-3806(98)00023-6. [DOI] [PubMed] [Google Scholar]