Abstract
HER family members are interdependent and functionally compensatory. Simultaneously targeting EGFR/HER2/HER3 by antibody combinations has demonstrated superior treatment efficacy over targeting one HER receptor. However, antibody combinations have their limitations, with high immunogenicity and high cost. In this study, we have developed a three-in-one nucleic acid aptamer-small interfering RNA (siRNA) chimera, which targets EGFR/HER2/HER3 in one molecule. This inhibitory molecule was constructed such that a single EGFR siRNA is positioned between the HER2 and HER3 aptamers to create a HER2 aptamer-EGFR siRNA-HER3 aptamer chimera (H2EH3). EGFR siRNA was delivered into HER2-expressing cells by HER2/HER3 aptamer-induced internalization. HER2/HER3 aptamers act as antagonist molecules for blocking HER2 and HER3 signaling pathways and also as tumor-targeting agents for siRNA delivery. H2EH3 enables down-modulation of the expression of all three receptors, thereby triggering cell apoptosis. In breast cancer xenograft models, H2EH3 is able to bind to breast tumors with high specificity and significantly inhibits tumor growth via either systemic or intratumoral administration. Owing to low immunogenicity, ease of production, and high thermostability, H2EH3 is a promising therapeutic to supplement current single HER inhibitors and may act as a treatment for HER2+ breast cancer with intrinsic or acquired resistance to current drugs.
Keywords: aptamer, siRNA, HER family, HER2, HER3, EGFR, breast cancer, trastuzumab resistance, non-immunogenicity, nontoxic
Introduction
20%–30% of breast cancer is characterized by HER2 overexpression that correlates with poor prognosis, high aggressiveness, and extensive drug resistance.1 Trastuzumab (Herceptin), a humanized antibody, is the first-line treatment for patients with HER2+ breast cancer. However, the majority of patients eventually develop resistance. Resistance to HER2-targeted therapies is associated with increased levels of other HER family receptors.2 The HER family consists of epidermal growth factor receptor (EGFR) (HER1), HER2, HER3, and HER4.3 It is well-known that the HER family is interdependent and displays functional redundancy in that the blockade of one HER receptor can often be compensated by another HER family member.4 EGFR, HER2, and HER3 contribute to the initiation and progression of many cancers and are well-recognized therapeutic targets. Ligand binding to HER receptors results in the formation of homo- and hetero-dimers. EGFR is one of the most studied HER family receptors and a key oncogenic driver in many epithelial cancers.5 Accumulating evidence shows that HER3 is a key node in many cancers and involved in the resistance against EGFR- and HER2-targeted therapies through activation of a compensatory PI3K-AKT survival pathway.6, 7 Resistance to trastuzumab in breast cancer cell lines is associated with upregulation of EGFR and HER3.2, 8 A HER2/HER3 heterodimer has been identified to function like an oncogene to support the proliferation of HER2-overexpressing tumor cells.7 The cross-talk and compensatory functionalities of HER family receptors provide strong rationales for co-targeting EGFR, HER2, and HER3 in HER-dependent cancer treatment. In line with the notion of combing treatment, bi-specific antibodies and pan-HER antibodies have been developed and show more efficacy than single receptor-targeting antibodies. Bispecific MM11 antibody targeting both HER2 and HER3 is able to suppress the proliferation of HER2-expressing tumor cells.9 Pan-HER with a six antibody mixture targeting EGFR, HER2, and HER3 has been developed and has displayed superior potency compared to agents targeting single receptors in preclinical studies.10
Because HER3 lacks intrinsic kinase activity, TKIs (tyrosine-kinase inhibitors) will be not effective in inhibiting HER3. Antibody combinations are the current strategies to inhibit the HER family signaling network.11, 12 However, most of these antibodies have a high cost in production and high immunogenicity. To address immunogenicity and the high cost of these antibodies, in our studies, we have engineered a nucleic-acid-based multiple function molecule with a small interfering RNA (siRNA) and two RNA aptamers. The new siRNA-aptamer chimera is able to target EGFR/HER2/HER3 simultaneously, with low toxicity. Recently, an aptamer-siRNA chimera, employing only RNA molecules, has become attractive for cell-type-specific gene silencing. Aptamers are single-stranded DNA (ssDNA) or RNA and selected through the in vitro enrichment process.13 Like antibodies, aptamers can bind to a target with high affinity and specificity. Due to small and oligonucleotide properties, aptamers offer many advantages over antibodies, including non-immunogenicity, high tissue penetration, thermostability, low cost, and ease of synthesis and modification.14, 15 Current cell-type-specific RNA aptamers have been used for targeting delivery of siRNA and drugs.16, 17
HER3 aptamer has been identified and is able to specifically bind to the extracellular domains of HER3.18 HER3 aptamer inhibits HRG-dependent tyrosine phosphorylation of HER2.19 With a similar generation approach, HER2 aptamer has been identified and synthesized. HER2 aptamer shows high specificity to HER2+ cancer cell lines but not HER2− cancer cell lines.20 In this study, we aim to target HER2/HER3/EGFR in one molecule using two aptamers and a siRNA. A new chimera was developed by fusing an EGFR siRNA between HER2 aptamer and HER3 aptamer. This structure will simultaneously block HER2 and HER3 signaling pathways and induce EGFR silencing in HER2-expressing cells. With a three-in-one structure, the new chimera will simplify the processes and cost in manufacturing and preclinical and clinical testing and be less complicated for patient administration. The new three-in-one design will provide a new therapeutic paradigm to address the HER network and overcome resistance to the therapies designed to target a single HER family protein. Furthermore, this novel design is nontoxic, simple, and cost effective compared with antibodies and small molecule inhibitors.
Results
HER2 Aptamer-EGFR siRNA-HER3 Aptamer (H2EH3) Chimera Was Constructed and Characterized
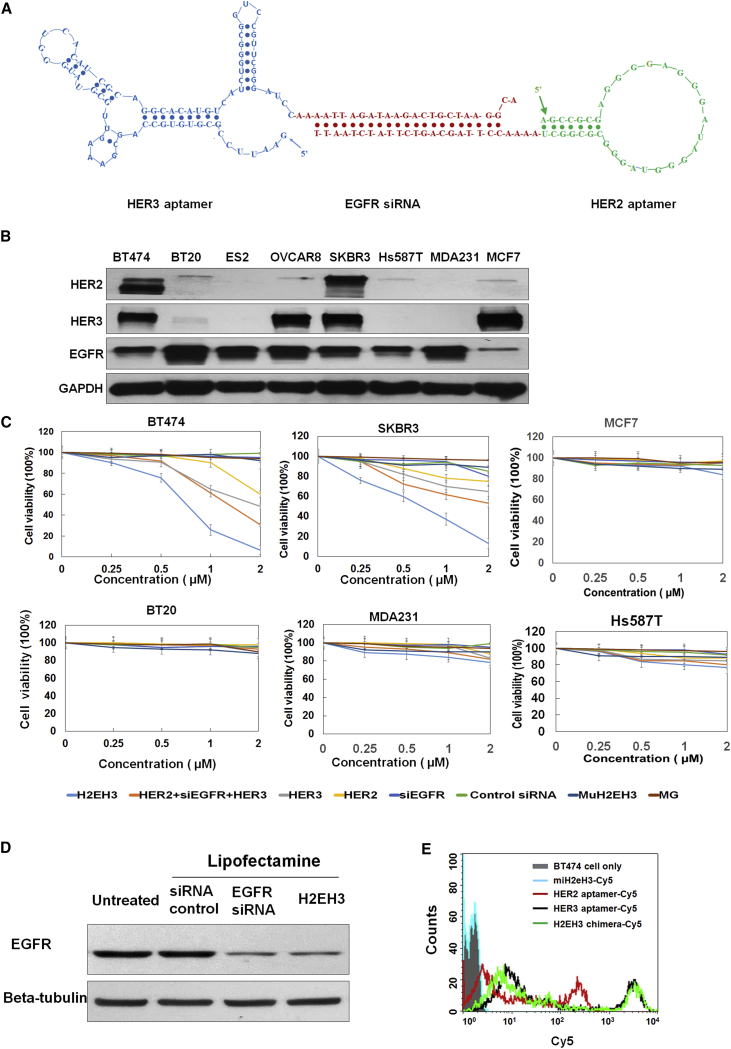
The 3′ terminus of HER3 aptamer (79 bases)18 was fused with the anti-sense strand of EGFR siRNA, and the 3′-terminus of HER2 aptamer (34 bases)20 was fused with the sense strand of EGFR siRNA. The dissociation constant (KD) value of HER2 aptamer is 3.49 nM, and the KD value of HER3 aptamer is 45 nM. Between an aptamer and a siRNA, 2–4 unpaired “A’s” are inserted to offer spatial flexibility to each aptamer (Figure 1A). Through in vitro transcription, 2′-fluoro pyrimidines were incorporated into two RNA chains to enhance serum stability. Two transcripts with 19-base complementing sequences (sense strand and anti-sense strand of EGFR siRNA) were annealed together by heating for 3 min at 95°C, followed by slowly cooling to room temperature within 1 hr. As shown in Figure 1A, the new chimera, with one HER2 aptamer (molecular weight [MW] 11.2 kDa), one EGFR siRNA, and one HER3 aptamer (MW 25.4 kDa), was annealed into one molecule with a molecular weight of 55.4 kDa, which is larger than each single aptamer and larger than the renal glomerulus cutoff mass (30–50 kDa), but smaller than an antibody (about 150 kDa). Thus, H2EH3 is expected to have a longer circulation time than a single aptamer alone. We have put the 3′ end of the anti-sense strand of EGFR siRNA with a 2-nt overhang, which will promote siRNA-RISC (RNA-induced silencing complex) formation.21
Figure 1.
Schematic Illustration of HER2 Aptamer-EGFR siRNA-HER3 Aptamer, H2EH3, and Characterization of H2EH3
(A) Structure of H2EH3. HER2 aptamer was conjugated with HER3 aptamer through 21 bases of EGFR siRNA and 2–4 unpaired base linkers. (B) Western blot detection of the expression levels of HER2, HER3, and EGFR in a panel of breast cancer cell lines. (C) Evaluation of the cytotoxicity of H2EH3 by CCK-8 assay. Breast cancer cell lines, including BT474, SKBR3, MCF7, MDA-MB-231, and Hs587T cells, were treated with varying concentrations of H2EH3 or controls for 72 hr, and cell viability was detected with the CCK-8 agent. Data are the mean ± SD from three independent experiments. (D) Evaluation of EGFR-silencing capability of H2EH3 using western blot. (E) Evaluation of H2EH3-binding capability compared with HER2 aptamer and HER3 aptamer.
Furthermore, we identified if H2EH3 possesses the activities of HER2 aptamer, EGFR siRNA, and HER3 aptamer. First, we evaluated the binding affinity of H2EH3 compared with HER2 aptamer and HER3 aptamer individually. In Figure 1E, H2EH3 showed a strong binding affinity comparable with HER2 aptamer and HER3 aptamer. To detect the EGFR-silencing activity of H2EH3, H2EH3 was transfected to BT474 cells using lipofectamine to evaluate EGFR knockdown. As shown in Figure 1D, H2EH3 kept a similar silencing effect as EGFR siRNA. In addition, H2EH3 has been incubated with a dicer for 12 hr. The digestion pattern is shown in Figure S1. The result showed that H2EH3 was digested with the dicer and released an RNA fragment with the same size as EGFR siRNA. This suggests that H2EH3 can be processed with RNA interference machinery.
H2EH3 Displayed Significant Cytotoxicity on HER2+HER3+ Cells
Next, we evaluated if H2EH3 shows cytotoxicity on breast cancer cell lines. We first probed the abundance of HER2, HER3, and EGFR in a panel of breast cancer and ovarian cancer cell lines, including BT474, BT20, ES2, OVCAR8, SKBR3, Hs587T, MDA-MB-231, and MCF7 by western blot analysis. As shown in Figure 1B, BT474 and SKBR3 cells express the highest levels of HER2 among all detected cell lines. ES2 and MDA-MB-231 cells are negative to HER2, whereas BT20, OVCAR8, and Hs587T cell lines just show slightly detectable HER2. BT474 and SKBR3 cells also express high levels of HER3. OVCAR8 and MCF7 show a high abundance of HER3, although with a trace amount of HER2. EGFR was expressed on all detected cell lines. MCF7 has a much lower expression of EGFR than other cell lines, whereas BT20 and MDA-MB-231, two triple-negative breast cancer cell lines, show the highest EGFR than other cell lines. BT474 and SKBR3 cells have a medium level of EGFR expression.
Next, we detected the growth inhibitory effect of H2EH3 on these cell lines using the Cell Counting Kit-8 (CCK-8). Cell lines were treated with increasing concentrations of H2EH3 or controls, including HER2 aptamer, HER3 aptamer, EGFR siRNA, mixture of HER2 aptamer, HER3 aptamer, and EGFR siRNA, non-targeting control aptamer MG (specific to Malachite Green), or mutant H2EH3. As shown in Figure 1C, the viability of BT474 and SKBR3 cells exposed to H2EH3 for 72 hr was significantly reduced in a dose-dependent manner. BT474 and SKBR3 cells only remain about 5%–10% viable upon treatment with 2 μM H2EH3. Upon treatment with 2 μM simply mixed HER2 aptamer, HER3 aptamer, and EGFR siRNA, the viability remains 30% for BT474 and 50% for SKBR3. The results suggest that three-in-one H2EH3 is superior to the simply mixed three individual components. HER3 aptamer and HER2 aptamer all show dose-dependent cytotoxicity for BT474 and SKBR3 cells. The efficacy of HER3 aptamer is more potent than that of HER2 aptamer. Upon treatment with 2 μM aptamers, the viability of BT474 is 48% with HER3 aptamer and 60% with HER2 aptamer; similarly, the viability of SKBR3 is 65% with HER3 aptamer and 75% with HER2 aptamer. EGFR siRNA only and Malachite green (MG) did not show cytotoxicity to SKBR3 and BT474. Mutant H2EH3 (muH2EH3) was constructed, which contained the same nucleic acid components as H2EH3 and the same EGFR siRNA as H2EH3 but is missing the original 3D conformations of HER2 and HER3 aptamers. muH2EH3 did not show cytotoxicity to any detected cell lines. MCF7 and BT20 cell lines do not have any response to H2EH3 treatment (Figure 1C). The viability of MDA-MB-231 and Hs578T cell lines shows only a 10%–20% decrease upon 2 μM H2EH3 treatments (Figure 1C, lower panel). Although MCF7 cells have high HER3 expression but no HER2 expression, HER3 aptamer and H2EH3 do not show inhibitory effects on MCF7 growth. Furthermore, we detected if MCF7 cell survival is dependent on HER3. We have silenced HRR3 in MCF7 cells and probed cell viability. As shown in Figure S2, cell viability only has a 7% decrease, which indicates the HER family is not the major survival pathway for MCF7 proliferation. BT20, MDA-231, and Hs578T are both HER2− and HER3−; thus, it is not surprising that their cell viabilities do not decrease significantly upon H2EH3 treatment. These results suggest that H2EH3 has a strong cell-type-specific targeting capability.
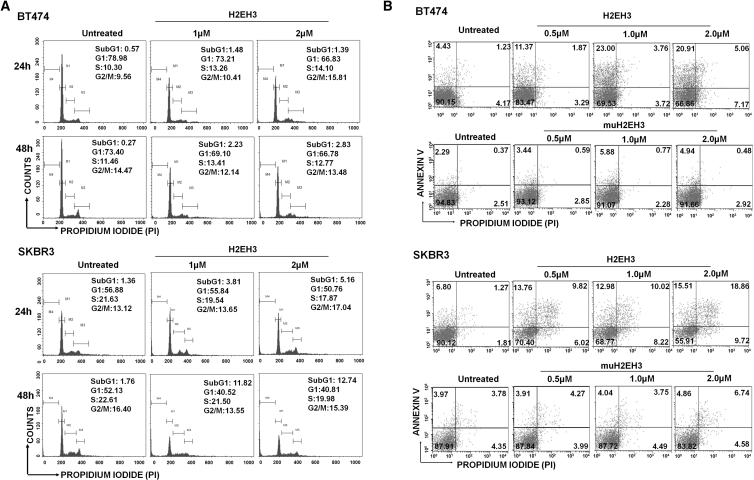
H2EH3 Induced Cell Cycle Arrest and Apoptosis in HER2+HER3+ Cell Lines
The decrease in cell viability may be attributed to reduced cell proliferation and/or increased cell death. To identify these effects, we first explored whether H2EH3 has interfered with cell cycle progression. BT474 and SKBR3 cells were treated with H2EH3 at different concentrations for 24 and 48 hr. As shown in Figure 2A, BT474 cells showed G2/M arrest in 24 hr and showed an increased subG1 population. from 0.27% of untreated cells to 2.83% of cells treated with 2 μM H2EH3. This suggests that the apoptosis of BT474 cells occurred after 48-hr treatment. SKBR3 cells showed slight G2/M arrest in 24 hr and significantly increased the subG1 population in 24 and 48 hr, indicating that SKBR3 cells underwent apoptosis within 24 hr.
Figure 2.
Analysis of Cell Cycle and Apoptosis
(A) Effects of H2EH3 on cell cycle progression of HER2+HER3+ cells. SKBR3 and BT474 cells were treated with H2EH3 (1 μM, 2 μM) for 24 and 48 hr. The cell cycle was then analyzed by flow cytometry. (B) Quantitative analysis of apoptosis after H2EH3 treatment by flow cytometry. BT474 and SKBR3 cells were treated with the different concentrations of H2EH3 for 72 hr, and cells were stained with Alexa Fluor 488 annexin-V-propidium iodide and analyzed by flow cytometry.
To further assess induction of apoptosis by H2EH3, SKBR3 cells and BT474 cells were treated with H2EH3 for 72 hr and stained with annexin V/PI (propidium iodide) for flow cytometry analyses. As shown in Figure 2B, for BT474 cells, early apoptosis (annexin V+/PI−) increased from untreated 4.43% to 11.37%, 23%, and 20.91% treated with H2EH3 at 0.5 μM, 1.0 μM, and 2.0 μM, respectively; late stage (annexin V+/PI+) increased from untreated 1.23% to 1.87%, 3.76%, and 5.06% treated with H2EH3 at 0.5 μM, 1.0 μM, and 2.0 μM, respectively. muH2EH3 at the 0.5–2.0 μM range has <6% early apoptosis and <5% late apoptosis. In SKBR3 cells, the most significant change occurred on the late apoptosis stage. Late apoptosis of SKBR3 cells increased from untreated 1.27% to 9.82%, 10.02%, and 18.86% when treated with H2EH3 at 0.5 μM, 1 μM, and 2 μM, respectively. muH2EH3 at the 0.5–2.0 μM range has <7% late apoptosis. These results from the analyses of cell cycle progression and apoptosis suggest that H2EH3 induces cell death through cell cycle arrest and apoptosis on HER2+HER3+ cell lines.
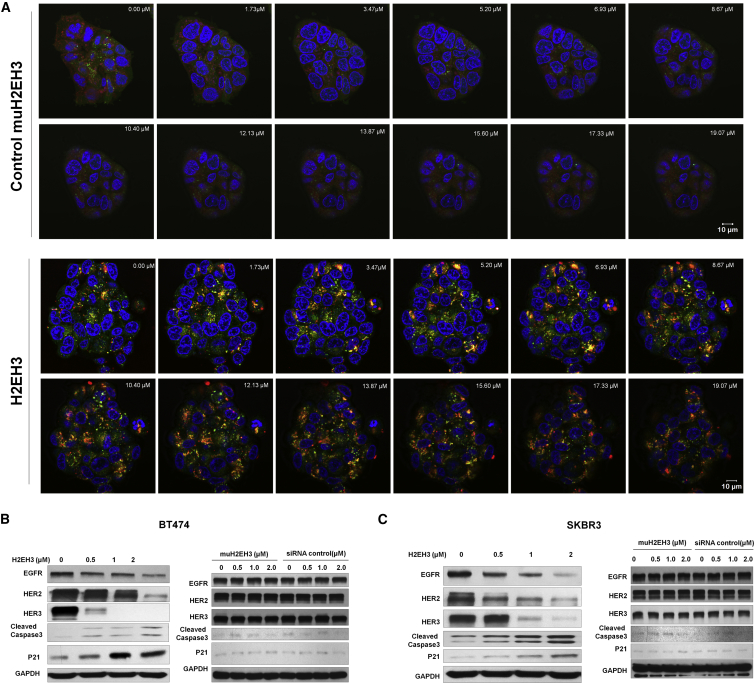
H2EH3 Reduced the Protein Expression Levels of EGFR, HER2, and HER3 and Upregulated Cleaved Caspase-3 and p21
Because EGFR siRNA was inserted between HER2 and HER3 aptamers, we expect to see EGFR gene knockdown. We hypothesize that H2EH3 binding to cell receptors of HER2 and HER3 will cross link two receptors and result in ligand-receptor complex internalization. The internalized H2EH3 may lead to EGFR silencing. First, we validated the internalization of H2EH3, which is the prerequisite of siRNA silencing. BT474 cells were treated with Cy5-H2EH3 and control Cy5-muH2EH3 aptamer. Confocal microscopy with z stack was performed to visualize the subcellular locations of H2EH3. Nuclei were stained with DAPI (blue), and endosome/lysosomes were revealed by Lysotracker (green). As shown in Figure 3A, after 12-hr incubation, H2EH3 has entered the cells and presented in the cytoplasm, and some H2EH2 has escaped from endosomes/lysosomes by showing red fluorescence, whereas some H2EH3 entrapped in endosomes showed a yellow signal. The result showed that HER2/HER3 dimerization can lead chimera internalization.
Figure 3.
Detection of H2EH3 Internalization by z Stack Confocal Microscopy and Evaluation of Protein Levels by Western Blot
(A) Cy5-labeled H2EH2 or Cy5-labeled muH2EH3 was added into BT474 cells for 12 hr at 37°C. LysoTracker Green was used to show lysosomes and endosomes. DAPI was used to display nucleus. Confocal laser scanning microscopy with z stack was performed to show cell binding and internalization. Scale bar, 10 μm. (B) Western blot analysis of total HER family receptors and apoptosis-associated molecules in BT474 cells. (C) Western blot analysis of total HER family receptors and apoptosis-associated molecules in SKBR3 cells.
After confirmation of H2EH3 internalization, we detected if EGFR siRNA has triggered gene silencing. EGFR expression was evaluated with western blot. As shown in Figure 3B, after treatment with H2EH3 for 72 hr, EGFR expression levels in both BT474 and SKBR3 cells were indeed significantly reduced at a dose-dependent manner but not treated with muH2EH3 or scrambled siRNA control.
Furthermore, we assessed if H2EH3 can reduce protein levels of HER2 and HER3. SKBR3 and BT474 cells were treated with the varying concentrations of H2EH3 for 72 hr, and HER2 and HER3 levels were quantified by western blot. As shown in Figure 3B, H2EH3 treatment resulted in a pronounced reduction of protein levels of HER2 and HER3 in SKBR3 and BT474 cells but not treatments with muH2EH3 or scrambled siRNA control. This indicates H2EH3 has led ligand-induced HER2 and HER3 receptor internalization and degradation, which results in the reduction of protein levels of HER2 and HER3. From previous studies, targeting a single HER family member often results in increased levels of other HER receptors, which will compensate the activity of the inhibited HER member and counteract the treatment efficacy.22, 23 In contrast, H2EH3 is able to significantly reduce the expression of all three receptors, thereby effectively blocking the compensatory network of HER members.
Next, we assessed whether H2EH3 has an impact on downstream apoptotic signaling. Upon treatment with H2EH3, BT474 and SKBR3 cells were probed for the expression of p21 and cleaved Caspase-3 by western blot. Cleaved Caspase-3 is regarded as a signature marker of apoptosis, and p21 upregulation is closely related to cell cycle arrest and apoptosis. As shown in Figure 3B, p21 and cleaved Caspase-3 in SKBR3 and BT474 cells were indeed significantly upregulated by H2EH3 at a dose-dependent manner. The results indicate that H2EH3 induces cell growth inhibition by activating the signaling pathways associated with cell cycle arrest and apoptosis.
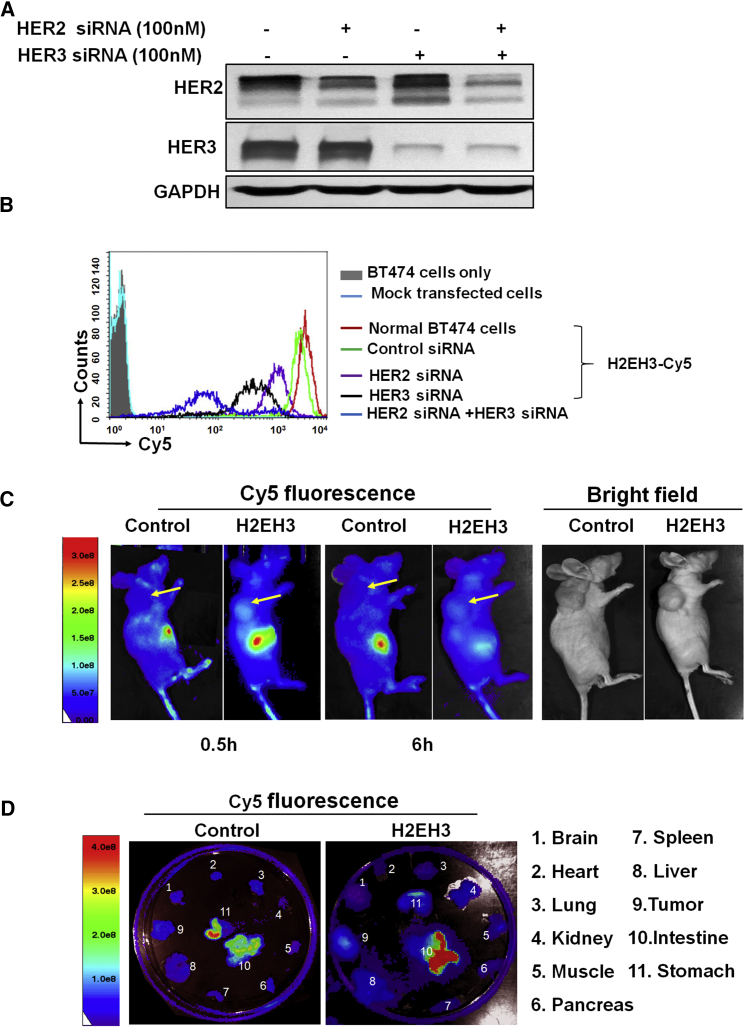
H2EH3 Possesses a High Binding Specificity to HER2 and HER3 Receptors and High Tumor-Targeting Capability In Vivo
To investigate whether H2EH3 indeed binds to HER2 and HER3 receptors, we used siRNAs to silence each receptor to assess binding specificity. We transfected HER2 and/or HER3 siRNAs via lipofectamine to BT474 cells, and then detected a binding pattern after receptor knockdown. We first identified HER2 and/or HER3 knockdown with western blot (Figure 4A). After treatment with 100 nM HER2 siRNA and/or HER3 siRNA, the protein levels of HER2 and HER3 are significantly reduced. Then, HER2- and/or HER3-silenced BT474 cells were compared with untreated BT474 on H2EH3 binding. BT474 cells (untreated or HER2/HER3 silenced) were first fixed with 1% paraformaldehyde-PBS and then incubated with Cy5-labeled H2EH3 or controls. As shown in Figure 4B, untreated BT474 cells have a strong fluorescence peak and high fluorescence intensity (red line). Upon knockdown of HER2 or HER3, the fluorescence peak of single positive HER3+ cells (purple line) or HER2+ cells (black line) has shifted to the left (lower intensity areas), which suggests reduced binding following HER2 or HER3 knockdown. Upon knockdown of both HER2 and HER3, most cells underwent dying. When we analyzed the remaining cells, the fluorescence intensity of double negative HER2-HER3-BT474 cells (blue line) shows the significant decrease compared with normal BT474. These results indicate that H2EH3 indeed possesses specificity toward HER2 and HER3 receptors.
Figure 4.
Evaluation of Binding Specificity and In Vivo Bio-distribution of H2EH3
(A) Western blot analysis of HER2 and/or HER3 gene knockdown. (B) Flow cytometry to quantitatively measure cell binding of H2EH3 upon HER2 and/or HER3 knockdown in BT474 cells. Untreated: solid gray; mock transfected cells: light blue; normal cells stained with H2EH3-Cy5: red line; control siRNA transfected cells stained with H2EH3: green line, HER2-silenced cells stained with H2EH3-Cy5: purple line; HER3-silenced cells stained with H2EH3-Cy5: black line; both HER2- and HER3-silenced cells stained with H2EH3: blue line. (C) Time course whole-body imaging to show the binding profile of H2EH3. Tumor-bearing mice were intravenously injected with Cy5-H2EH3 or non-targeting control aptamer. Cy5 fluorescence of whole body was captured at the time points of 2 and 6 hr with the Xenogen IVIS100 imaging system. (D) Ex vivo organ imaging. Following whole-body imaging, major organs were removed and detected with the Xenogen IVIS100 imaging system. The result is the representative of two independent experiments.
To explore tumor-targeting capability and bio-distribution in vivo, BT474 tumor-bearing mice were tail-vein injected with Cy5-H2EH3 or non-targeting control muH2EH3. As shown in Figure 4C, after a half-hour injection, in the reference bright-field images (Figure 4C, right), which show the locations of tumors, the tumors in H2EH3-treated mice with high fluorescence intensity could be clearly delineated from the surrounding background tissues with the Xenogen IVIS100 imaging system, but not the tumors in non-targeting control treated mice. The fluorescence signals can be detectable for 6 hr. After whole body imaging, organs were removed and ex vivo imaging was performed. As shown in Figure 4D, Cy5 fluorescence in H2EH3-treated tumors was detectable, but not in control muH2EH3-treated tumors. It is not surprising that strong fluorescence is shown in the intestines and stomach because those locations are the places for clearance of chimeras from bodies. Noteworthy, Cy5 fluorescence signals are not detectable in organs, including the brain, spleen, liver, heart, lung, kidney, muscle, and pancreas. These results suggest that H2EH3 can bind to tumors in vivo, with high specificity.
H2EH3 Treatment Led to Decreased Tumor Growth in Breast Cancer Xenografts
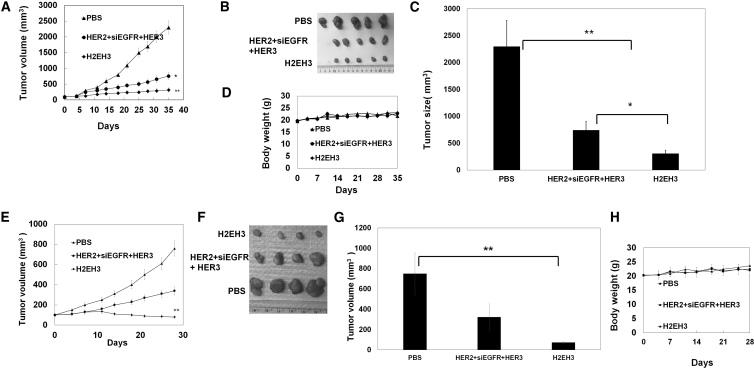
To investigate whether the in vitro anti-proliferative effect of H2EH3 could translate into in vivo efficacy, we tested H2EH3 in breast cancer subcutaneous and orthotopic xenografts. In the start test, BT474 cells were subcutaneously injected into 5- to 6-week-old female athymic nu/nu mice at the left flank. Upon the tumor reaching 100 mm3, mice were treated with PBS, mixture (HER2 aptamer, HER3 aptamer, and EGFR siRNA) (8 nmol of each component), or H2EH3 (8 nmol) by intra-tumor injection every other day for 5 weeks. As shown in Figures 5A–5C, H2EH3 treatment showed pronounced tumor growth inhibition and is superior to the mixture of HER2 aptamer, HER3 aptamer, and EGFR siRNA. Tumors in H2EH3-treated mice have a 6- to 7-fold reduction in size compared with PBS-treated controls and about a 1.5-fold reduction compared with those treated with the mixture of HER2 aptamer, HER3 aptamer, and EGFR siRNA. Next, mammary orthotopic xenografts were set up by injection of BT474 cells in mammary fat pads. Tumor (50 mm3) bearing mice were intravenously (via tail vein) treated with H2EH3 (8 nmol), mixture (HER2 aptamer, HER3 aptamer, and EGFR siRNA), or PBS every 3 days for 4 weeks. As shown in Figures 5E and 5F, H2EH3 treatment has achieved a 7- to 8-fold reduction in tumor sizes compared with the PBS control group, and a 3- to 4-fold reduction compared with those treated with the mixture of three components. Due to the limitation in available injection times through the tail vein, in this experiment, 4-week administration was taken. These results show H2EH3 is able to effectively inhibit HER2-expressing breast cancer growth through both intratumoral and intravenous (i.v.) injection. As a tumor control, MDA-MB-231 cells (HER2− and HER3−) were used to set up mammary orthotopic xenografts. Tumor-bearing mice were treated with H2EH3 and controls through i.v. injection. As shown in Figure S7, there is no significant treatment efficacy in the groups treated with H2EH3 or the mixture of three components compared with the PBS control group. The results proved the targeting capability of H2EH3 in vivo.
Figure 5.
H2EH3 Significantly Suppresses Tumor Growth in Breast Cancer Xenografts
(A) Inhibition of tumor growth by H2EH3 through intratumoral injection. Mice with subcutaneous tumors were intratumorally injected with H2EH3, a mixture of HER2 aptamer, HER3 aptamer, and EGFR siRNA, or PBS every other day for 5 weeks (n = 5 per group). (B) Dissected tumors after treatment though intratumoral injection (n = 5). (C) Quantitation of dissected tumor sizes from (B) (n = 5). (D) Body weight of mice treated with H2EH3 through intratumoral injection (n = 5). (E) Inhibition of tumor growth by H2EH3 through intravenous injection. Mice with orthotopic tumors were intratumorally injected with H2EH3, a mixture of HER2 aptamer, HER3 aptamer, and EGFR siRNA, or PBS every 3 days for 4 weeks (n = 4). (F) Dissected tumors after treatment through intravenous injection (n = 4). (G) Quantitation of dissected tumor sizes from (F) (n = 4). (H) Body weight of mice treated with H2EH3 through intravenous injection (n = 4). *p < 0.05; **p < 0.005. Data represent the mean ± SEM; n = 4–5 for each group.
H2EH3 Enabled Downregulation of HER2, HER3, and HER3 and Triggered Apoptosis In Vivo
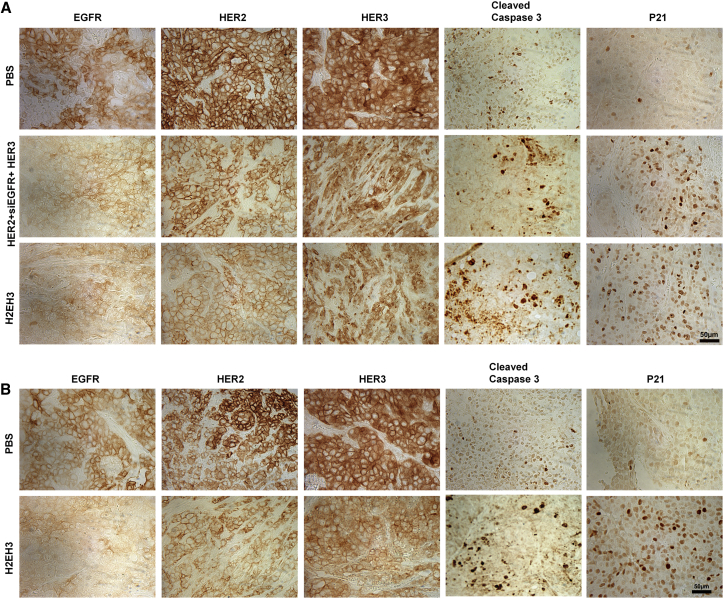
To determine the target specificity of our H2EH3, immunohistochemistry (IHC) assay was performed to validate gene knockdown and apoptosis in vivo. Tumor tissues from subcutaneous and orthotopic xenografts were collected and examined for the expression of HER2, HER3, EGFR, p21, and cleaved Caspase-3. As shown in Figure 6A, H2EH3 treatment has led to reduced protein levels of EGFR, HER2, and HER3 and increased cleaved Caspase-3 and p21 in subcutaneous tumors through intratumoral injection (Figure 6A). Similarly, the reduced expression of EGFR, HER2, and HER3 and the increase of cleaved Caspase-3 and p21 are visualized in orthotopic tumors through intravenous injection of H2EH3 (Figure 6B). We also proved EGFR knockdown in tumor tissues by western blot, as shown in Figure S6. These findings are consistent with the in vitro results in Figure 3B. The histology and western blot results suggest that H2EH3 enables intervention of EGFR/HER2/HER3 concomitantly and induces tumor cell-cycle arrest and apoptosis in vivo, which is translated into significant suppression of tumor growth in mouse xenograft models.
Figure 6.
Evaluation of Gene Expression and Apoptosis after H2EH3 Treatment In Vivo
Formalin-fixed paraffin-embedded sections of xenograft tumors were stained with antibodies targeting EGFR, HER2, HER3, P21, and cleaved caspase-3. (A) IHC analysis of subcutaneous tumors after intratumoral injection of H2EH3. (B) IHC analysis of orthotopic tumors after intravenous injection of H2EH3. Scale bar, 50 μm.
H2EH3 Did Not Show Detectable Toxicity
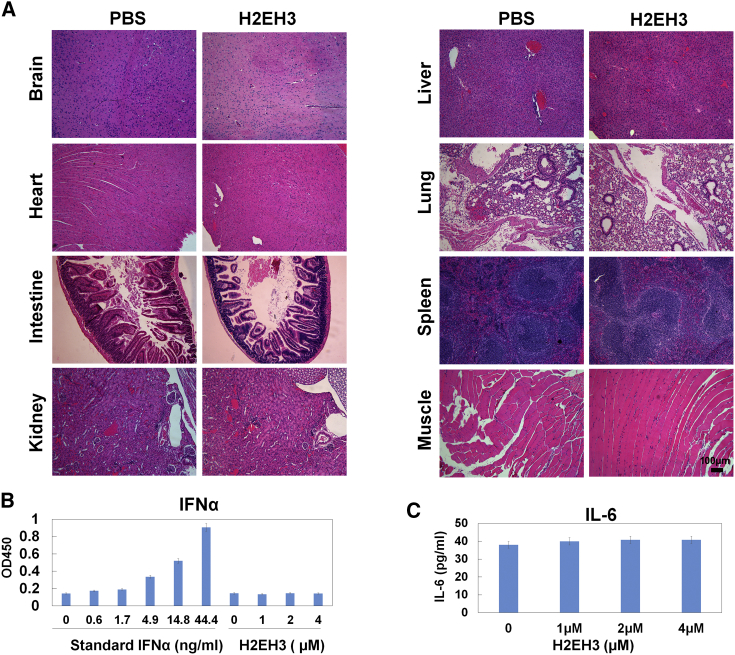
During treatment, the body weights of mice were measured. There was no significant difference in body weights between the PBS- and H2EH3-treated groups (Figures 5D and 5H). Furthermore, we examined the histopathology of major organs from mice that have experienced systemic drug administration by i.v. injection. As shown in Figure 7A, H&E staining revealed that there were no pathological changes observed in these organs of H2EH3-treated mice. Additionally, we have detected the possible innate immune response of H2EH3 by mixed lymphocyte reaction assay. It was reported that RNA synthesis by transcription may induce interferon-α (IFN-α) upregulation.24 We treated human peripheral mononuclear cells (PBMCs) with H2EH3 for 48 hr. IFN-α and pro-inflammatory cytokine interleukin-6 (IL-6) were measured with ELISA. As shown in Figure 7B, there was no detectable IFN-α at a concentration of up to 4 μM H2EH3. IL-6 levels in PBMCs treated with 1–4 μM H2EH3 did not show a statistical difference compared with untreated control (Figure 7B). Taken together, these results suggest that nucleic-acid-based H2EH3 has no discernible toxicity to the host and may not trigger an innate immune response.
Figure 7.
Evaluation of Toxicity
(A) Histological evaluation of the tissue damages. H&E staining of major organs. Compared with naive (no tumor implant), the organs from H2EH3-treated xenografts do not exhibit significant histological change. Scale bar, 100 μm. (B and C) Detection of potential immune response upon H2EH3 treatment. Human peripheral blood mononuclear cells were treated with different concentrations of H2EH3 for 48 hr. IFN-α (B) and IL-6 (C) in cell culture supernatants were quantified with ELISA kits. The results are the mean ± SD and represent three independent experiments.
Discussion
HER2-targeted antibodies, such as trastuzumab and pertuzumab, have revolutionized the treatment of HER+ breast cancer. However, 66%–88% of HER2+ metastatic breast cancer shows resistance to a single regimen of trastuzumab treatment.25 Combinations of trastuzumab with either paclitaxel or docetaxel have been used and provide improved treatment efficacy, but 15% of patients also develop resistance to these combination therapies.26 Moreover, these combinations are associated with significant cardiotoxicity and anaphylaxis.27 There is, therefore, an urgent need to develop improved combination therapeutics for HER2+ breast cancer patients.
Several laboratories have shown de novo expression of either EGFR and/or HER3 in trastuzumab-resistant HER2+ breast cancer cell lines, and these lines characteristically acquire sensitivity to either EGFR- or HER-targeted agents (e.g., gefitinib or cetuximab).2 Based on these observations, here we have developed an inhibitor that can simultaneously block EGFR, HER2, and HER3 signaling. Because HER4 expression has been associated with increased ER expression28 and low cell proliferation rates29 and has generally been correlated with a favorable outcome in breast cancer patients,30, 31 this HER family member was excluded as a target in these studies.
In our studies, to target heterodimer HER2/HER3, we propose that one multifunctional molecule will provide more spatial advantages to disable a heterodimer located in close proximity than two individual targeting molecules. Therefore, we fused HER2 aptamer and HER3 aptamer together with a double-stranded EGFR siRNA, which acts as a linker as well as an additional therapeutic component. 2–4 unpaired bases were put between each aptamer and EGFR siRNA to warrant aptamer flexibility for target binding. In our previous studies,32 we have proven that 2–4 unpaired bases are essential to maintain conjugated aptamer functionalities. HER2 and HER3 aptamers will direct EGFR siRNA to internalize to the HER2- and HER3-expressing cells through receptor-mediated endocytosis. We expect that double targeting on HER2 and HER3 will increase cargo delivery to breast cancer cells compared to targeting one receptor, and future studies are being designed to test this hypothesis in vivo. Our results showed that H2EH3 can significantly silence EGFR in vitro and in vivo. Therefore, when using H2EH3, both aptamers as well as the siRNA all have potent anti-tumor activities. Each aptamer has dual functions: as a receptor antagonist to block ligand binding and as a cell-type-specific siRNA delivery agent. Our studies demonstrate that H2EH3 is able to significantly reduce the protein levels of HER2 and HER3. Comparatively, Pan-HER antibodies can reduce total protein levels of HER2 and HER3,12 and HER3 antibody (KTN3379) is also able to reduce total HER3 protein.33 This indicates aptamer, like antibody, possesses the capability to induce ligand-receptor endocytosis, followed by degradation.
Our studies in vitro (Figure 1C) and in vivo (Figures 5A and 5E) demonstrated that the mixture of HER2 aptamer, HER3 aptamer, and EGFR siRNA has treatment efficacy but less than that of H2EH3. This is because HER2 aptamer and HER3 aptamer have played blocking functionalities. EGFR siRNA in the mixture cannot play silencing activity because free EGFR siRNA cannot enter cytoplasm without any delivery aid. In addition, a single HER2 aptamer is very small (11.2 kDa), which falls in the size of renal depletion (30–50 kDa). Shorter circulation time will result in the lower efficacy of unconjugated HER2 aptamer than the conjugated one. Due to a lack of EGFR siRNA activity and shorter circulation time of single aptamers, simply mixed three components have less efficacy than three-in-one H2EH3. As shown in Figures 1C, 5A, and 5E, H2EH3 has shown consistently better efficacy than the mixture of HER2 aptamer, HER3 aptamer, and EGFR siRNA.
Three-in-one chimera will significant increase overall drug size compared with each aptamer alone, suggesting that H2EH3 may have a longer circulation life time than each aptamer alone. In previous studies, polyethylene glycol (PEG; 20–40 kDa) has been added into chimeras for prolonging circulation life time in vivo.34, 35 In our design, conjugating two aptamers not only is amenable to reduce steric hindrance for ease of blocking hetero-dimerized two receptors, but can also increase structure size for longer circulation as compared with two single aptamers, and studies are ongoing to test this hypothesis.
Such a nucleic-acid-based aptamer-siRNA chimera is a unique class of targeting therapeutics that has potential for clinical translation. Vascular endothelial growth factor (VEGF) aptamer has been approved by the Food and Drug Administration (FDA) as a therapeutic for macular degradation.36 There are many aptamer-mediated therapeutics under development for the treatment of conventional undruggable diseases, in which aptamers act as antagonist drugs37 or delivery vehicles for small chemical drugs, proteins, siRNA, or even nanoparticles.38, 39 RNA-based aptamer-siRNA chimera can be generated by in vitro transcription and solid-phase chemical synthesis. Compared with antibodies, aptamer-siRNA chimeras provide ease of large-scale production, low immunogenicity, ease of modification, high thermostability, and a small size, whereas antibodies are cell-based products, associated with high immunogenicity, are sensitive to temperature, and have a high cost in manufacturing. Unmodified RNAs are sensitive to nucleases, which have higher degradation rates for pyrimidine residues. 2′-amino-pyrimidines and 2′-fluoro-pyridines have been successfully incorporated into aptamers and FDA-approved oligonucleotide drugs40, 41 to resist nuclease-induced degradation. We have proven that 2′-fluoro-modified aptamer-siRNA chimera can have over 24 hr of half-life in 50% serum-containing buffer.32, 42 It has been demonstrated that 2-fluoro modification of aptamer is safe and well-tolerated and also has increased binding affinity to the targets.43
Due to the accessibility of breast cancer, we tested the treatment efficacy of H2EH3 by intratumoral injection. For preparing to treat metastatic breast cancer, we tested the treatment efficacy of H2EH3 by intravenous injection. Through both administration routes, we have seen the potent inhibition of tumor growth. From the biodistribution study, through tail-vein injection of H2EH3, we did not find the nonspecific accumulation of H2EH3 in other major organs (brain, heart, lung, kidney, muscle, pancreas, spleen, and liver), except the intestine and stomach. Because we found non-targeting control aptamer also accumulated in the intestine and stomach, we may not say H2EH3 has an off-target effect by binding to other organs. Although HER2 and HER3 have been widely expressed in many organs, it is possible that there is a big difference of HER2/HER3 in the quantity at normal organs versus at breast tumors, thus creating a therapeutic window.
The new RNA therapeutic with low immunogenicity and low toxicity will provide a potent addition to HER-targeting therapeutics for HER2-expressing breast cancer. Our results show the new chimera can induce cell cycle arrest and apoptosis in HER2-expressing cells and significantly inhibits tumor growth in breast cancer xenografts. We envision that H2EH3 may be extended to treat other tumors that are dependent on the HER family for proliferation, such as lung and head and neck cancers.44, 45
Materials and Methods
Materials
Cell culture media were purchased from Life Technologies (Carlsbad, CA). Fetal bovine sera and Taq RNA polymerase were from Sigma-Aldrich (St. Louis, MO). Antibodies were from Cell Signaling Technology (Danvers, MA). Single-stranded DNAs were synthesized by IDT (Coralville, IA). Transcript Aid T7 High Yield Transcription Kits were ordered from Thermo Fisher Scientific. HER2 and HER3 siRNAs were purchased from Life Technologies. Alexa Fluor 488 Annexin V/Dead Cell apoptosis kits were from Invitrogen. ELISA kits for detection of IFN-α and IL-6 were obtained from RayBiotech (Norcross, GA). TUNEL assay kits were purchased from R&D systems (Minneapolis, MN). 2′-Fluoro-2′-deoxycytidine-5′-triphosphate, 2′-fluoro-2′-deoxyuridine-5′-triphosphate, and Cy5-labeled 2′-fluoro-labeled HER2 aptamer were purchased from TriLink Biotechnologies (San Diego, CA).
Cell Culture
All cell lines in this study were obtained from ATCC. Cell lines were authenticated by the manufacturer based on the following criteria: short tandem repeat profiling, cell morphology, and karyotyping. Cell lines were monitored for mycoplasma contamination and consistently found to be negative by qPCR kits. All cells except for MDA-MB-231 cells were cultured in RPMI-1640 medium. MDA-MB-231 cells were cultured in DMEM medium. All media were supplemented with 10% complete fetal bovine serum (FBS) with 100 U/mL penicillin and 0.1 mg/mL streptomycin. All cells were cultured in a humidified incubator with 5% CO2 at 37°C.
Mouse
All animal studies were approved by the Institutional Animal Care and Use Committee at Augusta University. Athymic nu/nu mice were purchased from Harlan Laboratories. The methods were carried out in accordance with the approved guidelines.
Synthesis of H2EH3 and Mutant H2EH3 Chimeras and Aptamers
The ssDNA templates and primers were synthesized from IDT.
For H2EH3 chimera synthesis,
RNA1 (HER3 aptamer-EGFR antisense siRNA): 5′-GAAUUCCGCGUGUGC CAGCGAAAGUUGCGUAUGGGUCACAUCGCAGGCACAUGUCAUCUGGGCGGUCCGUUCGGGAUC CAAAAUUAGAUAAGACUGCUAAGGCA-3′.
RNA1 PCR template: 5′-GAATTCCGCGTGTGCCAGCGAAAGTTGCGTATGGGTCACATCGCAGGC ACA T GTCATCTGGGCGGTCCGTTCGGGATCCAAAATTAGATAAGACTGCTAAGGCA-3′.
RNA1 5′ primer: 5′- TAATACGACTCACTATAGAATTCCGCGTGTGCCA-3′. The forward primer contains T7 RNA polymerase promoter site (bolded).
RNA1 3′ primer: 5′-TGCCTTAGCAGTCTTATCTAATTTTGGATCCCGA-3′.
RNA2 (HER2 aptamer-EGFR sense siRNA): 5′-AGCCGCGAGGGGAGGGAUAGGGUAGGGCGCGGC UAAAACCUUAGCAGUCUUAUCUAAUU-3′.
RNA2 PCR template: 5′-AGCCGCGAGGGGAGGGATAGGGTAGGGCGCGGCTAAAACCTTAGCAGTC TTATCTAATT-3′.
RNA2 5′ primer: 5′-TAATACGACTCACTATAAGCCGCGAGGGGAGGGA-3′. The forward primer contains T7 RNA polymerase promoter site (bolded).
RNA2 3′ primer: 5′-AATTAGATAAGACTGCTAAGGTTTTA-3′.
Two RNAs were generated by in vitro transcription, with PCR products as templates. The PCR products were put into T-A cloning pCR2.1 vector (Invitrogen) and sequenced. Transcription was performed with Transcript Aid T7 High Yield Transcription Kit following the manufacturer’s instruction. 2′ F-modified pyrimidines were incorporated into RNAs to replace CTP and UTP. The transcribed RNAs were purified following the methods described in our previous work.32, 42 Two RNAs were mixed at a molar ratio of 1:1 and annealed to form one entity by heating at 94°C for 3 min, followed by slowly cooling to room temperature within 1 hr.
For HER3 aptamer (RNA3) synthesis, RNA1 PCR template and RNA1 5′ primer will be used as the above sequences, and RNA3 3′ primer is 5′-GGAUCCCGAACGGACCGCCCA-3′.
For HER2 aptamer (RNA4) synthesis, RNA2 PCR template and RNA 2 5′ primer will be used as the above sequences, and RNA4 3′ primer is 5′-AGCCGCGCCCTACCCTATCCCT-3′.
For MG aptamer (RNA5) synthesis, RNA5: 5′- GGAUCCCGACUGGCGAGAGCCAGGUAACGAAUG GAUCC-3′. RNA5 PCR template: 5′- TAATACGACTCACTATAGGATCCCGACTGGCGAGAGCCAGG TAAC GAATGGATCC-3′.
RNA5 5′ primer: 5′- TAATACGACTCACTATAGGATCCCGACTGGC-3′.
RNA5 3′ primer: 5′-GGATCCATTCGTTACCT-3′.
The PCR products were transcribed into RNA3, RNA4, and RNA5, which were annealed into HER3, HER2, and MG aptamers individually.
For mutant H2EH3 synthesis, RNA6 (mutant HER3 aptamer-EGFR antisense siRNA). PCR template (IDT): 5′GAATTCCGCGTGTGCCAGCGAAAGTTGCGTATGGGTCACATCGCACAGGACGTTCATCTGGGCGGTCCGTTCGGGATCCAAAAUUAGAUAAGACUGCUAAGGCA-3′
RNA6 5′ primer: 5′-TAATACGACTCACTATAGAATTCCGCGTGTGCCA-3′
RNA6 3′ primer: 5′-TGCCTTAGCAGTCTTATCTAATTTTGGA-3′
RNA7 (mutant HER2 aptamer-EGFR sense siRNA). PCR template (IDT): 5′-AGCCAAA CGAGGGGGGAGAGGGTGGGGGCGCCTG AAAACCTTAGCAGTC TTATCTAATT-3′
RNA7 5′ primer: 5′-TAATACGACTCACTATAAGCCAAA CGAGGGGGGAGAGGGT-3′
RNA7 3′ primer: 5′-AATTAGATAAGACTGCTAAGGTTTTCA-3′
As the above procedures, the PCR products were transcribed into RNAs. RNA6 and RNA7 were annealed into mutant H2EH3. Cy5-labeled aptamers were synthesized by TriLink Biotechnologies.
Cytotoxicity Assay
Cellular cytotoxicity was quantified by measuring WST-8 formazan using CCK-8 (Dojindo, Japan). Cells were seeded in a 96-well plate at a density of 5 × 103 in a 5% CO2 incubator for 24 hr at 37°C. Cell lines were incubated with the varying concentrations of H2EH3 or controls for 72 hr without transfection reagents (e.g., lipofectamine). CCK-8 solution (10 μL) was added to each well and incubated at 37°C for 4 hr. Absorbance at 450 nm was measured using a microplate reader.
Western Blot
Cells were lysed in lysis buffer (M-PER Mammalian Protein Extraction Reagent, Thermo Fisher Scientific) containing 1x Protease Inhibitor Cocktails (Sigma-Aldrich). The cell lysates were centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was collected and the protein concentration was determined with Bio-Rad Protein Assay (Bio-Rad, Hercules, CA). An equal amount of proteins was mixed with 4x Laemmli sample buffer containing 5% β-mercaptoethanol and heated at 95°C for 10 min. Denatured samples was separated on 8%–10% SDS-PAGE and transferred to a polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with 5% non-fat milk overnight at 4°C and then incubated with primary antibodies overnight at 4°C, followed by incubation with horseradish-peroxidase-conjugated secondary antibodies for 2 hr at room temperature. After ECL Western Blotting Substrate (Pierce) was added onto the membrane, the signals were captured by the exposure to X-ray film.
Dicer Assay
H2EH3 (4 μg) was digested using human recombinant dicer enzyme (2 U) at 37°C for 12 hr following the manufacturer’s instructions (Genlantis, San Diego, CA). Reaction was quenched by adding dicer stop solution. The digestion pattern was analyzed on 3.5% agarose gel electrophoresis.
Cell Cycle Analysis
SKBR3 and BT474 cells were treated with the different concentrations of H2EH3 for 24 and 48 hr. Collected cells were fixed in cold 70% ethanol. After a fix for 30 min at 4°C, ethanol-fixed cells were centrifuged and washed once with PBS. The cells were treated with RNase A and stained with PI for 30 min at 37°C. Cellular DNA contents were measured by exciting PI at 488 nm and measuring the emission at 580 nm using a BD FACSCalibur flow cytometer.
Detection of Apoptosis by Flow Cytometry
SKBR3 and BT474 cells were treated with the different concentrations of H2EH3 for 72 hr. The cells were harvested and washed in cold PBS. Cells were stained with Alexa Fluor 488 annexin-V-PI solution for 1 hr at room temperature. Cells (1 × 104/sample) were acquired by BD FACSCalibur and analyzed using BD FACStation software.
Knockdown of HER2 and/or HER3
BT474 cells were plated in 6-well plates at a density of 5 × 105 cells/well for 24 hr. HER2 siRNA and/or HER3 siRNA or control siRNA were transfected into cells using Lipofectamine RNAi MAX (Life Technologies) according to the manufacturer’s instruction. Cells were harvested 72 hr post-transfection, and western blot was performed to confirm gene knockdown. HER2- and/or HER3-silenced cells were subjected to aptamer-binding assay by BD flow cytometry.
Bio-distribution Assay
To evaluate the bio-distribution properties of H2EH3, tumor-bearing mice (n = 3 per group) were intravenously administered Cy5-labeled H2EH3 (20 nmol, 200 μL) or an equal mole amount of Cy5-labeled muH2EH3. The whole-body images were obtained at 0.5 and 6 hr using the Xenogen IVIS100 imaging system by setting the wavelength at an excitation of 640 nm and emission at 710 nm. After 6-hr injection of Cy5-labeled chimeras, mice were euthanized with CO2 following whole-body imaging and organs (heart, lung, liver, spleen, kidney, muscle, brain, stomach, and intestine) were removed. The ex vivo imaging of Cy5 signal was performed using Xenogen IVIS100.
In Vivo Xenograft Treatment Study
4- to 5-week-old female athymic nu/nu mice were injected with tumor cells (BT474 or MDA-MB-231) (2 × 106) mixed with Matrigel (v/v 1:1) (Corning, NY) subcutaneously at the left flank or orthotopically at the mammary fat pads. Following the establishment of tumors (100 mm3), mice were randomly divided into three groups in subcutaneous xenografts, H2EH3 (8 nmol, 200 μL), a mixture of HER2 aptamer, EGFR siRNA, and HER3 aptamer (each 8 nmol, total of 200 μL), or PBS (200 μL) was intra-tumorally injected every other day for 5 weeks. In the orthotopic xenografts, upon the tumor reaching 50 mm3, mice were administrated with H2EH3 (8 nmol, 200 μL) or PBS every 3 days through tail-vein intravenous injection for 4 weeks. Tumor sizes and body weights were measured twice a week. The animals were euthanized 2 days after the last treatment. The tumors and organs (liver, spleen, kidney, brain, heart, muscle, blood, and intestine) were removed and fixed in 10% formalin buffer. The sections of tissues were analyzed by IHC.
Histology Assay
Tumors and organs (spleen, lung, kidney, intestine, heart, liver, and muscle) were collected from xenografts and fixed with 4% paraformaldehyde. Sections (6 μm) were cut and mounted on the slides and deparaffinized in xylene and ethyl alcohol. For IHC assay, sections were incubated in 3% normal goat serum for 2 hr and incubated with primary antibodies: caspase-3 (1:200), HER2 (1:800), HER3 (1:250), p21 (1:50), and EGFR (1:50). After washing, the sections were incubated with biotinylated secondary antibody (1:200) (Vector Labs, Burlingame, CA) for 1 hr. Following washing, the sections were incubated with VECTASTAIN ABC reagents for 30 min. The images were captured with a Nuance fluorescence microscope with a bright field imaging system.
ELISA Assay
Normal human PBMCs were separated with Ficoll-Plaque Plus (STEMCELL Technologies). Cells were seeded into 24-well plates at 2 × 106/well for 24 hr in RPMI-1640 medium containing 10% fetal bovine serum. H2EH3, with varying concentrations, was added into cells, and cells were incubated for 48 hr in a 5% CO2 incubator at 37°C. The cell culture supernatant was detected with human IFN-α and IL-6 ELISA kits following the manufacturer’s instruction.
Statistical Analysis
GraphPad Prism software was used to perform all statistical analyses. The data were analyzed using a two-tailed Student’s t test. The differences of p < 0.05 were considered statistically significant.
Author Contributions
H.Y.L. conceived and designed the experiments, H.Y.L., X.Y., H.L., L.X., S.Z., W.T., and L.Z. performed the experiments. H.Y.L., X.Y., and H.L. analyzed the data. H.Y.L., N.J.M., S.G., H.K., D.W., and S.-C.T. wrote the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
This work is supported by start-up funding of Augusta University and Department of Defense W81XWH-15-1-0333 (to H.Y.L.). H.Y.L. thanks the NIH for an F32 fellowship (CA150301).
Footnotes
Supplemental Information includes seven figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.015.
Supplemental Information
References
- 1.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 2.Narayan M., Wilken J.A., Harris L.N., Baron A.T., Kimbler K.D., Maihle N.J. Trastuzumab-induced HER reprogramming in “resistant” breast carcinoma cells. Cancer Res. 2009;69:2191–2194. doi: 10.1158/0008-5472.CAN-08-1056. [DOI] [PubMed] [Google Scholar]
- 3.Arteaga C.L., Engelman J.A. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olayioye M.A., Neve R.M., Lane H.A., Hynes N.E. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholson R.I., Gee J.M., Harper M.E. EGFR and cancer prognosis. Eur. J. Cancer. 2001;37(Suppl 4):S9–S15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- 6.Lee-Hoeflich S.T., Crocker L., Yao E., Pham T., Munroe X., Hoeflich K.P., Sliwkowski M.X., Stern H.M. A central role for HER3 in HER2-amplified breast cancer: implications for targeted therapy. Cancer Res. 2008;68:5878–5887. doi: 10.1158/0008-5472.CAN-08-0380. [DOI] [PubMed] [Google Scholar]
- 7.Holbro T., Beerli R.R., Maurer F., Koziczak M., Barbas C.F., 3rd, Hynes N.E. The ErbB2/ErbB3 heterodimer functions as an oncogenic unit: ErbB2 requires ErbB3 to drive breast tumor cell proliferation. Proc. Natl. Acad. Sci. USA. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritter C.A., Perez-Torres M., Rinehart C., Guix M., Dugger T., Engelman J.A., Arteaga C.L. Human breast cancer cells selected for resistance to trastuzumab in vivo overexpress epidermal growth factor receptor and ErbB ligands and remain dependent on the ErbB receptor network. Clin. Cancer Res. 2007;13:4909–4919. doi: 10.1158/1078-0432.CCR-07-0701. [DOI] [PubMed] [Google Scholar]
- 9.Kontermann R.E., Brinkmann U. Bispecific antibodies. Drug Discov. Today. 2015;20:838–847. doi: 10.1016/j.drudis.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen H.J., Poulsen T.T., Dahlman A., Kjær I., Koefoed K., Sen J.W., Weilguny D., Bjerregaard B., Andersen C.R., Horak I.D. Pan-HER, an antibody mixture simultaneously targeting EGFR, HER2, and HER3, effectively overcomes tumor heterogeneity and plasticity. Clin. Cancer Res. 2015;21:4110–4122. doi: 10.1158/1078-0432.CCR-14-3312. [DOI] [PubMed] [Google Scholar]
- 11.Hu S., Fu W., Xu W., Yang Y., Cruz M., Berezov S.D., Jorissen D., Takeda H., Zhu W. Four-in-one antibodies have superior cancer inhibitory activity against EGFR, HER2, HER3, and VEGF through disruption of HER/MET crosstalk. Cancer Res. 2015;75:159–170. doi: 10.1158/0008-5472.CAN-14-1670. [DOI] [PubMed] [Google Scholar]
- 12.Iida M., Bahrar H., Brand T.M., Pearson H.E., Coan J.P., Orbuch R.A., Flanigan B.G., Swick A.D., Prabakaran P.J., Lantto J. Targeting the HER family with Pan-HER effectively overcomes resistance to cetuximab. Mol. Cancer Ther. 2016;15:2175–2186. doi: 10.1158/1535-7163.MCT-16-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 14.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler L.A., Trifonova R., Vrbanac V., Basar E., McKernan S., Xu Z., Seung E., Deruaz M., Dudek T., Einarsson J.I. Inhibition of HIV transmission in human cervicovaginal explants and humanized mice using CD4 aptamer-siRNA chimeras. J. Clin. Invest. 2011;121:2401–2412. doi: 10.1172/JCI45876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neff C.P., Zhou J., Remling L., Kuruvilla J., Zhang J., Li H., Smith D.D., Swiderski P., Rossi J.J., Akkina R. An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice. Sci. Transl. Med. 2011;3:66ra6. doi: 10.1126/scitranslmed.3001581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen C.H.B., Chernis G.A., Hoang V.Q., Landgraf R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. USA. 2003;100:9226–9231. doi: 10.1073/pnas.1332660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi M., Wu X., Ho M., Chomchan P., Rossi J.J., Burnett J.C., Zhou J. High throughput sequencing analysis of RNA libraries reveals the influences of initial library and PCR methods on SELEX efficiency. Sci. Rep. 2016;6:33697. doi: 10.1038/srep33697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim M.Y., Jeong S. In vitro selection of RNA aptamer and specific targeting of ErbB2 in breast cancer cells. Nucleic Acid Ther. 2011;21:173–178. doi: 10.1089/nat.2011.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma J.B., Ye K., Patel D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler D.L., Huang S., Kruser T.J., Nechrebecki M.M., Armstrong E.A., Benavente S., Gondi V., Hsu K.T., Harari P.M. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–3956. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dua R., Zhang J., Nhonthachit P., Penuel E., Petropoulos C., Parry G. EGFR over-expression and activation in high HER2, ER negative breast cancer cell line induces trastuzumab resistance. Breast Cancer Res. Treat. 2010;122:685–697. doi: 10.1007/s10549-009-0592-x. [DOI] [PubMed] [Google Scholar]
- 24.Kim D.H., Longo M., Han Y., Lundberg P., Cantin E., Rossi J.J. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- 25.Vogel C.L., Cobleigh M.A., Tripathy D., Gutheil J.C., Harris L.N., Fehrenbacher L., Slamon D.J., Murphy M., Novotny W.F., Burchmore M. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J. Clin. Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- 26.Nahta R., Esteva F.J. Trastuzumab: triumphs and tribulations. Oncogene. 2007;26:3637–3643. doi: 10.1038/sj.onc.1210379. [DOI] [PubMed] [Google Scholar]
- 27.McKeage K., Perry C.M. Trastuzumab: a review of its use in the treatment of metastatic breast cancer overexpressing HER2. Drugs. 2002;62:209–243. doi: 10.2165/00003495-200262010-00008. [DOI] [PubMed] [Google Scholar]
- 28.Witton C.J., Reeves J.R., Going J.J., Cooke T.G., Bartlett J.M. Expression of the HER1-4 family of receptor tyrosine kinases in breast cancer. J. Pathol. 2003;200:290–297. doi: 10.1002/path.1370. [DOI] [PubMed] [Google Scholar]
- 29.Tovey S.M., Witton C.J., Bartlett J.M., Stanton P.D., Reeves J.R., Cooke T.G. Outcome and human epidermal growth factor receptor (HER) 1-4 status in invasive breast carcinomas with proliferation indices evaluated by bromodeoxyuridine labelling. Breast Cancer Res. 2004;6:R246–R251. doi: 10.1186/bcr783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barnes N.L., Khavari S., Boland G.P., Cramer A., Knox W.F., Bundred N.J. Absence of HER4 expression predicts recurrence of ductal carcinoma in situ of the breast. Clin. Cancer Res. 2005;11:2163–2168. doi: 10.1158/1078-0432.CCR-04-1633. [DOI] [PubMed] [Google Scholar]
- 31.Kew T.Y., Bell J.A., Pinder S.E., Denley H., Srinivasan R., Gullick W.J., Nicholson R.I., Blamey R.W., Ellis I.O. c-erbB-4 protein expression in human breast cancer. Br. J. Cancer. 2000;82:1163–1170. doi: 10.1054/bjoc.1999.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng J., Zhao S., Yu X., Huang S., Liu H.Y. Simultaneous targeting of CD44 and EpCAM with a bispecific aptamer effectively inhibits intraperitoneal ovarian cancer growth. Theranostics. 2017;7:1373–1388. doi: 10.7150/thno.17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao Z., Carrasco R.A., Schifferli K., Kinneer K., Tammali R., Chen H., Rothstein R., Wetzel L., Yang C., Chowdhury P. A potent HER3 monoclonal antibody that blocks both ligand-dependent and -independent activities: differential impacts of PTEN status on tumor response. Mol. Cancer Ther. 2016;15:689–701. doi: 10.1158/1535-7163.MCT-15-0555. [DOI] [PubMed] [Google Scholar]
- 34.Da Pieve C., Blackshaw E., Missailidis S., Perkins A.C. PEGylation and biodistribution of an anti-MUC1 aptamer in MCF-7 tumor-bearing mice. Bioconjug. Chem. 2012;23:1377–1381. doi: 10.1021/bc300128r. [DOI] [PubMed] [Google Scholar]
- 35.Dassie J.P., Liu X.Y., Thomas G.S., Whitaker R.M., Thiel K.W., Stockdale K.R., Meyerholz D.K., McCaffrey A.P., McNamara J.O., 2nd, Giangrande P.H. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ricklin D., Lambris J.D. Complement-targeted therapeutics. Nat. Biotechnol. 2007;25:1265–1275. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratico E.D., Sullenger B.A., Nair S.K. Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic Acid Ther. 2013;23:35–43. doi: 10.1089/nat.2012.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan L., Neoh K.G., Kang E.T., Choe W.S., Su X. PEGylated anti-MUC1 aptamer-doxorubicin complex for targeted drug delivery to MCF7 breast cancer cells. Macromol. Biosci. 2011;11:1331–1335. doi: 10.1002/mabi.201100173. [DOI] [PubMed] [Google Scholar]
- 39.Wengerter B.C., Katakowski J.A., Rosenberg J.M., Park C.G., Almo S.C., Palliser D., Levy M. Aptamer-targeted antigen delivery. Mol. Ther. 2014;22:1375–1387. doi: 10.1038/mt.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sundaram P., Kurniawan H., Byrne M.E., Wower J. Therapeutic RNA aptamers in clinical trials. Eur. J. Pharm. Sci. 2013;48:259–271. doi: 10.1016/j.ejps.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 41.Rettig G.R., Behlke M.A. Progress toward in vivo use of siRNAs-II. Mol. Ther. 2012;20:483–512. doi: 10.1038/mt.2011.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu H.Y., Yu X., Liu H., Wu D., She J.X. Co-targeting EGFR and survivin with a bivalent aptamer-dual siRNA chimera effectively suppresses prostate cancer. Sci. Rep. 2016;6:30346. doi: 10.1038/srep30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Behlke M.A. Chemical modification of siRNAs for in vivo use. Oligonucleotides. 2008;18:305–319. doi: 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- 44.Li C., Huang S., Armstrong E.A., Francis D.M., Werner L.R., Sliwkowski M.X., van der Kogel A., Harari P.M. Antitumor effects of MEHD7945A, a dual-specific antibody against EGFR and HER3, in combination with radiation in lung and head and neck cancers. Mol. Cancer Ther. 2015;14:2049–2059. doi: 10.1158/1535-7163.MCT-15-0155. [DOI] [PubMed] [Google Scholar]
- 45.Wang D., Qian G., Zhang H., Magliocca K.R., Nannapaneni S., Amin A.R.M.R., Rossi M., Patel M., El-Deiry M., Wadsworth J.T. HER3 targeting sensitizes HNSCC to cetuximab by reducing HER3 activity and HER2/HER3 dimerization: evidence from cell line and patient-derived xenograft models. Clin. Cancer Res. 2017;23:677–686. doi: 10.1158/1078-0432.CCR-16-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.