Abstract
The reciprocal interaction between influenza virus and host microRNAs (miRNAs) has been implicated in the regulation of viral replication and host tropism. However, the global roles of the cellular miRNA repertoire and the mechanisms of miRNA-mediated antiviral defense await further elucidation. In this study, we systematically screened 297 cellular miRNAs from human and mouse epithelial cells and identified five inhibitory miRNAs that efficiently inhibited influenza virus replication in vitro and in vivo. Among these miRNAs, hsa-mir-127-3p, hsa-mir-486-5p, hsa-mir-593-5p, and mmu-mir-487b-5p were found to target at least one viral gene segment of both the human seasonal influenza H3N2 and the attenuated PR8 (H1N1) virus, whereas hsa-miR-1-3p inhibited viral replication by targeting the supportive host factor ATP6V1A. Moreover, the number of miRNA binding sites in viral RNA segments was positively associated with the activity of host miRNA-induced antiviral defense. Treatment with a combination of the five miRNAs through agomir delivery pronouncedly suppressed viral replication and effectively improved protection against lethal challenge with PR8 in mice. These data suggest that the highly expressed miRNAs in respiratory epithelial cells elicit effective antiviral defenses against influenza A viruses and will be useful for designing miRNA-based therapies against viral infection.
Keywords: IAVs, miRNAs, antiviral defense, ATP6V1A, antiviral therapy
Introduction
Influenza A viruses (IAVs), as causative agents of an infectious febrile illness in humans, are widely spread among diverse hosts and cause recurrent epidemics and sporadic pandemics.1, 2 IAVs primarily infect epithelial cells of the upper and lower respiratory tracts, which may lead to primary viral pneumonia in humans with rapid progression to lung failure. Aside from humans, influenza viruses have a reservoir in bats, birds, and pigs, from which reassortment among these viruses may lead to the emergence of epidemic and pandemic viruses with the capability for interspecies transmission.3 This exerts a public health threat and an extensive economic burden on a global scale,4 and it has led to an urgent demand to dissect the mechanism of the host-virus interplay to design effective drugs. IAVs contain a single-stranded negative RNA genome consisting of eight segments. Each of these RNA segments is bound by viral RNA-dependent PA, PB1, and PB2 proteins to form ribonucleoprotein (vRNP) complexes. These vRNPs carry out transcription of viral genes and replication of the viral RNA genome, both of which are regulated by multiple host factors.5, 6, 7 Nevertheless, only a few host factors have been intensively investigated to determine their roles in IAV infection.8
MicroRNAs (miRNAs) belong to a class of genome-encoded, small non-coding RNAs, typically 20–22 nt in length, that post-transcriptionally regulate gene expression, mostly by repressing target transcripts containing partially complementary binding sites.9 It is estimated that approximately half of the human transcriptome is affected by miRNAs, leading to target mRNA degradation and translational repression. Thus, miRNAs have emerged as vital regulators of multiple biological processes, including viral infection, replication, and pathogenicity.10 Several lines of evidence indicate differential expression of miRNA signatures post-infection with various strains of IAVs, including avian H5N3 virus-infected chickens,11 H1N2 virus-infected pigs,12 a mouse-adapted and highly virulent avian H5N2 virus-infected mouse,13 and H1N1 virus-infected mouse lungs14, 15 and A549 cells.16 Under IAV infections, combinatorial cellular miRNAs may mediate a series of temporal and strain-specific host responses.17 These studies suggest that miRNAs are involved in the regulation of different stages in the viral life cycle via targeting of viral RNAs or host factors. For instance, hsa-miR-323-5p, hsa-miR-491-5p, and hsa-miR-654-5p directly bind to viral PB1 RNA, exerting an inhibitory effect on IAV replication in Madin-Darby canine kidney (MDCK) cells.18 Hsa-let-7c reduces viral replication by targeting and degrading M1 cRNA.19 In addition, hsa-miR-485-5p targets both retinoic acid-inducible gene I (RIG-I) and viral PB1 to prevent spurious activation of antiviral signaling and suppress influenza virus infection.20 To date, most studies have used miRNA target prediction for analyses of certain miRNAs that interact with IAV or with host mRNAs. As a consequence, the exact role of the miRNA repertoire has not been determined in IAV infection, and it is largely unclear whether the miRNA-mediated antiviral defense confers viral adaption to respiratory epithelial cells.
In this study, we systematically screened and identified candidate miRNAs from the total miRNA pools that are highly expressed in human and mouse respiratory epithelial cells to assess their regulation of IAV replication. We explored the underlying mechanisms of a set of miRNAs as host inhibitory factors to suppress viral expression and replication of human seasonal influenza virus H3N2 and A/Puerto Rico/8/1934 (PR8; H1N1). In addition, the co-relationship between number of miRNA targeting sites within viral sequences and miRNA-mediated suppression of viral replication was evaluated in response to selection pressure.
Results
High-Throughput Screen for Candidate miRNAs that Inhibit IAV Replication
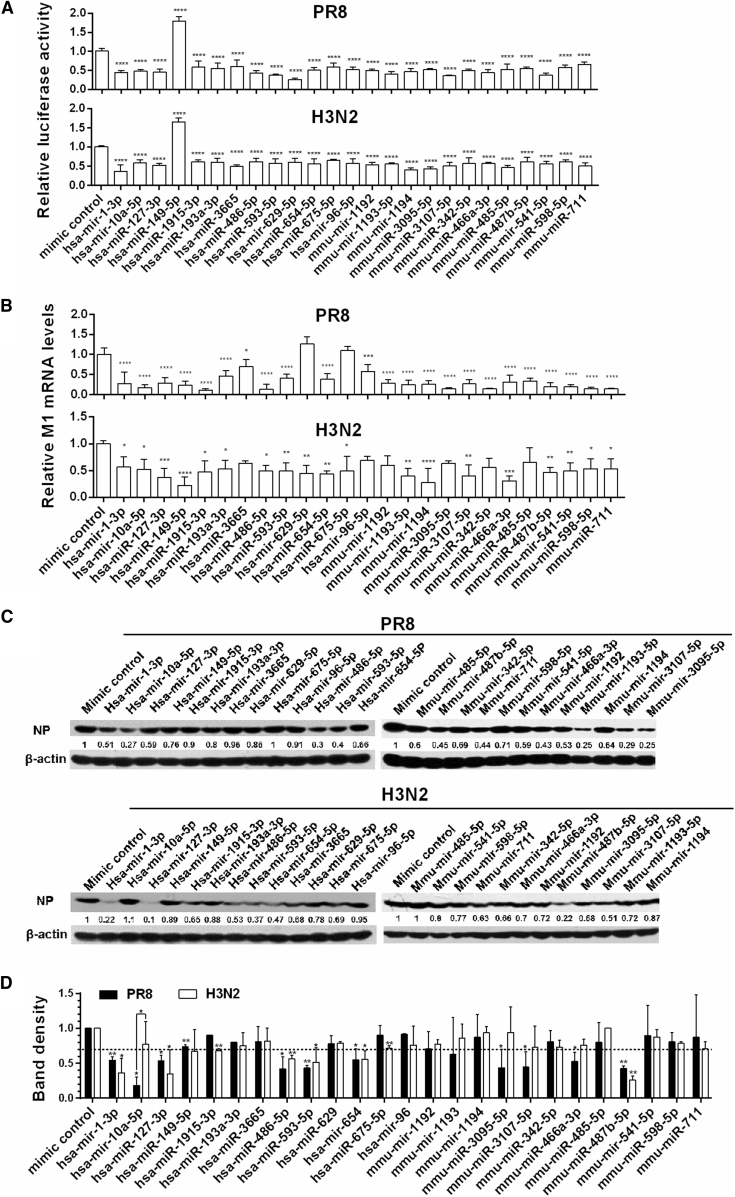
A plasmid carrying firefly luciferase gene with virus nonstructural gene promoter (vNS-Luc)21 that can be activated by viral RNA-dependent RNA polymerase (RdRp) was used for high-throughput screening of 297 host miRNAs expressed in either human or mouse epithelial cells (Table S1). These miRNAs were synthesized in mature form. HEK293T cells were chosen for high-throughput screening, because the cells are easily transfected with miRNAs and generally have relatively low abundance of the 297 selected miRNAs. HEK293T cells transfected with miRNAs and vNS-Luc were infected with two IAV subtypes, H1N1 (PR8) or H3N2 (A/Jiangxi/262/2005), and viral replication was determined by measurement of the luciferase reporter activity. The screen timeline and validation of the nonstructural (NS) luciferase reporter assay are shown in Figure S1. Renilla luciferase activity increases linearly with increasing MOI dose or infection time. Among the 297 miRNAs, 62 and 55 miRNAs exhibited regulatory (mostly inhibitory) effects on NS reporter activity in HEK293T cells infected with PR8 and H3N2, respectively (Figure S2; Table S2). Furthermore, 25 miRNAs affected the luciferase activities (mostly inhibitory) of the NS reporter in both IAV subtype-infected cells relative to mimic controls (all p < 0.05 or 0.01) (Figure 1A). Among these 25 miRNAs, only hsa-miR-149-5p facilitated IAV replication; all others inhibited IAV replication. Cell viability assays were performed to rule out the possibility that the observed differences were caused by altered cell growth and cytotoxicity (Figures S3A and S3B).
Figure 1.
Screen and Validation of miRNA Top Hits that Inhibit H1N1 and H3N2 Replication
(A) HEK293T cells were co-transfected with vNS-Luc, pRL-TK, and miRNA mimics or a randomized oligonucleotide as a control. At 24 hr post-transfection, cells were infected with PR8 or H3N2 at a MOI of 0.01 for 24 or 36 hr before measuring luciferase activities. The luciferase activity was normalized to Renilla luciferase. (B–D) A549 cells were infected with PR8 or H3N2 at a MOI of 0.1 24 hr post-transfection with 20 nM miRNA mimics or an equal amount of randomized oligonucleotide as a control. Total RNAs and proteins were prepared 24 hr post-infection of PR8 or 36 hr post-infection of H3N2. Viral M mRNA levels were measured by real-time PCR, with GAPDH as an internal control (B). NP protein levels were determined by immunoblotting, with β-actin as an internal control (C). The bands on western blots were scanned by densitometry, and the amounts of NP were quantified (D). The relative amounts of NP in control cells were arbitrarily taken as 1.0. Error bars are the mean ± SD from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control.
To validate the top 25 miRNAs from high-throughput screen, human A549 lung cells were transfected with each miRNA and then infected with PR8 or H3N2. Their regulatory effect on viral replication was confirmed by measuring viral M1 mRNA (Figure 1B) and NP protein levels (Figures 1C and 1D). The five miRNAs with the most potent inhibitory activity on PR8 and H3N2 replication, hsa-mir-1-3p, hsa-mir-127-3p, hsa-mir-486-5p, hsa-mir-593-5p, and mmu-mir-487b-5p (all p < 0.05, 0.01, or 0.001 compared to controls for M1 mRNA and NP protein levels), were chosen for further study.
To determine the efficiency and specificity of miRNA mimic transfections, expression levels of the validated targets of miRNAs were measured by real-time PCR in transfected A549 cells. As shown in Figure S3C, all target genes were significantly inhibited by the mimic transfection.
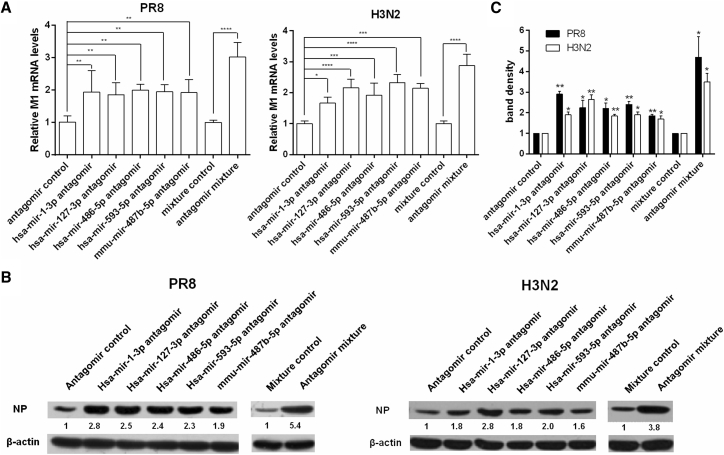
Conversely, miRNA suppression with specific antagomirs displayed enhancive effects on viral replication by analysis of M1 mRNA (Figure 2A) and NP protein levels (Figures 2B and 2C) in infected A549 cells, the adenocarcinomic human alveolar basal epithelial cells as the target cells for infection by IAV. This validates the inhibitory role of these endogenous miRNAs for IAVs.
Figure 2.
The Effect of the Five Selected miRNA Antagomirs on PR8 and H3N2 Replication
(A and B) A549 cells were transfected with 20 nM individual miRNA antagomirs, 20 nM mixture of the five miRNA antagomirs (each at a final concentration of 4 nM), or control antagomir. At 24 hr post-transfection, cells were infected with H1N1 or H3N2 at a MOI of 0.1 for 24 or 36 hr, respectively. Viral M1 mRNA (A) and NP protein (B) levels were determined by real-time PCR and immunoblotting analysis, respectively. (C) The bands of western blots were scanned by densitometry, and the amounts of NP were quantified. The relative amounts of NP in control cells were arbitrarily set as 1.0. Error bars are the mean ± SD from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control.
hsa-mir-149-5p promoted vNS-Luc reporter activity (Figure 1A) while repressing viral M1 transcripts (Figure 1B). The inconsistency may be due to both its promoting effect of vRNP activity and its direct targeting of viral M mRNAs, because there is a putative binding site in the M transcripts predicted by miRanda.
Identification of Shared miRNA Targets in the H1N1 and H3N2 Genomes
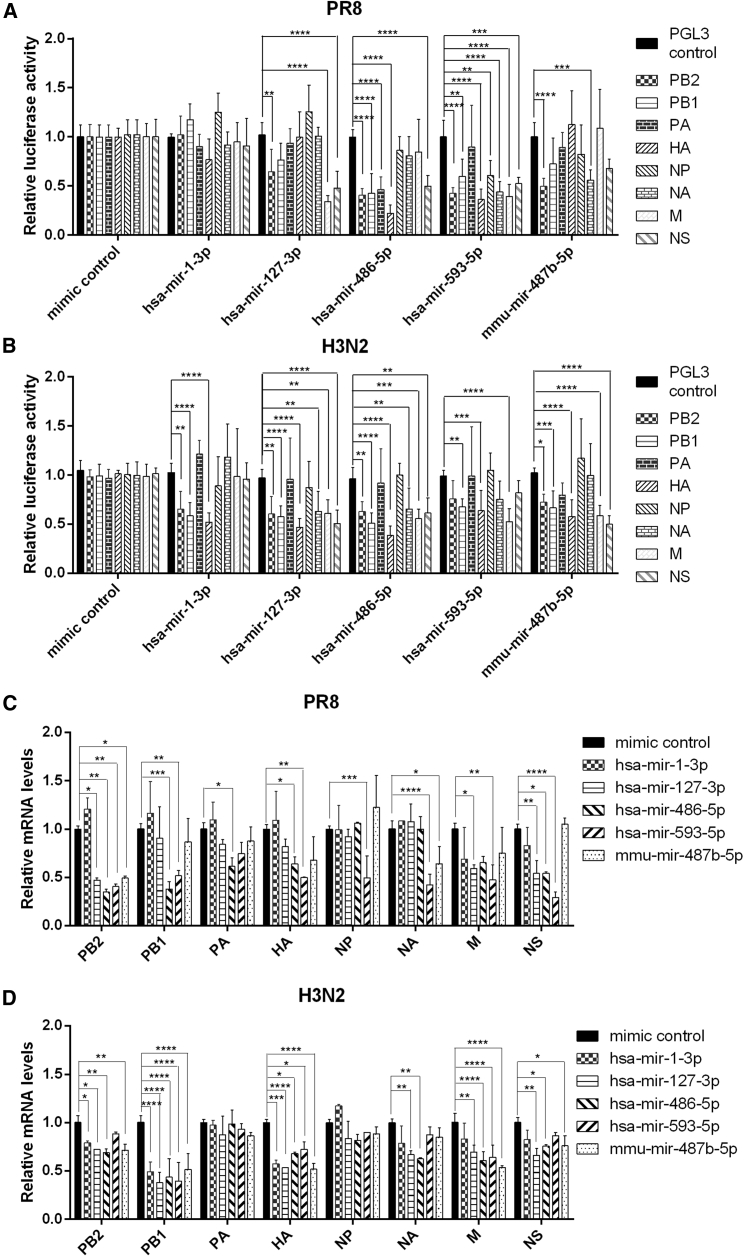
To explore the mechanisms of these five miRNAs in viral inhibition, luciferase reporters with each viral RNA segment in the 3′ UTR—HA, NA, M, PA, PB1, PB2, NP, and NS—were used to determine whether there were direct target sites in viral genes. Treatment with hsa-mir-1-3p, hsa-mir-127-3p, hsa-mir-486-5p, hsa-mir-593-5p, or mmu-mir-487b-5p mimics reduced the reporter activities of at least one PR8 (Figure 3A) and H3N2 (Figure 3B) RNA segment, except the PA and NP RNA segments of H3N2 (all p < 0.05, 0.01, or 0.001). In addition, levels of the corresponding viral transcripts of PR8 (Figure 3C) and H3N2 (Figure 3D) were significantly decreased in miRNAs-transfected A549 cells (all p < 0.05, 0.01, or 0.001). This indicates that these miRNAs directly target viral mRNAs.
Figure 3.
The Top miRNAs Target mRNAs of H1N1 or H3N2
(A and B) Eight RNA segments of H1N1 (A) and H3N2 (B) —HA, NA, M, PA, PB1, PB2, NP, and NS—were cloned into the 3′ UTR of the luciferase reporter vector pGL3. HEK293T cells were co-transfected with viral RNA segment luciferase reporter, Renilla luciferase vector pRL-TK, and 20 nM indicated miRNA mimics or 20 nM randomized oligonucleotide as a mimic control. At 48 hr post-transfection, the cells were lysed and subjected to luciferase assays, and the firefly luciferase activity was standardized to that of Renilla luciferase. (C and D) A549 cells were transfected with 20 nM indicated miRNA mimics or 20 nM randomized oligonucleotide as a control. At 24 hr post-transfection, the cells were infected with PR8 (C) or H3N2 (D) at a MOI of 0.1 for 14 hr. Then the cells were harvested to measure viral mRNA levels by real-time PCR. Data are presented as the mean ± SD from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control.
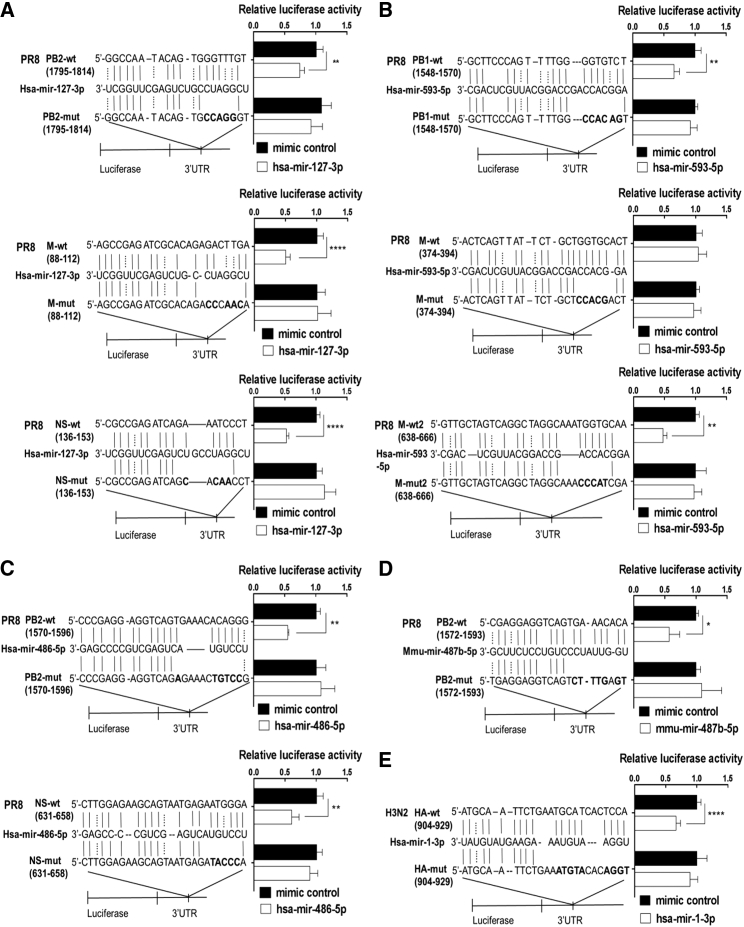
Because these five miRNAs inhibit luciferase activities of most viral RNA segments of both PR8 and H3N2, we further searched for shared miRNA binding sites in the two viral genomic sequences using Miranda software (Table S3). hsa-mir-127-3p has three shared binding sites located in the PB2, M, and NS RNA segments of both PR8 and H3N2. Mutations were made in the seed region of the hsa-mir-127-3p binding sites as controls. Treatment with hsa-mir-127-3p mimics reduced the reporter activity of the wild-type (WT), but not mutant (mut), 3′ UTR-containing luciferase reporter (Figures 4A and S4A). Similarly, hsa-mir-593-5p harbors three shared pairing regions located on PB1 and M RNAs (Figures 4B and S4B). The activities of two reporters constructed with two shared binding sites for hsa-mir-593-5p were decreased by treatment with hsa-mir-593-5p, while a suppressive effect for the reporter harboring a truncation from nucleotides 374–394 in the M segment was not observed. hsa-mir-486-5p has two shared binding sites, located on the PB2 and NS RNAs (Figures 4C and S4C), and mmu-mir-487b-5p only has one shared pairing region on the PB2 RNA (Figures 4D and S4D). All of these shared binding sites were verified by luciferase reporter assays. The limited differences between PR8 and H3N2 binding regions did not significantly alter inhibitory efficiency of luciferase reporter by miRNA mimics. In contrast, the targeting site for hsa-mir-1-3p was only found in the HA RNA of H3N2 (Figure 4E).
Figure 4.
Identification of Potential Target Sites of Top miRNAs in the H1N1 and H3N2 Genomes
The viral genomic fragments harboring the indicated miRNA pairing regions were cloned into the 3′ UTR of the luciferase reporter vector pGL3. HEK293T cells were co-transfected with viral RNA truncation luciferase reporter, Renilla luciferase vector pRL-TK, and each miRNA mimic or a randomized oligonucleotide as a mimic control. A dual-luciferase assay kit was used to detect the firefly luciferase activities 48 hr post-transfection, and the firefly luciferase activity was standardized to that of Renilla luciferase. (A–E) Perfect matches are indicated by a line between indicated viral RNA truncations and hsa-mir-127-3p (A), hsa-mir-593-5p (B), hsa-mir-486-5p (C), mmu-mir-487b-5p (D), or has-miR-1-3p (E). Mutations were made in the seed region of the miRNA binding site for the reporter gene assays. Data are presented as the mean ± SD from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control.
Aside from the shared targeting sites, a targeting site for hsa-mir-127-3p in the NA RNA was found in H3N2, but not in PR8 (Figure S4E). hsa-mir-486-5p, hsa-mir-593-5p, and mmu-mir-487b-5p were also found to target PA, HA, NP, and NA of PR8 (Figure S4F).
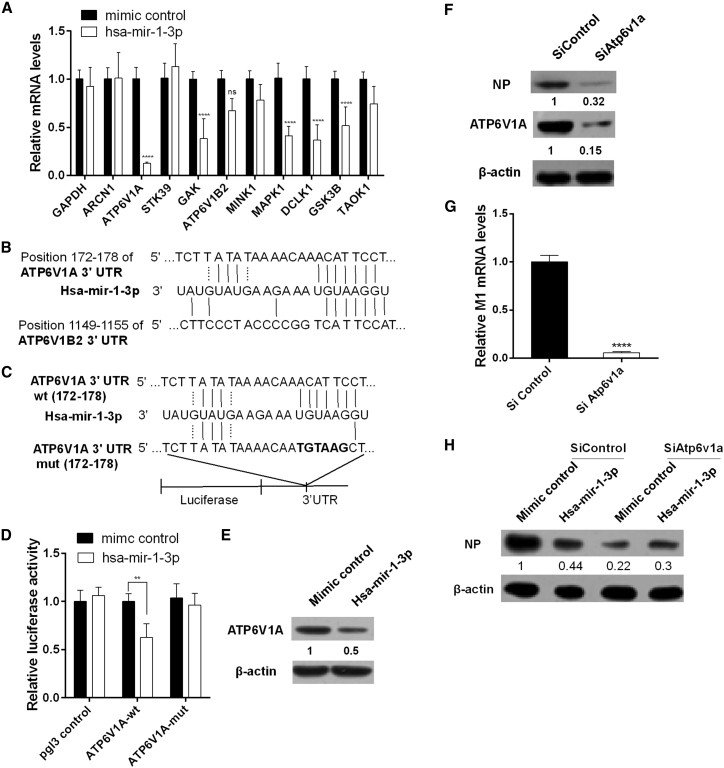
hsa-miR-1-3p Inhibits PR8 Replication Partially via Its Target ATP6V1A
Because hsa-miR-1-3p suppresses PR8 replication but has no effect on the 3′ UTR reporter activities of all viral RNA segments, we analyzed its predicted host targets that have been shown to affect IAV replication (Table S4)7 using TargetScan (http://www.mirbase.org/). Ten potential host factors (ARCN1, ATP6V1A, STK39, GAK, ATP6V1B2, MINK1, MAPK1, DCLK1, GSK3B, and TAOK1) were selected, and the reduction of ATP6V1A transcript was the most significant upon hsa-miR-1-3p treatment (Figure 5A). The discrepancy between ATP6V1A and ATP6V1B2 binding sites with miR-1-3p may account for the different targeting efficiency by miR-1-3p (Figure 5B). A putative hsa-miR-1-3p complementary region was found in the ATP6V1A mRNA 3′ UTR (Figure 5C) and verified by luciferase reporter assays (Figure 5D). A549 cells transfected with hsa-miR-1-3p displayed a decrease in ATP6V1A protein levels (Figure 5E). In addition, ATP6V1A knockdown by RNAi decreased viral M mRNA and NP protein levels (Figures 5F and 5G). ATP6V1A depletion by RNAi largely abrogated the inhibitory effect of hsa-miR-1-3p on NP expression (Figure 5H), suggesting that hsa-miR-1-3p inhibits H1N1 replication, at least partially, through ATP6V1A.
Figure 5.
hsa-miR-1-3p Targets ATP6V1A and Represses H1N1 Replication
(A) A549 cells were transfected with 20 nM hsa-mir-1-3p mimics or an equal amount of randomized oligonucleotide as a control, and the mRNA levels of the indicated potential targets were measured by real-time PCR 24 hr post-transfection. (B) The alignments of ATP6V1A and ATP6V1B2 binding sites with miR-1-3p. (C) Perfect matches are indicated by a line between 3′ UTR of ATP6V1A (ATP6V1A-WT) and miR-1-3p. Mutations (ATP6V1A-mut) were made in the seed region of the miR-1-3p binding site for the reporter assay. (D) HEK293T cells were co-transfected with 20 nM hsa-mir-1-3p mimic or an equal amount of randomized oligonucleotide as a control and the reporter plasmid ATP6V1A-WT, ATP6V1A-mut, or pGL3 as a control, together with pRL-TK. Cell lysates were harvested for dual-luciferase assays 48 hr post-transfection. (E) A549 cells were transfected with 20 nM hsa-mir-1-3p mimics or an equal amount of mimic control for 48 hr. Cell lysates were harvested for immunoblotting with rabbit anti-human ATP6V1A antibody. (F and G) A549 cells were transfected with 20 nM Atp6v1a-specific siRNA (SiAtp6v1a) or an equal amount of scrambled siRNA (SiControl) as a control. At 24 hr post-transfection, cells were infected with PR8 at a MOI of 0.1 and were harvested for immunoblotting against viral protein NP (F) and for measuring M1 mRNA by real-time PCR (G). (H) A549 cells were co-transfected with 20 nM hsa-mir-1-3p or an equal amount of randomized oligonucleotide as a control, along with 20 nM Atp6v1a-specific siRNA (SiAtp6v1a) or an equal amount of randomized oligonucleotide (SiControl), and then were infected with PR8 at a MOI of 0.1 24 hr post-transfection. Western blot analyses were subsequently performed for detection of viral protein NP 24 hr post-infection. Data are presented as the mean ± SD from three independent experiments; **p < 0.01 and ****p < 0.0001 compared to the control transfection.
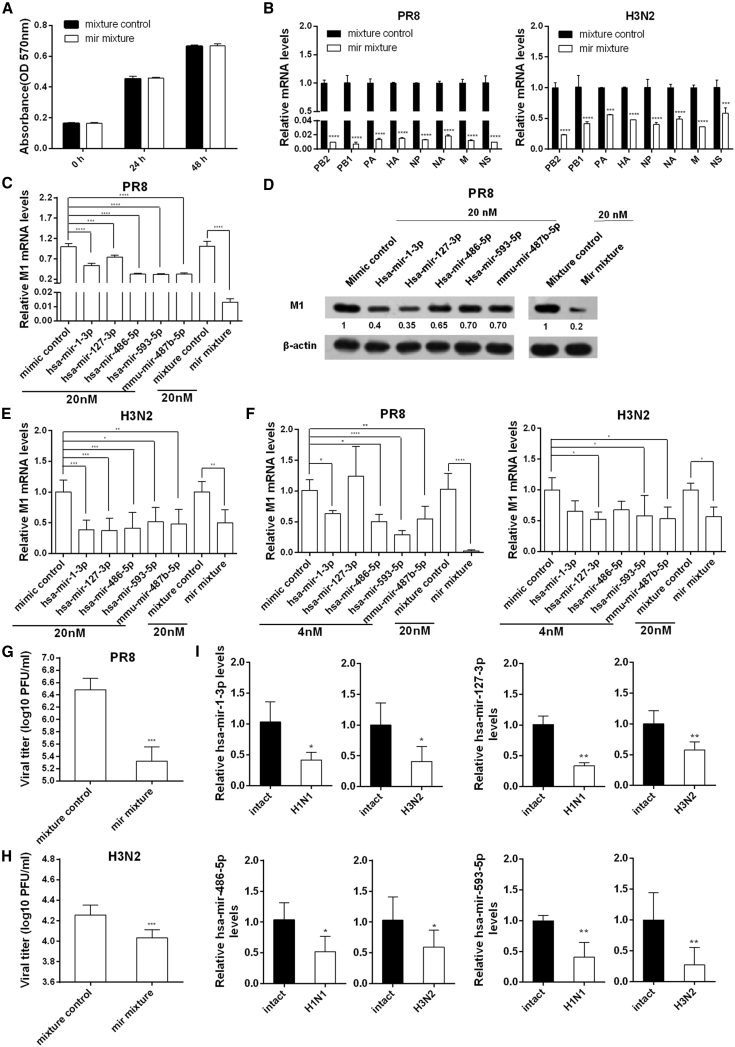
Synergistic Inhibition of PR8 Replication by the Combination of Five miRNAs
We found that hsa-mir-127-3p, hsa-mir-486-5p, hsa-mir-593-5p, and mmu-mir-487b-5p directly target viral RNA segments of H1N1 and H3N2 and that hsa-miR-1-3p targets host ATP6V1A, which positively regulates viral replication. Next, we investigated whether the combination of these five miRNAs would synergistically inhibit IAV. There was no obvious cytotoxicity for A549 cell viability under transfection with 20 nM five-mimic mixture (Figure 6A). A549 cells were transfected with 20 nM individual miRNAs or 20 nM five-mimic mixture and then infected with PR8 or H3N2. The results demonstrate that treatment with the miRNA combination significantly suppressed expression of all viral transcripts (Figure 6B) and had a higher inhibitory effect on PR8 than that of any individual miRNA by analysis of M1 mRNA (all p < 0.001) (Figure 6C) and protein levels (Figure 6D). Nevertheless, no such synergistic effect was observed for H3N2 (Figure 6E). Because the miRNA combination contained only 4 nM of each miRNA, we further compared the viral inhibitory effect of each miRNA at a 4-nM dose with that of the miRNA combination. We still found that the five miRNAs synergistically decreased viral M1 levels of PR8, but not H3N2 (Figure 6F). The difference may result from a higher number of virus gene segments harboring miRNA binding sites and total miRNA binding sites in the PR8 genome than in the H3N2 genome. The inhibitory effect of the five-miRNA combination on viral replication of PR8 (Figure 6G) and H3N2 (Figure 6H) was confirmed by plaque assays. As expected, the miRNA combination exhibited a higher suppression rate for PR8 (93.08%) than for H3N2 (39.87%). Meanwhile, infection of A549 cells with PR8 or H3N2 led to decreased levels of these inhibitory miRNAs (Figure 6I).
Figure 6.
Suppression of H1N1 and H3N2 Replication In Vitro by the Combination of the Five miRNAs
(A–H) A549 cells were transfected with 20 nM mixture of the five miRNAs, each at a final concentration of 4 nM; 20 nM indicated individual miRNA; or an equal amount of randomized oligonucleotide as control. MTT assays were carried out 0, 24, or 48 hr post-transfection (A). At 24 hr post-transfection, cells were infected with H1N1 or H3N2 at a MOI of 0.1 for 24 or 36 hr, respectively. Viral mRNA levels of eight gene segments were determined by real-time PCR (B). Viral M1 mRNA and protein levels were determined by real-time PCR (C, E, and F) and immunoblotting analysis (D), respectively. At 24 hr post-transfection, the cells were infected with H1N1 at a MOI of 0.5 (G) or H3N2 at a MOI of 1.0 (H) for 24 or 12 hr, respectively. Viral titers were determined by plaque assays. (I) A549 cells were infected with PR8 at a MOI of 0.1 for 24 hr or with H3N2 at a MOI of 1.0 for 10 hr and then harvested for measurement of indicated miRNAs by real-time PCR. Data are presented as the mean ± SD from three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control transfection.
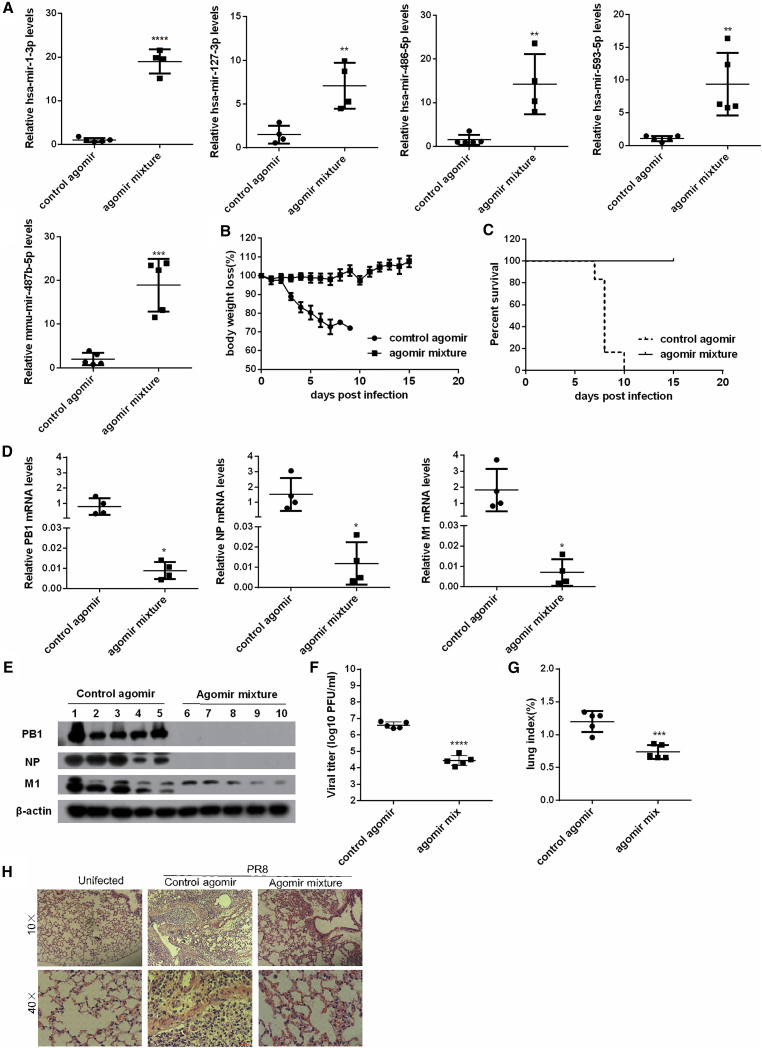
To assess whether the miRNA combination suppressed viral replication in vivo, we used cholesterylated stable miRNA mimics with two oxygen methylation modifications and sulfur-modified phosphate (i.e., agomirs) to deliver miRNAs to mouse lungs by intranasal administration. The delivery efficiency of agomirs was examined by measuring the levels of miRNA in mouse lungs 7 days post-intranasal administration of a mixture of the five agomirs. Expression levels of each miRNA in the lungs were increased by 4.5- to 31.6-fold (Figure 7A).
Figure 7.
Administration of the Five-Agomir Combination Protects Mice from H1N1 Infection
The 6- to 8-week-old female BALB/c mice (n = 5 per group) were intranasally administrated with the agomir mixture of hsa-mir-127-3p, hsa-mir-486-5p, hsa-mir-593-5p, mmu-mir-487b-5p, and hsa-miR-1-3p at 0.8 nmol each per mouse or the equivalent amount of randomized oligonucleotide agomir (control), and then the mice were intranasally challenged with 104 PFUs of H1N1 virus per mouse. (A) The agomir delivery efficiency was determined by measuring miRNA levels in mouse lungs 7 days post-intranasal administration of the mixture of agomirs. (B and C) The weight loss (B) and mortality rate (C) of infected mice were recorded daily, ending 15 days following infection. (D and E) The levels of viral PB1, NP, and M1 mRNA (D) and protein (E) in the mouse lungs were measured by real-time PCR and western blotting on day 4 post-challenge. Plaque assays were performed to measure viral titers in the diluted lung homogenates. (F) The titers are presented as log10 PFU/mL. (G) Lung index of mice was calculated by lung weight (g)/body weight (g) × 100%. (H) Photomicrographs of the lung tissue morphology with HE staining. Data show the mean ± SD of five mice from a representative experiment that was repeated two times with similar results; *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control.
BALB/c mice treated with the combination of the five agomirs or a randomized control agomir were intranasally challenged with 104 plaque-forming units (PFUs) of PR8 virus. By days 6–9 post-challenge, all control mice suffered typical influenza symptoms and displayed significant body weight loss and shiver (Figure 7B). All died at day 10 (Figure 7C). In contrast, mice treated with the combination of the five agomirs exhibited only moderate weight loss after H1N1 challenge and quickly restored weight from day 10 post-challenge. All agomir mixture-treated mice survived following the challenge with a lethal dose of H1N1 virus (Figure 7C).Viral replication and titers in mouse lungs were measured on day 4 after the challenge. Remarkable decreases in viral PB1, NP, and M1 mRNA (Figure 7D) and protein (Figure 7E) levels were observed in mice by agomir mixture treatment. As shown in Figure 7F, the lung viral titers of mice treated with the agomir mixture were approximately 100-fold lower than those of control mice (p < 0.001).
The lung index was used to monitor the degree of injury caused by influenza virus in mice. Mice treated with the agomir combination exhibited significantly lower lung indices than control mice (Figure 7G). H&E staining also showed that the alveoli, peribronchiolar, and perivascular regions were suffused with abundant inflammatory cells in the lungs of control mice, whereas inflammatory cells were barely seen in the alveoli of agomir combination-treated mice (Figure 7H). These results indicate that increased levels of inhibitory miRNAs in lungs can enhance resistance of mice to IAV infection.
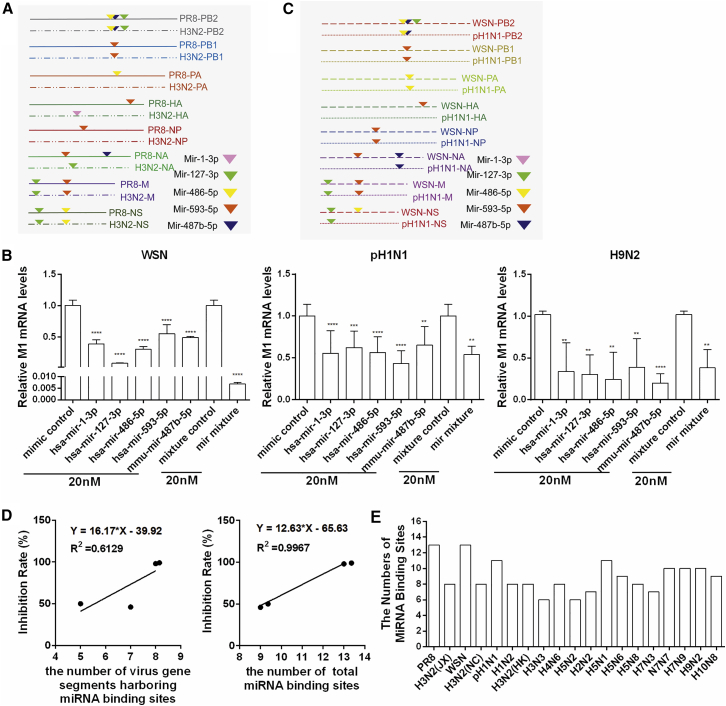
The miRNA-Mediated Antiviral Defense Is Associated with miRNA Binding Sites
To explore the mechanism of the different sensitivities of PR8 and H3N2 to endogenous miRNA-mediated inhibition, we compared the miRNA binding sites between the two IAV subtypes. As shown in Figure 8A, complementary regions of these five miRNAs were found in all eight RNA segments of PR8, whereas they were found in only six RNA segments of H3N2. We further tested whether the combination of the five miRNAs had synergistic inhibition of human virus WSN (H1N1), human pandemic (pH1N1), and avian-origin H9N2 IAVs. As shown in Figure 8B, the five miRNAs exhibited significant inhibitory activity on these IAVs. A synergistic effect was observed for WSN, but not for pH1N1 and H9N2. Only the WSN virus has binding sites for these five miRNAs in all eight RNA segments (Figures 8A and 8C). The inhibition rate of the IAVs by the miRNAs was positively associated with the number of virus gene segments harboring miRNA binding sites, as well as the number of total miRNA binding sites (Figure 8D). The complementary binding sites of the five inhibitory cellular miRNAs were also found in certain gene segments of aquatic bird influenza, domestic poultry influenza, swine influenza, and human seasonal influenza viruses (Figure 8E; Table S5). These data indicate that the five endogenous cellular miRNAs from respiratory epithelial cells identified in this study mainly act as host inhibitory factors against IAV replication, which may be involved in host defense mechanisms and interspecies transmission.
Figure 8.
Analysis of the Suppression of Other Viral Strains by the Combination of the Five miRNAs
(A) Summary of verified miRNA complementary regions located in viral RNAs of PR8 and H3N2. (B) Suppression of WSN, pH1N1, and H9N2 replication in vitro by the combination of the five miRNAs. A549 cells were transfected with 20 nM individual miRNA or with 20 nM mixture of five miRNAs, each at a final concentration of 4 nM. At 24 hr post-transfection, cells were infected with WSN, pH1N1, or H9N2 at a MOI of 0.1 for 24 hr. Viral M1 mRNA levels were determined by real-time PCR. (C) The shared miRNA binding sites located in the viral RNA of WSN and pH1N1. (D) Correlation of the inhibition rate of IAVs by miRNAs and the number of virus gene segments harboring miRNA binding sites (right) or the number of total miRNA binding sites (left). (E) Summary of the complementary binding sites of the five miRNAs in certain gene segments of different virus strains. Data are presented as the mean ± SD from three independent experiments; **p < 0.01, ***p < 0.001, and ****p < 0.0001 compared to the control.
Discussion
Cellular miRNAs play a key role in the control of virus infection and replication. However, the global role of the cellular miRNA repertoire in influenza virus infection has not been determined, and the mechanisms of antiviral defense and viral adaption await further elucidation. In this study, we screened a miRNA pool covering highly expressed miRNAs in human and mouse respiratory epithelial cells to unbiasedly identify endogenous cellular miRNAs that regulate two subtypes of IAV: PR8 and H3N2. With this approach, five miRNAs were identified, four of which directly targeted viral RNA segments, with the other one targeting supportive host ATP6V1A. Furthermore, agomir treatment by these five miRNAs synergistically suppressed PR8 replication in vitro and in vivo. Moreover, our findings demonstrate that miRNA-mediated antiviral defense correlates with miRNA binding sites in virus gene segments. Our work may therefore provide a comprehensive evaluation of miRNA-based antiviral therapeutic interventions in the context of viral escape of miRNA inhibition during IAV infection.
IAVs mainly target different regions of the respiratory tract, causing a respiratory disease in humans. In the present study, we identified 25 miRNAs highly expressed in respiratory epithelial cells that affect IAV replication, and among them, most exert suppression activities (Figure 1). In addition, a panel of the top five inhibitory miRNAs exhibited higher inhibitory activity against replication of PR8 and WSN strains that are well attenuated and adapted in mice and humans than other avian or swine IAV subtypes. Only the PR8 and WSN strains have the complementary binding sites of these five miRNAs in all eight gene segments, and miRNA-mediated inhibition of IAVs is correlated with the number of virus gene segments that have the miRNA binding sequences (Figure 7). This indicates the global inhibitory role of the cellular miRNA repertoire against influenza virus infection, which is involved in viral attenuation and adaption mechanisms. Dicer knockdown leads to enhancement of influenza virus replication in A549 cells that are hindered in endogenous miRNA production (Figure S5).22 In agreement with our results, other studies provide compelling evidence that the miRNA-based gene-silencing machinery can effectively control influenza virus replication. Perez et al.23 altered several bases in the viral NP gene to perfectly match the miRNA response elements of hsa-mir-93-5p, and the engineered virus displayed a greater than two-log reduction in mice mortality compared to WT viruses. The recombinant virus miRT-H1N1 engineered to be perfectly targeted by hsa-let-7b-5p was found to exert the desired replication restrictive effect and decreased infectivity in let-7b-rich-expressing tissues and organs.24 The modified miRT-H1N1 has a low virulence in mice, with the potential for the development of influenza vaccines.25 The engineered H5N1 viruses, whose genome was incorporated with miRNA target sites, exhibit attenuated pathogenesis in mice.26 In addition, an attenuated virus generated by inserting a miR-93 expression cassette into the viral NS segment, PR8-amiR-93NP, confers cross-protective immunity against heterologous influenza virus strains in mice.27 Altogether, we hypothesize that in IAV infection, highly expressed miRNAs in the respiratory tract constitute an effective antiviral defense against IAVs that harbor multiple miRNA binding elements in gene segments. In this specific selection setting, conditions may arise in which miRNA-associated viral sequence polymorphisms may occur in miRNA binding sites. Many viral and host factors determine interspecies IAV transmission. Aside from the receptor-binding specificity of the virus, host inhibitory factors are implicated in this event.28, 29 From our data, we speculate that miRNA-mediated defense may restrain the host switch, and this hypothesis deserves further study.
Reduced expression levels of inhibitory hsa-mir-1-3p, hsa-mir-127-3p, hsa-mir-486-5p, and hsa-mir-593-5p were observed during IAV infection (Figure 5G), which is consistent with the results of previous studies.15, 16, 30 Influenza virus infection possibly downregulates the transcription levels of these miRNAs by affecting related transcription factors31 and/or by sequestering endogenous miRNAs with viral RNAs harboring miRNA response elements.32 Conceivably, to establish infection, influenza viruses likely evolve to decrease host miRNAs that restrict viral infection or accumulate escape mutations within miRNA binding sites to avoid host miRNA targeting, which is supported by our observation that different strains of viruses have different numbers of total miRNA binding sites (Figure 8). Viruses, including IAVs, may have evolved RNAi suppressor activity under miRNA pressure,33 indicating the complexity of interaction between influenza viruses and their hosts.
Although influenza virus appears to be refractory to suppression by endogenous miRNAs at physiological levels,34 we believe that exogenous delivery with excessive inhibitory miRNA mimics or agomirs may provide a novel therapeutic approach against viral infection, because our data demonstrate that miRNA treatment effectively suppressed a range of IAV subtypes (Figure 7). There are several advantages to using endogenous cellular miRNAs as a therapeutic strategy against IAVs. First, our study reveals that IAV infection results in reduced expression levels of inhibitory cellular miRNAs. Increasing these miRNAs in infected cells may restore the compromised antiviral defense by host miRNAs. Second, the cellular miRNAs may target aquatic bird influenza, domestic poultry influenza, swine influenza, and human seasonal influenza viruses (Figure 8; Table S5), validating their potential utility against various viral subtypes and interspecies transmission. Third, these endogenous natural miRNAs may have fewer side effects as potential antiviral drugs, although their off-target effects cannot be totally avoided. Therefore, besides artificial miRNAs, small hairpin RNAs (shRNAs), or small interfering RNAs (siRNAs),24, 25, 26, 27, 35, 36 our study provides an alternative perspective for designing miRNA-based therapies against IAVs.
Some miRNAs that were previously reported to affect influenza virus infection and replication, including hsa-miR-323-5p, hsa-miR-491-5p, hsa-miR-654-5p,18 hsa-let-7c,19 hsa-miR-136-5p,37 mmu-miR-485-5p,20 hsa-miR-146a-5p,38 and hsa-miR-9-5p,39 were not selected in this study using the NS luciferase reporter for measuring viral RdRp activity. These discrepancies may be due to different influenza virus subtypes or different miRNA screening methods used, indirect regulation of viral replication by affecting the immune and inflammatory responses (e.g., retinoic acid-inducible gene I [RIG-I] or cytokines) by miRNAs, or low abundance of miRNAs in respiratory epithelial cells that were not included in the miRNA pool that we screened. In addition, due to miRNA target diversity, besides directly targeting viral RNA segments, it is possible that the defined inhibitory miRNAs may affect viral replication through regulation of host gene expression. This deserves further investigation.
In conclusion, we performed a systemic screening of a miRNA pool in respiratory epithelial cells and identified >20 cellular miRNAs that inhibit IAV infection. We further investigated the functions of five highly expressed suppressive miRNAs targeting either most viral RNA segments or the supportive host factor. Moreover, we provide evidence suggesting that the cellular miRNA-mediated antiviral defense plays an important role against influenza virus infection and interspecies transmission. Given the broad function of miRNAs in the influenza virus-host interplay, our work provides insight for a better understanding of miRNA-elicited antiviral responses against IAVs and raises potential interest in the delivery of exogenous miRNAs as a therapeutic approach for antiviral therapy.
Materials and Methods
Ethics Statement
The animal protocol used in this study was approved by the Research Ethics Committee of the Institute of Microbiology, Chinese Academy of Sciences (permit number PZIMCAS2012009). All animal experimental procedures were performed in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals approved by the State Council of People’s Republic of China.
Cells, Virus, and Chicken Embryos
Human embryonic kidney HEK293T cells (ATCC), human type II alveolar epithelial A549 cells (ATCC), and MDCK cells (ATCC) were propagated in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco) and antibiotics (100 IU/mL penicillin and 25 mg/mL streptomycin) at 37°C in a humidified 5% CO2 atmosphere.
Influenza viruses PR8 (H1N1), A/WSN/33 (H1N1, WSN), A/Jiangxi/262/2005 (H3N2), A/Swine/Fujian/0325/2008 (H1N1), and A/Chicken/Liaoning/1/100 (H9N2) were propagated in 9-day-old, specific-pathogen-free (SPF) chicken embryos, which were purchased from Vital River Laboratories, Beijing.
miRNA Library
miRNAs that are highly expressed in human and mouse respiratory epithelial cells were carefully searched from published papers and miRNA databases (Table S6), and 297 miRNAs (Table S1) were selected in this study. The miRNAs library was synthesized in mature form by RiboBio (Guangzhou, China).
Antibodies and siRNA
Mouse anti-M1 monoclonal antibody and rabbit anti-NP polyclonal antibody were generated as previously described.40 Rabbit anti-PB1 polyclonal antibody and rabbit anti-ATP6V1A polyclonal antibody were purchased from GeneTex (USA). Mouse anti-β-actin monoclonal antibody and secondary antibodies conjugated with horseradish peroxidase were from Zhongshan Goldenbridge Biotechnology (Beijing, China).
siRNA for ATP6V1A was synthesized by RiboBio (Guangzhou, China). The ATP6V1A siRNA sequences7 were as follows: 5′-GAGCUUGAAUUUGAAGGUGUAdTdT-3′ (sense) and 5′-UACACCUUCAAAUUCAAGCUCdTdT-3′ (antisense).
Plasmid Construction
The plasmid carrying an influenza virus-like RNA encoding firefly luciferase (vNS-Luc) was used to monitor influenza virus replication.21 The PB2, PB1, PA, HA, NP, NA, M, and NS gene segments of PR8 or H3N2 were amplified and cloned into the 3′ UTR of the luciferase gene in the luciferase expression vector pGL3 (Promega). The miRNA pairing regions in the viral RNAs were cloned into the pGL3 vector, and mutations at the seed pairing region were induced by site-directed mutagenesis.
The QuikChange Site-Directed Mutagenesis Kit (Stratagene, United States and Canada) was used to make point mutations. The mutagenic oligonucleotide primers were designed individually according to the desired mutation. The two primers were extended during temperature cycling by high-fidelity DNA polymerase. Following temperature cycling, the PCR product was recovered, purified, and treated with Dpn I endonuclease (target sequence: 5′-Gm6ATC-3′). The digested products were transformed into DH5α supercompetent cells. The mutation constructs were extracted and validated by sequencing.
The ATP6V1A 3′ UTR (wide-type and mut) harboring the hsa-mir-1-3p binding site was amplified by PCR and cloned into pGL3 as set forth. The accuracy of all DNA constructs was validated by sequencing. The sequences of all primers used are listed in Table S7.
Cell Transfection
Transfections of HEK293T or A549 cells were performed using Lipofectamine 2000 reagent or Lipofectamine 3000 (Invitrogen, USA), respectively. In brief, HEK293T cells were seeded in 24-well or 48-well culture plates. Forward transfections were performed with 20 nM miRNA mimics and 0.1 μg (for 24-well) or 0.05 μg (48-well) plasmids when cells were approximately 60%–70% confluent. A549 cells were plated in 6-well or 12-well culture plates and grown 60%–70% confluent for transfection with 20 nM miRNA mimics. Each treatment was performed at least three times.
Virus Infection
HEK293T or A549 cells were used for infection with IAVs (PR8, H3N2, WSN, SWINE, or H9N2). Cells were washed and incubated with virus at the indicated MOI for 1 hr with medium containing 1 μg/mL TPCK (L-1-tosylamido-2-phenylethyl chloromethyl ketone)-treated trypsin (Sigma-Aldrich, USA). After adsorption, the supernatant was removed, and the cells were washed twice before culturing in medium contain 1% FBS (Gibco) for the indicated time.
Virus Titers Assays
MDCK cells plated in 12-well plates were washed twice with PBS and then inoculated with 0.5 mL serial 10-fold dilutions of virus. After a 2-hr incubation at 37°C, inoculums were removed and washed four times with PBS. The cells were overlaid with 1% low-melting-point agarose in DMEM with 2 μg/mL TPCK-treated trypsin and cultured for 3 days. The visible plaques were then counted, and virus titers were calculated after a 72-hr incubation at 37°C. All data are expressed as the mean of triplicate samples.
MTT Assays
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed to evaluate the cytotoxicity of miRNAs. The transfected cells were seeded in a 96-well culture plate. At 24 or 48 hr post-transfection, the cells were incubated with MTT (M2128; Sigma-Aldrich, St. Louis, MO, USA) solution for 4 hr, and the resulting formazan was solubilized with DMSO (D2650; Sigma-Aldrich, St. Louis, MO, USA). The absorbance of each well was measured at 570 nm using a spectrophotometer.
Luciferase Assays
HEK293T cells were co-transfected with a firefly luciferase reporter plasmid, pRL-TK (as an internal control), and the corresponding miRNA mimic or a randomized oligonucleotide as a control. At 48 hr post-transfection, the relative luciferase activities were measured using a dual-luciferase assay kit (Promega). Each set of assays was performed in triplicate.
Real-Time qPCR
Total RNA was extracted with Trizol reagent and quantified using SYBR Green Premix Reagent (Takara Bio, Shiga, Japan) and a TaqMan miRNA kit (Applied Biosystems) according to the manufacturer’s protocols. The endogenous control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 was used for normalization. The primers used are described in Table S8. Relative expression was quantified using the comparative threshold cycle (Ct) method.
Immunoblotting Assay
Immunoblotting analysis was performed according to our previous description.29
Mouse Experiments
Six- to eight-week-old female BALB/c mice, weighing 16–20 g at study initiation, were purchased from Vital River Laboratories, Beijing. Mice were intraperitoneally (i.p.) anaesthetized with trichloroacetaldehyde hydrate (375 mg/kg body weight). Under anesthesia, the mice were administrated intranasally with the five-miRNA mixture or an equivalent randomized oligonucleotide as a control and then intranasally challenged with 104 PFUs of PR8 in 50 μL saline per mouse. Mice were observed daily for mortality and weight loss for 14 days, or they were dissected at day 4 post-infection. Left lung tissues were prepared for western blotting, real time PCR, and viral titers assays. The right lungs were fixed with 4% neutral buffered formalin (NBF) for at least 24 hr and then stained with H&E for histological analysis.
Lung Index
On day 4 post-infection, mice were sacrificed. The lung tissues were washed with 0.9% normal saline and dried with filter paper. The lung index was calculated as below.41
Histopathology
For histological analysis, the lung tissues fixed in 4% NBF were dehydrated and embedded in paraffin. Tissue sections (5 μm thick) were stained with H&E and then examined using a Zeiss Axio Imager M1 microscope equipped with an AxioCam HRc camera under control of AxioVision 4 software.
Statistical Analysis
Differences between groups were determined by two-tailed Student’s t tests. A value of p < 0.05 was considered significant. Association between variables was assessed by Spearman’s nonparametric correlation.
Author Contributions
M.F. and S.M. conceived the original idea and designed and supervised the studies and analysis, and S.M. wrote the manuscript. S.P. designed the studies, performed most experiments with PR8, analyzed the data, and wrote the manuscript. J.W. performed most experiments with H3N2, pH1N1, and H9N2 and analyzed miRNA binding sites in different viral strains. S.W. performed dual-luciferase assay experiments with the preliminary screen for H3N2. C.L. and K.Z. established the miRNA library. J.H., X.Y., and J.Y. participated in analysis of the data and gave advice. X.Y., J.Y., W.L., and G.F.G. provided the vNS-Luc vector, virus strains, antibodies of NP and M1, and other assistance.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank past and present members of the Meng laboratory and Fang laboratory, particularly Dr. Bao Zhao, for technical assistance and experimental suggestions. We also thank Dr. Xuyu Zhou and his lab for the use of Single-Sample Luminometers (Promega). This work is supported by the funding from the Major State Basic Research Development Program of China (2014CB542602 and 2015CB910503) and grants from the National Natural Science Foundation of China (81761128002, 31230026, 81621091, 81471960, and 81672815) the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB0304).
Footnotes
Supplemental Information includes five figures and eight tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2017.12.016.
Contributor Information
Min Fang, Email: fangm@im.ac.cn.
Songdong Meng, Email: mengsd@im.ac.cn.
Supplemental Information
References
- 1.Mc Mahon A., Martin-Loeches I. The pharmacological management of severe influenza infection—“existing and emerging therapies.”. Expert Rev. Clin. Pharmacol. 2017;10:81–95. doi: 10.1080/17512433.2017.1255550. [DOI] [PubMed] [Google Scholar]
- 2.WHO. (2016). Fact sheet on influenza. http://www.who.int/mediacentre/factsheets/fs211/en/.
- 3.Shi Y., Wu Y., Zhang W., Qi J., Gao G.F. Enabling the “host jump”: structural determinants of receptor-binding specificity in influenza A viruses. Nat. Rev. Microbiol. 2014;12:822–831. doi: 10.1038/nrmicro3362. [DOI] [PubMed] [Google Scholar]
- 4.Nair H., Brooks W.A., Katz M., Roca A., Berkley J.A., Madhi S.A., Simmerman J.M., Gordon A., Sato M., Howie S. Global burden of respiratory infections due to seasonal influenza in young children: a systematic review and meta-analysis. Lancet. 2011;378:1917–1930. doi: 10.1016/S0140-6736(11)61051-9. [DOI] [PubMed] [Google Scholar]
- 5.Ando T., Yamayoshi S., Tomita Y., Watanabe S., Watanabe T., Kawaoka Y. The host protein CLUH participates in the subnuclear transport of influenza virus ribonucleoprotein complexes. Nat. Microbiol. 2016;1:16062. doi: 10.1038/nmicrobiol.2016.62. [DOI] [PubMed] [Google Scholar]
- 6.Kedzierski L., Tate M.D., Hsu A.C., Kolesnik T.B., Linossi E.M., Dagley L., Dong Z., Freeman S., Infusini G., Starkey M.R. Suppressor of cytokine signaling (SOCS)5 ameliorates influenza infection via inhibition of EGFR signaling. eLife. 2017;6:6. doi: 10.7554/eLife.20444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.König R., Stertz S., Zhou Y., Inoue A., Hoffmann H.H., Bhattacharyya S., Alamares J.G., Tscherne D.M., Ortigoza M.B., Liang Y. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Te Velthuis A.J.W., Fodor E. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat. Rev. Microbiol. 2016;14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 10.Trobaugh D.W., Klimstra W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Brahmakshatriya V., Lupiani B., Reddy S.M., Soibam B., Benham A.L., Gunaratne P., Liu H.C., Trakooljul N., Ing N. Integrated analysis of microRNA expression and mRNA transcriptome in lungs of avian influenza virus infected broilers. BMC Genomics. 2012;13:278. doi: 10.1186/1471-2164-13-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skovgaard K., Cirera S., Vasby D., Podolska A., Breum S.O., Dürrwald R., Schlegel M., Heegaard P.M.H. Expression of innate immune genes, proteins and microRNAs in lung tissue of pigs infected experimentally with influenza virus (H1N2) Innate Immun. 2013;19:531–544. doi: 10.1177/1753425912473668. [DOI] [PubMed] [Google Scholar]
- 13.Choi E.J., Kim H.B., Baek Y.H., Kim E.H., Pascua P.N., Park S.J., Kwon H.I., Lim G.J., Kim S., Kim Y.I., Choi Y.K. Differential microRNA expression following infection with a mouse-adapted, highly virulent avian H5N2 virus. BMC Microbiol. 2014;14:252. doi: 10.1186/s12866-014-0252-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bao Y., Gao Y., Jin Y., Cong W., Pan X., Cui X. MicroRNA expression profiles and networks in mouse lung infected with H1N1 influenza virus. Mol. Genet. Genomics. 2015;290:1885–1897. doi: 10.1007/s00438-015-1047-1. [DOI] [PubMed] [Google Scholar]
- 15.Chockalingam A.K., Hamed S., Goodwin D.G., Rosenzweig B.A., Pang E., Boyne M.T., 2nd, Patel V. The effect of oseltamivir on the disease progression of lethal influenza A virus infection: plasma cytokine and miRNA responses in a mouse model. Dis. Markers. 2016;2016:9296457. doi: 10.1155/2016/9296457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buggele W.A., Johnson K.E., Horvath C.M. Influenza A virus infection of human respiratory cells induces primary microRNA expression. J. Biol. Chem. 2012;287:31027–31040. doi: 10.1074/jbc.M112.387670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loveday E.-K., Svinti V., Diederich S., Pasick J., Jean F. Temporal- and strain-specific host microRNA molecular signatures associated with swine-origin H1N1 and avian-origin H7N7 influenza A virus infection. J. Virol. 2012;86:6109–6122. doi: 10.1128/JVI.06892-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song L., Liu H., Gao S., Jiang W., Huang W. Cellular microRNAs inhibit replication of the H1N1 influenza A virus in infected cells. J. Virol. 2010;84:8849–8860. doi: 10.1128/JVI.00456-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma Y.-J., Yang J., Fan X.-L., Zhao H.-B., Hu W., Li Z.-P., Yu G.-C., Ding X.-R., Wang J.-Z., Bo X.-C. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J. Cell. Mol. Med. 2012;16:2539–2546. doi: 10.1111/j.1582-4934.2012.01572.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ingle H., Kumar S., Raut A.A., Mishra A., Kulkarni D.D., Kameyama T., Takaoka A., Akira S., Kumar H. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015;8:ra126. doi: 10.1126/scisignal.aab3183. [DOI] [PubMed] [Google Scholar]
- 21.Gao S., Wu J., Liu R.-Y., Li J., Song L., Teng Y., Sheng C., Liu D., Yao C., Chen H. Interaction of NS2 with AIMP2 facilitates the switch from ubiquitination to SUMOylation of M1 in influenza A virus-infected cells. J. Virol. 2015;89:300–311. doi: 10.1128/JVI.02170-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matskevich A.A., Moelling K. Dicer is involved in protection against influenza A virus infection. J. Gen. Virol. 2007;88:2627–2635. doi: 10.1099/vir.0.83103-0. [DOI] [PubMed] [Google Scholar]
- 23.Perez J.T., Pham A.M., Lorini M.H., Chua M.A., Steel J., tenOever B.R. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat. Biotechnol. 2009;27:572–576. doi: 10.1038/nbt.1542. [DOI] [PubMed] [Google Scholar]
- 24.Shen X., Sun W., Shi Y., Xing Z., Su X. Altered viral replication and cell responses by inserting microRNA recognition element into PB1 in pandemic influenza A virus (H1N1) 2009. Mediators Inflamm. 2015;2015:976575. doi: 10.1155/2015/976575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan M., Sun W., Feng C., Xia D., Shen X., Ding Y., Liu Z., Xing Z., Su X., Shi Y. The microRNA-let-7b-mediated attenuated strain of influenza A (H1N1) virus in a mouse model. J. Infect. Dev. Ctries. 2016;10:973–981. doi: 10.3855/jidc.6861. [DOI] [PubMed] [Google Scholar]
- 26.Tundup S., Kandasamy M., Perez J.T., Mena N., Steel J., Nagy T., Albrecht R.A., Manicassamy B. Endothelial cell tropism is a determinant of H5N1 pathogenesis in mammalian species. PLoS Pathog. 2017;13:e1006270. doi: 10.1371/journal.ppat.1006270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Arévalo M.T., Diaz-Arévalo D., Chen Y., Choi J.-G., Zeng M. Generation of a safe and effective live viral vaccine by virus self-attenuation using species-specific artificial microRNA. J. Control. Release. 2015;207:70–76. doi: 10.1016/j.jconrel.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimakami T., Yamane D., Jangra R.K., Kempf B.J., Spaniel C., Barton D.J., Lemon S.M. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. USA. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang S., Qiu L., Yan X., Jin W., Wang Y., Chen L., Wu E., Ye X., Gao G.F., Wang F. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1)-modulated P53 activity. Hepatology. 2012;55:730–741. doi: 10.1002/hep.24809. [DOI] [PubMed] [Google Scholar]
- 30.Tambyah P.A., Sepramaniam S., Mohamed Ali J., Chai S.C., Swaminathan P., Armugam A., Jeyaseelan K. microRNAs in circulation are altered in response to influenza A virus infection in humans. PLoS ONE. 2013;8:e76811. doi: 10.1371/journal.pone.0076811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z.P., Wu H., Zhu J., Miao H. Systematic identification of transcriptional and post-transcriptional regulations in human respiratory epithelial cells during influenza A virus infection. BMC Bioinformatics. 2014;15:336. doi: 10.1186/1471-2105-15-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C., Hu J., Hao J., Zhao B., Wu B., Sun L., Peng S., Gao G.F., Meng S. Competitive virus and host RNAs: the interplay of a hidden virus and host interaction. Protein Cell. 2014;5:348–356. doi: 10.1007/s13238-014-0039-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Vries W., Haasnoot J., Fouchier R., de Haan P., Berkhout B. Differential RNA silencing suppression activity of NS1 proteins from different influenza A virus strains. J. Gen. Virol. 2009;90:1916–1922. doi: 10.1099/vir.0.008284-0. [DOI] [PubMed] [Google Scholar]
- 34.Bogerd H.P., Skalsky R.L., Kennedy E.M., Furuse Y., Whisnant A.W., Flores O., Schultz K.L.W., Putnam N., Barrows N.J., Sherry B. Replication of many human viruses is refractory to inhibition by endogenous cellular microRNAs. J. Virol. 2014;88:8065–8076. doi: 10.1128/JVI.00985-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waring B.M., Sjaastad L.E., Fiege J.K., Fay E.J., Reyes I., Moriarity B., Langlois R.A. MicroRNA-based attenuation of influenza virus across susceptible hosts. J. Virol. 2018;92 doi: 10.1128/JVI.01741-17. e01741-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu K., Ren Y., Zhang X., Qiao T., Liu Z., Shi J., Wang Y. shRNA-mediated NP knockdown inhibits the apoptosis of cardiomyocytes induced by H1N1pdm2009 influenza virus. Mol. Med. Rep. 2017;16:1376–1382. doi: 10.3892/mmr.2017.6728. [DOI] [PubMed] [Google Scholar]
- 37.Zhao L., Zhu J., Zhou H., Zhao Z., Zou Z., Liu X., Lin X., Zhang X., Deng X., Wang R. Identification of cellular microRNA-136 as a dual regulator of RIG-I-mediated innate immunity that antagonizes H5N1 IAV replication in A549 cells. Sci. Rep. 2015;5:14991. doi: 10.1038/srep14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Terrier O., Textoris J., Carron C., Marcel V., Bourdon J.-C., Rosa-Calatrava M. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. J. Gen. Virol. 2013;94:985–995. doi: 10.1099/vir.0.049528-0. [DOI] [PubMed] [Google Scholar]
- 39.Dong C., Sun X., Guan Z., Zhang M., Duan M. Modulation of influenza A virus replication by microRNA-9 through targeting MCPIP1. J. Med. Virol. 2017;89:41–48. doi: 10.1002/jmv.24604. [DOI] [PubMed] [Google Scholar]
- 40.Liu X., Sun L., Yu M., Wang Z., Xu C., Xue Q., Zhang K., Ye X., Kitamura Y., Liu W. Cyclophilin A interacts with influenza A virus M1 protein and impairs the early stage of the viral replication. Cell. Microbiol. 2009;11:730–741. doi: 10.1111/j.1462-5822.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 41.Hu X.P., Shao M.M., Song X., Wu X.L., Qi L., Zheng K., Fan L., Liao C.H., Li C.Y., He J. Anti-influenza virus effects of crude phenylethanoid glycosides isolated from ligustrum purpurascens via inducing endogenous interferon-γ. J. Ethnopharmacol. 2016;179:128–136. doi: 10.1016/j.jep.2015.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.