Abstract
Glioblastoma multiforme (GBM) is the most prevalent and lethal malignant intracranial tumor in the brain, with very poor prognosis and survival. The epidermal growth factor receptor variant III (EGFRvIII) contributes to increased oncogenicity that does not occur through binding EGFR ligands and instead occurs through constitutive activation, which enhances glioma tumorigenicity and resistance to targeted therapy. Aptamers are nucleic acids with high affinity and specificity to targets selected by systematic evolution of ligands by exponential enrichment (SELEX), and are usually developed as antagonists of disease-associated factors. Herein, we generated a DNA aptamer U2, targeting U87-EGFRvIII cells, and demonstrated that U2 alters the U87-EGFRvIII cell growth, radiosensitivity, and radiotherapy of glioblastoma cells. We detected U2 and U87-EGFRvIII cells by flow cytometry and confocal microscopy to explore the binding ability of U2 to U87-EGFRvIII cells. Then, we found that aptamer U2 inhibits the proliferation, migration, invasion, and downstream signaling of U87-EGFRvIII cells. Moreover, the U2 aptamer can increase the radiosensitivity of U87-EGFRvIII in vitro and has a better antitumor effect on 188Re-U2 in vivo. Therefore, the results revealed the promising potential of the U2 aptamer to be a new type of drug candidate and aptamer-targeted drug delivery system for glioblastoma therapy.
Keywords: aptamer, EGFRvIII, glioblastoma, proliferation, radiosensitivity
Introduction
Glioblastoma multiforme (GBM) is one of the most common and lethal malignant intracranial tumors in the brain. The tumor cells have infiltrative growth in vivo, with a high degree of malignancy, complex clinical manifestations, and poor prognosis. The effect of traditional treatments, including surgery, radiotherapy, chemotherapy, and a combination of radiotherapy and chemotherapy, is extremely limited. GBM is still difficult to cure. The median survival time of GBM patients is no longer than 1.5 years. Early diagnosis and prognosis have been unsuccessful. The primary reasons that GBM causes patient death are the abnormal activation and migration invasion of tumor cells as well as resistance to chemotherapy and radiotherapy.1 Therefore, finding a drug that is capable of inhibiting GBM cell growth, proliferation, migration, and invasion and increasing radioresistance is quite significant for developing an effective treatment.
Epidermal growth factor receptor (EGFR), which is one of the most frequently altered proteins, is amplified in half of GBM patients. EGFR is a type I receptor tyrosine kinase (RTK) that plays an important role in the signal transduction of cell growth and proliferation. EGFR is overexpressed and amplified in a variety of malignancies, which is related to the degree of malignancy.2, 3 There are various EGFR mutations, such as EGFRvI, EGFRvII, and EGFRvIII.4 The most common mutation is EGFR variant III (EGFRvIII, EGFR type III, or ΔEGFR) in half of the EGFR amplification of GBM patients. Compared to wild-type EGFR (EGFRwt), EGFRvIII is a gain-of-function mutation that includes a genomic deletion of exons 2–7. It cannot bind any ligands, but it produces lower constitutive signals of EGFRwt, which is very important to its tumorigenicity.5, 6, 7 Previous studies have demonstrated a correlation between the EGFRvIII expression and the poor prognosis, chemoresistance, and radioresistance in patients.8, 9, 10, 11 EGFRvIII, which enhances the tumorigenic behavior and increases malignancy, is commonly found in many tumors, such as GBM, non-small cell lung cancer (NSCLC), colorectal cancer, and colon cancer. It is specifically expressed in cancer cells but not in normal cells; therefore, it is a good target for cancer therapy.12, 13
Aptamers are single-stranded oligonucleotides that are selected from a large capacity random single-stranded DNA (ssDNA) or RNA library by systematic evolution of ligands by exponential enrichment (SELEX).6, 14 Aptamers are capable of binding to specific target molecules, with high affinity and specificity. They have the following attractive features as molecular probes compared to conventional antibodies: (1) low molecular weight and long-term stability; (2) low immunogenicity and toxicity; (3) high affinity and specificity; and (4) quick, reproducible synthesis and modification.8, 9 Aptamers can be effectively used on targeted therapy, detection, and diagnosis of cancer. These advantages make aptamers an excellent alternative as a molecular probe and drug for clinical applications.10, 11
In our previous work, we acquired DNA aptamer U2 with a high affinity (Kd = 6.27 ± 1.40 nM), with overexpression of EGFRvIII protein in U87 cells (U87-EGFRvIII) by cell SELEX. The U2 aptamer could specifically bind U87-EGFRvIII cells; then, we radiolabeled U2 with 188Re (188Re U2) to serve as a molecule imaging probe that could significantly target U87-EGFRvIII xenografts in nude mice.12 RNA aptamers targeting the EGFRwt/EGFRvIII protein that affect the migration and proliferation of glioblastoma cells have been observed.13, 14
To explore the binding ability of U2 to U87-EGFRvIII cells and the effects of inhibiting U87-EGFRvIII cells, we detected the U2 aptamer and U87-EGFRvIII cells by flow cytometry (FCM) and confocal microscopy. Then, we found that U2 can affect the growth, proliferation, apoptosis, migration, invasion, and downstream signaling of U87-EGFRvIII cells. The U2 aptamer can increase the radiosensitivity of U87-EGFRvIII in vitro and have a better antitumor effect of 188Re-U2. Our results revealed the promising potential of U2 to be a new type of drug candidate for glioblastoma therapy.
In the current study, we investigated whether U2 treatment might affect the proliferation, migration, invasion, and apoptosis of U87-EGFRvIII cells and the involvement of relevant signaling pathways. Furthermore, we examined whether the U2 aptamer can increase the radiosensitivity of U87-EGFRvIII cells in vitro and improve the antitumor effect of 188Re-U2. Our findings revealed the promising potential of U2 to be a new type of drug candidate for glioma therapy.
Results
U2 Specifically Binds to the U87-EGFRvIII Cells
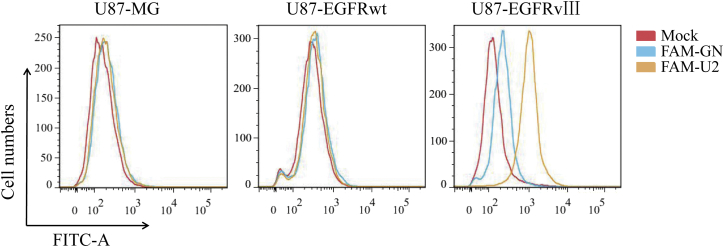
U2 is a DNA aptamer obtained by cell SELEX technology using U87-EGFRvIII cells. To investigate the specificity of U2 for the different glioblastoma cells, including U87MG, U87-EGFRwt, and U87-EGFRvIII cells, we applied an FCM binding assay using the 5′ end FAM-labeled U2 aptamers, and the FAM-labeled original library GN was used as a control. According to the FCM findings, FAM-U2 was bound to U87-EGFRvIII at a higher extent than FAM-GN bound to U87-EGFRvIII, whereas FAM-U2 shows no different significant binding to FAM-GN in U87MG and U87-EGFRwt cells (Figure 1). U2 binding to U87-EGFRvIII cells but not to U87-EGFRwt cells or U87MG cells confirmed its specificity for U87-EGFRvIII cells. Besides, we added other four primary GBM cell lines to confirm the specificity of U2 and the results showed that the average rate of aptamer U2 binding to the four cell lines is less than 3% (Figure S1A).
Figure 1.
The Binding Relatives of FAM-U2 or FAM-GN with U87MG cells, U87-EGFRwt cells, and U87-EGFRvIII Cells Obtained by Flow Cytometry
U87MG cells, U87-EGFRwt cells, and U87-EGFRvIII cells bind with FAM-U2 and FAM-GN detected by flow cytometry. ***p < 0.001.
Subcellular Localization of U2 Aptamer
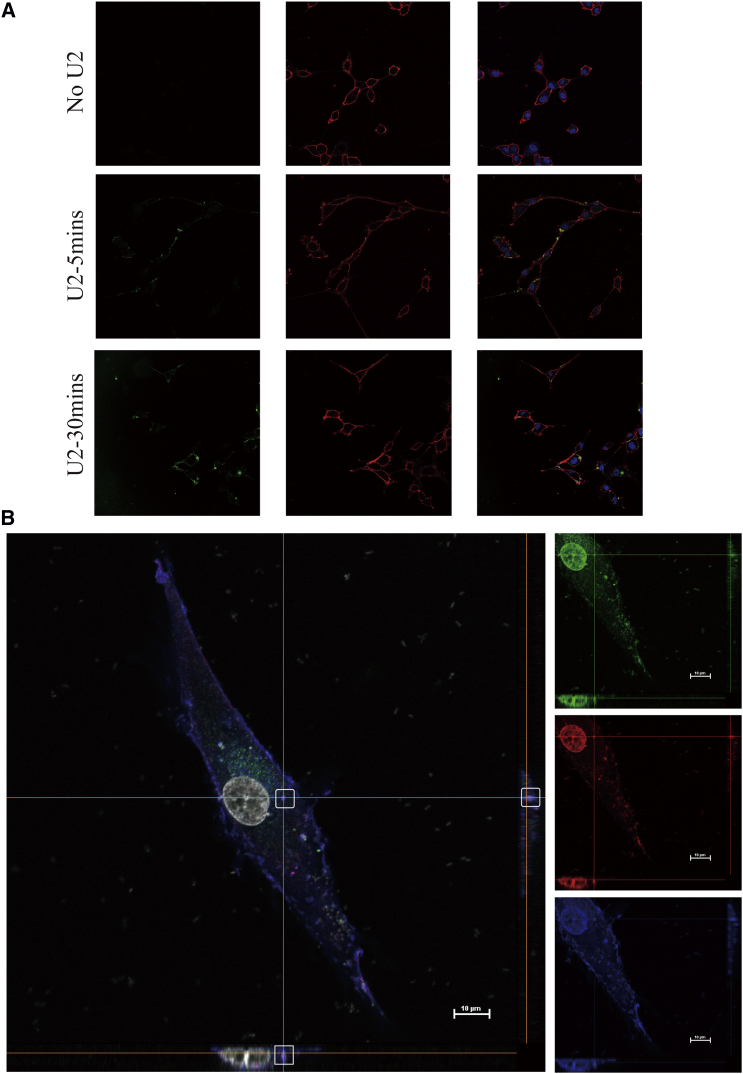
Consistent with the results by FCM, confocal microscopy on U87EGFRvIII cells with FAM-labeled U2 showed that cells with FAM-labeled aptamer for 5 min were combined with staining via a specific EGFR antibody (targeting to the extracellular EGFR domain). A wide overlap of EGFR antibody and FAM-U2 fluorescent signals was detected on the membrane, indicating clear co-localization of the aptamer and antibody on the receptor expressed on the cell surface (Figure 2A). Due to the phenomenon of FAM-U2 incubation after 20 min, overlap signals appeared in the cell and the next objective was to validate the uptake mechanism for an anti-EGFR-aptamer complex. Consistently, after co-localization experiments of FAM-U2 with endocytosis markers, early endosome antigen 1 (EEA1) was confirmed by using z stack processing. After incubation for 30 min and then fixing and staining with anti-EGFR antibody and anti-EEA1 antibody, FAM-U2 and EGFR were co-localized inside the cells (Figure 2B), suggesting that U87EGFRvIII cells internalize the compounds through the endosome recycling pathway.
Figure 2.
U2 Can Internalize into U87-EGFRvIII Cells
(A) U87-EGFRvIII cells were treated with 2 μM FAM-U2 for 5 and 20 min. Cells were fixed and labeled with anti-EGFR antibody targeting on the cell membrane without permeabilization. Green: fluorescence labeling FAM-U2; blue: cell nucleus (staining by DAPI); red: anti-EGFR antibody. (B) Z stack of U87-EGFRvIII cells incubated with 2 μM FAM-U2 for 30 min. Scale bar, 10 μm. Cells were fixed, permeabilized, and labeled with anti-EGFR and anti-EEA1 antibodies. Green: fluorescence labeling FAM-U2; red: anti-EEA1 antibody; blue: anti-EGFR antibody.
Induction of Apoptosis and Inhibition of Proliferation in U87-EGFRvIII Cells with U2 Aptamer
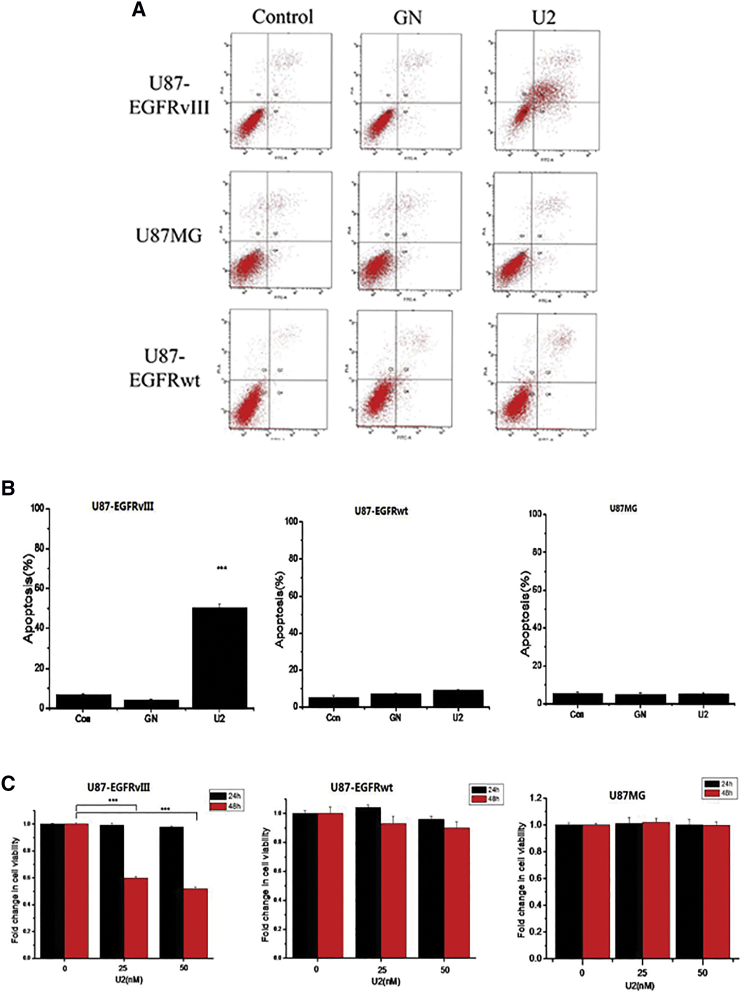
To determine whether U2 treatment could lead to apoptosis of U87-EGFRvIII cells, we performed Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) experiments. We observed that U2 significantly increased the apoptosis rate of U87-EGFRvIII cells but not U87MG cells or U87-EGFRwt cells (Figures 3A and 3B). We performed Cell Counting Kit-8 (CCK8) experiments to determine whether long-term U2 treatment alters the cell viability of EGFRvIII-expressing GBM cells. A significant decrease (p < 0.001) in cell viability was observed in U87-EGFRvIII cells treated for 24 hr with 25 and 50 nM U2 (fold change viability rates of 59% and 51%, respectively), whereas there were no effects on U87MG and U87-EGFRwt cells (Figure 3C). We found that the U2 aptamer reduced the survival of U87-EGFRvIII cells, whereas it did not affect the U87MG cells and U87EGFRwt cells. The above results confirm that U2 aptamer caused a time- and dose-dependent inhibition of U87-EGFRvIII cell viability.
Figure 3.
Apoptosis and Inhibition of Cell Growth in Glioblastoma Cells
(A) Effect of U2 on the apoptosis of U87-EGFRvIII cells, U87MG cells, and U87-EGFRwt cells. (B) Analysis of the apoptosis rate of each glioblastoma cell. ***p < 0.001 compared to control. (C) Fold change in the cell viability of U87 glioma cells treated with of U2 (25 or 50 nM) for 24 and 48 hr detected by the CCK8 assay in U87-EGFRvIII, U87MG, and U87-EGFRwt cells.
U2 Inhibits U87-EGFRvIII Cell Migration and Invasion
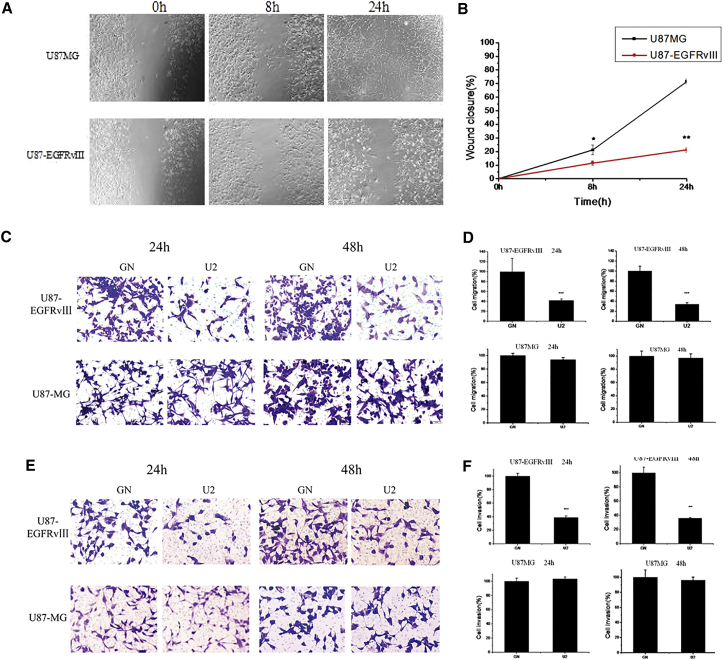
To evaluate whether U2 could affect the migration and invasion of U87-EGFRvIII cells, we first used a scratch-wound assay that measures the cell motility. We observed that U87-EGFRvIII cells treated with U2 still had a wide gap at 8 hr after the scratch was created. The U87MG cell wound treated by U2 was barely visible. These data have statistical significance (p = 0.048). At 24 hr after the scratch, the U87MG wound was almost closed compared with U87-EGFRvIII cells treated with U2. However, the U87-EGFRvIII cells treated with U2 significantly delayed the wound closure compared to control cells treated with U2 (Figures 4A and 4B) but did not show any statistical significance after treating with GN or mock (Figure S2).
Figure 4.
U2 Inhibits the Migration and Invasion of U87-EGFRvIII
(A) U2 inhibits U87-EGFRvIII cell migration obtained by scratch assays to measure cell migration. U87-EGFRvIII cells treated for 8 and 24 hr. Microscopy images were taken at the indicated times. (B) The extent of wound closure was calculated. *p < 0.05 and **p < 0.01. (C) A transwell migration assay was performed in the presence of U2 or GN for 24 and 48 hr. Photographs of a representative experiment are shown. (D) Data from the transwell migration assay are presented as the percentage of migrated cells in the presence of U2 compared with GN control. Each determination represents the average of three individual experiments, and error bars represent the SD. ***p < 0.001 relative to GN. (E) Invasion of U87MG and U87-EGFRvIII cells through matrigel was evaluated in the presence of U2 or GN for the indicated times. Photographs of a representative experiment are shown. (F) Data from the invasion assay are presented as the percentage of invaded cells in the presence of U2 compared with GN control. ***p < 0.001 relative to GN.
Furthermore, migration was analyzed by a “transwell migration assay” that assesses the chemotactic cell capacity. The benchmark is the number of cells, treated by GN, that travel through the transwell membrane. After treatment for 24 hr, compared to U87MG cells treated with U2, the percentage of migrated U87-EGFRvIII cells was reduced to 41.94% ± 3.0000 (50 nM). Additionally, the percentage of migrated cells was 34% ± 3.0000 U87-EGFRvIII cells and 96.946% ± 6.4100 U87MG cells at 48 hr, respectively (Figures 4C and 4D). According to the above findings, U2 is capable of reducing the migration ability of U87-EGFRvIII cells.
Then, we examined the effect of U2 on the capacity of the cells to invade through a matrigel-coated membrane by a “transwell invasion assay,” which has been reported to mimic the entire process of cell invasion through basement membranes. Using this assay, we observed that the invasion rate of U87-EGFRvIII cells in the presence of U2 treatment for 24 hr was significantly decreased compared with U87MG cells. There were 36.1027% ± 0.5160 U87-EGFRvIII cells and 96.206% ± 4.0890 U87MG cells at 48 hr, respectively (Figures 4E and 4F). These results emphasize the use of the U2 aptamer to affect U87-EGFRvIII migration and invasion.
Effects of Aptamer U2 on the EGFR Signaling Pathway
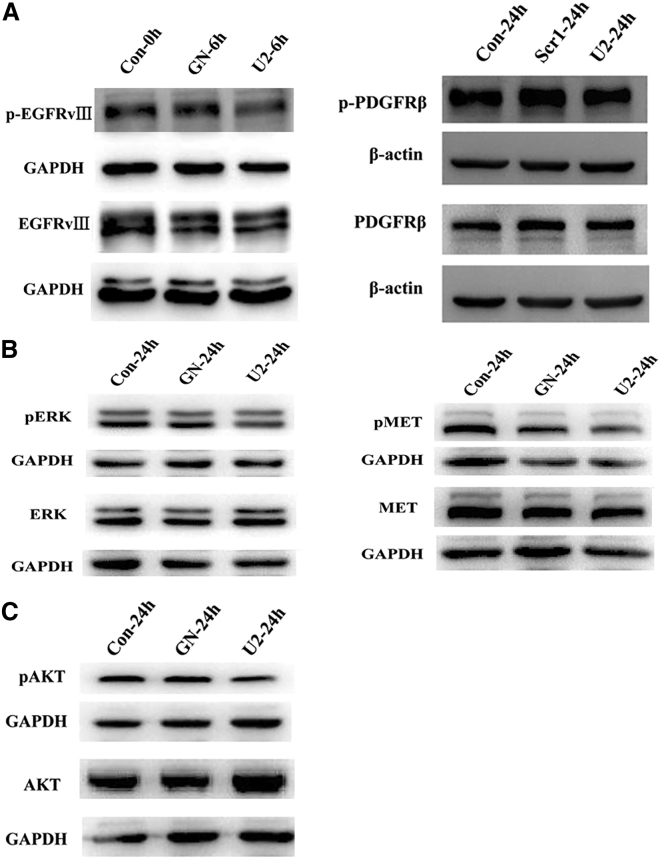
We analyzed the mechanism of U2 inhibition on the proliferation of GBM cells. For this purpose, we first targeted the autophosphorylation activity of EGFRvIII in U87-EGFRvIII cells. To measure the cell cycle at the same stage, the cells were treated with medium containing 2% fetal bovine serum (FBS) for starvation treatment and then with 200 nM GN or U2 for another 6 hr. Then, we collected the lysates and immunoblotted with antibodies as indicated. The results showed that after therapy with U2, the level of phosphorylation was significantly decreased compared to that of the other two groups, but there was no change in total EGFRvIII. Based on the previous report, the expression of platelet-derived growth factor receptor β (PDGFRβ) contributed to the alteration of EGFRvIII;13 therefore, we tested the PDGFRβ level. The expression of PDGFRβ does not changed, whereas the phosphorylation level of EGFRvIII was attenuated (Figure 5B). Due to the decrease of phospho-EGFRvIII, we asked whether the U2 aptamer could affect the tyrosine kinase activation of multiple downstream signaling pathways. U2 treatment could consistently reduce the extent of phospho-MET and phospho-ERK1/2, and the total protein levels of MET and ERK1/2 remained unchanged (Figure 5C). However, there was no significant decrease in phospho-AKT or AKT activity, which is consistent with the observation that U87MG cells lack the phosphatase and tensin homolog (PTEN) gene. The above results indicate that U2, because of binding to U87-EGFRvIII cells, interferes with activation of the tyrosine kinase receptor and its downstream signaling, representing a promising inhibitor candidate.
Figure 5.
U2 Inhibits the Activation of EGFRvIII
(A) U87-EGFRvIII cells were started in 2% FBS-containing medium for 6 hr and then treated with 200 nM GN or U2 for another 6 hr; then, the lysates were collected and immunoblotted with anti-pEGFR and anti-EGFR. U87-EGFRvIII cells were started in 2% FBS-containing medium for 24 hr and then treated with 200 nM GN or U2 for another 24 hr and the cells lysates were collected and immunoblotted with anti-pPDGFRβ and anti-PDGFRβ antibodies. Setting the values of the relative ratio of untreated cells to 100%, the values below the blot indicate the ratio of pEGFR to total EGFR signal levels after normalization with the β-actin signal level. *p < 0.05. (B) U87-EGFRvIII cells were treated with 2% FBS-containing medium for 6 hr and then treated with 200 nM GN or U2 for another 24 hr. They were immunoblotted with anti-pERK, anti-pMET, and anti-pAKT antibodies, and the values below the blot indicate the ratio of the phosphorylated protein to the total protein levels after normalization with the β-actin level. *p < 0.05. (C) The cells were treated as in (B), and the lysates were collected and immunoblotted with anti-pAKT and anti-AKT antibodies. p > 0.05.
U2 Inhibits the Repair of DNA Damage after Radiation
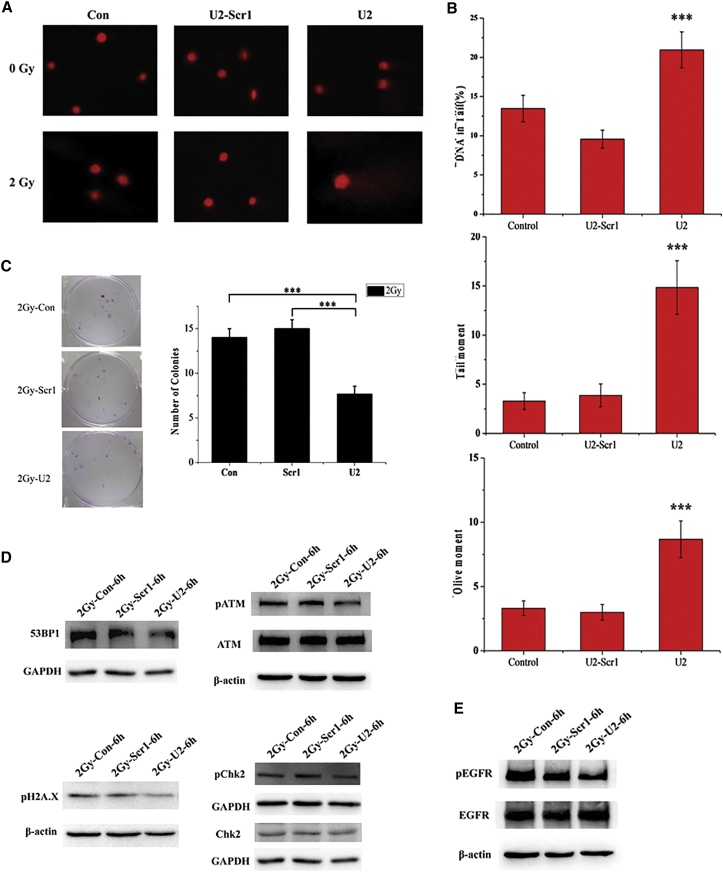
In the above study, we found that U2 can interfere with the MAPK/ERK pathway and has anti-U87-EGFRvIII cell proliferation activity. We designed this part of the study to determine whether U2 can increase glioma cell radiation sensitivity. We used a comet assay to evaluate the effects of U2 combined with radiation on DNA damage in target cells. With U2 treatment for 24 and 3 hr after 2 Gy radiation, cells in the U2 group had a high percentage of constant DNA in the tail, which is a marker for the degree of DNA damage (Figure 6A). Additionally, we found that the measurement of the tail DNA content and olive tail moment confirmed that cells in the co-treated group have significantly more DNA fragments than cells in the other two groups (Figure 6B). Moreover, we performed the colony formation assay on U87-EGFRvIII cells after exposure to irradiation with a single dose of 2 Gy. We observed that cells after U2 treatment formed fewer colonies than the other two groups (Figure 6C). These results suggest that U2 inhibits the repair of DNA damage after radiation, which could be explained by the decrease in the activation of the repair process.
Figure 6.
U2 Inhibits DNA Damage Repair after Radiation
(A) U87-EGFRvIII cells were incubated with U2-Scr1 or U2 (200 nmol/L). After 24 hr, the monolayer cells were exposed to a single dose of irradiation with 2 Gy and harvested 3 hr post irradiation for the comet assay. (B) The cells were treated as in (A). The tail DNA content, tail moment, and olive tail moment were calculated by software to assess the severity of DNA damage under each condition after irradiation therapy. ***p < 0.001. (C) Picture of U87EGFRvIII cells treated with U2 or U2-Scr1 (200 nmol/L), with exposure to a single dose of 2 Gy. The cells were then submitted to a clonogenic assay after radiotherapy. ***p < 0.001. (D) U87-EGFRvIII cells were starved in 2% FBS-containing medium for 6 hr and then incubated with 200 nM U2-Scr1 or U2 for 24 hr. After exposure to irradiation (2 Gy), cells were harvested for 6 hr post irradiation and the lysates were immunoblotted for phosphorylated and total markers related to DNA damage repair, as indicated. *p < 0.05. (E) The cells were treated as in (D), and the lysates were collected and immunoblotted with anti-pEGFRvIIIand anti-EGFRvIII antibodies. *p < 0.05.
Because of the results of the comet and clonogenic assays, we asked whether the high percentage of DNA fragments and reduction of colonies after U2 treatment played a role in the activation of the DNA break repair process. Therefore, we detected the level of the major effectors in the repair of DNA damage after radiation. The cells were also starved as before and exposed to irradiation (2 Gy); then, they were harvested at 6 hr post irradiation and the lysates were immunoblotted as indicated. Consistent with the results in the comet and congenic assays, DNA damage occurred because of irradiation, but the level of phosphorylation of ATM (ataxia telangiectasia mutated) and H2AX decreased after U2 treatment compared to the other two control groups (Figure 6D). To better understand this pathway in the DNA break repair process, we examined the effect of 53BP1 and Chk2 on the activation as upstream and downstream targets of ATM and H2AX, respectively. Furthermore, U2 still inhibits the phosphorylation of EGFRvIII after irradiation (Figure 6E). Therefore, U2 decreases the phosphorylation of the main DNA repair effectors to inhibit the DNA damage repair caused by radiation and U2 can enhance the radiosensitivity of U87-EGFRvIII cells.
In Vivo Antitumor Effects of 188Re-U2
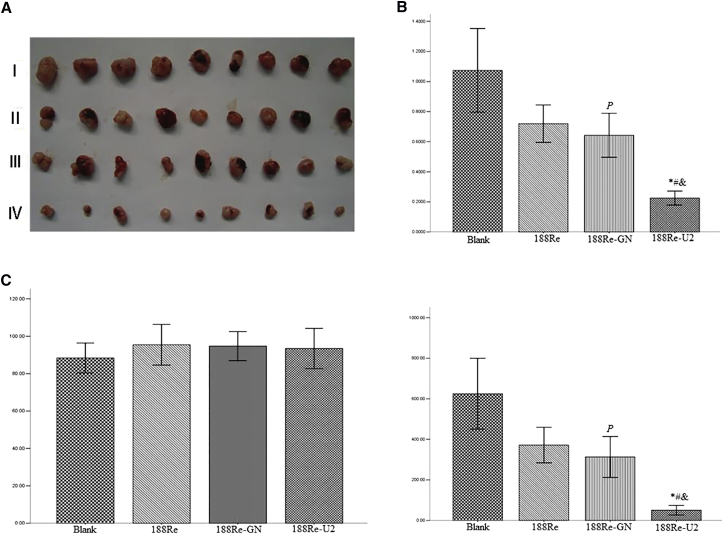
We assumed that 188Re-U2 has more powerful antitumor activity based on its targeting ability and increased radiosensitivity of U87-EGFRvIII cells. We established a nude mouse model bearing U87-EGFRvIII cells and then injected different drugs into the tumor. 188Re-U2 can dramatically inhibit the tumor volume and weights and showed significant antitumor effects compared to the other groups, whereas saline or 188Re alone showed no antitumor effects (Figure 7A). The 188Re-GN group had a slight significant difference compared to the blank group, which may due to the antitumor effect of random short nucleic acids (Figures 7B and 7C).
Figure 7.
In Vivo Antitumor Effects of 188Re-U2
(A) The in vivo effectiveness of 188Re-U2 was evaluated in xenograft murine models bearing tumors originating from U87-EGFRvIII cells. (I) Blank control group. (II) 188Re: dissociate radionuclide 188Re group. (III) 188Re-GN: 188Re-labeled original library GN group. (IV) 188Re-U2: 188Re-labeled aptamer U2 group. (B) The tumor size before and after injecting drugs. (C) The tumor weight that had been stripped (n = 9). *p < 0.01 compared with the blank group; #p < 0.01 compared with the 188Re radionuclide group; &p < 0.01 compared with the 188Re-GN group; and P, p < 0.05 compared with the blank group.
Discussion
GBM is the most lethal tumor, with little advancement treatment over the last 20 years. Overexpression and mutations of EGFR, some of most significant factors responsible for the development of gliomas, usually co-exist with overexpression with EGFRvIII, the most common mutant of EGFR in GBM. EGFR tyrosine kinase inhibitors are the first-line therapy for patients with colorectal cancer,15 non-small-cell lung cancer,16 and other tumors. However, it was reported that erlotinib poorly inhibits glioma-specific EGFR mutants;17 therefore, we focused on a targeted molecule: EGFRvIII. In our study, we investigated the effects of a DNA aptamer, U2, on U87-EGFRvIII cells at the cellular level, animal model, and with radiotherapy.
We found that U2 can specifically bind to U87-EGFRvIII cells and be internalized into the cells through the endosome recycling pathway by FCM and immunofluorescence methods. Then, we looked at the effects of U2 on U87MG/EGFRvIII cells. It was reported that suppressing the expression of EGFR inhibits tumor cell proliferation, migration, and invasion.13, 18, 19 Consistent with previous studies, we demonstrated that 24 hr of U2 treatment could significantly reduce U87-EGFRvIII cell proliferation, migration, and invasion as well as promote cell apoptosis. Moreover, the oncogenic role of EGFR has been functionally validated at both the cellular level and in animal models.4, 20 In agreement with these findings, we confirmed that after U2 treatment, the phosphorylation level of EGFRvIII is consistently markedly decreased and accompanied by a reduction in the phosphorylation level of MAPK/ERK (MEK) downstream signaling. Nevertheless, U2 does not change the activity of AKT in our study, which may be attributed to a defective PTEN gene in the U87 cell line, which agrees with previous observations.13, 21, 22
It has been clear for quite some time that the expression or activation of EGFR is relative to radioresistance.23 Moreover, EGFRvIII plays an important role in the response to ionizing radiation24, 25 and contributes radioresistance by promoting the hyperactivation of the AKT and ERK signaling pathways26 as well as DNA damage repair in GBM.27 Therefore, it is urgently necessary to reduce the effect of radiotherapy resistance of GBM and explore the effective radiotherapy sensitizer in the treatment of GBM. In the last few years, aptamers have begun to be studied in radiotherapy in combination with small interfering RNA (siRNA)28 or small hairpin RNA (shRNA).29 However, no study has been performed to evaluate radiotherapy in combination with an aptamer to treat glioma. In this report, we tentatively put forward that U2 inhibits the growth of U87-EGFRvIII cells, enhancing the radiosensitivity of human glioma cell lines for the first time. Importantly, using the comet and colony formation assays, our results indicated that U2 inhibits DNA damage repair after radiation, which is in line with previous data, indicating that inhibition of EGFR by gefitinib significantly suppresses congenic survival.30 It has been widely accepted that DNA damage repair proteins, such as 53BP1 (53 binding protein1), ATM, and phosphorylated histone H2AX, are critical to the response to DNA double-strand breaks after ionizing radiation.31, 32 We found that the phosphorylation level of pH2AX and total protein level of 53BP1 distinctly decrease after U2 treatment and radiation. Then, we observed that the activity of check-point protein Chk2, the major downstream target of ATM in response to DNA damage, also declines. Meanwhile, the phosphorylation of EGFRvIII remarkably decreased after radiation, agreeing with inhibition of EGFR inducing the radiosensitivity of GBM cells by siRNA33 and microRNA.34
It is noteworthy that inhibition of the DNA damage response has great potential for radiosensitization in cancers,35, 36 especially inhibition of ATM.37 The results in our manuscript demonstrated that U2 can promote the radiosensitivity of GBM cells by altering the phosphorylation of EGFRvIII, affecting the MET and ATM signaling pathways. It has been reported that inhibition of EGFR inhibits ATP level recovery, which is essential to the DNA repair process after radiation, resulting in radiosensitivity in A549 cells.38 Afterward, Huang et al.39 showed that downregulation of the mitochondrial ATP-sensitive potassium channels and MEK reinforces radiosensitivity in GBM. Moreover, it was published that targeting MET promotes radiosensitivity in tumor cells40 and glioblastoma stem-like cells.41 From the above statements, we speculated that increased radiosensitivity with U2 treatment in GBM cells might occur through decreasing the ATP supply and inhibiting the signaling molecules in the common pathways induced by EGFRvIII and MET.
In previous studies, researchers reported that aptamers are potential candidates for molecular imaging applications because of a number of attractive characteristics, such as rapid blood clearance and tumor penetration.42, 43 Jacobson et al.44 demonstrated that 18F-labeled aptamer Sgc8 targeting protein tyrosine kinase-7 had high specificity and affinity in positron emission tomography (PET) imaging in HCT116 cells. In light of these findings, we have previously published that U2 with 188Re significantly radiolabeled targeted EGFRvIII overexpressing GBM xenografts in mice.12 To further evaluate the radiotherapy, we applied different drug treatments to the tumor xenografts and observed that U2 with 188Re labeled significantly shrinks the tumor as well as decreases the tumor weight. The result implies that U2 labeling with radionuclide not only serves as a molecular imaging probe, but also shows the tumor suppression effects to GBM in vivo.
In conclusion, aptamer U2 was observed to exhibit outstanding amplifications in vitro and in vivo to GBM. Based on specific targeting of U87-EGFRvIII cells, our findings highlight the potential value of U2 as a multifunctional therapeutic strategy for increasing the sensitivity to radiotherapy and killing the tumor cells while acting as a molecular imaging probe and inhibiting tumor growth. In combination with ionizing radiation, treatment with U2 might be a promising, efficient clinical approach to overcome resistance to GBM treatment. Further studies will explore the biological interactions between the EGFR and DNA damage repair pathways, reinforcing the alteration of the cell cycle with U2 incubation after radiation and indicating the therapeutic effects of U2 in orthotropic brain tumor xenografts.
Materials and Methods
Cell Lines and Animals
The human GBM U87MG cell line was acquired from the American Type Culture Collection Company. The U87MG cell line was maintained in DMEM supplement with 10% fetal FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin. U87-EGFRvIII and U87-EGFRwt cell lines45 (kindly provided by Dr. Webster Cavenee, Ludwig Cancer Institute, San Diego, CA) were grown in DMEM supplement with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 200 mg/mL G418 (Sigma-Aldrich, St. Louis, MO). All cells were cultured in 95% air/5% CO2 at 37°C.
Male BALB/c nude mice at 4–6 weeks of age were supplied by Laboratory Animal Centre, Southern Medical University, to construct the tumor model. Approximately 2 × 106 U87-EGFRvIII cells re-suspended in 100 μL modified RPMI-1640 (Invitrogen, Carlsbad, CA) culture medium were subcutaneously injected into the right rump of each nude mouse. The formation of tumors was observed 2 weeks later. The animal experiments were conducted according to ethical committee approved protocols and regulations.
Aptamer Synthesis
The GN screening library containing a central random region of 40 nt was subjected to the cell-SELEX process, and U2 is one of the aptamers for U87-EGFRvIII cells.12 GN library and U2scr1 were used as the negative controls of the U2 aptamer. All sequences and FAM-labeled ssDNA were synthesized by Invitrogen (Carlsbad, CA). Before each treatment in the following experiment, the aptamers were first incubated at 90°C for 5 min, cooled on ice for 5 min, and treated at 37°C for 5 min.
Binding Assay of FCM and Confocal Microscopy
Binding of the U2 aptamer to the three cell lines was performed as described.12 Trypsinized and washed three times and then resuspended in Dulbecco’s PBS (DPBS), the cells were incubated with 1 μM FAM-U2 in 500 μL binding buffer (10 mg yeast tRNA and 5 mg salmon sperm DNA in 100 mL serum-free media) at 37°C for 30 min in darkness. The cells were washed, suspended with DBPS, and analyzed using a FACS cytometry assay (Becton Dickinson, Franklin Lakes, NJ).
The binding assay by confocal microscopy was performed as previously described.13 After overnight culture, the U87-EGFRvIII cells were treated with 2 μM FAM-U2 for 5 and 20 min. Cells were fixed in DPBS/4% PFA and labeled with anti-EGFR antibody targeting on the cell membrane without permeabilization. To assay the internalization of cells, the cells were fixed and permeabilized with 0.2% Triton X-100 for 10 min, which was followed by incubation with anti-EGFR and anti-EEA1 antibodies. After washing in DPBS, the cells were incubated with fluorescent secondary antibody. All cells were incubated in 1.5 μM DAPI to visualize the nuclei using a laser scanning confocal microscope (Leica).
Cell Proliferation Analysis
Cell proliferation was analyzed using a CCK8 (Dojindo Molecular Technologies, Gaithersburg, MD) test. Cells were seeded at 104 per well with a 96-well plate and incubated with six replicates for each condition. The cells were incubated for 24 hr at 37°C prior to transfection with different concentrations of U2 aptamer or GN library (0, 25, and 50 nM). The cells infected with GN library or U2 were incubated for 24 or 48 hr and quantification of cell viability was performed with a colorimetric assay using CCK8. The cell viability was expressed as the mean ± SD in percentage of the control viability (the percentage of control cell viability is 100%).
Apoptosis Assays by Annexin V-FITC/PI Double Staining
For apoptosis analysis, 2 × 105 cells were seeded on 6-well plates 24 hr prior to transfection with 50 nM aptamer U2 or 50 nM GN library as described above and were maintained at 37°C for 48 hr. Cells were trypsinized, centrifuged, and washed twice with PBS; then, they were detected by a FITC/Annexin V Apoptosis Kit (Abcam, Cambridge, MA). The cells were suspended in the binding buffer; then, FITC-conjugated Annexin V and PI were added at 50 μg/mL. Cells were analyzed using a FACS cytometry assay (Becton Dickinson, Franklin Lakes, NJ)
Cell Migration and Invasion
Cells were plated in 6-well plates and grown to confluence for using the healing assay.3, 13 Cells were serum starved for 24 hr and then scraped to induce a wound after adding 50 nM U2 aptamer or GN library. The wounds were quantitatively measured, and the remaining wound areas were calculated using ImageJ software (NIH).
The migration ability was also determined using a transwell migration assay as previously described.13 Cells were incubated in serum-free medium for 24 hr and then in the presence of 50 nM U2 or GN library on a 24-well transwell plate (Corning Incorporate, Corning, NY). The invasion assays on cells was performed using transwell filters coated with Matrigel (BD Biosciences, NJ) diluted 1:5 in serum medium and then incubated at 37°C for 30 min. Other procedures were performed as in the migration assay.
Western Blot Analysis
Cells were lysed in the RIPA buffer (P89901, Thermo Scientific) supplemented with protease inhibitors (P1862209, Thermo Scientific). The protein concentration was detected using a BCA protein assay kit (Thermo Scientific). For western blot, 40 μg protein was separated by 8% SDS-PAGE gels and blotted onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA). The membranes were incubated with the following specific antibodies: anti-pEGFR (tyr1038), anti-Akt, anti-pAkt (Ser 473), anti-pMET (Y1234/1235), anti-ERK, anti-pERK (T202/Y204), anti-PDGFRβ, anti-ATM, anti-53BP1, anti-pH2AX (S139), anti-pChk2 (T68), and anti-Chk2 (Cell Signaling Technology, Danvers, MA); and anti-EGFR, anti-Met (C-12) (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-GAPDH (Sigma-Aldrich, St. Louis, MO); then, a secondary horseradish peroxidase (HRP)-conjugate antibody was used for immunodetection. The protein bands were visualized using an ECL western blotting kit (Millipore, Bedford, MA) and densitometry analysis with AlphaEaseFC software.
Colony Formation Assay
The U87-EGFRvIII cells were seeded into 6-well plates at 250 cells per well. After 24 hr of culture in the presence of 200 nM U2 or U2-Scr1, the cells were exposed to radiation with doses of 2 Gy. After treatment, the cells were cultured for 10–12 days depending on the cell line. The colonies were fixed with 4% paraformaldehyde solution and stained with crystal violet. Colonies with more than 50 cells were counted and analyzed.
Comet Assay
All reagents used in the comet assay were prepared as previously described.46 Briefly, cells were seeded at 1 × 105 cells/well and cultured overnight. After treatment with U2 and U2Scr1 at a concentration of 200 nM for 24 hr, respectively, cells were exposed to irradiation with 2 Gy. Cultured for another 3 hr, the cells were collected and suspended in PBS. The cell suspension was mixed with 0.75% low temperature melting agarose (LMA) in PBS at 37°C and pipetted onto fully frosted slides, which were precoated with 0.5% normal temperature melting agarose (NMA). The coverslips were removed and the slides were placed in precooled lysis buffer at 4°C for 120 min and rinsed in double distilled water thrice. Electrophoresis was performed for 25 min at 25 V in an alkaline electrophoresis solution after unwinding for 25 min. After rinsing with neutral buffer thrice, slides were stained with PI for 10 min and covered by a coverslip. Stained cells were photographed using a fluorescence microscope with at least 100 cells per slide. The results were analyzed with the Comet Assay Software Project (CASP). Measurement of DNA in the tail, olive tail moment (OTM), and tail moment was used to quantify the extent of DNA damage.
In Vivo Radiotherapy of Tumor-Bearing Mice by 188Re-U2
Radiolabeling of aptamer or GN library with 188Re was prepared as previously described.12 188Re-U2 or 188Re-GN was purified with a C-18 Sep-Pak reverse-phase column; the radiochemical purity was determined by a paper chromatogram, and its radioactivity was measured using a scintillation γ-counter.
The in vivo experiment was approved by the Animal Care and Use Committee of Southern Medical University. The male nude mice were 18–20 g in weight and 4–6 weeks old. To establish a U87-EGFRvIII animal model, 5 × 105 U87-EGFRvIII cells in 0.1 mL PBS were subcutaneously injected in the back of BALB/c nude mice, and tumors were grown for 14 days and then randomized into the following four groups: blank group, free 188Re group, 188Re-labeled original library GN (88Re-GN) group, and 188Re-labeled aptamer U2 (188Re-U2) group. Mice were intratumorally injected with 10 μL saline or 10 μL drugs (200 pmol), respectively. All mice were sacrificed after 10 days, and the tumors were carefully dissected; then, the volumes and weights of the tumor were measured.
Statistical Analysis
Statistical analyses were performed with the SPSS 20.0 statistical software (IBM). A p value < 0.05 was considered significant. Comparison between group means was performed using one-way ANOVA; comparisons between two groups were performed using Student’s t test unless otherwise indicated. All experiments were performed at least 3 times.
Author Contributions
X.Z. conceived the study, supervised experiments, and participated in manuscript writing; L.P. supervised experiments, analyzed the data, and wrote the manuscript; Z.L. performed data analysis of western blot and flow cytometry assays; Z.K. supervised experiments in primary cell assays; Y.C. performed data analysis of western blot and G.S. performed the data and figures processing; X.L., Y.L., and F.W. participated in manuscript writing; Y.S. performed the radiotherapeutic assays and participated in manuscript writing. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81272509, 81471388, and 81771484), Guangdong Natural Science Foundation (9151051501000053 and 2014A030313351), President Foundation of Nanfang Hospital (2015Z007), and Program for Changjiang Scholars and Innovative Research Team in University (IRT_16R37).
Footnotes
Supplemental Information includes two figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.01.001.
Contributor Information
Xingmei Zhang, Email: zxmray@hotmail.com.
Yusheng Shi, Email: shiyush@mail.nfyy.com.
Supplemental Information
References
- 1.Cloughesy T.F., Cavenee W.K., Mischel P.S. Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 2.Brennan C.W., Verhaak R.G., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S., Chakravarty D., Sanborn J.Z., Berman S.H., TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi B.D., Kuan C.T., Cai M., Archer G.E., Mitchell D.A., Gedeon P.C., Sanchez-Perez L., Pastan I., Bigner D.D., Sampson J.H. Systemic administration of a bispecific antibody targeting EGFRvIII successfully treats intracerebral glioma. Proc. Natl. Acad. Sci. USA. 2013;110:270–275. doi: 10.1073/pnas.1219817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang P.H., Xu A.M., White F.M. Oncogenic EGFR signaling networks in glioma. Sci. Signal. 2009;2:re6. doi: 10.1126/scisignal.287re6. [DOI] [PubMed] [Google Scholar]
- 5.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 7.Bastien J.I., McNeill K.A., Fine H.A. Molecular characterizations of glioblastoma, targeted therapy, and clinical results to date. Cancer. 2015;121:502–516. doi: 10.1002/cncr.28968. [DOI] [PubMed] [Google Scholar]
- 8.Ni X., Castanares M., Mukherjee A., Lupold S.E. Nucleic acid aptamers: clinical applications and promising new horizons. Curr. Med. Chem. 2011;18:4206–4214. doi: 10.2174/092986711797189600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ilgu M., Nilsen-Hamilton M. Aptamers in analytics. Analyst (Lond.) 2016;141:1551–1568. doi: 10.1039/c5an01824b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimjee S.M., White R.R., Becker R.C., Sullenger B.A. Aptamers as therapeutics. Annu. Rev. Pharmacol. Toxicol. 2017;57:61–79. doi: 10.1146/annurev-pharmtox-010716-104558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X., Liang H., Tan Y., Yuan C., Li S., Li X., Li G., Shi Y., Zhang X. Cell-SELEX aptamer for highly specific radionuclide molecular imaging of glioblastoma in vivo. PLoS One. 2014;9:e90752. doi: 10.1371/journal.pone.0090752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camorani S., Crescenzi E., Colecchia D., Carpentieri A., Amoresano A., Fedele M., Chiariello M., Cerchia L. Aptamer targeting EGFRvIII mutant hampers its constitutive autophosphorylation and affects migration, invasion and proliferation of glioblastoma cells. Oncotarget. 2015;6:37570–37587. doi: 10.18632/oncotarget.6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esposito C.L., Passaro D., Longobardo I., Condorelli G., Marotta P., Affuso A., de Franciscis V., Cerchia L. A neutralizing RNA aptamer against EGFR causes selective apoptotic cell death. PLoS One. 2011;6:e24071. doi: 10.1371/journal.pone.0024071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivatsa S., Paul M.C., Cardone C., Holcmann M., Amberg N., Pathria P., Diamanti M.A., Linder M., Timelthaler G., Dienes H.P. EGFR in tumor-associated myeloid cells promotes development of colorectal cancer in mice and associates with outcomes of patients. Gastroenterology. 2017;153:178–190.e10. doi: 10.1053/j.gastro.2017.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosell R., Dafni U., Felip E., Curioni-Fontecedro A., Gautschi O., Peters S., Massutí B., Palmero R., Aix S.P., Carcereny E., BELIEF collaborative group Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): an international, multicentre, single-arm, phase 2 trial. Lancet Respir. Med. 2017;5:435–444. doi: 10.1016/S2213-2600(17)30129-7. [DOI] [PubMed] [Google Scholar]
- 17.Vivanco I., Robins H.I., Rohle D., Campos C., Grommes C., Nghiemphu P.L., Kubek S., Oldrini B., Chheda M.G., Yannuzzi N. Differential sensitivity of glioma- versus lung cancer-specific EGFR mutations to EGFR kinase inhibitors. Cancer Discov. 2012;2:458–471. doi: 10.1158/2159-8290.CD-11-0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J., Deng Q., Zhou J., Zou J., Zhang Y., Tan P., Zhang W., Cui H. CSN6 controls the proliferation and metastasis of glioblastoma by CHIP-mediated degradation of EGFR. Oncogene. 2017;36:1134–1144. doi: 10.1038/onc.2016.280. [DOI] [PubMed] [Google Scholar]
- 19.Yu X., Li W., Deng Q., You S., Liu H., Peng S., Liu X., Lu J., Luo X., Yang L. Neoalbaconol inhibits angiogenesis and tumor growth by suppressing EGFR-mediated VEGF production. Mol. Carcinog. 2017;56:1414–1426. doi: 10.1002/mc.22602. [DOI] [PubMed] [Google Scholar]
- 20.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu W., Zhou X., Hong B., Liu J., Yue Z. Suppression of invasion in human U87 glioma cells by adenovirus-mediated co-transfer of TIMP-2 and PTEN gene. Cancer Lett. 2004;214:205–213. doi: 10.1016/j.canlet.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 22.Zhu Y., Shah K. Multiple lesions in receptor tyrosine kinase pathway determine glioblastoma response to pan-ERBB inhibitor PF-00299804 and PI3K/mTOR dual inhibitor PF-05212384. Cancer Biol. Ther. 2014;15:815–822. doi: 10.4161/cbt.28585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyati M.K., Morgan M.A., Feng F.Y., Lawrence T.S. Integration of EGFR inhibitors with radiochemotherapy. Nat. Rev. Cancer. 2006;6:876–885. doi: 10.1038/nrc1953. [DOI] [PubMed] [Google Scholar]
- 24.Lammering G., Hewit T.H., Valerie K., Contessa J.N., Amorino G.P., Dent P., Schmidt-Ullrich R.K. EGFRvIII-mediated radioresistance through a strong cytoprotective response. Oncogene. 2003;22:5545–5553. doi: 10.1038/sj.onc.1206788. [DOI] [PubMed] [Google Scholar]
- 25.Lammering G., Hewit T.H., Holmes M., Valerie K., Hawkins W., Lin P.S., Mikkelsen R.B., Schmidt-Ullrich R.K. Inhibition of the type III epidermal growth factor receptor variant mutant receptor by dominant-negative EGFR-CD533 enhances malignant glioma cell radiosensitivity. Clin Cancer Res. 2004;10:6732–6743. doi: 10.1158/1078-0432.CCR-04-0393. [DOI] [PubMed] [Google Scholar]
- 26.Golding S.E., Morgan R.N., Adams B.R., Hawkins A.J., Povirk L.F., Valerie K. Pro-survival AKT and ERK signaling from EGFR and mutant EGFRvIII enhances DNA double-strand break repair in human glioma cells. Cancer Biol. Ther. 2009;8:730–738. doi: 10.4161/cbt.8.8.7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mukherjee B., McEllin B., Camacho C.V., Tomimatsu N., Sirasanagandala S., Nannepaga S., Hatanpaa K.J., Mickey B., Madden C., Maher E. EGFRvIII and DNA double-strand break repair: a molecular mechanism for radioresistance in glioblastoma. Cancer Res. 2009;69:4252–4259. doi: 10.1158/0008-5472.CAN-08-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni X., Zhang Y., Zennami K., Castanares M., Mukherjee A., Raval R.R., Zhou H., DeWeese T.L., Lupold S.E. Systemic administration and targeted radiosensitization via chemically synthetic aptamer-siRNA chimeras in human tumor xenografts. Mol. Cancer Ther. 2015;14:2797–2804. doi: 10.1158/1535-7163.MCT-15-0291-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni X., Zhang Y., Ribas J., Chowdhury W.H., Castanares M., Zhang Z., Laiho M., DeWeese T.L., Lupold S.E. Prostate-targeted radiosensitization via aptamer-shRNA chimeras in human tumor xenografts. J. Clin. Invest. 2011;121:2383–2390. doi: 10.1172/JCI45109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang K.B., Zhu C., Wong Y.L., Gao Q., Ty A., Wong M.C. Gefitinib radiosensitizes stem-like glioma cells: inhibition of epidermal growth factor receptor-Akt-DNA-PK signaling, accompanied by inhibition of DNA double-strand break repair. Int. J. Radiat. Oncol. Biol. Phys. 2012;83:e43–e52. doi: 10.1016/j.ijrobp.2011.11.037. [DOI] [PubMed] [Google Scholar]
- 31.Goldberg M., Stucki M., Falck J., D’Amours D., Rahman D., Pappin D., Bartek J., Jackson S.P. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature. 2003;421:952–956. doi: 10.1038/nature01445. [DOI] [PubMed] [Google Scholar]
- 32.Weber A.M., Ryan A.J. ATM and ATR as therapeutic targets in cancer. Pharmacol. Ther. 2015;149:124–138. doi: 10.1016/j.pharmthera.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Palumbo S., Tini P., Toscano M., Allavena G., Angeletti F., Manai F., Miracco C., Comincini S., Pirtoli L. Combined EGFR and autophagy modulation impairs cell migration and enhances radiosensitivity in human glioblastoma cells. J. Cell. Physiol. 2014;229:1863–1873. doi: 10.1002/jcp.24640. [DOI] [PubMed] [Google Scholar]
- 34.Lee K.M., Choi E.J., Kim I.A. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother. Oncol. 2011;101:171–176. doi: 10.1016/j.radonc.2011.05.050. [DOI] [PubMed] [Google Scholar]
- 35.Lundholm L., Hååg P., Zong D., Juntti T., Mörk B., Lewensohn R., Viktorsson K. Resistance to DNA-damaging treatment in non-small cell lung cancer tumor-initiating cells involves reduced DNA-PK/ATM activation and diminished cell cycle arrest. Cell Death Dis. 2013;4:e478. doi: 10.1038/cddis.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gil del Alcazar C.R., Hardebeck M.C., Mukherjee B., Tomimatsu N., Gao X., Yan J., Xie X.J., Bachoo R., Li L., Habib A.A. Inhibition of DNA double-strand break repair by the dual PI3K/mTOR inhibitor NVP-BEZ235 as a strategy for radiosensitization of glioblastoma. Clin Cancer Res. 2014;20:1235–1248. doi: 10.1158/1078-0432.CCR-13-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furgason J.M., Bahassi M. Targeting DNA repair mechanisms in cancer. Pharmacol. Ther. 2013;137:298–308. doi: 10.1016/j.pharmthera.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Dittmann K., Mayer C., Rodemann H.P., Huber S.M. EGFR cooperates with glucose transporter SGLT1 to enable chromatin remodeling in response to ionizing radiation. Radiother. Oncol. 2013;107:247–251. doi: 10.1016/j.radonc.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 39.Huang L., Li B., Tang S., Guo H., Li W., Huang X., Yan W., Zou F. Mitochondrial KATP channels control glioma radioresistance by regulating ROS-induced ERK activation. Mol. Neurobiol. 2015;52:626–637. doi: 10.1007/s12035-014-8888-1. [DOI] [PubMed] [Google Scholar]
- 40.De Bacco F., Luraghi P., Medico E., Reato G., Girolami F., Perera T., Gabriele P., Comoglio P.M., Boccaccio C. Induction of MET by ionizing radiation and its role in radioresistance and invasive growth of cancer. J. Natl. Cancer Inst. 2011;103:645–661. doi: 10.1093/jnci/djr093. [DOI] [PubMed] [Google Scholar]
- 41.De Bacco F., D’Ambrosio A., Casanova E., Orzan F., Neggia R., Albano R., Verginelli F., Cominelli M., Poliani P.L., Luraghi P. MET inhibition overcomes radiation resistance of glioblastoma stem-like cells. EMBO Mol. Med. 2016;8:550–568. doi: 10.15252/emmm.201505890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt K.S., Borkowski S., Kurreck J., Stephens A.W., Bald R., Hecht M., Friebe M., Dinkelborg L., Erdmann V.A. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 2004;32:5757–5765. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang D.W., Ko H.Y., Lee J.H., Kang H., Ryu S.H., Song I.C., Lee D.S., Kim S. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. J Nucl Med. 2010;51:98–105. doi: 10.2967/jnumed.109.069880. [DOI] [PubMed] [Google Scholar]
- 44.Jacobson O., Weiss I.D., Wang L., Wang Z., Yang X., Dewhurst A., Ma Y., Zhu G., Niu G., Kiesewetter D.O. 18F-labeled single-stranded DNA aptamer for PET imaging of protein tyrosine kinase-7 expression. J Nucl Med. 2015;56:1780–1785. doi: 10.2967/jnumed.115.160960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang P.H., Mukasa A., Bonavia R., Flynn R.A., Brewer Z.E., Cavenee W.K., Furnari F.B., White F.M. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. USA. 2007;104:12867–12872. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olive P.L., Banáth J.P. The comet assay: a method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.