Abstract
Citrullination is the post-translational hydrolysis of peptidyl-arginines to form peptidyl-citrulline, a reaction that is catalyzed by the protein arginine deiminases (PADs), a family of calcium-regulated enzymes. Aberrantly increased protein citrullination is associated with a slew of autoimmune diseases (e.g., rheumatoid arthritis (RA), multiple sclerosis, lupus, and ulcerative colitis) and certain cancers. Given the clear link between increased PAD activity and human disease, the PADs are therapeutically relevant targets. Herein, we report the development of next generation cell permeable and “Clickable” probes (BB-Cl-Yne and BB-F-Yne) for covalent labeling of the PADs both in vitro and in cell-based systems. Using advanced chemoproteomic technologies, we also report the off targets of both BB-Cl-Yne and BB-F-Yne. The probes are highly specific for the PADs, with relatively few off targets, especially BB-F-Yne, suggesting the preferential use of the fluoroacetamidine warhead in next generation irreversible PAD inhibitors. Notably, these compounds can be used in a variety of modalities, including the identification of off-targets of the parent compounds and as ABPPs in target engagement assays to demonstrate the efficacy of PAD inhibitors.
INTRODUCTION
Aberrantly upregulated protein citrullination is associated with a slew of autoimmune diseases (e.g., rheumatoid arthritis (RA), multiple sclerosis, lupus, and ulcerative colitis), as well as certain cancers.1, 2 Given these disease links, the protein arginine deiminases (PADs), the enzymes that catalyze this reaction, have garnered significant recent interest. The most deeply investigated disease associated with aberrantly increased PAD activity is RA, where these patients produce autoantibodies targeting numerous citrullinated proteins (e.g., citrullinated keratin, fibrin, vimentin and enolase).3–7 Importantly, the presence of these autoantibodies is the most specific diagnostic test available for RA. Moreover, these autoantibodies are present in patients’ sera 4-5 years before clinical onset, and higher titers are associated with a more severe clinical outcome.7–9 Thus, the presence of these anti-citrullinated protein antibodies (i.e., ACPA) is highly predictive of both disease incidence and severity. In addition to ACPAs, PAD2 and PAD4 are released by immune cells into the synovial joints of patients with RA where they remain active and citrullinate proteins. Within the joint, these citrullinated proteins then bind ACPAs,7, 10, 11 thereby setting up a classic positive feedback loop that recruits additional immune cells into the joint, the release of additional PAD isozymes into the synovium and enhanced protein citrullination and consequent inflammation.7
Although the specific cells that release PAD isozymes into the joints of RA patients is still debated, one likely source is neutrophils. Neutrophils are the predominant white blood cell in humans and are generally the first responders to signs of infection/inflammation. Depending on the environmental cues, a subset of these cells will undergo a novel form of cell death known as Neutrophil Extracellular Trap formation (NET) or NETosis.12–14 During this process, the chromatin decondenses and histones and other proteins are hypercitrullinated ultimately resulting in the ejection of chromatin fibers from the cell to form a web like structure that can trap pathogens (e.g. bacteria, fungi, viruses) as well as promote the formation of blood clots.12–14 Notably, neutrophils have long been known to be important players in the development and progression of RA as they are a predominant cell type in the synovial fluid of RA patients.15, 16 Enhanced NETosis, as is observed in RA,12 also results in the exposure of citrullinated autoantigens, which is key to the progression of RA, and is additionally thought to be the source of extracellular PADs.4, 12 How the PADs contribute to other inflammatory diseases is less well defined, but characteristic features include enhanced citrullination in the inflamed regions, suggesting that aberrant NETosis may contribute to these diseases as well. In addition, since the PADs are histone modifying enzymes that contribute to the epigenetic control of gene expression, there is emerging evidence to suggest that enhanced PAD activity promotes an inflamed state by altering the expression and/or activity of key cytokines and chemokines.17, 18
The role that the PADs play in these diseases is further highlighted by the efficacy of several PAD inhibitors in a variety of pre-clinical disease models. Specifically, the first-generation irreversible inhibitor Cl-amidine (1, Figure 1) has demonstrated efficacy in animal models of rheumatoid arthritis, lupus, ulcerative colitis, breast cancer, and atherosclerosis.12, 19–27 The therapeutic importance of the PADs was further highlighted by the second generation inhibitor, BB-Cl-amidine (3), which has shown enhanced efficacy over Cl-amidine (1) in animal models of lupus and RA.17, 28, 29 Moreover, the allosteric inhibitor GSK199 also shows efficacy in an RA model.30 Together, these findings have validated the PADs as viable therapeutic targets for a wide range of inflammatory conditions.
Figure 1.
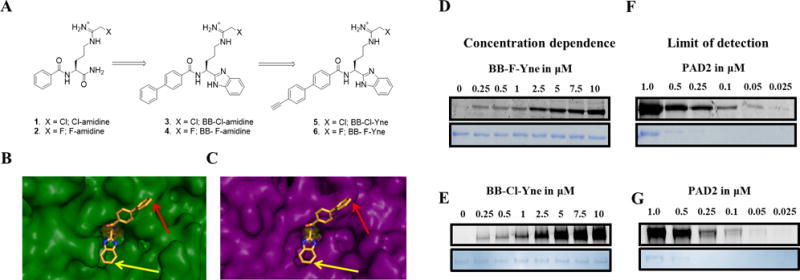
(A and B) Development of Benzimidazole-Based ABPPs. (A) Progression of inhibitor design to current probe design. (B) Co-crystal structure of BB-F-amidine (4) bound to PAD4. (C) PAD2 (PDB: 4N2C) overlay with PAD4/BB-F-amidine (4) co-crystal structure in PyMOL. (D and E) Concentration dependent labeling of recombinant PAD2 with (D) BB-F-Yne and (E) BB-Cl-Yne. PAD2 was treated with increasing concentrations of BB-F-Yne and BB-Cl-Yne for 1 h followed by click chemistry with TAMRA-N3. (F and G) The limit of detection for PAD2. Decreasing concentrations of PAD2 were treated with (F) BB-F-Yne and (G) BB-Cl-Yne for 1 h followed by click chemistry with TAMRA-N3.
The PAD reaction involves the hydrolysis of the guanidinium group of arginine to generate citrullinated proteins.1, 31, 32 This reaction, variably termed citrullination or deimination, is a calcium-dependent process wherein calcium binding triggers a conformational change that moves a nucleophilic cysteine residue into the active site, resulting in a >10,000-fold increase in PAD activity.33, 34 There are five PAD isozymes (PADs 1-4 and 6)1, 31, 32 with unique cellular and tissue distribution patterns, however, only PADs 1-4 are catalytically active. It is also important to note that the PADs are uniquely distributed both within the cell and throughout the body. Specifically, all the PADs can be found in the cytoplasm while only PADs 1, 2, and 4 are also localized in the nucleus.35–37 When it comes to distribution throughout the body, PAD2 is expressed in most tissues and cell types, PAD1 and PAD3 are principally expressed in the skin and hair follicles, PAD6 in oocytes, and PAD4 primarily in neutrophils and other myeloid derived cells.31 While PAD2 and PAD4 play a significant role in the development of RA,3, 38 PAD2 is also overexpressed in luminal breast cancer cells and its expression correlates with the level of the HER2 protooncogene.27, 36, 39
Cl-amidine (1) and the related PAD inhibitor F-amidine (2, Figure 1) irreversibly inhibit the PADs by covalently modifying an active site cysteine (Cys645 and 647 in PAD4 and PAD2, respectively). Taking advantage of the conserved nature of this catalytic cysteine, we previously reported the development of activity-based protein profiling (ABPP) probes targeting the PADs. These probes include Rhodamine-conjugated F-amidine (RFA)40 which labels all 4 active PAD isozymes (PADs 1-4) and was used to identify novel inhibitors of both PAD2 and PAD4.41, 42 “Clickable” versions of this probe were also developed and demonstrated their ability to label and isolate the PADs under a variety of conditions.43
Despite these early advances, the peptidic nature of these first-generation inhibitors and ABPPs possessed a number of limitations including their susceptibility to proteolysis and poor membrane permeability.28 This prompted the development of second-generation inhibitors predicated on improving metabolic stability and membrane permeability by substituting the C-terminal carboxamide with a benzimidazole moiety.28 This progression led to the discovery of BB-Cl-amidine (3) which has shown enhanced efficacy in a number of cell-based assays as well as animal models.17, 28, 29, 44 Specifically, BB-Cl-amidine (3) demonstrated an ability to block NETosis while improving efficacy in animal models of lupus and RA.17, 28, 29 In addition to further validating the PADs as important therapeutic targets, this scaffold also provided a novel platform for the development of next generation of ABPPs.
Herein we report the development of BB-Cl-Yne (5) and BB-F-Yne (6) for labeling the PADs both in vitro and in cell-based systems (Figure 1). These probes covalently modify the catalytic cysteine residue conserved in all PAD isozymes and present a handle for a subsequent copper catalyzed azide-alkyne cycloaddition reaction (CuAAC)45 with either TAMRA-N3 or Biotin-N3. The probes described herein are highly specific with relatively few off targets. Using advanced chemoproteomics technology we also report for the first time the off targets of both BB-Cl-Yne and BB-F-Yne. Notably, these compounds can be used in a variety of modalities, including the identification of off-targets of the parent compounds and as ABPPs in target engagement assays to demonstrate the efficacy of PAD inhibitors. Moreover, we expect that these probes will aid in furthering our understanding of PAD biology by detecting PADs in complex biological systems as well as ultimately identifying novel proteins and signaling pathways that interact with and/or regulate PAD activity.
RESULTS AND DISCUSSION
Probe Design
Three key features that we considered during the design of our next generation PAD-targeted ABPPs were: 1) cell permeability; 2) metabolic stability; and 3) alkyne position for optimal CuAAC. The first two features were implemented by utilizing the next generation PAD inhibitors, BB-Cl-amidine (3) and BB-F-amidine (4) as the core scaffold; this scaffold shows improved cell permeability and metabolic stability as compared to the first-generation counterparts, Cl-amidine (1) and F-amidine (2) (Figure 1A). Considering the structure of BB-Cl-amidine (3) and BB-F-amidine (4), we hypothesized that incorporation of an alkyne on either the benzimidazole moiety or the biphenyl group would optimally facilitate the copper catalyzed azide-alkyne cycloaddition reaction (CuAAC) between the probe and either TAMRA-N3 or Biotin-N3.
Key to developing these “Clickable” probes was the determination of the co-crystal structure of BB-F-amidine (4) bound to PAD4 (Figure 1B).46 Close examination of this co-crystal structure (Figure 1B) shows that the benzimidazole moiety binds deep within the active site. By contrast, the biphenyl group orients towards the solvent. Figure 1C illustrates the potential binding of this benzimidazole-based scaffold to active PAD2 through an overlay of the PAD4-BB-F-amidine (4) co-crystal structure with holoPAD2 (PDB: 4N2C).47 This overlay indicates that BB-F-amidine (4) binds to PAD2 in a similar fashion to PAD4 where the biphenyl group is solvent exposed and the benzimidazole moiety is buried within the substrate binding pocket. These data suggested that placement of the alkyne at the 4-position of the biphenyl moiety would allow for optimal CuAAC after covalent modification of the catalytic cysteine conserved in all active PAD isozymes; PAD6 is not catalytically active.48
Probe Synthesis
The synthesis of these clickable probes (Scheme 1) commenced with the generation of key intermediate 16 through a previously published method.28, 49 Briefly, Fmoc-Orn(Boc)-OH underwent HBTU-mediated amide coupling with phenylenediamine, followed by acid-catalyzed cyclization to form the benzimidazole moiety. Subsequent treatment with piperidine resulted in decarboxylation of the Fmoc protecting group giving 16. This intermediate was then subjected to HBTU-mediated amide coupling with biphenyl carboxylic acid 13 to give Boc-protected intermediate 17. Subsequent acid catalyzed decarboxylation of the side chain Boc group followed by installation of either the chloroacetamidine or fluoroacetamidine or warhead gave desired probes 5 (BB-Cl-Yne) and 6 (BB-F-Yne) in good yield.
Scheme 1.
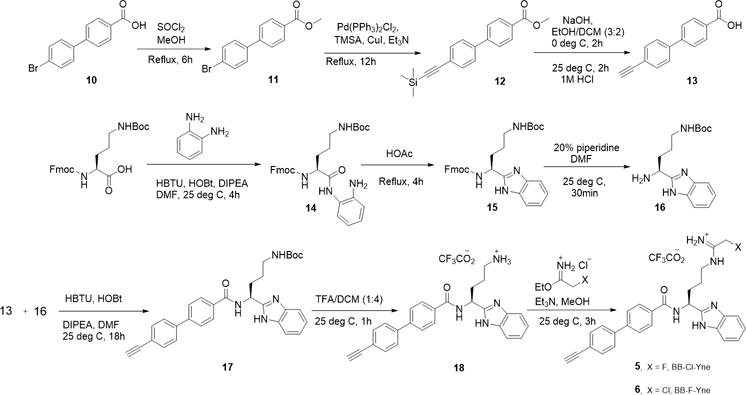
Synthesis of BB-F-Yne and BB-Cl-Yne
Probe Validation – Potency and in vitro labeling
To initially establish the utility of these “clickable” probes, we first determined kinact/KI values to assess their ability to inhibit PADs 1, 2, 3, and 4 (Table 1, Figures S1-S2, and Table S1). Importantly, the kinact/KI values for BB-Cl-Yne (5) and BB-F-Yne (6) are within 3-fold of the parent compounds BB-Cl-amidine and BB-F-amidine indicating that the additional alkyne moiety does not significantly impact potency, consistent with our modeling (Figure 1B-C). Next, we evaluated the ability of BB-F-Yne and BB-Cl-Yne to covalently modify recombinant PADs and undergo a subsequent click reaction with TAMRA-N3. Figures 1D-E and Figures S3-S4 illustrate the ability of these probes to effectively label PADs 1-4 in a dose-dependent manner down to probe concentrations as low as 250 nM. Notably, the sensitivity of the probes is as low as 375 fmol with BB-F-Yne (Figure 1F) and 756 fmol with BB-Cl-Yne (Figure 1G). Despite the two probes exhibiting similar potencies against PAD2, the slightly lower limit of detection may be attributable to the higher selectivity of BB-F-Yne compared to BB-Cl-Yne (see below).
Table 1.
Inhibition of PAD isozymes by second generation inhibitors.
| Compound | PAD1 kinact/KI (M−1min−1) |
PAD2 kinact/KI (M−1min−1) |
PAD3 kinact/KI (M−1min−1) |
PAD4 kinact/KI (M−1min−1) |
|---|---|---|---|---|
| BB-Cl-amidine (3) | 16100a | 4100a | 6800a | 13300a |
| BB-F-amidine (4) | 900b | 1200b | 3400b | 3750b |
| BB-Cl-Yne (5) | 6400 | 3600 | 10800 | 4900 |
| BB-F-Yne (6) | 2050 | 1700 | 1100 | 3100 |
kinact/KI was determined from a linear fit.
A single kobs was determined.
Probe validation – Labeling in cells
Having demonstrated their ability to label recombinant PADs, we next evaluated their ability to label the PADs in cell-based systems. Specifically, PAD2-overexpressing HEK293T cell lines were treated with BB-F-Yne and BB-Cl-Yne in the presence of a calcium ionophore (i.e., ionomycin) (Figure 2A). Cells were then harvested, lysed by sonication, and then “Clicked” with TAMRA-N3. Notably, BB-F-Yne labeled a single prominent band at the correct molecular weight for PAD2 (Figure 2B), even when used at concentrations as high as 25 μM. This level of selectivity is remarkable. In agreement with our in vitro results, BB-Cl-Yne did not label PAD2 as efficiently as BB-F-Yne (Figure 2 C), again indicating the superior selectivity of BB-F-Yne. The diminished labeling ability of BB-Cl-Yne may be due to the increased reactivity of the chloro-warhead, compared to the fluoro-warhead,41, 43 which increases the number of off targets such that a smaller amount of the probe is available to label PAD2.
Figure 2.

(A) Workflow showing probe labeling of mammalian cells by BB-F-Yne or BB-Cl-Yne followed by TAMRA-N3 mediated click chemistry. HEK293T/PAD2 cells were treated with increasing concentrations of BB-F-Yne (B) or BB-Cl-Yne (C), 5μM ionomycin and 1 mM CaCl2 and incubated at 37 °C for 1 h. The cells were sonicated, and the probe labeled proteins were tagged with TAMRA-N3 to facilitate visualization after SDS-PAGE. The visualized gel was transferred to a PVDF membrane and anti-PAD2 antibody was used to visualize PAD2 by western blot (shown in the bottom). (D) Workflow showing probe labeling of mammalian cells by BB-F-Yne or BB-Cl-Yne followed by biotin-N3 coupling. Labeled proteins were selectively separated using streptavidin beads. (E) HEK293T/PAD2 cells were treated with increasing concentrations of BB-F-Yne or (F) BB-Cl-Yne, 5 μM ionomycin and 1 mM CaCl2 and incubated at 37 °C for 1 h. The cells were sonicated and the probe labeled proteins were tagged with Biotin-N3. Biotin tagged proteins were then isolated on streptavidin agarose and the eluted proteins were probed for PAD2 by western blot. The full gel is shown in supporting information (Figure S9).
To demonstrate their utility in assessing target engagement, BB-F-Yne and BB-Cl-Yne were subjected to competition experiments. Briefly, PAD2-ovexpressing HEK293T cell lysates were incubated with increasing concentrations of either BB-F-amidine or BB-Cl-amidine in the presence of CaCl2 and were then treated with a fixed concentration BB-F-Yne or BB-Cl-Yne. Labeled lysates were “Clicked” with TAMRA azide and probe modified proteins were visualized by fluorescence. Both BB-F-amidine and BB-Cl-amidine dose dependently decrease the labeling of PAD2 (Figure S6 and S7) by BB-F-Yne and BB-Cl-Yne, thereby demonstrating their ability to assess target engagement.
To further illustrate the versatility of these probes, PAD2-ovexpressing HEK293T cell lysates were treated with either BB-F-Yne or BB-Cl-Yne and then “Clicked” with Biotin-N3 (Figure 2D). These lysates were then incubated with streptavidin-agarose beads, washed and probed for PAD2 or biotinylated proteins as shown in Figure 2D. Both BB-F-Yne and BB-Cl-Yne show a similar ability to label and isolate PAD2 (Figure 2E and F). The small amount of PAD2 isolated in the untreated control may be due to the ability of streptavidin-agarose beads to isolate autocitrullinated PAD2; the urea moiety of citrulline is structurally related to biotin and we consistently observe streptavidin binding to autocitrullinated PADs (data not shown). Note that PAD2 is expressed at ~0.8% of total proteins in our PAD2 overexpressing cell line (Figure S8).
Chemoproteomics experiments
Next, we employed a chemoproteomic approach to identify the targets of these two probes. Specifically, HEK293T PAD2 overexpressing cell lysates were treated with BB-F-Yne (25 μM), BB-Cl-Yne (2.5 μM), or DMSO for one hour (Figure 3A). These concentrations were chosen because they correspond to the EC50 values of the parent compounds in cellular citrullination experiments.46 Labeled proteomes were then “Clicked” with Biotin-N3 in the presence of TBTA and CuSO4. The clicked lysates were then isolated on streptavidin agarose beads followed by on-bead trypsin digestion and subsequent LC/LC-MS/MS. Protein identities were determined by database searches using the SEQUEST algorithm. Relative quantitation of probe labeled proteins in probe and DMSO treated samples was achieved by spectral counting.
Figure 3.
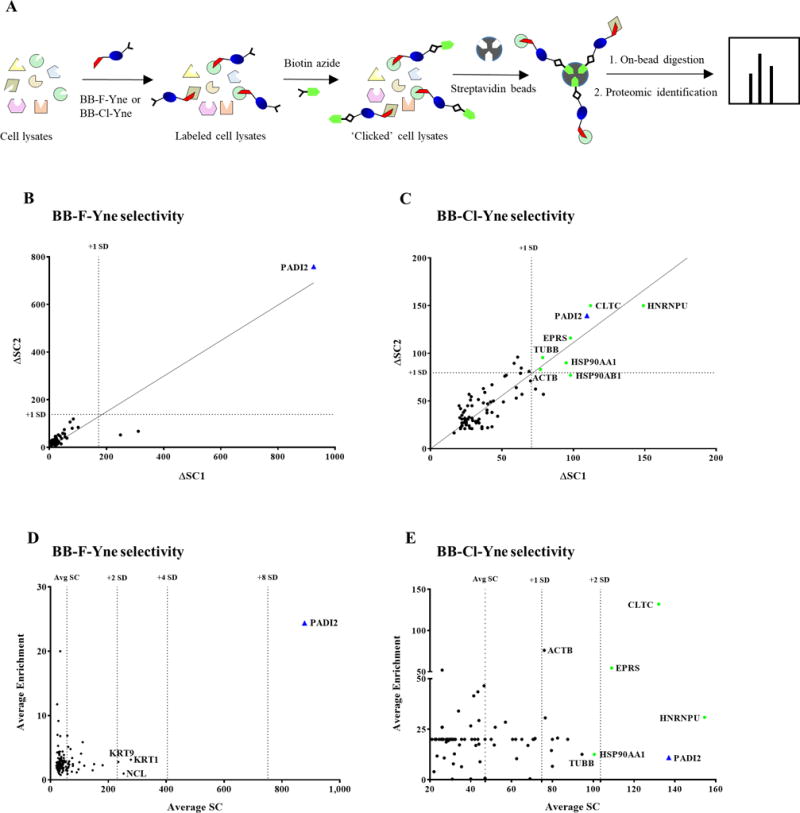
(A) Workflow showing probe labeling of cell lysates by BB-F-Yne (25 μM) or BB-Cl-Yne (2.5 μM) followed by coupling with biotin-N3. Labeled proteins were selectively separated using streptavidin agarose beads. (B and C) Plot showing the difference in spectral counts between the sample and the control (Δ SC) for the experimental replicates. (D and E) A plot of average enrichment versus average spectral counts for BB-F-Yne and BB-Cl-Yne labeled cell lysates from proteomics analysis.
Consistent with our Tamra-N3 labeling data, BB-F-Yne selectively enriched PAD2 with few off targets (Figure 3B and 3D). Figure 3B depicts the difference in spectral counts (ΔSC) between DMSO and BB-F-yne-treated samples for proteins identified in two biological replicates. Proteins along the diagonal are consistently enriched in the BB-F-yne sample and those above one standard deviation (1 SD) are considered statistically significant. Figure 3D depicts the average spectral count data plotted against the probe/DMSO spectral count ratio, which provides a relative measure of probe selectivity. Those proteins with high spectral counts and high probe/DMSO ratios are significantly enriched. Both plots demonstrate the remarkable selectivity of BB-F-Yne.
By contrast, BB-Cl-Yne reacted with PAD2 and a modest number of off targets in this in vitro experiment. These proteins include Heterogeneous Nuclear Ribonucleoprotein U (HNRPU), Clathrin heavy chain 1 (CLTC), Bifunctional glutamate/proline-tRNA ligase (EPRS), Tubulin beta chain (TUBB) and Actin beta chain (ACTB). Most of these proteins are highly abundant, contain reactive cysteines that covalently react with cysteine-targeted electrophiles, and have been found in other proteomic screens.50 Nevertheless, it is important recognize that PAD2 is amongst the most prominent proteins labeled by BB-Cl-Yne (Figure 3C and 3E), thereby indicating the preferential targeting of this protein by both this probe and likely the parent compound BB-Cl-amidine. The lower fold enrichment relates to background binding of PAD2 to the streptavidin beads, which, as noted above, relates to the ability of autocitrullinated PADs to bind weakly to streptavidin-agarose.
Because the proteomic experiments were performed with cell lysates, we next wished to confirm that the identified proteins were labeled in cells (Figure 4A). We focused on HNRPU, CLTC), EPRS), β-tubulin, and β-Actin because they were prominent off targets in our chemoproteomic screen. GAPDH and RPL were included as negative controls because they were slightly elevated but the data were not statistically significant. For these experiments, HEK cells overexpressing PAD2 were treated with BB-Cl-Yne (2.5 μM) and BB-F-Yne (25 μM) and then clicked to biotin azide. These concentrations were chosen because they correspond to the EC50 values for BB-F-amidine and BB-Cl-amidine in cellular citrullination experiments.46 Proteins were then isolated using streptavidin beads and the bound proteins were detected by western blotting after SDS-PAGE. In parallel we performed similar experiments in cell lysates. Consistent with our proteomic and labeling studies (see above), BB-F-Yne selectively isolated PAD2. Remarkably, BB-Cl-Yne was also selective and failed to isolate β-tubulin, β-Actin, clathrin, HSP90, GAPDH, and EPRS. By contrast, we did confirm that BB-Cl-Yne modifies β-tubulin, β-Actin, clathirin, HSP90 and EPRS but not GAPDH and RPL3 when cell lysates are treated with BB-Cl-Yne at 2.5 μM (Figure 4B). When cells were treated with a higher concentration of BB-Cl-Yne (10 μM), we did detect some labeling of β-tubulin and HSP90, but still failed to label β-Actin and GAPDH (Figure 4C). Overall, these data indicate that the selectivity of BB-Cl-Yne in cells is higher than that in cell lysates. The enhanced selectivity is likely due to a combination of factors including cell permeability which for low permeable compounds would adversely impact the cellular concentration of the probe and thereby improve cellular selectivity. Additionally, it is important to recognize that proteins like actin and tubulin adopt complex structures in cells, as compared to lysates, and as such their reactive cysteines may not be susceptible to modification.
Figure 4.
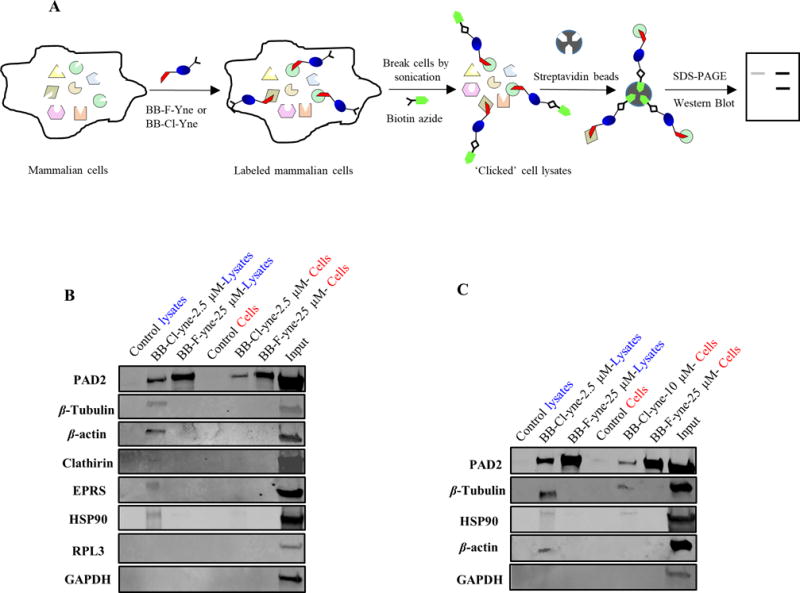
(A) Workflow showing probe labeling of mammalian cells by BB-F-Yne or BB-Cl-Yne followed coupling to biotin-N3. Labeled proteins were selectively separated using streptavidin beads. (B) HEK293T/PAD2 cells were treated with BB-Cl-Yne (2.5 μM), BB-F-Yne (25 μM), or DMSO. (C) HEK293T/PAD2 cells were treated with BB-Cl-Yne (10 μM), BB-F-Yne (25 μM) or DMSO, along with 5 μM ionomycin and 1 mM CaCl2 and incubated at 37 °C for 1h. Cells were sonicated and the probe labeled proteins were tagged with Biotin-N3. Biotin tagged proteins were then isolated on streptavidin agarose and the eluted proteins were probed for specific antibodies. Full image of the western blot along with more experimental details are given in the supporting information. See Figure S10 and S11.
Conclusions
In summary, we describe the development of two new PAD targeted ABPPs, BB-F-Yne (6) and BB-Cl-Yne (5), that show enhanced cellular uptake and efficient labeling of the PADs in cells. We predict that these compounds will be used in a variety of modalities, including as ABPPs in target engagement assays to demonstrate the efficacy of PAD inhibitors. Moreover, we expect that these probes will aid in furthering our understanding of PAD biology by detecting PADs in complex biological systems as well as ultimately identifying novel proteins and signaling pathways that interact with and/or regulate PAD activity. Our chemoproteomic data also highlights several features of the haloacetamidine warheads present in BB-F-amidine, BB-Cl-amidine, and related compounds. First, the fluoroacetamidine warhead is fairly non-reactive and in the context of the BB-F-amidine scaffold facilitate the isolation of only a single protein, i.e. PAD2, despite being tested at 25 μM. Such selectivity is remarkable and unprecedented for thiol reactive warheads. Second, BB-Cl-amidine reacts with a wider array of proteins and this feature likely explains the generally higher toxicity of chloroacetamidine containing compounds.46 Nevertheless, BB-Cl-Yne still shows high selectivity when applied to intact cells. This feature, along with its greater cell permeability, likely explains why BB-Cl-amidine has shown remarkable efficacy in animal models of RA and lupus.17, 28, 29 Projecting forward, we suggest that a primary focus should be placed on developing fluoroacetamidine-based irreversible inhibitors of the PADs due to the inherent specificity conferred by this warhead.
METHODS
Materials
PADs 1, 2, 3 and 4 were purified as reported.22, 51 HEK293T and HEK293T cells stably expressing human PAD2 (HEK293T/PAD2) were cultured as previously described.41 Bioin-TEV-N3 was synthesized as previously reported.52 TAMRA-N3 was obtained from Lumiprobe.
Inactivation Kinetics
Inactivation kinetic parameters were determined by incubating PAD1, 2, or 4 (2.0 μM) or PAD3 (5.0 μM) in a pre-warmed (10 min, 37 °C) inactivation mixture (50 mM HEPES pH 7.6, 10 mM CaCl2, and 2 mM DTT, with a final volume of 60 μL) containing various concentrations of inhibitor. Aliquots for were removed at various time points and added to a pre-warmed (10 min, 37 °C) reaction mixture (50 mM HEPES pH 7.6, 50 mM NaCl, 10 mM CaCl2, 2 mM DTT, and 10 mM BAEE or 10 mM BAA in the case of PAD3). The final dilution from the pre-incubation assay to the activity assay was 10-fold. After 15 min, reactions were quenched in liquid nitrogen and citrulline production quantified using the COLDER assay.34, 53 Data were plotted as a function of time and fit to eq 1,
| eq 1, |
using GraFit version 5.0.11, where v is velocity, vo is initial velocity, k (equal to kobs) is the pseudo-first order rate constant of inactivation, and t is time. When saturation was reached upon plotting kobs versus inactivator concentration, the data were fit to eq 2,
| eq 2, |
using GraFit version 5.0.11, where kinact corresponds to the maximal rate of inactivation and KI is the concentration of inhibitor that gives half-maximal inactivation. If the plot of kobs versus [I] was linear and did not saturate, then the value for kinact/KI equaled the slope of the line.
In vitro labeling of PADs dose dependently with BB-F-Yne (6) and BB-Cl-Yne (5)
Increasing concentrations of BB-F-Yne (6) or BB-Cl-Yne (5) (0 to 10 μM) were incubated with recombinant PADs (1 μM) in the presence of CaCl2 (2 mM) in 1× PBS at 37 ºC for 1 h. The probe labeled enzymes were coupled to TAMRA-N3 (20 μM) in the presence of 1X TBTA (0.31 mM), sodium ascorbate (2 mM) and freshly prepared CuSO4 (1 mM). The tubes were gently tumbled for 1 h. The reactions were quenched with 6× SDS loading buffer and separated by SDS-PAGE (12.5% gel). The bands were visualized by scanning the gel in a typhoon scanner (approximate excitation/emission maxima ~546/579, respectively).
In vitro labeling of PAD2 with BB-F-Yne (6) and BB-Cl-Yne (5) to determine their limit of detection (LOD)
BB-F-Yne (6) or BB-Cl-Yne (5) (10 μM) were incubated with decreasing concentrations of recombinant PAD2 (1.0 to 0.025 μM) in the presence of CaCl2 in 1× PBS (2 mM) at 37 ºC for 1 h. The probe labeled enzymes were coupled to TAMRA-N3 (20 μM) in the presence of 1X TBTA (0.31 mM), sodium ascorbate (2 mM) and freshly prepared CuSO4 (1 mM). The tubes were gently tumbled for 1 h. The reactions were quenched with 6× SDS loading buffer and separated by SDS-PAGE (12.5% gel). The bands were visualized by scanning the gel in a typhoon scanner (approximate excitation/emission maxima ~546/579, respectively).
Cellular labeling of PAD2 with BB-F-Yne and BB-Cl-Yne in ionomycin stimulated HEK293T/PAD2 cells
HEK293T cells stably expressing human PAD2 (HEK293T PAD2) were cultured as previously described.41 Cells were grown to ~80% confluence (8 × 106 cells), trypsinized, and trypsin activity quenched with complete media. The cells were harvested by centrifugation at 1000 × g for 2 min and washed 4× with 1× PBS. Cells were resuspended in PBS at 8 × 106 cells/mL and 4 × 105 cells were added to 0.65 mL tubes for subsequent assays.
HEK293T PAD2 cells were treated with increasing concentrations of BB-F-Yne or BB-Cl-Yne (0 to 50 μM) in the presence of CaCl2 (2 mM) at 37 ºC. After 30 min, ionomycin (5 μM) was added and the cells incubated for 1 h followed by sonication at 4 ºC for 1 h. Lysates were cleared by centrifugation at 21,000 × g for 15 min. The soluble protein fraction was isolated and quantified by the Detergent Compatible assay (DC assay, Biorad). Lysates (2 μg/μL, 50 μL total) were “Clicked” with TAMRA-N3 (20 μM), 1X TBTA (0.3 mM), sodium ascorbate (2 mM) and freshly prepared CuSO4 (1 mM). The tubes were gently tumbled for 2 h. The reactions were quenched by the addition of 6× SDS loading buffer and separated by SDS-PAGE (12.5% gel). The bands were visualized by scanning the gel in a typhoon scanner (approximate excitation/emission maxima ~546/579, respectively).
Lysates (2 μg/μL, 500 μL total) were also “Clicked” with Biotin azide (100 μM), 1X TBTA (0.3 mM), sodium ascorbate (2 mM) and freshly prepared CuSO4 (1 mM). The tubes were gently tumbled for 2 h. The cloudy solution was transferred to a microconcentrator (10 kDa molecular weight cutoff) and centrifuged at 10,000 × g for 5 min at 4 ºC to remove the excess biotin azide. The protein was then resolubilized in 1X PBS with 0.2% SDS. Resolubilized protein samples were incubated with 100 μL streptavidin-agarose beads (Thermo Scientific) at 4 °C for 16 h. The solutions were then incubated at room temperature for 1 h. The beads were washed with 0.2% SDS/PBS, PBS, and water. The beads were pelleted by centrifugation (1,600 × g, 3 min) between washes. To the washed beads, 2X SDS loading buffer was added and heated in a 95 °C heat block for 15 min. The resolubilized protein was separated by SDS-PAGE (4 – 20 % gel) and transferred to PVDF membranes (Biorad) at 150 V for 60 min. The membranes were analyzed for PAD2 and for other proteins.
Competition experiments
HEK293T PAD2 cell lysates (100 μg) were treated with increasing concentrations of BB-Cl-amidine or BB-F-amidine (0 – 200 μM) in the presence of CaCl2 (1 mM) at 37 ºC. After 15 minutes, BB-Cl-Yne or BB-F-Yne (25 μM) was added and the lysates were incubated for 1 h. Lysates (100 μg, 50 μL total) were “Clicked” with TAMRA-N3 (20 μM), 1X TBTA (0.3 mM), and freshly prepared CuSO4 (1 mM). The tubes were gently tumbled for 2 h. The reactions were quenched by the addition of 6× SDS loading buffer and separated by SDS-PAGE (4-20 % gel). The bands were visualized by scanning the gel in a typhoon scanner (approximate excitation/emission maxima ~546/579, respectively). Western blot analysis of the lysates was performed to detect the presence of PAD2.
Chemoproteomic analysis of cellular PAD2 labeled with BB-F-yne and BB-Cl-yne
Cell lysates (2 mg lysate per tube) prepared from HEK293T PAD2 expressing cells were labeled with BB-F-yne (25 μM) and BB-Cl-yne (1 μM) by incubating at 37 °C for 1h. The probes were ‘Clicked’ to biotin azide (100 μM) in the presence of 1X TBTA (0.3 mM) and CuSO4 (1 mM) for 2 h at room temperature. Samples were mixed well by vortexing them every 15 min. The reaction mixture was then centrifuged and the pellet was washed with methanol and resuspended in 1.2% SDS in PBS (w/v) followed by sonication and heating (5 min, 80 °C). The SDS-solubilized samples were diluted with PBS (5 mL) for a final SDS concentration of 0.2%. The solutions were incubated with 100 μL of streptavidin agarose beads (Thermo Fisher Scientific) at 4 °C for 16 h. The solutions were then incubated at room temperature for 3 h. The beads were washed with 6M urea in PBS (2 × 5 mL), 0.2% SDS/PBS (2 × 5 mL), PBS (3 × 5 mL), and water (3 × 5 mL). The beads were pelleted by centrifugation (1,400 × g, 3 min) between washes. The washed beads were suspended in 6 M urea in PBS (500 μL) and 10 mM dithiothreitol (from 20× stock in water) and placed in a 65 °C heat block for 15 min. Iodoacetamide (20 mM, from 20× stock in water) was then added to the samples and allowed to react at 37 °C for 30 min. Following reduction and alkylation, the beads were pelleted by centrifugation (1,400 × g, 3 min) and resuspended in a premixed solution of 2 M urea in PBS (200 μL), 100 mM CaCl2 in water (2 μL), and trypsin (4 μL of 20 mg reconstituted in 40 μL of trypsin buffer). The digestion was allowed to proceed overnight at 37 °C. The digested peptides were separated from the beads by centrifugation and the beads were washed twice with 50 μL of H2O. Formic acid (15 μL) was added to the samples, and the samples were stored at −80 °C until MS analysis.
LC/LC-MS/MS and Data Processing
Mass Spectrometry was performed using a Thermo Fisher LTQ Orbitrap Discovery mass spectrometer in conjunction with Agilent 1200 series HPLC. Enriched and digested peptide samples were pressure-loaded onto 250 μm fused silica desalting column packed with 4 cm of Aqua C18 reverse phase resin (Phenomenex). Peptides were eluted onto a biphasic 100 μm fused silica column with a 5 μm tip. The column was packed with 3 cm Partisphere strong cation exchange resin (SCX, Whatman) followed by 10 cm of C18 resin. Elution took place using a gradient from 5% Buffer A (95% water, 5% acetonitrile, 0.1% formic acid), to 100% Buffer B (20% water, 80% acetonitrile, 0.1% formic acid). The peptides were detached from the SCX onto the C18 resin and into the mass spectrometer using distinct salt elutions (95% high-purity water, 5% Optima-grade acetonitrile, 0.1% formic acid and 500 mM ammonium acetate)52. The flow rate through the column was 250 nL/min, with a spray voltage of 2.75 kV. With dynamic exclusion enabled, each full MS1 scan (400-1800 MW) was followed by 8 high-resolution, data-dependent scans of the nth most intense ion.
The tandem MS data from the multidimensional LC/LC-MS/MS protocol (MudPIT) was analyzed by the SEQUEST algorithm54 using a static Cys modification (+57 Da). Data was searched against a human reverse-concatenated non-redundant FASTA database applying Uniprot identifiers. MS2 spectra was assembled using DTASelect 2.055, with trypstat option applied.
Pulldown experiments using biotin azide
HEK293T cells stably expressing human PAD2 (HEK293T PAD2) were cultured as previously described.41 Cells were grown to ~80% confluence (8 × 106 cells), trypsinized, and trypsin activity quenched with complete media. The cells were harvested by centrifugation at 1000 × g for 2 min and washed 4× with 1× PBS. Cells were resuspended in PBS at 8 × 106 cells/mL and 4 × 105 cells (0.6 mL) were added to 1.5 mL tubes and were treated with BB-F-Yne (25 μM) or BB-Cl-Yne (2.5 μM and 10 μM) in the presence of CaCl2 (2 mM) at 37 ºC. After 30 min, ionomycin (5 μM) was added and the cells incubated for 1 h followed by sonication at 4 ºC for 1 h. Lysates were cleared by centrifugation at 21,000 × g for 15 min. The soluble protein fraction was isolated and quantified by the Detergent Compatible assay (DC assay, Biorad). Probe labeled proteins present in lysates (2 mg per tube) were then coupled to biotin azide using the click chemistry protocol described below. HEK cell lysates (2 mg lysate per tube) were separately labeled with BB-F-yne (25 μM) and BB-Cl-yne (2.5 μM) for 1 h at 37 °C. Click chemistry was performed on all samples using biotin azide (100 μM), 1X TBTA (0.3 mM) and CuSO4 (1 mM) and incubating at room temperature for 2 h. Samples were mixed well by vortexing them every 15 min. The reaction mixture was then centrifuged and the pellet was washed with methanol and re-suspended in 1.2% SDS in PBS (w/v) sonication and heating (5 min, 80 °C). The SDS-solubilized samples were diluted with PBS (5 mL) for a final SDS concentration of 0.2%. The solutions were incubated with 100 μL of streptavidin agarose beads (Thermo Fisher Scientific) at 4 °C for 16 h. The solutions were then incubated at room temperature for 3 h. The beads were washed with PBS (2 × 5 mL), and water (5 mL). The beads were pelleted by centrifugation (1,400 × g, 3 min) between washes. The washed beads were suspended in 8 M urea in PBS with 1.2% SDS (with 1X loading dye) and heated at 95 °C for 30 min and centrifuged. The supernatant was separated by SDS-PAGE (4 – 20 % gel) and transferred to PVDF membranes (Biorad) at 150 V for 60 min. The membranes were analyzed for the presence of PAD2 and other proteins of interest using the following antibodies: anti-clathrin heavy chain antibody (Abcam, ab21679), anti-beta Tubulin antibody (Abcam, ab604), anti-beta actin antibody (Abcam, ab8227), anti-GAPDH antibody (Abcam ab9484), anti-RPL3 antibody (Abcam, ab154882), anti-Hsp90 antibody (Abcam, ab13495), anti-glutamyl prolyl tRNA synthetase antibody (Abcam, ab85447), PADI2 antibody (Protein Tech, Cat No: 12110-1-AP).
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant GM109767 (P.R.T.) and GM117004 and GM118431 (E.W.).
ABBREVIATIONS
- RA
Rheumatoid arthritis
- PAD
Protein arginine deiminase
- ACPA
Anti-citrullinated protein antibodies
- NET
Neutrophil extracellular trap
- ABPP
Activity-based protein profiling
- RFA
Rhodamine-conjugated F-amidine
- CuAAC
Copper catalyzed azide-alkyne cycloaddition
- TAMRA-N3
Tetramethylrhodamine azide
- Biotin-N3
Biotin azide
- Orn
Ornithine
- LOD
Limit of detection
Footnotes
Supporting information. Full synthetic procedures, Table S1, and Figures S1-S11. This information is available free of charge via the Internet at http://pubs.acs.org.
ORCID ID
Paul R. Thompson: 0000-0002-1621-3372
Eranthie Weerapana: 0000-0002-0835-8301
References
- 1.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, Thompson PR. Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr Opin Drug Discov Devel. 2009;12:616–627. [PMC free article] [PubMed] [Google Scholar]
- 2.Bicker KL, Thompson PR. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers. 2013;99:155–163. doi: 10.1002/bip.22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Steendam K, Tilleman K, Deforce D. The relevance of citrullinated vimentin in the production of antibodies against citrullinated proteins and the pathogenesis of rheumatoid arthritis. Rheumatology. 2011;50:830–837. doi: 10.1093/rheumatology/keq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puszczewicz M, Iwaszkiewicz C. Role of anti-citrullinated protein antibodies in diagnosis and prognosis of rheumatoid arthritis. Arch Med Sci. 2011;7:189–194. doi: 10.5114/aoms.2011.22067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Boekel MA, Vossenaar ER, van den Hoogen FH, van Venrooij WJ. Autoantibody systems in rheumatoid arthritis: specificity, sensitivity and diagnostic value. Arthritis Res. 2002;4:87–93. doi: 10.1186/ar395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, Nogueira L, Vincent C, Senshu T, Serre G. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the alpha- and beta-chains of fibrin. J Immunol. 2001;166:4177–4184. doi: 10.4049/jimmunol.166.6.4177. [DOI] [PubMed] [Google Scholar]
- 7.Burska AN, Hunt L, Boissinot M, Strollo R, Ryan BJ, Vital E, Nissim A, Winyard PG, Emery P, Ponchel F. Autoantibodies to posttranslational modifications in rheumatoid arthritis. Mediators Inflamm. 2014;2014:492873. doi: 10.1155/2014/492873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berlyne GM. Carbamylated proteins and peptides in health and in uremia. Nephron. 1998;79:125–130. doi: 10.1159/000045013. [DOI] [PubMed] [Google Scholar]
- 9.Swaminathan S, Shah SV. Novel inflammatory mechanisms of accelerated atherosclerosis in kidney disease. Kidney Int. 2011;80:453–463. doi: 10.1038/ki.2011.178. [DOI] [PubMed] [Google Scholar]
- 10.van Gaalen F, Ioan-Facsinay A, Huizinga TW, Toes RE. The devil in the details: the emerging role of anticitrulline autoimmunity in rheumatoid arthritis. J Immunol. 2005;175:5575–5580. doi: 10.4049/jimmunol.175.9.5575. [DOI] [PubMed] [Google Scholar]
- 11.Wright HL, Moots RJ, Edwards SW. The multifactorial role of neutrophils in rheumatoid arthritis. Nat Rev Rheumatol. 2014;10:593–601. doi: 10.1038/nrrheum.2014.80. [DOI] [PubMed] [Google Scholar]
- 12.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, Friday S, Li S, Patel RM, Subramanian V, Thompson P, Chen P, Fox DA, Pennathur S, Kaplan MJ. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5:178ra140. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li P, Li M, Lindberg MR, Kennett MJ, Xiong N, Wang Y. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med. 2010;207:1853–1862. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 15.Ottonello L, Cutolo M, Frumento G, Arduino N, Bertolotto M, Mancini M, Sottofattori E, Dallegri F. Synovial fluid from patients with rheumatoid arthritis inhibits neutrophil apoptosis: role of adenosine and proinflammatory cytokines. Rheumatology. 2002;41:1249–1260. doi: 10.1093/rheumatology/41.11.1249. [DOI] [PubMed] [Google Scholar]
- 16.Weissmann G, Korchak H. Rheumatoid arthritis. The role of neutrophil activation. Inflammation. 1984;8(Suppl):S3–14. doi: 10.1007/BF00915708. [DOI] [PubMed] [Google Scholar]
- 17.Ghari F, Quirke AM, Munro S, Kawalkowska J, Picaud S, McGouran J, Subramanian V, Muth A, Williams R, Kessler B, Thompson PR, Fillipakopoulos P, Knapp S, Venables PJ, La Thangue NB. Citrullination-acetylation interplay guides E2F1 activity during the inflammatory response. Sci Adv. 2016;2:e1501257. doi: 10.1126/sciadv.1501257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun B, Dwivedi N, Bechtel TJ, Paulsen JL, Muth A, Bawadekar M, Li G, Thompson PR, Shelef MA, Schiffer CA, Weerapana E, Ho IC. Citrullination of NF-kappaB p65 promotes its nuclear localization and TLR-induced expression of IL-1beta and TNFalpha. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aal3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Li P, Wang S, Hu J, Chen XA, Wu J, Fisher M, Oshaben K, Zhao N, Gu Y, Wang D, Chen G, Wang Y. Anticancer peptidylarginine deiminase (PAD) inhibitors regulate the autophagy flux and the mammalian target of rapamycin complex 1 activity. J Biol Chem. 2012;287:25941–25953. doi: 10.1074/jbc.M112.375725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chumanevich AA, Causey CP, Knuckley BA, Jones JE, Poudyal D, Chumanevich AP, Davis T, Matesic LE, Thompson PR, Hofseth LJ. Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol. 2011;300:G929–938. doi: 10.1152/ajpgi.00435.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange S, Gogel S, Leung KY, Vernay B, Nicholas AP, Causey CP, Thompson PR, Greene ND, Ferretti P. Protein deiminases: New players in the developmentally regulated loss of neural regenerative ability. Dev Biol. 2011;355:205–214. doi: 10.1016/j.ydbio.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Causey CP, Jones JE, Slack JL, Kamei D, Jones LE, Subramanian V, Knuckley B, Ebrahimi P, Chumanevich AA, Luo Y, Hashimoto H, Sato M, Hofseth LJ, Thompson PR. The development of N-alpha-(2-carboxyl)benzoyl-N(5)-(2-fluoro-1-iminoethyl)-l-ornithine amide (o-F-amidine) and N-alpha-(2-carboxyl)benzoyl-N(5)-(2-chloro-1-iminoethyl)-l-ornithine amide (o-Cl-amidine) as second generation protein arginine deiminase (PAD) inhibitors. J Med Chem. 2011;54:6919–6935. doi: 10.1021/jm2008985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight JS, Kaplan MJ. Lupus neutrophils: ‘NET’ gain in understanding lupus pathogenesis. Curr Opin Rheumatol. 2012;24:441–450. doi: 10.1097/BOR.0b013e3283546703. [DOI] [PubMed] [Google Scholar]
- 24.Knight JS, Zhao W, Luo W, Subramanian V, O’Dell AA, Yalavarthi S, Hodgin JB, Eitzman DT, Thompson PR, Kaplan MJ. Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest. 2013;123:2981–2993. doi: 10.1172/JCI67390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CK, Vivekanandan-Giri A, Tang C, Knight JS, Mathew A, Padilla RL, Gillespie BW, Carmona-Rivera C, Liu X, Subramanian V, Hasni S, Thompson PR, Heinecke JW, Saran R, Pennathur S, Kaplan MJ. Neutrophil extracellular trap-derived enzymes oxidize high-density lipoprotein: an additional proatherogenic mechanism in systemic lupus erythematosus. Arthritis Rheumatol. 2014;66:2532–2544. doi: 10.1002/art.38703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight JS, Luo W, O’Dell AA, Yalavarthi S, Zhao W, Subramanian V, Guo C, Grenn RC, Thompson PR, Eitzman DT, Kaplan MJ. Peptidylarginine deiminase inhibition reduces vascular damage and modulates innate immune responses in murine models of atherosclerosis. Circ Res. 2014;114:947–956. doi: 10.1161/CIRCRESAHA.114.303312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McElwee JL, Mohanan S, Griffith OL, Breuer HC, Anguish LJ, Cherrington BD, Palmer AM, Howe LR, Subramanian V, Causey CP, Thompson PR, Gray JW, Coonrod SA. Identification of PADI2 as a potential breast cancer biomarker and therapeutic target. BMC Cancer. 2012;12:500. doi: 10.1186/1471-2407-12-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight JS, Subramanian V, O’Dell AA, Yalavarthi S, Zhao W, Smith CK, Hodgin JB, Thompson PR, Kaplan MJ. Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann Rheum Dis. 2015;74:2199–2206. doi: 10.1136/annrheumdis-2014-205365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawalkowska J, Quirke AM, Ghari F, Davis S, Subramanian V, Thompson PR, Williams RO, Fischer R, La Thangue NB, Venables PJ. Abrogation of collagen-induced arthritis by a peptidyl arginine deiminase inhibitor is associated with modulation of T cell-mediated immune responses. Sci Rep. 2016;6:26430. doi: 10.1038/srep26430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willis VC, Banda NK, Cordova KN, Chandra PE, Robinson WH, Cooper DC, Lugo D, Mehta G, Taylor S, Tak PP, Prinjha RK, Lewis HD, Holers VM. Protein arginine deiminase 4 inhibition is sufficient for the amelioration of collagen-induced arthritis. Clin Exp Immunol. 2017;188:263–274. doi: 10.1111/cei.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vossenaar ER, Zendman AJ, van Venrooij WJ, Pruijn GJ. PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 32.Stone EM, Schaller TH, Bianchi H, Person MD, Fast W. Inactivation of two diverse enzymes in the amidinotransferase superfamily by 2-chloroacetamidine: dimethylargininase and peptidylarginine deiminase. Biochemistry. 2005;44:13744–13752. doi: 10.1021/bi051341y. [DOI] [PubMed] [Google Scholar]
- 33.Liu YL, Chiang YH, Liu GY, Hung HC. Functional role of dimerization of human peptidylarginine deiminase 4 (PAD4) PLoS One. 2011;6:e21314. doi: 10.1371/journal.pone.0021314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kearney PL, Bhatia M, Jones NG, Yuan L, Glascock MC, Catchings KL, Yamada M, Thompson PR. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry. 2005;44:10570–10582. doi: 10.1021/bi050292m. [DOI] [PubMed] [Google Scholar]
- 35.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Bolt M, Guertin MJ, Chen W, Zhang S, Cherrington BD, Slade DJ, Dreyton CJ, Subramanian V, Bicker KL, Thompson PR, Mancini MA, Lis JT, Coonrod SA. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci U S A. 2012;109:13331–13336. doi: 10.1073/pnas.1203280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, Liu X, Zhang M, Li T, Muth A, Thompson PR, Coonrod SA, Zhang X. Peptidylarginine deiminase 1-catalyzed histone citrullination is essential for early embryo development. Sci Rep. 2016;6:38727. doi: 10.1038/srep38727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Damgaard D, Senolt L, Nielsen MF, Pruijn GJ, Nielsen CH. Demonstration of extracellular peptidylarginine deiminase (PAD) activity in synovial fluid of patients with rheumatoid arthritis using a novel assay for citrullination of fibrinogen. Arthritis Res Ther. 2014;16:498. doi: 10.1186/s13075-014-0498-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohanan S, Cherrington BD, Horibata S, McElwee JL, Thompson PR, Coonrod SA. Potential role of peptidylarginine deiminase enzymes and protein citrullination in cancer pathogenesis. Biochem Res Int. 2012;2012:895343. doi: 10.1155/2012/895343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knuckley B, Luo Y, Thompson PR. Profiling Protein Arginine Deiminase 4 (PAD4): a novel screen to identify PAD4 inhibitors. Bioorg Med Chem. 2008;16:739–745. doi: 10.1016/j.bmc.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewallen DM, Bicker KL, Madoux F, Chase P, Anguish L, Coonrod S, Hodder P, Thompson PR. A FluoPol-ABPP PAD2 high-throughput screen identifies the first calcium site inhibitor targeting the PADs. ACS Chem Biol. 2014;9:913–921. doi: 10.1021/cb400841k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones JE, Slack JL, Fang P, Zhang X, Subramanian V, Causey CP, Coonrod SA, Guo M, Thompson PR. Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors. ACS Chem Biol. 2012;7:160–165. doi: 10.1021/cb200258q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Slack JL, Causey CP, Luo Y, Thompson PR. Development and use of clickable activity based protein profiling agents for protein arginine deiminase 4. ACS Chem Biol. 2011;6:466–476. doi: 10.1021/cb1003515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horibata S, Vo TV, Subramanian V, Thompson PR, Coonrod SA. Utilization of the Soft Agar Colony Formation Assay to Identify Inhibitors of Tumorigenicity in Breast Cancer Cells. J Vis Exp. 2015:e52727. doi: 10.3791/52727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speers AE, Adam GC, Cravatt BF. Activity-based protein profiling in vivo using a copper(i)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 46.Muth A, Subramanian V, Beaumont E, Nagar M, Kerry P, McEwan P, Srinath H, Clancy K, Parelkar S, Thompson PR. Development of a Selective Inhibitor of Protein Arginine Deiminase 2. J Med Chem. 2017;60:3198–3211. doi: 10.1021/acs.jmedchem.7b00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slade DJ, Fang P, Dreyton CJ, Zhang Y, Fuhrmann J, Rempel D, Bax BD, Coonrod SA, Lewis HD, Guo M, Gross ML, Thompson PR. Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design. ACS Chem Biol. 2015;10:1043–1053. doi: 10.1021/cb500933j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen PA, Drijfhout JW, Pruijn GJ. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol. 2007;367:1118–1129. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- 49.Xiang Y, Wang Q, Wang G, Li X, Zhang D, Jin W. Synthesis and coordination of star-shaped electron-deficient hexaheteroarylbenzene derivatives containing three pyrimidylbenzene derivatives. Tetrahedron. 2016;72:2574–2580. [Google Scholar]
- 50.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, Gonzalez-Paez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, Wolan DW, Cravatt BF. Proteome-wide covalent ligand discovery in native biological systems. Nature. 2016;534:570–574. doi: 10.1038/nature18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Knuckley B, Causey CP, Jones JE, Bhatia M, Dreyton CJ, Osborne TC, Takahara H, Thompson PR. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weerapana E, Speers AE, Cravatt BF. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)–a general method for mapping sites of probe modification in proteomes. Nat Protoc. 2007;2:1414–1425. doi: 10.1038/nprot.2007.194. [DOI] [PubMed] [Google Scholar]
- 53.Knipp M, Vasak M. A colorimetric 96-well microtiter plate assay for the determination of enzymatically formed citrulline. Anal Biochem. 2000;286:257–264. doi: 10.1006/abio.2000.4805. [DOI] [PubMed] [Google Scholar]
- 54.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spec. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 55.Tabb DL, McDonald WH, Yates JR. DTASelect and Contrast: Tools for Assembling and Comparing Protein Identifications from Shotgun Proteomics. J Prot Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


