Abstract
Purpose
Tendon injuries are clinically challenging due to poor healing. A better understanding of the molecular events that regulate tendon differentiation would improve current strategies for repair. The mouse model system has been instrumental to tendon studies and several key molecules were initially established in mouse. However, the study of gene function has been limited by the absence of a standard in vitro tendon system for efficiently testing multiple mutations, physical manipulations, and mis-expression. The purpose of this study is therefore to establish such a system.
Methods
We adapted an existing design for generating 3D tendon constructs for use with mouse progenitor cells harboring the ScxGFP tendon reporter and the Rosa26-TdTomato Cre reporter. Using these cells, we optimized parameters for construct formation, inducing tenogenesis via TGFβ2, and genetic recombination via an adenovirus encoding Cre recombinase. Finally, for proof of concept, we used Smad4 floxed cells and tested the robustness of the system for gene knockdown.
Results
We found TGFβ2 treatment induced a tenogenic phenotype depending on timing of initiation. Addition of TGFβ2 after 3D ‘tensioning’ enhanced tendon differentiation. Interestingly, while TGFβ2-induced proliferation depended on Smad4, tenogenic parameters such as ScxGFP expression and fibril diameter were independent of Smad4.
Conclusions
Our results demonstrate the feasibility of this optimized system for harnessing the power of mouse genetics for in vitro applications.
Keywords: tendon biology, tendon tissue engineering, mouse embryonic fibroblasts, TGFβ signaling, Smad signaling
Introduction
Tendons are dense connective tissues that transmit muscle-generated forces to the skeleton. Tendon function is enabled by an extracellular matrix (ECM) primarily composed of aligned type I collagen fibers, which confer the bulk of tensile resistance (1). During embryonic development, tendons are formed as highly cellular structures with small collagen fibrils of uniform diameter. After birth, the ECM undergoes rapid maturation as the collagen matrix transforms from an assembly of small and uniform fibrils to a heterogeneous distribution of large and small fibers by the end of tendon growth (2). Tendon development from embryonic differentiation to postnatal maturation is orchestrated by the resident tenocytes, which in the mature tendon comprise only ~20% of the total tissue volume (3). Although tendon and ligament injuries are extremely common, tissue repair remains a challenge. Surgical intervention remains the standard of care, however clinical outcomes are variable (4–7). A better understanding of the molecular events that regulate tendon differentiation and maturation may enable new therapies that induce innate regenerative healing responses or advance strategies for engineering tissue replacements.
To date, the biological processes underlying tendon differentiation and maturation remain largely unknown. Several of the known regulators of tendon cell fate were originally identified and tested using genetic mouse models (8–11). The ability to engineer the mouse genome, the wide range of genetic resources currently available (including null and conditional mutants), and the presence of long-range tendons anatomically analogous to human makes the mouse a powerful system for developmental and regenerative tendon biology (12–15). In recent years, a number of mouse reagents have been generated for tendon, facilitated by the discovery of key transcription factors for tendon such as Scleraxis (Scx) and Mohawk (9–11, 16). The generation of a robust tendon reporter mouse, ScxGFP, has been especially useful for studies of tendon differentiation, as it enables easy detection of tendon cells (17). Despite these advances, studies of gene function in tendon remain severely limited by the absence of a standard in vitro system to facilitate experiments that combine multiple mutations, physical manipulations and nimble gene mis-expression strategies. One common method to test gene function in cell culture is RNAi, however, efficient delivery of siRNA depends on a variety of factors including cell type, siRNA molecule size and mode of delivery (18). Although transfection is frequently used, transfection of primary mammalian cells can be challenging. Inhibition by siRNA is also transient (limiting culture durations), potentially non-specific, and depending on transcript turnover, knockdown may be incomplete or variable (19). These challenges may be mitigated by the Cre-lox system, which has been widely used to manipulate the mouse genome. This system is attractive due to the availability of existing mouse conditional mutants that have already been generated using this system (20, 21). Cre mediated recombination is also permanent, and recombined cells can be readily distinguished using a variety of robust Cre reporters, such as the Ai14 Rosa26-TdTomato reporter (RosaT) (22).
Although in vitro gene deletions have been carried out using primary cells isolated from conditional mouse mutants to assess tenogenesis, most of these studies tested cells cultured under 2D conditions (23). For tendon, conventional culture systems using 2D plastic adherence is undesirable since tenocytes maintained in 2D conditions rapidly lose expression of characteristic markers (24). More promising are 3D culture systems that mimic the natural mechanical environment of tendons in vivo. In the field of tissue engineering, a number of platforms have emerged for generating such 3D environments (24–26). Although the biomaterial scaffolds and cell types used may vary (27–30), the central conceptual design is fairly consistent. Generally, a linear construct with high cell density is formed such that the construct is anchored between two fixed points and held under tension. In this study, we have adapted a previously established and widely used method from the Kadler lab for generating 3D tendon-like structures (31–34), for use with primary mouse progenitor cells derived from ScxGFP and RosaT embryos. The overall purpose of this study is to establish a tendon system to study gene function using available mouse genetic tools, thereby harnessing the strengths of the mouse system for in vitro studies. Toward this end, we optimized tenogenic differentiation of mouse embryonic fibroblasts (MEFs) in this 3D culture system by TGFβ supplementation, and developed parameters to direct in vitro recombination of conditional mutant cells using an adenovirus encoding Cre recombinase. Using Smad4f/f MEFs as a proof of concept study, we demonstrated efficacy of our optimized platform for testing gene function and showed that while TGFβ-induced proliferation depends on downstream Smad signaling, the tenogenic effects of TGFβ may be independent of Smads.
Methods
Mice
Existing mouse lines used for these studies were previously described: ScxGFP tendon reporter (17), Ai14 Rosa26-TdTomato (RosaT) Cre reporter (22), Scxf/f (9), and Smad4f/f (35). All mice were crossed with ScxGFP to enable visualization of tendon differentiation. Embryos were collected from timed matings to generate cells and control tissues for immunostains. All animal procedures were approved by the Institutional Animal Care and Use Committee and are consistent with animal care guidelines.
Cell isolation and expansion
Mouse embryonic fibroblasts (MEFs) were isolated from E13.5 embryos as previously described (36). Briefly, the head and all viscera were removed from the embryo, followed by fine dicing of the remaining tissues using sterile instruments. The tissue slurry was then incubated in 0.25% Trypsin-EDTA for 10 min at 37°C and plated in culture medium (high glucose DMEM containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS)). After 1–2 passages, MEFs were incubated for 24 hours in culture medium supplemented with 20 ng/mL TGFβ2 (US Biological), and ScxGFP+ cells were isolated by Fluorescence-Activated Cell Sorting (BD FACSVantage Cell Sorter, BD Biosciences). ScxGFP+ sorted cells were then expanded in culture medium for an additional two passages prior to construct formation (Figure 1A). Note that ScxGFP expression in sorted cells is lost after 2D expansion. Embryos from each litter were pooled (including male and female embryos), and experiments were carried out using pooled cells from distinct litters.
Figure 1. MEFs form viable 3D constructs and spontaneously re-express ScxGFP in 3D culture.
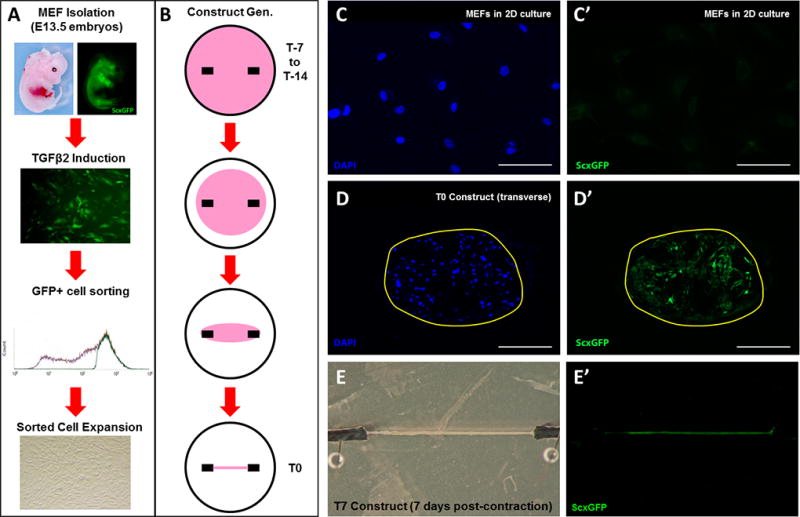
(A) Schematic showing MEF isolation from E13.5 embryos, cell sorting, and expansion. (B) Schematic showing contraction of cell-fibrin gels over a 7 to 14 day period until a linear construct is formed around stationary anchors at T0. (C, C’) MEFs cultured under 2D conditions do not express ScxGFP while (D, D’) transverse section and (E, E’) whole mount images show that MEFs under 3D culture spontaneously re-express ScxGFP. Scalebars: 100 µm.
Construct formation
3D linear constructs were formed as previously established by the Kadler lab (33). Briefly, MEFs (700,000 cells in 400 µL media) were mixed with 83 µL of fibrinogen (20 mg/mL, Sigma), crosslinked with 10 µL thrombin (200 U/mL) and cast into prepared 35 mm plates containing two suture anchors pinned onto Sylgard. Gels were then incubated at 37°C and 5% CO2 for 5 minutes to fully set the gel, scored around the edge to release the gel from the plate walls, and flooded gently with 3 mL of media. Media were changed every 2–3 days and the edges of each gel scored daily until full contraction was achieved (T0 timepoint) and a linear construct formed between the two suture anchors. Since contraction time was generally 7 or 14 days, the time of gel casting is indicated as the T-7 or T-14 timepoint (Figure 1B).
Experimental culture conditions
In Study 1, MEFs isolated from ScxGFP embryos were used to generate 3D constructs or plated on 2D tissue culture plastic. 3D constructs were cultured for 7 additional days following full contraction of the gel (T7 timepoint). To evaluate the effect and timing of TGFβ2 supplementation on construct formation and tenogenic differentiation, constructs were incubated with 10 ng/mL of TGFβ2 (US Biological) prior to construct formation (from T-14 to T10), or after construct formation (from T0 to T10).
In Study 2, effective in vitro recombination of MEFs using an adenovirus for Cre recombinase (Ad-Cre, Vector Biolabs) was evaluated. To determine optimal dosing of Ad-Cre, MEFs were isolated from RosaT embryos, plated on 2D tissue culture plastic and incubated in Ad-Cre at doses ranging from Multiplicity of Infection (MOI) 0–200. Media were changed after 2 days and cells fixed and stained with DAPI at 4 days; cells were then imaged at 5× and efficiency of recombination quantified via cell counting (Tomato+ and total DAPI+ cells within a defined field) with ImageJ Cell Counter. To confirm effective recombination, MEFs were also generated from Scxf/f embryos and Scx gene deletion was carried out using Ad-Cre at MOI 0, 10, and 100. To determine optimal timing of Ad-Cre deletion, Ad-Cre (MOI 10) was added at T-7 or T0 and constructs assessed for Tomato expression at T7. For all experiments in Study 2, cells were incubated with Ad-Cre for 2 days followed by fresh media change.
Finally, in Study 3, a proof of concept study using MEFs isolated from Smad4f/f; RosaT; ScxGFP embryos was carried out to determine whether the effects of TGFβ supplementation on construct growth and differentiation is mediated by Smad4. Using the dose and timing parameters established in Studies 1 and 2, Ad-Cre was added at T-14 and constructs were maintained in 10 ng/mL TGFβ2 from T0 to T14. Constructs were harvested at T0, T3, T7, and T14.
For all studies, constructs and cells were maintained in DMEM containing 10% FBS and 1% PS. Tenogenic differentiation was determined by ScxGFP expression, immunostaining, histology, in situ hybridization and transmission electron microscopy (TEM) analysis.
Immunofluorescence and histology
Constructs were fixed overnight at 4°C in 4% paraformaldehyde (PFA), rinsed in phosphate buffered saline and embedded in 2% w/v agarose to facilitate visibility and positioning of sample during embedding and sectioning. The construct/agarose block was embedded in OCT medium and 12 µm cryosections were generated for immunofluorescence staining and histology. Immunostaining was carried out using the Mouse on Mouse kit (Vector Laboratories) according to manufacturer’s directions. Antibodies for collagen type I (AB765P, Millipore, 1:40 dilution) and collagen type II (II-II6B3, Developmental Hybridoma Bank, 1:100 dilution) were used to distinguish fibrous and cartilaginous collagens, respectively. The secondary antibody used for detection was streptavidin-conjugated Cy3 (016-160-084, Jackson Laboratories, 1:400 dilution) and DAPI counterstaining was used to visualize cell nuclei. Cell number was determined by counting DAPI+ nuclei in transverse sections using the ImageJ Cell Counter plugin tool. Cross-sectional area was also determined from transverse sections by measuring the major and minor radii and calculating the area for an ellipse. To minimize variability due to section location, four sections were counted for each construct and then averaged. Quantitative measurements are reported for n=3–4 constructs at T0 and T14. To stain for glycosaminoglycans, cryosections were stained with Alcian Blue (pH 1.0). To identify apoptotic cells, TUNEL assays were performed on cryosections using the In Situ Cell Death Detection kit (Roche) according to manufacturer’s instructions.
Fluorescence microscopy
Fluorescence imaging of cryosections was carried out using the Zeiss AxioImager with optical sectioning by Apotome and the Zeiss Zen software. Whole mount imaging of constructs was carried out after overnight fixation in 4% PFA at 4°C. Whole mount images were captured using either a Leica stereomicroscope fitted with GFP filter, or using laser scanning confocal microscopy (Zeiss LSM780 with Zen software). Cell images were captured using a Leica DMIL LED inverted fluorescence microscope with Texas Red and UV filters. Images were captured at 5×, 10×, or 20× magnification.
In situ hybridization
In situ hybridization was performed on cryosections as previously described (37). Briefly, cryosections were subjected to proteinase K treatment, post-fixation in PFA and acetylation. Hybridization with digoxygenin-labeled probe against Col1a1 RNA was performed overnight at 65°C. Post-hybridization, slides were rinsed in a 5× saline-sodium citrate buffer (SSC) at 65°C, washed with 1× SSC/50% formamide, and unbound probe was digested using RNase A. Additional washes in 2× and 0.2× SSC buffers were carried out at 65°C, and bound probe detected using NBT/BCIP.
Transmission electron microscopy
Constructs were prepared for electron microscopy by immersion in 1.5% glutaraldehyde/1.5% PFA (Tousimis Research Corporation, Rockville, MD) in Dulbecco’s serum-free media (SFM) containing 0.05% tannic acid. Constructs were further washed in SFM followed by post-fixation in 1% OsO4 and further washing in SFM. Samples were then gradually dehydrated in ethanol to 100%, rinsed in propylene oxide, and infiltrated in Spurrs epoxy and polymerized at 70°C over 18 hours. Micrographs were obtained in transverse and longitudinal sections to visualize collagen fibril diameter, alignment, and density. For morphometric analysis, fibril diameters were measured using Image J (n=200 fibrils/group).
Statistical analysis
Results are presented as mean ± standard deviation. All statistical analyses were carried out using one way ANOVA with Bonferroni posthoc testing. Significant differences were detected at p<0.05.
Results
Purified ScxGFP MEFs generated constructs with tendon-like features under 3D culture
To evaluate whether MEFs are able to form viable 3D constructs, we isolated MEFs from E13.5 ScxGFP embryos and encapsulated the cells into fibrin gels as described by the Kadler Lab (33). Interestingly, the MEFs were unable to form a viable, linear construct; most gels were completely torn within 2–3 days of casting, likely due to uneven contraction (not shown). Despite their name, MEFs are a heterogeneous population of cells comprising not only fibroblastic cells, but also cells from diverse lineages, including myogenic, chondrogenic, neurogenic and vascular lineages. To restrict the population of cells to only those with tenogenic potential, we induced expression of the tendon reporter, ScxGFP, in responsive cells using TGFβ2, and isolated the GFP+ fraction by FACS (Figure 1A, B). TGFβ ligand was chosen as it is well established that TGFβ signaling is essential for tendon formation, as deletion of either the ligands (TGFβ2 and TGFβ3) or the receptor (TβR2) results in a complete loss of tendons early in embryonic development (8). TGFβ signaling is also a strong inducer of the tendon cell fate, which has been shown both in limb organ culture and in mesenchymal stem cells (8, 38, 39). After sorting, this purified population of MEFs reproducibly formed linear constructs, requiring 7 to 14 days for full contraction around the anchors. Compared to cells cultured in parallel on 2D tissue culture plastic (in which ScxGFP expression of sorted cells is lost after 2D expansion), MEFs within 3D constructs spontaneously expressed higher levels of ScxGFP at T0, although the level of ScxGFP expression was not consistent between all cells within the construct (Figure 1C–D’). Whole mount images showed that ScxGFP expression was observed throughout the construct length and was not localized to any specific location (Figure 1E, E’). For all subsequent studies, we therefore induced and enriched for ScxGFP+ MEFs via FACS, prior to experimentation.
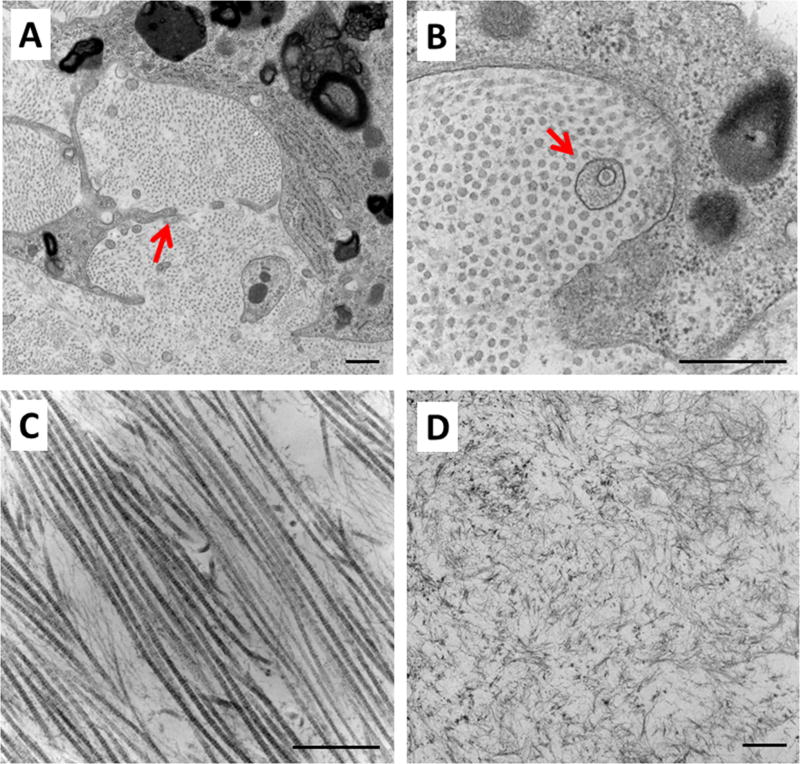
TEM analysis of constructs at T7 revealed several hallmarks of tendon cell identity, including cell protrusions resembling those of native tenocytes, as well as aligned collagen fibrils and the presence of fibropositors (Figure 2A–C). However, the collagen fibrils were uniformly small in diameter, indicating an immature state. There were also regions of disorganized collagen, suggesting possible local, aberrant differentiation or non-uniform loading through the construct (Figure 2D).
Figure 2. MEFs in 3D constructs deposit tendon-like matrix.
TEM images revealed several features characteristic of embryonic tendon, including (A) cell protrusions surrounding collagen fibrils, (B) collagen fibrils of small uniform size, fibropositors, and (C) aligned collagen fibrils organized in the direction of tension. (D) However, there were also regions of disorganized collagen. Red arrow in (A) and (B) highlight cell protrusion and fibropositor, respectively. Scalebars: 500 nm.
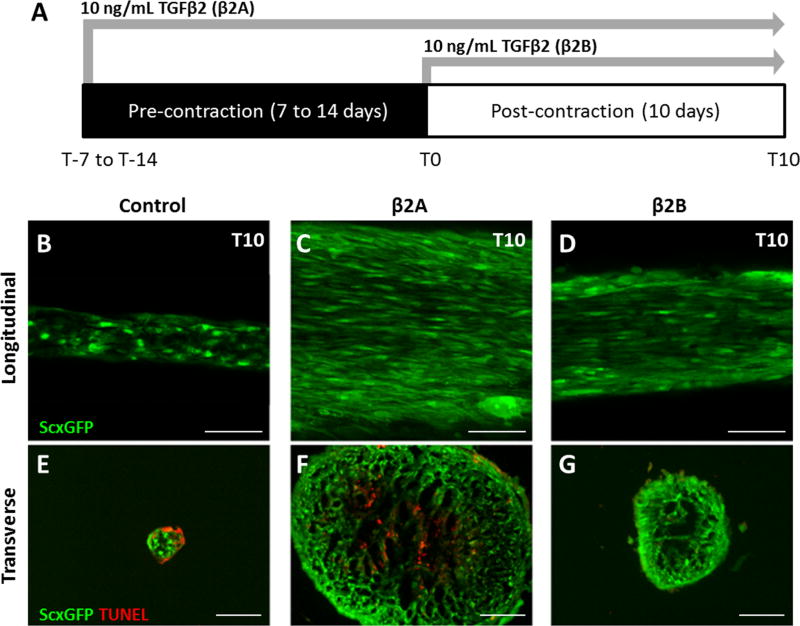
Study 1: TGFβ2 enhanced tenogenic differentiation of MEFs in 3D culture depending on time of initiation
To enhance tenogenic differentiation of MEFs in 3D culture, we tested the tenogenic capacity of TGFβ in this context, and initiated TGFβ2 supplementation either immediately after gel casting at T-7 (group β2A) or after construct formation at T0 (group β2B) (Figure 3A). Ten days after construct formation (T10), whole mount and transverse sections showed that all TGFβ2 treated constructs appeared qualitatively larger compared to non-treated control constructs, although the β2A group was the most robust in size. Whole mount imaging further showed highly aligned and elongated cells with strong expression of ScxGFP in both TGFβ2 treatment groups (Figure 3B–D). In contrast, ScxGFP expression in the control construct was less uniform and cells appeared relatively rounded. Transverse sections stained with TUNEL for apoptotic cells showed cell death in both the control and β2A groups, with no evidence of cell death in the β2B group (Figure 3E–G). Interestingly, while apoptotic cells were observed in the periphery of control constructs, TUNEL positive cells were largely localized to the center of β2A constructs.
Figure 3. TGFβ2 supplementation improves tenogenic differentiation.
(A) Schematic of study design. Whole mount longitudinal images of (B) control, (C) immediate β2A, and (D) delayed β2B constructs showed enhanced ScxGFP expression, aligned cells, and elongated cell morphology in the presence of TGFβ2 relative to control, regardless of timing. Tunel staining of transverse sections showed cell death in (E) control and (F) β2A constructs at T10, but not (G) β2B constructs. While apoptotic cells were localized to the periphery of control constructs, apoptotic cells were localized in the center of β2A constructs. Scalebars: 100 µm.
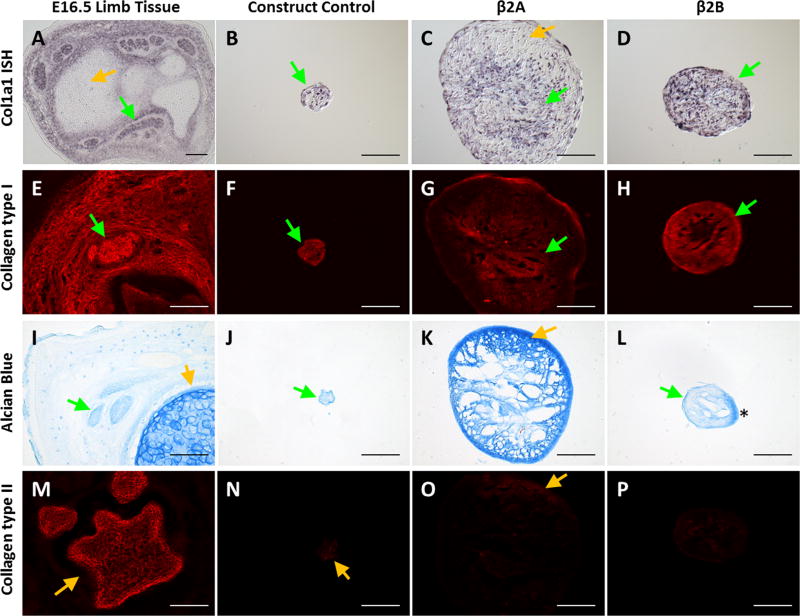
In situ hybridization for Col1a1 showed intense and homogeneous expression in β2B constructs, compared to the more sporadic expression in controls. While Col1a1 expression was detected in cells throughout β2A constructs, expression was highly localized to the center regions, in contrast to ScxGFP expression, which was detected mostly in the periphery (Figure 4A–D). Regional specificity was confirmed by immunostaining for type I collagen, which showed a similar distribution. While immunostaining for type I collagen was robust throughout β2B constructs, staining was again centrally localized in β2A (Figure 4E–H). Since TGFβ ligands are well known to be strong chondrogenic inducers in vitro, we also evaluated potential differentiation toward chondrogenesis using Alcian Blue staining for proteoglycans and immunostaining for type II collagen. Alcian Blue staining of E16.5 embryonic tissue was strong in cartilage (orange arrow), but relatively weak in tendons (green arrow, Figure 4I). Interestingly, intense Alcian Blue staining was observed in β2A constructs, at levels comparable to embryonic cartilage. Staining was localized to outer regions that were coincident with low type I collagen staining and Col1a1 expression. Alcian Blue staining was uniformly light in both controls and β2B constructs, at levels similar to embryonic tendon (Figure 4I–L). However, while type II collagen immunostaining was intense in embryonic cartilage, there was little to no staining in any of the construct groups (Figure 4M–P).
Figure 4. Delayed application of TGFβ2 results in improved tenogenic phenotype.
(A–D) Col1a1 in situ hybridization, (E–H) Collagen type I immunostaining, (I–L) Alcian Blue staining, and (M–P) Collagen type II immunostaining of transverse cryosections taken from E16.5 embryonic limbs, control constructs, immediate β2A constructs, and delayed β2B constructs. While control and β2B constructs showed minimal Alcian blue staining and uniform Collagen type I immunostaining, and Col1a1 expression, β2A constructs showed intense Alcian Blue staining comparable to embryonic cartilage; Collagen type I immunostaining and Col1a1 expression was primarily localized to the center of β2A constructs. Minimal Collagen type II immunostaining was observed for all constructs. Green arrows highlight tendon or tendon-like staining while yellow arrows highlight cartilage or cartilage-like staining. * shows artificial staining due to section folding artifact. Scalebars: 100 µm.
Collectively, this data indicates that initiation of TGFβ2 supplementation directly after gel casting resulted in constructs with tenogenic features, but high proteoglycan content and centralized cell death. In contrast, initiation of TGFβ2 at T0 (after cells have formed a linear construct) resulted in constructs with homogeneous tenogenesis, low proteoglycan staining and no cell death. TGFβ2 was therefore initiated at T0 for subsequent studies.
Study 2: Optimization and validation of adenovirus Cre-mediated gene deletion in 2D and 3D culture
Having established an effective protocol for tenogenesis, we next tested parameters (dose and timing) necessary to achieve effective recombination of cells in 3D culture using an adenovirus expressing Cre recombinase (Ad-Cre). For this experiment, MEFs were isolated from RosaT embryos, so that recombined cells could be easily detected by visualization of the Tomato reporter. As expected, Cre-mediated recombination of cells in 2D culture depended on the dose of Ad-Cre. Since no cells were recombined in the absence of Ad-Cre (MOI 0), quantification was carried out for MOI 0.5 and above. We found a progressive increase in the number of Tomato+ cells with increasing Ad-Cre dose (MOI 0.5–200), with the highest recombination rate observed at MOI 100 (p<0.001, Figure 5A, B). Recombination rate was not improved with MOI 200 (p>0.1 vs MOI 100); therefore, MOI 100 was chosen for subsequent studies. In addition, Ad-Cre did not affect cell viability since total cell numbers were similar across all samples (Figure 5C). To further confirm functional deletion by Ad-Cre, we also used MEFs derived from Scxf/f embryos and evaluated recombination by PCR amplification for Scx−/− DNA. Consistent with the cell counting results, we did not detect any Scx−/− PCR product in the absence of Ad-Cre (MOI 0). However weak bands (of comparable size to Scx−/− control tissue) were detected at the low dose (MOI 10) while strong bands were detected at the high dose (MOI 100), indicating functional, dose-dependent recombination of the floxed Scx gene by Ad-Cre (Figure 6A).
Figure 5. Optimizing dosage for Ad-Cre mediated recombination in MEFs.
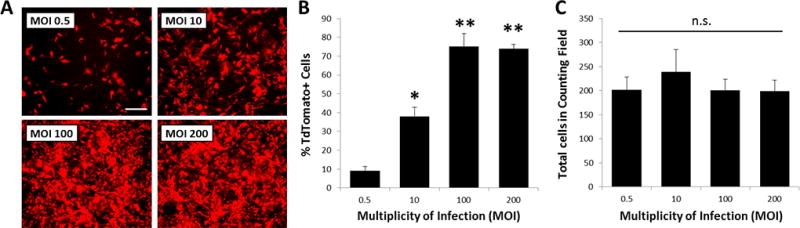
(A, B) Increasing Ad-Cre MOI on RosaT MEFs results in dose-dependent increase in number of Tomato expressing cells, with maximum recombination achieved at MOI 100. (C) Total DAPI+ cell number was not affected with increasing Ad-Cre dose. * indicates p<0.05 compared to all; ** indicates p<0.05 compared to MOI 0.5 and 10; n.s. indicates p>0.1 for all comparisons (n=6/group). Cell images taken at 5× magnification. Scalebar: 100 µm.
Figure 6. Optimizing timing for Ad-Cre mediated recombination in fibrin gels.
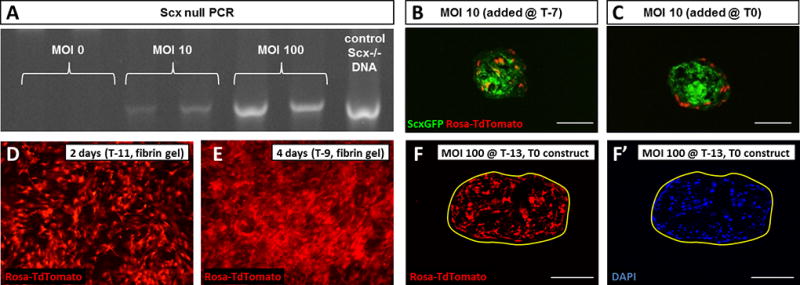
(A) Increasing Ad-Cre MOI on Scxf/f MEFs results in dose-dependent increase in Scx−/− DNA, indicating functional recombination by Ad-Cre. (B) Immediate addition of Ad-Cre results in uniform distribution of recombined cells within constructs compared to (C) delayed addition after construct formation. MEFs express the Tomato reporter (D) 2 and (E) 4 days after Ad-Cre induction with greater numbers of recombined cells observed at 4 day compared to 2 day. (F) Tomato expression and (F’) DAPI staining showed excellent recombination of cells within construct with no adverse effect on construct formation by Ad-Cre. Cell images were taken at 4× magnification. Scalebars: 100 µm.
To determine optimal timing, we used a low dose of Ad-Cre (MOI 10) and initiated recombination directly after gel casting (T-7) or after construct formation (T0). The localization and distribution of Tomato+ cells was visualized after an additional 7 days of culture (T7). When cells were recombined with Ad-Cre directly after gel casting, Tomato+, recombined cells were detected throughout the construct at T7. In contrast, when Ad-Cre was added at T0, only the cells in the construct periphery were Tomato+, suggesting inadequate penetration by Ad-Cre after construct formation (Figure 6B, C).
Finally, we tested our optimized parameters for dose (MOI 100) and timing (T-7) on MEFs in 3D culture, to evaluate recombination efficiency and construct formation under high Ad-Cre exposure. Since Ad-Cre is non-replicating, exposure of the virus was transient (2 days), however new cells expressing the Tomato reporter were detected up to 4 days (Figure 6D, E). This was likely due to the lag time between infection and subsequent Cre expression, recombination, and Tomato transcription. Construct formation was not impaired by high Ad-Cre exposure as all constructs formed successfully, within a standard contraction time of 7 to 14 days. Transverse sections of T0 constructs showed a large number of Tomato+ cells throughout the construct (Figure 6F, F’), further confirming effective recombination by Ad-Cre using the optimized parameters.
Study 3: Smad4 deletion abolished TGFβ-induced cell proliferation but did not affect tendon differentiation
In Study 1, we showed that delayed initiation of TGFβ2 resulted in constructs with larger diameter and enhanced expression of tenogenic markers (ScxGFP and Col1a1). Since canonical TGFβ signaling is mediated by two receptor Smads (Smad2 and Smad3) and the common Smad (Smad4), we decided to delete Smad signaling as a proof of concept study to test the efficacy of our system for interrogating gene function. Since Smad2 and Smad3 may be functionally redundant, we chose to use MEFs derived from homozygous Smad4f/f embryos for simplicity.
Combining our optimized parameters for TGFβ supplementation and Ad-Cre dose and timing, we formed constructs using Smad4f/f MEFs, and cultured the constructs under the following conditions: control media (Wild Type, WT), TGFβ2 supplementation (TGFβ2), Ad-Cre only (Mutant, Mut), and Ad-Cre with TGFβ2 supplementation (Mut-TGFβ2). TGFβ2 was used to induce tenogenesis and Ad-Cre was used to generate Smad4−/− mutant cells. Both WT and Mut groups formed viable constructs within 14 days and transverse sections at T0 showed that WT and Mut constructs were indistinguishable. Constructs were then cultured for an additional 14 days after construct formation, and harvested at T3, T7 and T14. Surprisingly, we found a dramatic reduction in the size of WT constructs from T0 to T14. To evaluate whether this reduction was due to contraction of cells or loss of cells, we counted cell number in representative transverse sections using DAPI to highlight cell nuclei and found a significant reduction in cell number by T14 (Figure 7). In contrast, supplementation with TGFβ2 resulted in increased cell number at T14 (p=0.001 vs WT) (Figure 8A). While ScxGFP levels remained uniform across timepoints for WT constructs, there was a gradual increase in ScxGFP expression in TGFβ2 constructs from T0 to T7. Although DAPI staining revealed the presence of cells in the central regions at T14, intense ScxGFP expression was largely restricted to outer regions (Figure 8B–G).
Figure 7. Construct cell number declines with time in the absence of TGFβ2.
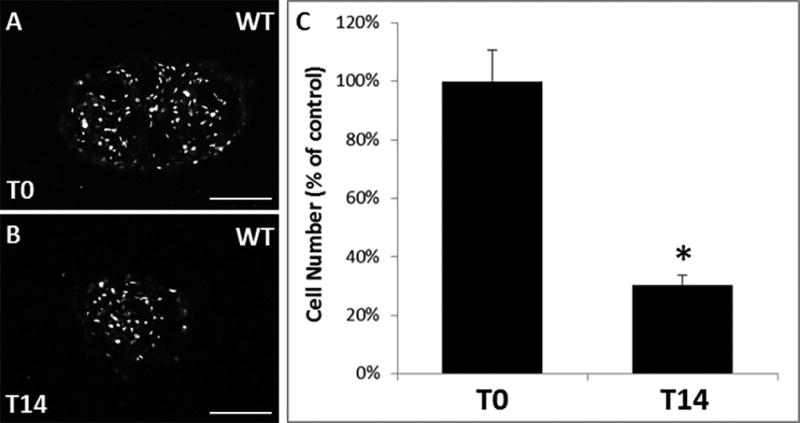
DAPI staining of (A) WT control construct at T0 and (B) WT control construct at T14. (C) Average cell number decreases from T0 to T14 in WT constructs (percentage of T0 control). * indicates p<0.05 compared to T0 control. Scalebars: 100 µm.
Figure 8. Smad4 mutant cells lose proliferative capacity in the presence of TGFβ2 but maintain enhanced ScxGFP expression.
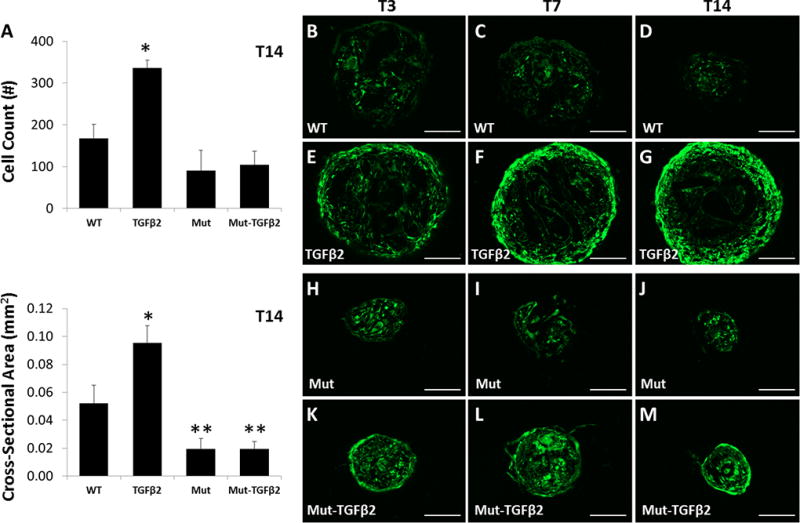
(A) Cell number and cross-sectional area measurements for constructs at T14. ScxGFP expression of transverse sections from (B–D) WT control, (E–G) WT TGFβ2, (H–J) Smad4 Mut control, and (K–M) Smad4 Mut TGFβ2 constructs from T0 to T14. ScxGFP expression was enhanced at T7 and T14 compared to respective cell control groups. * indicates significant increase compared to all other groups, ** indicates significant decrease compared to WT control (p<0.001) (n=3–4 group). Scalebars: 100 µm.
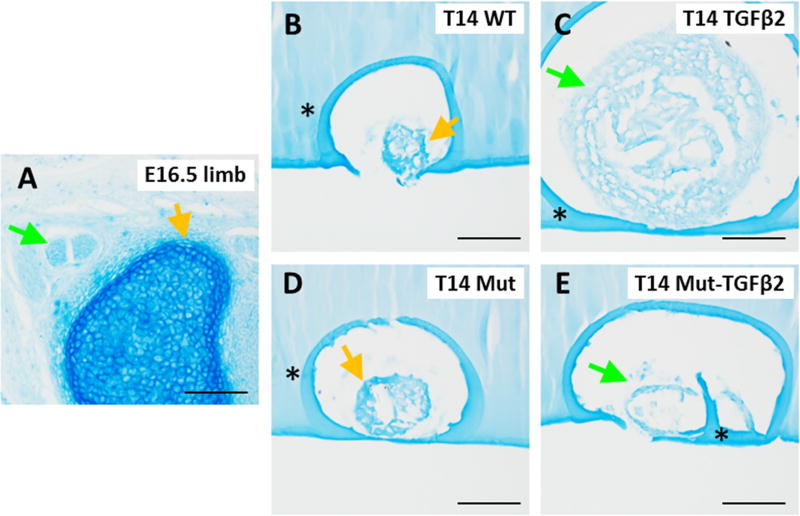
Interestingly, Smad4 Mut constructs failed to grow in response to TGFβ2 stimulation, since cell number of Mut-TGFβ2 were comparable to WT and Mut control groups (p>0.05), indicating that the proliferative activity of TGFβ2 in this context is likely mediated by Smad signaling (Figure 8A). Although cell number was not significantly different for any group except TGFβ2-treated, the cross-sectional area was sigificantly lower for both Mut and Mut-TGFβ2 relative to control, which may suggest reduced matrix deposition with loss of Smad4 function (Figure 8A). Despite the loss of cell proliferation, ScxGFP expression was enhanced in Mut-TGFβ2 compared to Mut constructs, and the peripheral cells were organized circumferentially around the construct, a pattern which was also observed in TGFβ2 constructs (Figure 8B–M). Alcian blue staining at T14 revealed stronger staining in the non-treated WT and Mut groups compared to either TGFβ treated group (Figure 9A–E).
Figure 9. TGFβ2 inhibits proteoglyan deposition.
Alcian Blue staining of transverse cryosections from (A) E16.5 embryonic limb, (B, C) WT and Smad4 mutant (D, E) constructs showed less intense staining in TGFβ2 treated constructs compared to non-treated controls. Green arrows highlight tendon or tendon-like staining while yellow arrows highlight cartilage or cartilage-like staining. * indicates background staining of agarose used to embed constructs. Scalebars: 100 µm.
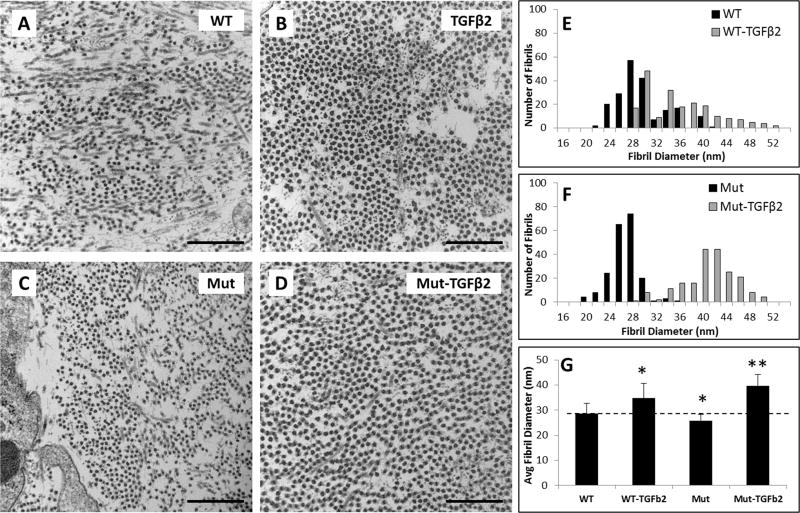
To evaluate matrix deposition, TEM imaging was carried out for T14 samples. Transverse micrographs showed that collagen fibrils were relatively homogeneous within each group, indicative of an immature matrix phenotype, however fibril density appeared higher in TGFβ2 treated groups (Figure 10A–D). Quantification of fibril diameter revealed increased diameter with TGFβ2 treatment for both WT and Mut groups relative to controls. Interestingly, while control Mut fibrils were smaller in size compared to control WT (p<0.001), Mut-TGFβ2 fibrils were significantly larger compared to WT TGFβ2 fibrils (p<0.001) (Figure 10E–G).
Figure 10. TGFβ2 improves collagen fibril diameter in both WT and Smad4 mutant constructs.
TEM images of (A) WT, (B) TGFβ2, (C) Smad4 Mut, and (D) Smad4 Mut-TGFβ2 constructs showed increased collagen fibril density in TGFβ2 treated groups and larger fibril diameter. Histograms show collagen fibril diameter distribution in (E) WT and (F) Mut groups in response to TGFβ2. (G) Average fibril diameter increased for both TGFβ2 groups, but was highest in Mut-TGFβ2 constructs. * indicates p<0.05 compared to WT, ** indicates p<0.05 compared to all groups. Scalebars: 500 nm.
Discussion
In this study, we established an in vitro system for testing tendon differentiation and maturation that combines the strengths of the mouse model system with design principles from tissue engineering. Using a previously described method for generating linear cell-fibrin constructs (33), we optimized conditions for the use of mouse progenitor cells derived from embryos containing alleles for ScxGFP and RosaT and tested parameters for inducing tenogenesis by TGFβ2 and Smad4 gene deletion by Ad-Cre.
The capacity of TGFβ to induce both chondrogenic and tenogenic differentiation is supported by numerous in vivo and in vitro reports in the literature. In vivo, the loss of TGFβ signaling via deletion of either ligands (TGFβ2 and TGFβ3) or the type II receptor (TβR2) results in a complete loss of tendons (8). Interestingly, recent studies indicate that TGFβ signaling induces an early pool of progenitors that expresses both Sox9 (chondrogenic) and Scx (tenogenic) and that subsequent allocation of these progenitors to cartilage or tendon depends on signals that suppress either Sox9 or Scx in these cells (40, 41). In vitro, TGFβ has long been used to induce chondrogenesis; in this context, cell morphology plays a crucial role and chondrogenic protocols are typically carried out in high density cultures where cells are maintained in a rounded morphology (42, 43). When cells are allowed to adopt an elongated morphology, TGFβ induces differentiation toward fibrocartilage (44). Although type II collagen staining was not apparent in any constructs, proteoglycan staining was enhanced when TGFβ was added immediately after gel casting, suggesting potential differentiation toward chondrogenesis. Although we found minimal proteoglycan staining with delayed addition of TGFβ2, it may be that longer culture durations will eventually result in mixed phenotypes. Future studies will therefore evaluate the effect of long-term culture duration and apply cyclic mechanical loading to further improve tenogenesis (24, 45). Since we also observed centralized cell death once constructs reached a certain size (although diameter and cross-sectional areas were not measured in this study), mechanical stimulation may also improve nutrient diffusion and enable sustained construct growth. Interestingly, cell death was also observed at the periphery of control constructs after 10 days of culture (T10) in the absence of TGFβ2 supplementation. We also found that there was a significant loss of cells from the time of construct formation at T0 to T14 of culture. The smaller size of control constructs and peripheral location of cell death suggests that inadequate nutrient diffusion is likely not the cause of cell death. It may be that TGFβ2 supplementation exerts a protective effect on the cells, since cell death was not observed in the delayed TGFβ2 condition. TGFβ2 also induced a strong proliferative and matrix synthesis response regardless of timing, which is consistent with other in vitro studies using TGFβ ligands (28, 29, 46, 47).
To optimize parameters for gene knockdown using the Cre/lox system, we used MEFs derived from RosaT embryos (to facilitate detection of cell recombination by Tomato expression) and induced recombination by Ad-Cre. We found that the proportion of wild type and mutant cells can be controlled by Ad-Cre dose and that Ad-Cre dose correlated with functional recombination of the Scx gene when Scxf/f cells were used. While the focus of this study was to generate the largest number of mutant cells with minimal toxicity, the ability to form constructs with mixed mutant and wild type populations could be a powerful tool for studying cell-cell interactions in vitro. Finally, we also tested the robustness of our system for gene function studies by applying our optimized parameters for inducing tenogenesis and recombination to MEFs generated from Smad4f/f embryos. While TGFβ2 induced cell proliferation in wild type cells, increases in cell number were not observed in Smad4 mutant constructs, indicating that the proliferative effects of TGFβ2 supplementation is mediated by Smad4 signaling. Further, cell numbers in Mut-TGFβ2 constructs at T14 were comparable to control groups, which may suggest that the protective effect of TGFβ2 also depends on Smad4.
Surprisingly, the tenogenic effects of TGFβ2 treatment were not affected by loss of Smad4. ScxGFP expression remained enhanced in Smad4 mutant constructs with TGFβ2 treatment, proteoglycan staining was low, and collagen fibril diameter was enhanced. Although canonical TGFβ signaling is mediated by Smads, TGFβ can also activate a number of non-Smad pathways (48). Our data indicates that the tenogenic activity of TGFβ may be mediated by a non-canonical pathway. In addition, loss of Smad4 may also inhibit BMP signaling since Smad4 is also required in this context. It’s been shown that BMP inhibits tenogenesis and loss of Smad4 in embryonic limbs results in expansion of the Scx expression domain, which is likely due to loss of BMP signaling (16, 49). The extent to which these in vitro findings will correlate to in vivo biology must be confirmed through parallel loss of function experiments in vivo; however this is a limitation with all in vitro culture systems. Despite this caveat, our in vitro system holds enormous potential for identifying new molecules that regulate critical aspects of tendon differentiation and maturation.
One important limitation with our system is the poor penetration by Ad-Cre once a linear construct had formed. Since the ideal system would permit gene deletion at any point during construct culture, future studies will test strategies to overcome this limitation. One solution may be mechanical loading to facilitate penetration of Ad-Cre, however we can also test inducible systems that utilize smaller molecules such as 4-hydroxytamoxifen (6–10 nm compared to Ad-Cre which is 100 nm), by incorporating a ubiquitously expressed, inducible CreERT2 allele into the background of cells. The current study also focused primarily on gene knockdown, however the ability to mis-express genes would enable a wider range of experiments. Toward that end, utilizing MEFs that express the avian tumor virus receptor A (TVA) would allow for infection of mouse cells by chick retroviral RCAS vectors. Since a number of TVA expressing mice have already been generated, adapting the current system for use with TVA expressing MEFs should be relatively straightforward and is the focus of current efforts. Finally, although MEFs were used here, other cell types with greater clinical potential (such as mesenchymal stem cells or induced pluripotent cells) can also be derived from the same mutant lines and tested within this system (32, 34).
Acknowledgments
Funding Sources
This work was supported by funding from NIH/NIAMS (R01 AR069537) and the Icahn School of Medicine at Mount Sinai to A.H.H.
Acknowledgements and Funding Sources
We thank Dr. Ronen Schweitzer and Dr. Karl Kadler for helpful discussions, advice, and support. The type II collagen antibody developed by T. F. Lisenmayer was obtained from the Developmental Studies Hybridoma Bank.
Footnotes
Declaration of Interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- 1.Zhang G, Young BB, Ezura Y, Favata M, Soslowsky LJ, Chakravarti S, Birk DE. Development of tendon structure and function: regulation of collagen fibrillogenesis. J Musculoskelet Neuronal Interact. 2005;5(1):5–21. [PubMed] [Google Scholar]
- 2.Benjamin M, Kaiser E, Milz S. Structure-function relationships in tendons: a review. J Anat. 2008;212(3):211–28. doi: 10.1111/j.1469-7580.2008.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benjamin M, Ralphs JR. The cell and developmental biology of tendons and ligaments. Int Rev Cytol. 2000;196:85–130. doi: 10.1016/s0074-7696(00)96003-0. [DOI] [PubMed] [Google Scholar]
- 4.United States Bone and Joint Initiative. The Burden of Musculoskeletal Disease in the United States, Second Edition. Rosemont, IL: American Academy of Orthopaedic Surgeons; 2011. [Google Scholar]
- 5.Thompson S, Salmon L, Waller A, Linklater J, Roe J, Pinczewski L. Twenty-Year Outcomes of a Longitudinal Prospective Evaluation of Isolated Endoscopic Anterior Cruciate Ligament Reconstruction With Patellar Tendon Autografts. Am J Sports Med. 2015;43(9):2164–74. doi: 10.1177/0363546515591263. [DOI] [PubMed] [Google Scholar]
- 6.Negrin LL, Nemecek E, Hajdu S. Extensor mechanism ruptures of the knee: Differences in demographic data and long-term outcome after surgical treatment. Injury. 2015 doi: 10.1016/j.injury.2015.06.042. [DOI] [PubMed] [Google Scholar]
- 7.Shon MS, Koh KH, Lim TK, Kim WJ, Kim KC, Yoo JC. Arthroscopic Partial Repair of Irreparable Rotator Cuff Tears: Preoperative Factors Associated With Outcome Deterioration Over 2 Years. Am J Sports Med. 2015;43(8):1965–75. doi: 10.1177/0363546515585122. [DOI] [PubMed] [Google Scholar]
- 8.Pryce B, Watson S, Murchison N, Staverosky J, Dünker N, Schweitzer R. Recruitment and maintenance of tendon progenitors by TGFbeta signaling are essential for tendon formation. Development (Cambridge, England) 2009;136(8):1351–61. doi: 10.1242/dev.027342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murchison N, Price B, Conner D, Keene D, Olson E, Tabin C, Schweitzer R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development (Cambridge, England) 2007;134(14):2697–708. doi: 10.1242/dev.001933. [DOI] [PubMed] [Google Scholar]
- 10.Ito Y, Toriuchi N, Yoshitaka T, Ueno-Kudoh H, Sato T, Yokoyama S, Nishida K, Akimoto T, Takahashi M, Miyaki S, Asahara H. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci U S A. 2010;107(23):10538–42. doi: 10.1073/pnas.1000525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu W, Watson S, Lan Y, Keene D, Ovitt C, Liu H, Schweitzer R, Jiang R. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Molecular and cellular biology. 2010;30(20):4797–807. doi: 10.1128/MCB.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang AH, Lu HH, Schweitzer R. Molecular regulation of tendon cell fate during development. J Orthop Res. 2015 doi: 10.1002/jor.22834. [DOI] [PubMed] [Google Scholar]
- 13.Watson S, Riordan T, Pryce B, Schweitzer R. Tendons and muscles of the mouse forelimb during embryonic development. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238(3):693–700. doi: 10.1002/dvdy.21866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glaser S, Anastassiadis K, Stewart AF. Current issues in mouse genome engineering. Nat Genet. 2005;37(11):1187–93. doi: 10.1038/ng1668. [DOI] [PubMed] [Google Scholar]
- 15.Huang A, Riordan T, Wang L, Eyal S, Zelzer E, Brigande J, Schweitzer R. Repositioning forelimb superficialis muscles: tendon attachment and muscle activity enable active relocation of functional myofibers. Developmental cell. 2013;26(5):544–51. doi: 10.1016/j.devcel.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schweitzer R, Chyung JH, Murtaugh LC, Brent AE, Rosen V, Olson EN, Lassar A, Tabin CJ. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development. 2001;128(19):3855–66. doi: 10.1242/dev.128.19.3855. [DOI] [PubMed] [Google Scholar]
- 17.Pryce B, Brent A, Murchison N, Tabin C, Schweitzer R. Generation of transgenic tendon reporters, ScxGFP and ScxAP, using regulatory elements of the scleraxis gene. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236(6):1677–82. doi: 10.1002/dvdy.21179. [DOI] [PubMed] [Google Scholar]
- 18.Reischl D, Zimmer A. Drug delivery of siRNA therapeutics: potentials and limits of nanosystems. Nanomedicine. 2009;5(1):8–20. doi: 10.1016/j.nano.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9(7):554–66. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 20.Argos P, Landy A, Abremski K, Egan JB, Haggard-Ljungquist E, Hoess RH, Kahn ML, Kalionis B, Narayana SV, Pierson LS, 3rd, et al. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5(2):433–40. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14(4):381–92. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 22.Madisen L, Zwingman T, Sunkin S, Oh S, Zariwala H, Gu H, Ng L, Palmiter R, Hawrylycz M, Jones A, Lein E, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13(1):133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wada S, Ideno H, Shimada A, Kamiunten T, Nakamura Y, Nakashima K, Kimura H, Shinkai Y, Tachibana M, Nifuji A. H3K9MTase G9a is essential for the differentiation and growth of tenocytes in vitro. Histochem Cell Biol. 2015;144(1):13–20. doi: 10.1007/s00418-015-1318-2. [DOI] [PubMed] [Google Scholar]
- 24.Kuo CK, Tuan RS. Mechanoactive tenogenic differentiation of human mesenchymal stem cells. Tissue engineering Part A. 2008;14(10):1615–27. doi: 10.1089/ten.tea.2006.0415. [DOI] [PubMed] [Google Scholar]
- 25.Legant W, Pathak A, Yang M, Deshpande V, McMeeking R, Chen C. Microfabricated tissue gauges to measure and manipulate forces from 3D microtissues. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(25):10097–102. doi: 10.1073/pnas.0900174106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler D, Juncosa-Melvin N, Boivin G, Galloway M, Shearn J, Gooch C, Awad H. Functional tissue engineering for tendon repair: A multidisciplinary strategy using mesenchymal stem cells, bioscaffolds, and mechanical stimulation. Journal of orthopaedic research : official publication of the Orthopaedic Research Society. 2008;26(1):1–9. doi: 10.1002/jor.20456. [DOI] [PubMed] [Google Scholar]
- 27.Bayer ML, Yeung CY, Kadler KE, Qvortrup K, Baar K, Svensson RB, Magnusson SP, Krogsgaard M, Koch M, Kjaer M. The initiation of embryonic-like collagen fibrillogenesis by adult human tendon fibroblasts when cultured under tension. Biomaterials. 2010;31(18):4889–97. doi: 10.1016/j.biomaterials.2010.02.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breidenbach AP, Dyment NA, Lu Y, Rao M, Shearn JT, Rowe DW, Kadler KE, Butler DL. Fibrin Gels Exhibit Improved Biological, Structural, and Mechanical Properties Compared with Collagen Gels in Cell-Based Tendon Tissue-Engineered Constructs. Tissue Eng Part A. 2014 doi: 10.1089/ten.tea.2013.0768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farhat Y, Al-Maliki A, Chen T, Juneja S, Schwarz E, O'Keefe R, Awad H. Gene expression analysis of the pleiotropic effects of TGF-β1 in an in vitro model of flexor tendon healing. PloS one. 2012;7(12) doi: 10.1371/journal.pone.0051411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calve S, Dennis RG, Kosnik PE, 2nd, Baar K, Grosh K, Arruda EM. Engineering of functional tendon. Tissue Eng. 2004;10(5–6):755–61. doi: 10.1089/1076327041348464. [DOI] [PubMed] [Google Scholar]
- 31.Kalson NS, Holmes DF, Herchenhan A, Lu Y, Starborg T, Kadler KE. Slow stretching that mimics embryonic growth rate stimulates structural and mechanical development of tendon-like tissue in vitro. Dev Dyn. 2011;240(11):2520–8. doi: 10.1002/dvdy.22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapacee Z, Yeung CY, Lu Y, Crabtree D, Holmes DF, Kadler KE. Synthesis of embryonic tendon-like tissue by human marrow stromal/mesenchymal stem cells requires a three-dimensional environment and transforming growth factor beta3. Matrix Biol. 29(8):668–77. doi: 10.1016/j.matbio.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalson NS, Holmes DF, Kapacee Z, Otermin I, Lu Y, Ennos RA, Canty-Laird EG, Kadler KE. An experimental model for studying the biomechanics of embryonic tendon: Evidence that the development of mechanical properties depends on the actinomyosin machinery. Matrix Biol. 2010;29(8):678–89. doi: 10.1016/j.matbio.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerquin M-J, Charvet B, Nourissat G, Havis E, Ronsin O, Bonnin M-A, Ruggiu M, Olivera-Martinez I, Robert N, Lu Y, Kadler K, Baumberger T, Doursounian L, Berenbaum F, Duprez D. Transcription factor EGR1 directs tendon differentiation and promotes tendon repair. The Journal of clinical investigation. 2013;123(8):3564–76. doi: 10.1172/JCI67521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X, Li C, Herrera PL, Deng CX. Generation of Smad4/Dpc4 conditional knockout mice. Genesis. 2002;32(2):80–1. doi: 10.1002/gene.10029. [DOI] [PubMed] [Google Scholar]
- 36.Xu J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol. 2005 doi: 10.1002/0471142727.mb2801s70. Chapter 28:Unit 28 1. [DOI] [PubMed] [Google Scholar]
- 37.Murtaugh LC, Chyung JH, Lassar AB. Sonic hedgehog promotes somitic chondrogenesis by altering the cellular response to BMP signaling. Genes Dev. 1999;13(2):225–37. doi: 10.1101/gad.13.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Zhang C, Zhu S, Lu P, Zhu T, Gong X, Zhang Z, Hu J, Yin Z, Heng BC, Chen X, Ouyang HW. Mohawk Promotes the Tenogenesis of Mesenchymal Stem Cells through Activation of the TGFbeta Signaling Pathway. Stem Cells. 2014 doi: 10.1002/stem.1866. [DOI] [PubMed] [Google Scholar]
- 39.Brown JP, Galassi TV, Stoppato M, Schiele NR, Kuo CK. Comparative analysis of mesenchymal stem cell and embryonic tendon progenitor cell response to embryonic tendon biochemical and mechanical factors. Stem Cell Res Ther. 2015;6:89. doi: 10.1186/s13287-015-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blitz E, Sharir A, Akiyama H, Zelzer E. Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development. 2013;140(13):2680–90. doi: 10.1242/dev.093906. [DOI] [PubMed] [Google Scholar]
- 41.Eyal S, Blitz E, Shwartz Y, Akiyama H, Ronen S, Zelzer E. On the development of the patella. Development. 2015 doi: 10.1242/dev.121970. [DOI] [PubMed] [Google Scholar]
- 42.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 43.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238(1):265–72. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 44.Baker BM, Mauck RL. The effect of nanofiber alignment on the maturation of engineered meniscus constructs. Biomaterials. 2007;28(11):1967–77. doi: 10.1016/j.biomaterials.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Juncosa-Melvin N, Matlin KS, Holdcraft RW, Nirmalanandhan VS, Butler DL. Mechanical stimulation increases collagen type I and collagen type III gene expression of stem cell-collagen sponge constructs for patellar tendon repair. Tissue Eng. 2007;13(6):1219–26. doi: 10.1089/ten.2006.0339. [DOI] [PubMed] [Google Scholar]
- 46.Huang AH, Stein A, Tuan RS, Mauck RL. Transient exposure to TGF-beta3 improves the mechanical properties of MSC-laden cartilage constructs in a density dependent manner. Tissue Eng Part A. 2009;15(11):3461–72. doi: 10.1089/ten.tea.2009.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagerty P, Lee A, Calve S, Lee CA, Vidal M, Baar K. The effect of growth factors on both collagen synthesis and tensile strength of engineered human ligaments. Biomaterials. 2012;33(27):6355–61. doi: 10.1016/j.biomaterials.2012.05.045. [DOI] [PubMed] [Google Scholar]
- 48.Mu Y, Gudey SK, Landstrom M. Non-Smad signaling pathways. Cell Tissue Res. 2011 doi: 10.1007/s00441-011-1201-y. [DOI] [PubMed] [Google Scholar]
- 49.Benazet JD, Pignatti E, Nugent A, Unal E, Laurent F, Zeller R. Smad4 is required to induce digit ray primordia and to initiate the aggregation and differentiation of chondrogenic progenitors in mouse limb buds. Development. 2012;139(22):4250–60. doi: 10.1242/dev.084822. [DOI] [PubMed] [Google Scholar]