Abstract
The complex lipids present in the cell wall of Mycobacterium tuberculosis (Mtb) act as major effector molecules that actively interact with the host, modulating its metabolism and stimulating the immune response, which in turn affects the physiology of both, the host cell and the bacilli. Lipids from the host are also nutrient sources for the pathogen and define the fate of the infection by modulating lipid homeostasis. Although new technologies and experimental models of infection have greatly helped understanding the different aspects of the host-pathogen interactions at the lipid level, the impact of this interaction in the Mtb lipid regulation is still incipient, mainly because of the low background knowledge in this area of research.
Introduction
Tuberculosis (TB) is a lung infection disease caused by Mtb that has afflicted humans for thousands of years, and still remains a major health emergency provoking more than a million deaths each year (World Health Organization, 2016). The fact that approximately one third of the world’s population is infected with Mtb demonstrates a remarkable well-adapted long-term interaction between this pathogen and its host. Mtb is transmitted to a new individual via inhalation and within the lung bacteria are ingested by alveolar macrophages, the first line of the innate immune system [1]. However, due to the effectiveness of this pathogen at subverting many of the host immune defenses, instead of being cleared by the immune system, Mtb often resist degradation by arresting phagosome maturation creating a permissive niche, in which it can persist or replicate to ultimately trigger the formation of a granuloma [2,3]. Furthermore, it is well established now that a fraction of the Mtb population can produce phagosomal rupture and escape to the cytosol allowing bacterial replication and inducing host cell death involving necrosis [4–6]. In order for Mtb to succeed and actively replicate causing an acute TB, or to survive within the granuloma for long periods of time, in an asymptomatic state, Mtb has evolved a wide array of specific lipids and related metabolisms that actively interact with the immune response and the lipid metabolism in the host [7,8]. This review will focus in the latest understanding of the adaptation and regulation of the different lipid related pathways in Mtb and in the interactions between the bacteria and the host lipid metabolisms during the infection process.
Lipid Metabolism and its regulation in the fate of Mtb infection
The complexity of lipid biosynthesis in Mtb
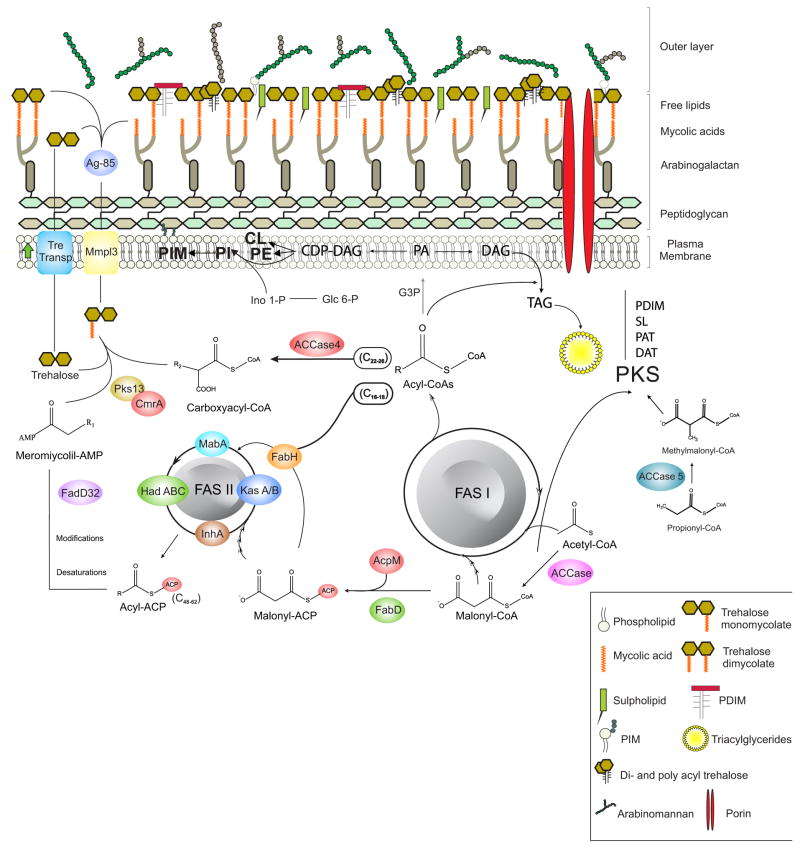
The composition and complexity of the mycobacterial cell envelope is the most distinctive feature of the Mycobacterium genus. Cryotransmission electron microscopy (EM) data provided direct evidence for a multilayered cell wall organization in mycobacteria and confirmed that the outermost layer of mycobacteria is an outer membrane with a bilayer structure [9,10]. The most recent model divides the mycobacterial cell envelope in three entities: an outermost layer, also called capsule in the case of pathogenic species, a cell wall core and a conventional plasma membrane [11]. The cell wall core consists of the mycomembrane and peptidoglycan (PG) covalently linked to arabinogalactan (AG). The mycomembrane exhibits an asimetric bilayer organization: the inner leaflet is made of α-alkyl, β-hydroxy long-chain mycolic acids linked to AG and the outer leaflet is composed of free, non covalently bound to the cell, lipids and glycolipids like trehalose monomycolates (TMM), trehalose dimycolates (TDM), glycerol monomycolates, glucose monomycolates, phthiocerol dimycocerosates (PDIM), poly-acylated threaloses (PAT), sulfolipids (SL), phosphatidylinositol mannosides (PIM), phenolic glycolipids (PGL) and mannose-capped lipoarabinomannans (Man-LAM). Genomics, bioinformatics, proteomics as well as advances in genetic manipulation of mycobacteria have resulted in a thorough understanding of the enzymes involved in the biosynthesis of the main cell envelope constituents. A remarkable characteristic of mycobacteria is the use of a multifunctional fatty acid synthase (FAS) system, FAS I, for de novo synthesis of medium and long chain acyl-CoAs [12,13]. The extraordinary diversity of lipids synthesized by Mtb is directly related with the unusual level of complexity regarding the fate of fatty acids, as they are substrates of all the different lipid biosynthetic pathways. The complex interplay that exists between these pathways is schematized in Figure 1.
Figure 1.
Schematic representation of lipid biosynthesis pathways and their interactions in M. tuberculosis.
Role of Lipids in Mtb entry and phagocytosis mechanisms
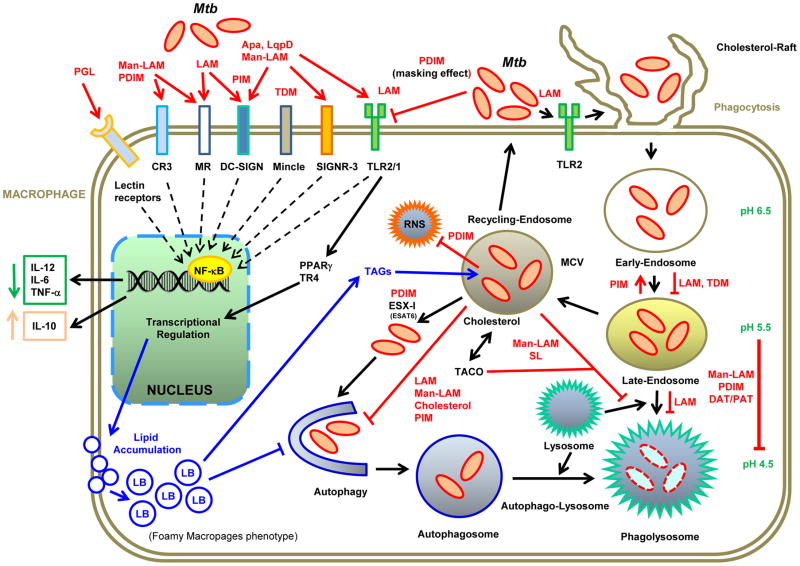
Several studies have demonstrated the relevance of Mtb and the host lipids in their interaction and in the early events of phagocytosis (Figure 2) [7,8,14]. The entry of Mtb into the macrophage occurs through receptor mediated phagocytosis and in this process the most relevant components are the pathogen-associated molecular patterns (PAMPs) [15], which are recognized by macrophages and dendritic cells (DCs) through pattern recognition receptors (PRRs). This recognition triggers the internalization of bacilli and the lipid specific signaling cascades that differentially modulate the host immune response [reviewed in 15,16] (Figure 2). A recent publication in this area demonstrated that sulfoglycolipids contribute to Mtb virulence by inhibiting the human innate immune responses acting as competitive antagonists of TLR2 receptors [17].
Figure 2.
Role of the host-mycobacterial lipids interactions during macrophages infection.
The cell envelope of mycobacteria comprise a wealth of unique glyco-lipids that act as PAMPs and are recognized by macrophages and DCs through PRRs such as Toll-like receptors (TLRs), Nod-like receptors (NLRs), and CLRs. In this scheme, we highlight the main host-mycobacterial lipid interactions and their consequences. Mtb lipids prevent the phagosome maturation, acidification and fusion with lysosomes; they also inhibit autophagy, in order to create a permissive niche that allows bacteria to resist degradation, and eventually replicate within the macrophage. During infection, macrophages accumulate lipids that can be used for Mtb as carbon and energy sources. Apa and LpqH: surface glyco- and lipoproteins, TACO/coronin 1: actin-binding host protein, Mincle: macrophage-inducible C-type lectin, RNS: reactive nitrogen species, MR: C-type lectins Mannose Receptor, DC-SIGN: dendritic cell-specific ICAM-grabbing non-integrin (only in DCs), CR3: complement receptor type 3, SIGNR3: DC-SIGN homologue in mice.
Lipids and modulation of phagosome-maturation
Once mycobacteria is delivered into a phagosome, it could be eliminated by the cellular lysosomal machinery; however, virulent mycobacteria have evolved unique strategies to modulate host endocytic pathways, including immune evasion [18,19]. Cell wall lipids may have multiple, overlapping functions [8] and this is the case for Man-LAM, TDM and PDIM, which besides their role in phagocytosis they also mediate the intracellular trafficking and the vacuole maturation arrest induced by Mtb. While most wild type Mtb are found in non-acidic phagosomes, PDIM-deficient mutants are principally found in acidic phagosomes, demonstrating the relevance of PDIM in the arrest of phagosomal acidification [20]. Furthermore, the characterization of mutants attenuated for intracellular survival revealed that ESX-1 and PDIM deficient mutants shared similar characteristics of attenuation (Figure 2). Building upon the shared phenotype of these mutants, it was found that PDIM production and export are required for phagosomal permeabilization and for production of the macrophage type I interferon response [21,22]. The characterization of an Mtb mutant in the transcriptional repressor Rv3167c demonstrated that increased PDIM levels correlated well not only with an increased capacity of Mtb to escape from the phagosome but also to induce host cell necrosis; attributing a new role to PDIM in intracellular host-cell modulation [23]. TDM also modulate phagosome maturation but by not fully understood mechanisms [24]. Only recently, it was shown that IgG-opsonized TDM-coated particles could recruit Mincle and FcγRIIB to induce signals that delay FcγR-mediated phagosome maturation [25]. The contribution of SL, DAT and PAT to bacterial survival, is relevant only in a PDIM mutant strain [8].
Lipids and granuloma biogenesis
The vacuole that contains Mycobacterium (MCV) encloses lipids moieties of the mycobacterial cell wall [26], which are trafficked out of MCV to associate with several intracellular organelles, representing one possible mechanism by which Mycobacterium influences the environment within the infection foci [27]. These lipids were also found extracellularly in small vesicles where they can elicit proinflammatory cytokines from macrophages, contributing to the granulomatous response [28]. Although Mtb survives and grow, albeit slowly, inside vacuoles, this bacterium can also escape from the MCV to the cytosol and induce a non-apoptotic mechanism [4–6], supporting mycobacterial growth and the recruitment of additional host phagocytes which leads to infection spread. Mtb aggregates can also be internalized by macrophages directly into the cytosol, where they actively replicate and kill the host cell. Dead cells full of active bacteria can then be phagocytosed by other macrophages triggering a cell death cascade, which might be the dynamics of necrosis and bacterial proliferation in lung granulomas of active TB [29].
Lipid metabolism and regulation within the host
Mtb resides or replicates in a very nutritionally-defined environment, either a macrophage, a DC, or a granuloma, relying on specific metabolic pathways to use host-derived nutrients [30]. The different carbon and nitrogen sources, as well as the varying oxygen tension that Mtb encounters during infection are known to impact the lipid composition of the envelope, i.e. the changes observed in the typical acid-fast staining of Mtb during infection [31]. Although it has been shown in different models that Mtb uses different carbon sources at different stages of the infection process [32–35], it is generally accepted that host lipids are the primary carbon source for Mtb in vivo. Furthermore, Mtb infected macrophages induce the formation of foamy macrophages (FM) by the accumulation of lipid bodies (LB) which mainly contain cholesterol esters and TAG (Figure 2). The formation of FM is a clear consequence of a bacilli manipulation of the host metabolism promoting the accumulation of neutral lipids [36,37]. Within the FM, Mtb-containing phagosomes progressively surround and engulf the LB, which then serve as nutrient for the microorganism [38]. Under these conditions Mtb faces important physiological changes that result in the accumulation of TAG within intracytosolic lipid inclusions (ILI), reduced growth and lower metabolic activity and resistance to antibiotics; which represent a hallmark of persistent and non-dividing bacilli [34]. Interestingly, TAG accumulation can be prevented by transporting them out of the cell through the putative efflux pump LprG-Rv1410 which is essential for Mtb growth in mouse [39].
The characterization of several mutant strains, demonstrated that genes involved in metabolizing the products of FA oxidation through the glyoxylate shunt and the gluconeogenesis had a strong impact in the Mtb life cycle in different infection models [40–42]. Mutants in the cholesterol transport system Mce4 provided the first evidences that this lipid was not required for establishing infection but was essential for persistence in the lungs of chronically infected animals and for growth within the IFN-γ-activated macrophages [43]. Later on, mutants unable to metabolize cholesterol also showed important defects in intracellular growth or survival [44]. Furthermore, a whole cell-based drug screen against Mtb in macrophages found that a large fraction of hit compounds inhibited cholesterol related processes, indicating that cholesterol plays a central role for Mtb during infection [45]. However, it has been demonstrated that Mtb co-metabolizes simple carbon substrates in vitro [46], suggesting that this bacterium uses both cholesterol and FAs during infection. Supporting the simultaneous use of FAs and cholesterol, it was shown that the metabolic pressure experienced by Mtb inside the host macrophage by the use of cholesterol can be balanced by the utilization of host FAs which increase the acetyl-CoA pool and allows the utilization of propionyl-CoA in the synthesis of methyl-branched lipids [30]. Furthermore, a protein named LucA was found to facilitate the simultaneous uptake of FA and cholesterol by stabilizing protein subunits of the Mce1 and Mce4 transporters. These studies also revealed that the Mce1 complex transports fatty acids and that LucA is essential for full virulence in vivo [47]. Altogether, these results suggest that although FAs cannot substitute for cholesterol during intra-cellular growth, they are needed in order to prevent or relieve the toxicity of propionyl-CoA. It will be interesting to obtain mutants unable to β-oxidize FAs to rigorously test the essentiality of this catabolic pathway. Likewise, it is still not known if exogenous FAs could overcome the absence of de novo FA biosynthesis.
As shown in Figure 1, FAS I provides the substrates for several other lipids biosynthesis pathways in Mtb, suggesting that a highly complex network of regulation between all these pathways should exist in order to maintain lipid homeostasis under control. However, the components and the molecular mechanisms involved in the regulation and interaction of all these pathways, is only starting to emerge. Regulatory proteins and transcriptional regulators, that directly control the transcription of lipids biosynthesis genes, have been described. The first transcriptional regulator of the main fasII operon (fabD-acpM-kasA-kasB) that was characterized is MabR [48]. Genetic studies showed that mabR is essential for M. smegmatis survival and biochemical analysis carried out in a mabR conditional mutant strain showed alterations in mycolic acid and in de novo FA biosynthesis, demonstrating for the first time the existence of a crosstalk between the two FAS systems and confirming MabR as one of the key modulator of lipid homeostasis in Mycobacterium [48,49]. A second non-essential transcription factor, FadR, that represses the fasII operon expression has also been described [50]. FadR is induced upon starvation, leading to reduced fasII expression under those conditions [51]. The hadABC genes, coding for the dehydratase complexes of the FAS II system, are part of a seven-gene operon together with four genes involved in translation, and they all respond to the alarmone (p)ppGpp which leads to dramatic reprogramming of cell transcription [52]. This suggests that the coordination of the expression of the complete set of FAS II genes is really complex and indicates that an eventual interplay exists between different regulatory pathways. The transcription of the fas-acpS operon (encoding for the multidomain FAS I and the phosphopantetheinyl transferase AcpS) is regulated by FasR [53]. Analysis of a conditional mutant in M. smegmatis showed that FasR is a transcriptional activator and proved its essential role in mycobacteria viability. Interestingly, the activity of all these transcription factors (MabR, FadR and FasR) is modulated by long chain acyl-CoAs, the products of the FAS I system, highlighting a key role for these molecules in the modulation of lipid biosynthesis in mycobacteria [49,50,53]. An important difference between MabR and FasR with other bacteria transcriptional regulators of FA biosynthesis is that these two proteins are essential for bacterial survival while all the others are not. This suggests that the coordination and modulation of the two FAS systems is highly relevant in order to maintain lipid homeostasis in mycobacteria. Different approaches also revealed that genes involved in complex lipid biosynthesis (SL, DAT/PAT and PDIM) are up regulated upon infection [54,55]. However, only some global transcriptional regulators of those pathways have been described. EspR, a nucleoid-associated protein with architectural and regulatory roles, impact cell wall functions and pathogenesis through the transcriptional regulation of multiple genes, particularly those involved in PDIM biosynthesis [56]. The global transcriptional regulator PhoP has also been involved in the EspR mediated response [57]. Disruption of PhoP leads to the absence of SL, DAT, and PAT [58] and global transcription assays demonstrated that PhoP is an activator of the genes required for SL and DAT/PAT biosynthesis [59,60]. More recently, Quigley et al (2017) analyzed the regulon of the transcription factor Rv3167c [23], previously characterized as a repressor of Mtb virulence [61], and found that it is enriched for genes involved in the synthesis and transport of PDIM. However, this regulation appears to be indirect and the ligands and regulators involved in the molecular mechanism of this response remains to be elucidated.
The impact of having reduced levels of de novo FA biosynthesis was studied in a fas conditional mutant in M. smegmatis [62]. As expected, fas was essential for survival and in the non-permissive conditions the reduction of FAS I activity led to the accumulation of the FAS I substrates (acetyl-CoA and malonyl-CoA) and to a strong reduction of C12–18 acyl-CoAs. Unexpectedly, even when de novo FA biosynthesis was impaired, the fas mutant was still able to synthesize mycolic acids at the expense of TAG, suggesting that storage lipids could be an intracellular reservoir of FAs for the biosynthesis of complex lipids in mycobacteria. Understanding the interaction between FAS I and the metabolic pathways that rely on FAS I products is a key step to better understand how lipid homeostasis is regulated in Mtb and how this regulation could play a role during infection in pathogenic mycobacteria.
Conclusions
The extensive exchange of information between Mtb and its environment during infection induces a reciprocal modulation of both, host and bacteria metabolisms. Many of these interactions are mediated by lipids, opening the question about how different lipids or lipids derive molecules interact and modulate host and pathogen metabolisms for the outcome of an Mtb infection. On this regard, identifying and characterizing the proteins, the signal molecules and the mechanisms involved in the complex network that regulate lipid homeostasis in Mtb, in the context of the host environment, could help us identify essential regulatory elements for maintaining an efficient and successful interaction of the pathogen with the host. These key regulatory components could be potential drug-targets for the development of conceptually new anti-mycobacterial agents.
Lipids from Mycobacterium tuberculosis and the host play essential roles in almost every step of the infection cycle of this pathogen.
Mtb lipid metabolism is highly complex and the regulatory network that preserves lipid homeostasis in this organism is now starting to be understood.
Host lipid metabolism has strong impact in Mtb lipid homeostasis
The regulatory proteins involved in the regulation of Mtb lipid metabolism are potential therapeutic targets.
Acknowledgments
This work was supported by NIH (1R01AI095183-01), ANPCyT PICT-2012-0168 and PICT 2015-2022 to HG; PICT 2015-0796 to GG and PICT 2014-1454 to LD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
◆ of special interest
◆◆ of outstanding interest
- 1.Awuh JA, Flo TH. Molecular basis of mycobacterial survival in macrophages. Cell Mol Life Sci. 2016;74:1625–1648. doi: 10.1007/s00018-016-2422-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbues A, Lugo-Villarino G, Neyrolles O, Guilhot C, Astarie-Dequeker C. Playing hide-and-seek with host macrophages through the use of mycobacterial cell envelope phthiocerol dimycocerosates and phenolic glycolipids. Front Cell Infect Microbiol. 2014;4:1–7. doi: 10.3389/fcimb.2014.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ◆◆3.Cambier CJ, Takaki KK, Larson RP, Hernandez RE, Tobin DM, Urdahl KB, Cosma CL, Ramakrishnan L. Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature. 2014;505:218–222. doi: 10.1038/nature12799. Demonstrates that Mtb preferentially recruits and infect permissive macrophages evading microbicidal ones. This immune evasion is accomplished through cell surface associated PDIM. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Wel N, Hava D, Houben D, Fluitsma D, van Zon M, Pierson J, Brenner M, Peters PJ. M. tuberculosis and M. leprae Translocate from the Phagolysosome to the Cytosol in Myeloid Cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 5.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog. 2012:8. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simeone R, Sayes F, Song O, Gröschel MI, Brodin P, Brosch R, Majlessi L. Cytosolic Access of Mycobacterium tuberculosis: Critical Impact of Phagosomal Acidification Control and Demonstration of Occurrence In Vivo. PLoS Pathog. 2015;11:1–24. doi: 10.1371/journal.ppat.1004650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neyrolles O, Guilhot C. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis. 2011;91:187–195. doi: 10.1016/j.tube.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Passemar C, Arbues A, Malaga W, Mercier I, Moreau F, Lepourry L, Neyrolles O, Guilhot C, Astarie-Dequeker C. Multiple deletions in the polyketide synthase gene repertoire of mycobacterium tuberculosis reveal functional overlap of cell envelope lipids in host-pathogen interactions. Cell Microbiol. 2014;16:195–213. doi: 10.1111/cmi.12214. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci U S A. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sani M, Houben ENG, Geurtsen J, Pierson J, De Punder K, Van Zon M, Wever B, Piersma SR, Jiménez CR, Daffé M, et al. Direct visualization by Cryo-EM of the mycobacterial capsular layer: A labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010:6. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiaradia L, Lefebvre C, Parra J, Marcoux J, Burlet-Schiltz O, Etienne G, Tropis M, Daffé M. Dissecting the mycobacterial cell envelope and defining the composition of the native mycomembrane. Sci Rep. 2017;7:12807. doi: 10.1038/s41598-017-12718-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloch K. Fatty acid synthases from Mycobacterium phlei. Methods Enzymol. 1975;35:84–90. doi: 10.1016/0076-6879(75)35141-0. [DOI] [PubMed] [Google Scholar]
- 13.Zimhony O, Vilchèze C, Jacobs WR. Characterization of Mycobacterium smegmatis expressing the Mycobacterium tuberculosis fatty acid synthase I (fas1) gene. J Bacteriol. 2004;186:4051–4055. doi: 10.1128/JB.186.13.4051-4055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vromman F, Subtil A. Exploitation of host lipids by bacteria. Curr Opin Microbiol. 2014;17:38–45. doi: 10.1016/j.mib.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa E, Mori D, Yamasaki S. Recognition of Mycobacterial Lipids by Immune Receptors. Trends Immunol. 2017;38:66–76. doi: 10.1016/j.it.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 16.Thirunavukkarasu S, de Silva K, Plain KMJ, Whittington R. Role of host- and pathogen-associated lipids in directing the immune response in mycobacterial infections, with emphasis on Mycobacterium avium subsp. paratuberculosis. Crit Rev Microbiol. 2016;42:262–275. doi: 10.3109/1040841X.2014.932327. [DOI] [PubMed] [Google Scholar]
- 17.Blanc L, Gilleron M, Prandi J, Song O, Jang M-S, Gicquel B, Drocourt D, Neyrolles O, Brodin P, Tiraby G, et al. Mycobacterium tuberculosis inhibits human innate immune responses via the production of TLR2 antagonist glycolipids. Proc Natl Acad Sci. 2017 doi: 10.1073/pnas.1707840114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cambier CJ, Falkow S, Ramakrishnan L. Host evasion and exploitation schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012;12:581–591. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 20.Astarie-Dequeker C, Le Guyader L, Malaga W, Seaphanh FK, Chalut C, Lopez A, Guilhot C. Phthiocerol dimycocerosates of M. tuberculosis participate in macrophage invasion by inducing changes in the organization of plasma membrane lipids. PLoS Pathog. 2009:5. doi: 10.1371/journal.ppat.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barczak AK, Avraham R, Singh S, Luo SS, Zhang WR, Bray MA, Hinman AE, Thompson M, Nietupski RM, Golas A, et al. Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog. 2017;13:1–27. doi: 10.1371/journal.ppat.1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ◆22.Augenstreich J, Arbues A, Simeone R, Haanappel E, Wegener A, Sayes F, Le Chevalier F, Chalut C, Malaga W, Guilhot C, et al. ESX-1 and phthiocerol dimycocerosates of Mycobacterium tuberculosis act in concert to cause phagosomal rupture and host cell apoptosis. Cell Microbiol. 2017 doi: 10.1111/cmi.12726. Demonstrates, by the use of several knock-out and/or knock-in mutants of Mtb or M. bovis BCG strains and cell biological assays, that ESX-1 and PDIM act in concert to induce phagosomal membrane damage and rupture in infected macrophages, ultimately leading to the host cell apoptosis. [DOI] [PubMed] [Google Scholar]
- ◆◆23.Quigley J, Hughitt VK, Velikovsky CA, Mariuzza RA, El-Sayed NM, Briken V. The cell wall lipid PDIM contributes to phagosomal escape and host cell exit of Mycobacterium tuberculosis. MBio. 2017;8:1–12. doi: 10.1128/mBio.00148-17. Demonstrates the relevance of PDIM in the scape of M. tuberculosis from its intracellular vacuole. The PDIM-dependent increased release of M. tuberculosis into the cytosol leads to increased host cell necrosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Axelrod S, Oschkinat H, Enders J, Schlegel B, Brinkmann V, Kaufmann SHE, Haas A, Schaible UE. Delay of phagosome maturation by a mycobacterial lipid is reversed by nitric oxide. Cell Microbiol. 2008;10:1530–1545. doi: 10.1111/j.1462-5822.2008.01147.x. [DOI] [PubMed] [Google Scholar]
- 25.Patin EC, Geffken AC, Willcocks S, Leschczyk C, Haas A, Nimmerjahn F, Lang R, Ward TH, Schaible UE. Trehalose dimycolate interferes with FcgammaR-mediated phagosome maturation through Mincle, SHP-1 and FcgammaRIIB signalling. PLoS One. 2017;12:e0174973. doi: 10.1371/journal.pone.0174973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhoades E, Hsu FF, Torrelles JB, Turk J, Chatterjee D, Russell DG. Identification and macrophage-activating activity of glycolipids released from intracellular Mycobacterium bovis BCG. Mol Microbiol. 2003;48:875–888. doi: 10.1046/j.1365-2958.2003.03473.x. [DOI] [PubMed] [Google Scholar]
- 27.Beatty WL, Rhoades ER, Ullrich H-J, Chatterjee D, Heuser JE, Russell DG. Trafficking and Release of Mycobacterial Lipids from Infected Macrophages. Traffic. 2000;1:235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- 28.van den Elzen P, Garg L, León L, Brigl M, Leadbetter E, Gumperz J, Dascher C, Cheng T, Sacks F, Illarionov P, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- ◆◆29.Mahamed D, Boulle M, Ganga Y, Mc Arthur C, Skroch S, Oom L, Catinas O, Pillay K, Naicker M, Rampersad S, et al. Intracellular growth of Mycobacterium tuberculosis after macrophage cell death leads to serial killing of host cells. Elife. 2017;6:1–26. doi: 10.7554/eLife.22028. Live cell imaging to track Mtb infection showed that bacteria aggregates can be internalized into the macrophage cytosol where they replicate and kill the cell host. Dead infected cells can be internalized by other macrophages which will then die leading to a cell death cascade. This is one way pathogen virulence can be achieved. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee W, VanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryan GJ, Hoff DR, Driver ER, Voskuil MI, Gonzalez-Juarrero M, Basaraba RJ, Crick DC, Spencer JS, Lenaerts AJ. Multiple M. tuberculosis phenotypes in mouse and guinea pig lung tissue revealed by a dual-staining approach. PLoS One. 2010:5. doi: 10.1371/journal.pone.0011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouzy A, Larrouy-Maumus G, Wu T-D, Peixoto A, Levillain F, Lugo-Villarino G, Gerquin-Kern J-L, de Carvalho LPS, Poquet Y, Neyrolles O. Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat Chem Biol. 2013;9:674–6. doi: 10.1038/nchembio.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trujillo C, Blumenthal A, Marrero J, Rhee KY, Schnappinger D, Ehrt S. Triosephosphate isomerase is dispensable in vitro yet essential for Mycobacterium tuberculosis to establish infection. MBio. 2014;5:1–11. doi: 10.1128/mBio.00085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011:7. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, Sassetti CM. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7:1–9. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ◆◆36.Singh V, Jamwal S, Jain R, Verma P, Gokhale R, Rao KVS. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe. 2012;12:669–681. doi: 10.1016/j.chom.2012.09.012. Demonstrate that Mtb induces foamy phenotype via targeted manipulation of host cellular metabolism directing the glycolytic pathway towards the synthesis of ketone bodies and the activation of the anti-lypolitic receptor GPR1094. This change ends up in the reduction of cellular cAMP and the accumulation of lipids bodies. [DOI] [PubMed] [Google Scholar]
- 37.Singh V, Kaur C, Chaudhary VK, Rao KVS, Chatterjee S. M tuberculosis Secretory Protein ESAT-6 Induces Metabolic Flux Perturbations to Drive Foamy Macrophage Differentiation. Sci Rep. 2015;5:12906. doi: 10.1038/srep12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffé M, Emile JF, Marchou B, Cardona PJ, et al. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:1–14. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ◆39.Martinot AJ, Farrow M, Bai L, Layre E, Cheng T-Y, Tsai JH, Iqbal J, Annand JW, Sullivan ZA, Hussain MM, et al. Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis. PLOS Pathog. 2016;12:e1005351. doi: 10.1371/journal.ppat.1005351. TAG accumulation restricts bacterial growth and here is shown that LprG-Rv11410 can mediate the transport of TAG into the cell wall, helping to release growth inhibition by acting as a sink of excess of free fatty acids. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci U S A. 2010;107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muñoz-Elias E, McKinney J. M. tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mckinney JD, Bentrup KHZ, Muñoz-elías EJ, Miczak A, Chen B, Chan W-T, Swenson DL, Sacchettini JC, Jacobs WR, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 43.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem Biol. 2012;19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.VanderVen BC, Fahey RJ, Lee W, Liu Y, Abramovitch RB, Memmott C, Crowe AM, Eltis LD, Perola E, Deininger DD, et al. Novel Inhibitors of Cholesterol Degradation in Mycobacterium tuberculosis Reveal How the Bacterium’s Metabolism Is Constrained by the Intracellular Environment. PLoS Pathog. 2015;11:1–20. doi: 10.1371/journal.ppat.1004679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Carvalho LPS, Fischer SM, Marrero J, Nathan C, Ehrt S, Rhee KY. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem Biol. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Nazarova EV, Montague CR, La T, Wilburn KM, Sukumar N, Lee W, Caldwell S, Russell DG, VanderVen BC. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife. 2017;6:1–22. doi: 10.7554/eLife.26969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salzman V, Mondino S, Sala C, Cole ST, Gago G, Gramajo H. Transcriptional regulation of lipid homeostasis in mycobacteria. Mol Microbiol. 2010;78:64–77. doi: 10.1111/j.1365-2958.2010.07313.x. [DOI] [PubMed] [Google Scholar]
- 49.Tsai YT, Salzman V, Cabruja M, Gago G, Gramajo H. Role of long chain acyl-CoAs in the regulation of mycolic acid biosynthesis in mycobacteria. Open Biol. 2017;7:170087. doi: 10.1098/rsob.170087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biswas RK, Dutta D, Tripathi A, Feng Y, Banerjee M, Singh BN. Identification and characterization of Rv0494: A fatty acid-responsive protein of the GntR/FadR family from Mycobacterium tuberculosis. Microbiology. 2013;159:913–923. doi: 10.1099/mic.0.066654-0. [DOI] [PubMed] [Google Scholar]
- 51.Yousuf S, Angara R, Vindal V, Ranjan A. Rv0494 is a starvation-inducible, auto-regulatory FadR-like regulator from Mycobacterium tuberculosis. Microbiology. 2015;161:463–476. doi: 10.1099/mic.0.000017. [DOI] [PubMed] [Google Scholar]
- 52.Jamet S, Quentin Y, Coudray C, Texier P, Laval F, Daffé M, Fichant G, Cam K. Evolution of mycolic acid biosynthesis genes and their regulation during starvation in Mycobacterium tuberculosis. J Bacteriol. 2015;197:3797–3811. doi: 10.1128/JB.00433-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mondino S, Gago G, Gramajo H. Transcriptional regulation of fatty acid biosynthesis in mycobacteria. Mol Microbiol. 2013;89:372–387. doi: 10.1111/mmi.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graham JE, Clark-Curtiss JE. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS) Proc Natl Acad Sci U S A. 1999;96:11554–9. doi: 10.1073/pnas.96.20.11554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodríguez JE, Ramírez AS, Salas LP, Helguera-Repetto C, Gonzalez-y-Merchand J, Soto CY, Hernández-Pando R. Transcription of Genes Involved in Sulfolipid and Polyacyltrehalose Biosynthesis of Mycobacterium tuberculosis in Experimental Latent Tuberculosis Infection. PLoS One. 2013:8. doi: 10.1371/journal.pone.0058378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, Pojer F, Cole ST. Virulence Regulator EspR of Mycobacterium tuberculosis Is a Nucleoid-Associated Protein. PLoS Pathog. 2012;8:e1002621. doi: 10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar VA, Goyal R, Bansal R, Singh N, Sevalkar RR, Kumar A, Sarkar D. Espr-dependent ESAT-6 protein secretion of Mycobacterium tuberculosis requires the presence of virulence regulator phoP. J Biol Chem. 2016;291:19018–19030. doi: 10.1074/jbc.M116.746289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalo-Asensio JG, Maia C, Ferrer NL, Barilone N, Laval F, Soto CY, Winter N, Daffe M, Gicquel B, Martín C, et al. The virulence-associated two-component PhoP-PhoR system controls the biosynthesis of polyketide-derived lipids in Mycobacterium tuberculosis. J Biol Chem. 2006;281:1313–1316. doi: 10.1074/jbc.C500388200. [DOI] [PubMed] [Google Scholar]
- 59.Walters SB, Dubnau E, Kolesnikova I, Laval F, Daffe M, Smith I. The Mycobacterium tuberculosis PhoPR two-component system regulates genes essential for virulence and complex lipid biosynthesis. Mol Microbiol. 2006;60:312–330. doi: 10.1111/j.1365-2958.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 60.Solans L, Gonzalo-Asensio J, Sala C, Benjak A, Uplekar S, Rougemont J, Guilhot C, Malaga W, Martín C, Cole ST. The PhoP-Dependent ncRNA Mcr7 Modulates the TAT Secretion System in Mycobacterium tuberculosis. PLoS Pathog. 2014:10. doi: 10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Srinivasan L, Gurses SA, Hurley BE, Miller JL, Karakousis PC, Briken V. Identification of a Transcription Factor That Regulates Host Cell Exit and Virulence of Mycobacterium tuberculosis. PLOS Pathog. 2016;12:e1005652. doi: 10.1371/journal.ppat.1005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ◆62.Cabruja M, Mondino S, Tsai YT, Lara J, Gramajo H, Gago G. A conditional mutant of the fatty acid synthase unveils unexpected cross talks in mycobacterial lipid metabolism. Open Biol. 2017:7. doi: 10.1098/rsob.160277. Detailed analysis of the impact of reduced de novo fatty acid biosynthesis in the overall composition of M. smegmatis cell envelope. Demonstrates that storage lipids could be an intracellular reservoir of fatty acids for the biosynthesis of complex lipids. [DOI] [PMC free article] [PubMed] [Google Scholar]