Abstract
Small molecules help intestinal pathogens navigate the complex human gastrointestinal tract to exploit favorable microhabitats. These small molecules provide spatial landmarks for pathogens to regulate synthesis of virulence caches and are derived from the host, ingested plant and animal material, and the microbiota. Their concentrations and fluxes vary along the length of the gut and provide molecular signatures that are beginning to be explored through metabolomics and genetics. However, while many small molecules have been identified and are reviewed here, there are undoubtedly others that may also profoundly affect how enteric pathogens infect their hosts.
Introduction
Enterohemorrhagic Escherichia coli (EHEC) is a non-invasive intestinal pathogen that adeptly senses small molecules to infect the human large intestine. After consuming contaminated food or water, EHEC causes hemorrhagic colitis, and in severe cases, hemolytic uremic syndrome or death [1]. EHEC is a problematic, common pathogen because of its low infectious dose (< 100 cells) [1]. In addition, EHEC skillfully utilizes a plethora of small molecules to tightly regulate expression of its type III secretion system (T3SS) encoded on the locus of enterocyte effacement (LEE), a pathogenicity island that harbors 41 genes the majority of them being organized on five operons [2–4]. The LEE is needed for EHEC to colonize the gut by forming attaching and effacing (AE) lesions on enterocytes. AE lesions are associated with the dynamic remodeling of the host’s cytoskeleton leading to the formation of a pedestal-like structure underneath the adherent bacterium. Expression of the LEE is energetically costly, and thus, tightly regulated to be deployed in microhabitats where it can help EHEC compete for a niche. Recent data about the type and role of these small molecules in EHEC pathogenesis will be explored in this review (summarized in Figure 1).
Figure 1.
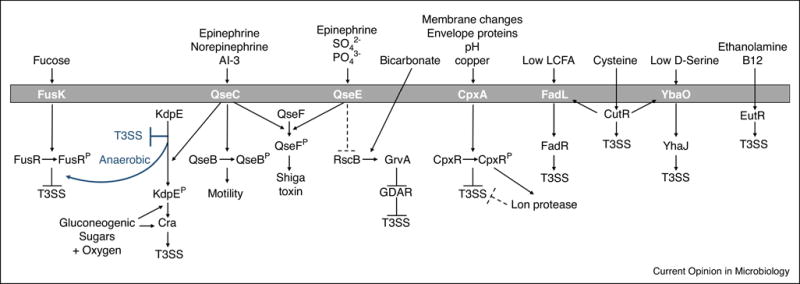
Summary of small molecules sensed by EHEC to regulate expression of the Type III secretion system (T3SS). Diagrams in blue are under anaerobic, gluconeogenic conditions. In general, molecules that are found near the epithelial surface such as oxygen, epinephrine, norepinephrine, gluconeogenic carbon sources (pyruvate, N-acetyl-galactosamine, N-acetyl-glucosamine, N-acetylneuraminic acid, mannose, galacturonic acid, gluconic acid, and glucuronic acid), and ethanolamine activate T3SS. Fucose, which is found in the lumen, inhibits T3SS. Cysteine concentrations increase with infection and can activate T3SS. Low concentrations of long chain fatty acids (LCFA) or D-Serine also activate the T3SS. Bicarbonate activates GrvA in an RscB dependent manner to inhibit the Glutamate-dependent acid resistance (GDAR) pathway, thereby activating T3SS.
Host and microbiota signal: Ethanolamine
Bacterial and host membranes constantly release ethanolamine into the intestines, providing a reliable gut signal. Ethanolamine is useful as both a carbon or nitrogen source. EHEC uses ethanolamine as a nitrogen source to compete with the microbiota and colonize their host [5,6]. However, ethanolamine also activates expression of the LEE through the EutR transcription factor, a known ethanolamine receptor, independently of activating the eut operon [5,6] (Figure 1). Ethanolamine and choline also promote expression of several fimbrial operons which helps EHEC attach to cells [7]. These data suggest sensing of ethanolamine and choline is important for the initial stages of EHEC adherence to epithelial cells.
Host signals: Epinephrine and Norepinephrine
The host-derived signals epinephrine and norepinephrine play important roles in gut physiology and motility [8]. The host inactivates epinephrine and norepinephrine using glucuronidation; however, the gut microbiota encoded enzymes to cleave glucuronic acid from epinephrine and norepinephrine, thus making them biologically active in the lumen [9]. To sense these neurotransmitters, EHEC uses two bacterial adrenergic histidine sensor kinases, QseC and QseE. Upon sensing these signals, these kinases initiate a signal cascade phosphorylating three response regulators (RRs). QseE only phosphorylates the QseF RR, while QseC phosphorylates QseF, KdpE and QseB [10–15] (Figure 1). Phosphorylated KdpE then works in concert with the catabolite repressor activator (Cra), a global regulator of genes involved in gluconeogenesis, to activate expression of the LEE [14,16]. Recent data in the EHEC murine surrogate model, Citrobacter rodentium, highlight how qseC, qseE, and qseEC mutants are attenuated for murine infection because they fail to correctly sense epinephrine and norepinephrine [9]. In addition, in mice lacking dopamine β-hydroxylase, which do not produce epinephrine or norepinephrine, C. rodentium has colonization defects and reduced expression of the LEE [9]. In an infant rabbit model of infection, EHEC mutants in qseC [12] or the qseEC double mutant are attenuated [9]. It is important to note that QseC also senses the microbiota-produced signal autoinducer-3 [10,17] and QseE senses SO4 and PO4 [11].
Host and microbiota signals: Mucin and diet derived sugars
Exploitation of non-preferred carbon sources helps pathogens gain a niche advantage. In the gut, E. coli prefers monosaccharides that feed into the Embden-Meyerhof-Parnas pathway (classical glycolysis), but also metabolizes sugars using the pentose phosphate pathway and the Entner-Doudoroff (ED) pathway [18–22]. These sugars are liberated by glycophagic bacteria, such as Bacteroides thetaiotaomicron, degrading ingested food as well as mucus [23] (Figure 2). The GI mucus layer is composed of mucins, glycoproteins composed of 80% carbohydrates, and provides a barrier between the microbiota and host epithelial cells [23]. Recently, it has been shown in C. rodentium infections that diet affects the amount of mucus degradation in the intestine [24] (Figure 2). Mice colonized with a synthetic human microbiota and fed diets devoid of fiber had eroded mucus layers compared to mice fed a fiber-rich diet. In addition, the fiber deprived mice, with diminished mucus layers, were more susceptible to C. rodentium infection and had more aggressive colitis [24]. In another study, mice colonized with B. thetaiotaomicron exacerbates C. rodentium infections by increasing gluconeogenic substrates such as succinate [25]. C. rodentium uses the gluconeogenic master regulator Cra to metabolize succinate and activate expression of the LEE [25]. B. thetaiotaomicron also enhances EHEC infection using another sugar utilization pathway. B. thetaiotaomicron liberates fucose from mucin, which is sensed by the histidine sensor kinase FusK that phosphorylates its response regulator FusR [26] (Figure 1). FusR represses LEE expression to avoid assembling the T3SS in the intestinal lumen where the microbiota would be cleaving terminal mucin sugars [26].
Figure 2.
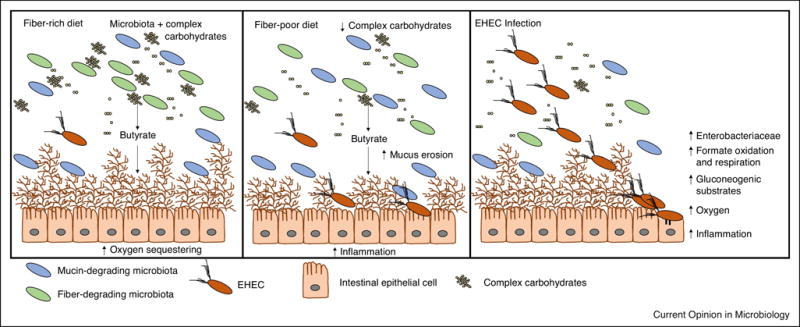
EHEC hosts have varied diets and environmental conditions. In the left panel, the host has a fiber-rich diet where the microbiota digest complex carbohydrates to short chain fatty acids such as butyrate. These hosts have a normal mucus layer to keep infecting pathogens away from gut epithelial cells. In the center panel, the host has a fiber-poor diet. Fewer complex carbohydrates are available, and thus the proportion of mucin-degrading microbiota members increases. There is enhanced mucus erosion and an increase in inflammation that makes it easier for enteric pathogens to infect gut epithelial cells. In the right panel, EHEC is infecting the host by sensing different small molecules. The infection causes a bloom in Enterobacteriaceae, an increase in formate oxidization and respiration, gluconeogenic substrates such as succinate, oxygen, and inflammation.
The mucus layer and the diet provide nutrients for the microbiota but also signals for EHEC (Figure 2). Mucin-derived sugars available for E. coli to utilize in the gut include: glucose, fucose, galactose, N-acetyl-galactosamine, N-acetyl-glucosamine, N-acetylneuraminic acid, fructose, xylose, and mannose [27,28]. Some diet derived sugars in the large intestine include those involved in pectin degradation. Pectin is constructed of long chains of α-1,4-glycoside-linked D-galacturonic acid which can be decorated with other terminal sugars such as rhamnose, D-xylose, L-fucose, D-glucuronic acid, and others [29]. Expression of the T3SS is affected when EHEC is grown with these mucin-derived and pectin-derived sugars as a sole carbon source. The ED pathway sugars galacturonic acid, glucuronic acid, and gluconic acid all increase secretion of EspB, which is needed to form the pore in mammalian cells for the needle apparatus of the T3SS to inject effectors into the cytosol [16]. N-acetyl-galactosamine, N-acetyl-glucosamine, N-acetylneuraminic acid, and mannose also increased secretion of EspB [16]. These sugars are important building blocks in O-linked and N-linked glycosylation of attached and secreted mucins in the large intestine [30,31]. Pyruvate, a glucoenogenic sugar, also increased secretion of EspB as well as increased expression of the LEE [16]. This increased secretion is not from a growth advantage, except potentially in the case of galacturonic acid where EHEC grows almost twice as quickly as on glucose, but suggests EHEC is regulating its virulence genes via catabolite repression [16] (Figure 2).
Host and microbiota signals: Electron acceptors
In the GI tract, E. coli use different electron acceptors depending on the location of their environmental niche, health of the host, and presence of the microbiota. Both aerobic respiration of oxygen (via cytochrome bd oxidase) and anaerobic respiration of nitrate and fumarate are needed for EHEC colonization in the intestine [32,33]. Oxygen and nitrate are both limited in the large intestine, but fumarate is more readily available and helps with longer term colonization [32]. Recent data suggest there is a radial oxygen gradient in the large intestine that varies depending on atmospheric pressure, the host’s ability to sequester oxygen, and the aerotolerant members of the microbiota that consume oxygen in the outer mucosal layer [34–36]. While the lumen is predominately anaerobic, near the epithelial surface microaerobic conditions exist because of diffusion across the microvillus capillary network [34,36–38]. If the microbiota is absent or in dysbiosis, then oxygen levels can increase further [39–41]. If there is a breach in the epithelium and blood infiltrates into the large intestine, as is common in EHEC infections, then the amount of oxygen can dramatically increase [42] (Figure 2).
New data exploring the role of oxygen on Enterobacteriaceae blooms and how oxygen regulates expression of EHEC virulence factors shed light on the dynamic processes taking place in the gut. For EHEC and C. rodentium, oxygen plays a strong role in regulating infection [40,42]. In mice with an inflamed gut, expression of E. coli genes involved in respiratory pathways are overrepresented [41,42]. Microbiota-derived formate concentrations also increase during dysbiosis and E. coli use it as an electron donor and oxygen as an electron acceptor to compete and proliferate in this environment [41,42] (Figure 2). For EHEC, oxygen availability also affects LEE expression. When oxygen is present, LEE expression is activated through Cra and KdpE, and when oxygen is absent, KdpE and FusR strongly repress the LEE [16] (Figure 1). However, if oxygen and pyruvate are present, then FusR represses the LEE [16]. Together, EHEC uses these three transcriptional regulators to express the T3SS when oxygen is present, which is consistent with EHEC being near the epithelial surface [16].
Host and microbiota signals: Short and long chain fatty acids, bicarbonate, and pH
Short chain fatty acids (SCFAs) vary throughout the gut and can regulate EHEC virulence genes. Concentrations of SCFAs acetate, propionate, and butyrate are affected by the microbiota and the diet (reviewed in [43]). Butyrate, a preferred food source for colonocytes, also affects EHEC colonization. In mice fed high-fiber diets with floras producing more butyrate, EHEC colonizes the mice better compared to their low fiber diet counterparts, presumably because fewer Escherichia species are present to outcompete incoming pathogens [44] (Figure 2). EHEC senses butyrate through the leucine-responsive regulatory (LRP) protein, which promotes transcription of pchA, a positive regulator of the LEE [39]. SCFAs also affect EHEC flagella and motility. Low concentrations of SCFAs, characteristic of the large intestine where EHEC forms AE lesions, reduce flagella gene fliC expression and motility [45].
SCFAs production affects the overall pH of the large intestine. The host uses bicarbonate to neutralize the proton byproducts of SCFAs production. EHEC can sense bicarbonate through rscB and grvA [46,47]. RscB forms a heterodimer with GadE to activate the glutamate-dependent acid resistance (GDAR) pathway and repress LEE expression when EHEC is in acidic environments such as the stomach [47]. However, once EHEC is in the intestine, it senses bicarbonate through GrvA [47]. GrvA represses the GDAR pathway, promoting expression of the LEE and increased adherence to epithelial cells [46–48] (Figure 1). EHEC also uses CpxA to sense pH changes, copper, alterations in the membrane, and overexpression of envelope proteins such as NlpE [49]. If such stimuli are present, the histidine kinase sensor CpxA phosphorylates CpxR, which modulates transcription of LEE activators ler and grlA [49]. CpxR activates rpoH, which then activates transcription of the lon protease (Figure 1). Mutants in cpxA or lon are attenuated in a Galleria mellonella infection model [49].
Long chain fatty acids (LCFAs), cysteine, and D-serine also affect EHEC virulence (Figure 1). The cysteine utilization regulator, CutR, was recently found to sense exogenous cysteine and promote LEE expression (Pifer submitted). CutR also activates expression of the D-serine transporter, YhaO, and long chain fatty acid transporter, FadL (Pifer submitted). D-serine concentrations are low in the large intestine, but high in other body locations [50,51]. When D-serine concentrations are low, YhaO can promote YhaJ to bind to the LEE1 promoter and activate LEE transcription and repress the LEE when D-serine concentrations are high [50,51]. If LCFAs are unavailable, FadL can signal through the transcription factor FadR to promote fatty acid synthesis; FadR can also bind upstream of the LEE1 promoter to repress LEE transcription (Pifer submitted). In a C. rodentium model, both cutR and fadR were attenuated (Pifer submitted).
Conclusions and Future Directions
Pathogens need information on their environments to control their virulence armamentariums. Through recently published data in EHEC and its mouse surrogate C. rodentium, it is clear enteric pathogens like EHEC must sense multiple small molecules to avoid constructing its T3SS in unfavorable niches. In addition, the advancements in visualizing intact mucus layers and infection environments in vivo will no doubt show the effect of other host, microbiota, or diet derived signals on EHEC virulence. Regardless of how healthy or inflamed the host environment is, EHEC can outwit the host defense systems to colonize, grow, and eventually relocate to other microhabitats.
Highlights.
Enterohemorrhagic Escherichia coli (EHEC) tightly controls expression of its type III secretion system to compete for favorable microhabitats in the large intestine.
EHEC co-opts metabolites derived from the host or the microbiota found in the gut to promote attachment to epithelial cells.
EHEC senses oxygen concentrations to regulate virulence gene expression, which may be indicative of gut dysbiosis.
Different combinations of small molecules can reflect the health of the host and potential competition from the microbiota.
Highlights.
Enterohemorrhagic E. coli (EHEC) exploits microbiota and host derived signals and metabolites to recognize the gut environment and properly regulate expression of its virulence repertoire.
The host hormones epinephrine and norepinephrine acts through QseC and QseE to increase EHEC virulence.
Several host and microbiota-derived metabolites such as SCFAs, succinate, ethanolamine, fucose and aminoacids are also sensed as signals to regulate virulence gene expression
Acknowledgments
We would like to thank our funding sources.
Funding Sources
This work was funded through National Institutes of Health grants T32AI007520, AI053067, and AI05135.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Both authors, Kimberly Carlson-Banning and Vanessa Sperandio have no conflicts of interest
References and Recommended Reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nature Reviews Microbiology. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Elliott SJ, Hutcheson SW, Maria S, Mellies JL, Wainwright LA, Batchelor M, Frankel G, Knutton S, Kaper JB. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Molecular microbiology. 1999;33:1176–1189. doi: 10.1046/j.1365-2958.1999.01559.x. [DOI] [PubMed] [Google Scholar]
- 3.Elliott SJ, Wainwright LA, Timothy K, Jarvis KG, Deng Y, Lai L, Mcnamara BP, Michael S, Kaper JB. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Molecular microbiology. 1998;28:1–4. doi: 10.1046/j.1365-2958.1998.00783.x. [DOI] [PubMed] [Google Scholar]
- 4.Mellies JL, Barron AMS, Carmona AM. Enteropathogenic and enterohemorrhagic Escherichia coli virulence gene regulation. Infection and immunity. 2007;75:4199–4210. doi: 10.1128/IAI.01927-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kendall MM, Gruber CC, Parker CT, Sperandio V. Ethanolamine controls expression of genes encoding components involved in interkingdom signaling and virulence in enterohemorrhagic Escherichia coli O157:H7. mBio. 2012;3 doi: 10.1128/mBio.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luzader DH, Clark DE, Gonyar LA, Kendall MM. EutR is a direct regulator of genes that contribute to metabolism and virulence in enterohemorrhagic Escherichia coli O157:H7. Journal of bacteriology. 2013;195:4947–4953. doi: 10.1128/JB.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonyar LA, Kendall MM. Ethanolamine and choline promote expression of putative and characterized fimbriae in enterohemorrhagic Escherichia coli O157: H7. Infection and Immunity. 2014;82:193–201. doi: 10.1128/IAI.00980-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grenham S, Clarke G, Cryan JF, Dinan TG. Brain-gut-microbe communication in health and disease. Frontiers in Physiology. 2011 Dec 2;:1–15. doi: 10.3389/fphys.2011.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreira CG, Russell R, Mishra AA, Narayanan S, Ritchie JM, Waldor MK, Curtis MM, Winter SE, Weinshenker D, Sperandio V. Bacterial adrenergic sensors regulate virulence of enteric pathogens in the gut. mBio. 2016;7:1–14. doi: 10.1128/mBio.00826-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reading NC, Rasko DA, Torres AG, Sperandio V. The two-component system QseEF and the membrane protein QseG link adrenergic and stress sensing to bacterial pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5889–5894. doi: 10.1073/pnas.0811409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Njoroge J, Sperandio V. Enterohemorrhagic Escherichia coli virulence regulation by two bacterial adrenergic kinases, QseC and QseE. Infection and immunity. 2012;80:688–703. doi: 10.1128/IAI.05921-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Njoroge JW, Gruber C, Sperandio V. The interacting Cra and KdpE regulators are involved in the expression of multiple virulence factors in enterohemorrhagic Escherichia coli. Journal of bacteriology. 2013;195:2499–2508. doi: 10.1128/JB.02252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Njoroge JW, Nguyen Y, Curtis MM, Moreira CG, Sperandio V. Virulence meets metabolism: Cra and KdpE gene regulation in enterohemorrhagic Escherichia coli. mBio. 2012;3:e00280–00212. doi: 10.1128/mBio.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes DT, Clarke MB, Yamamoto K, Rasko DA, Sperandio V. The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC) PLoS pathogens. 2009;5:e1000553. doi: 10.1371/journal.ppat.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlson-Banning KM, Sperandio V. Catabolite and Oxygen Regulation of Enterohemorrhagic Escherichia coli Virulence. mBio. 2016;7:1–11. doi: 10.1128/mBio.01852-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamada N, Kim Y-G, Sham HP, Vallance BA, Puente JL, Martens EC, Núñez G. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science. 2012;336:344–347. doi: 10.1126/science.1222195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabich AJ, Jones SA, Chowdhury FZ, Cernosek A, Anderson A, Smalley D, Mchargue W, Hightower GA, Smith JT, Autieri SM, et al. Comparison of Carbon Nutrition for Pathogenic and Commensal Escherichia coli Strains in the Mouse Intestine. Infection and Immunity. 2008;76:1143–1152. doi: 10.1128/IAI.01386-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bertin Y, Chaucheyras-Durand F, Robbe-Masselot C, Durand A, de la Foye A, Harel J, Cohen PS, Conway T, Forano E, Martin C. Carbohydrate utilization by enterohaemorrhagic Escherichia coli O157:H7 in bovine intestinal content. Environmental microbiology. 2013;15:610–622. doi: 10.1111/1462-2920.12019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snider TA, Fabich AJ, Conway T, Clinkenbeard KD. E. coli O157:H7 catabolism of intestinal mucin-derived carbohydrates and colonization. Veterinary microbiology. 2009;136:150–154. doi: 10.1016/j.vetmic.2008.10.033. [DOI] [PubMed] [Google Scholar]
- 22.Conway T, Cohen PS. Commensal and Pathogenic Escherichia coli Metabolism in the Gut. Microbiology Spectrum. 2015:1–15. doi: 10.1128/microbiolspec.MBP-0006-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: Bacterial consumption of host glycans in the gut. Glycobiology. 2013;0:1–9. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24••.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell. 2016;167:1339–1353.e1321. doi: 10.1016/j.cell.2016.10.043. Using gnotobiotic mice colonized by a synthetic human microbiota, this study reveals how the microbiotia consume secreted glycoproteins needed for colonic integrity when dietary fiber is deficient. Depletion of the colonic mucus layer from reduced fiber intake leaves the host suscpetible to Citrobacter rodentium infection.This study demonstrates that entertic pathogens can exploit hosts with poor diets. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis Meredith M, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The Gut Commensal Bacteroides thetaiotaomicron Exacerbates Enteric Infection through Modification of the Metabolic Landscape. Cell Host & Microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pacheco AR, Curtis MM, Ritchie JM, Munera D, Waldor MK, Moreira CG, Sperandio V. Fucose sensing regulates bacterial intestinal colonization. Nature. 2012;492:113–117. doi: 10.1038/nature11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johansson MEV, Ambort D, Pelaseyed T, Schütte A, Gustafsson JK, Ermund A, Subramani DB, Holmén-Larsson JM, Thomsson KA, Bergström JH, et al. Composition and functional role of the mucus layers in the intestine. Cellular and molecular life sciences: CMLS. 2011;68:3635–3641. doi: 10.1007/s00018-011-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robbe C, Capon C, Coddeville B, Michalski J-C. Structural diversity and specific distribution of O-glycans in normal human mucins along the intestinal tract. The Biochemical journal. 2004;384:307–316. doi: 10.1042/BJ20040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schols HA. Complex Pectins: Structure elucidation using enzymes. Progress in Biotechnology. 1996;14:3–19. [Google Scholar]
- 30.Konopka JB. N-acetylglucosamine (GlcNAc) functions in cell signaling. Scientifica. 2012;2012:631–632. doi: 10.6064/2012/489208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature reviews Microbiology. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones SA, Gibson T, Maltby RC, Chowdhury FZ, Stewart V, Cohen PS, Conway T. Anaerobic respiration of Escherichia coli in the mouse intestine. Infection and immunity. 2011;79:4218–4226. doi: 10.1128/IAI.05395-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang D, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, et al. Carbon nutrition of Escherichia coli in the mouse intestine. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147:1055–1063.e1058. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host and Microbe. 2015;17:662–671. doi: 10.1016/j.chom.2015.03.005. This study demonstrates in mice the importance of microbiota produced byturate to limit oxygen availability in the gut and promote intestinal barrier function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nature communications. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng L, Kelly CJ, Colgan SP. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A Review in the Theme: Cellular Responses to Hypoxia. American journal of physiology Cell physiology. 2015;309:C350–360. doi: 10.1152/ajpcell.00191.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schüller S, Phillips AD. Microaerobic conditions enhance type III secretion and adherence of enterohaemorrhagic Escherichia coli to polarized human intestinal epithelial cells. Environmental microbiology. 2010;12:2426–2435. doi: 10.1111/j.1462-2920.2010.02216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakanishi N, Tashiro K, Kuhara S, Hayashi T, Sugimoto N, Tobe T. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology. 2009;155:521–530. doi: 10.1099/mic.0.023499-0. [DOI] [PubMed] [Google Scholar]
- 40.Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 41••.Hughes ER, Winter MG, Duerkop BA, Spiga L, Furtado de Carvalho T, Zhu W, Gillis CC, Büttner L, Smoot MP, Behrendt CL, et al. Microbial Respiration and Formate Oxidation as Metabolic Signatures of Inflammation-Associated Dysbiosis. Cell Host and Microbe. 2017;21:208–219. doi: 10.1016/j.chom.2017.01.005. This study demonstrates how mice with chemically induced dysbiosis of the microbiota have elevated formate concentrations in the gut. Enterobacteriaceae use formate as an electron donor and oxygen as a terminal electron acceptor to bloom, showing that aerobic respiration is a metabolic signature for inflammation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42••.Lopez CA, Miller BM, Rivera-chávez F, Velazquez EM, Byndloss MX, Chávez-Arroyo A, Lokken KL, Tsolis RM, Winter SE, Bäumler AJ. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science. 2016;353:1249–1253. doi: 10.1126/science.aag3042. This study demonstrates that Citrobacter rodentium uses its T3SS in mice to induce colonic crypt hyperplasia and increase oxygenation at the mucosal surface. C. rodentium then uses aerobic respiration to bloom in the colon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud D-J, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. The Journal of Lipid Research. 2013;54:2325–2340. doi: 10.1194/jlr.R036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zumbrun SD, Melton-Celsa AR, Smith MA, Gilbreath JJ, Merrell DS, O’Brien AD. Dietary choice affects Shiga toxin-producing Escherichia coli (STEC) O157:H7 colonization and disease. Proceedings of the National Academy of Sciences. 2013;110:E2126–E2133. doi: 10.1073/pnas.1222014110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lackraj T, Kim JI, Tran SL, Barnett Foster DE. Differential modulation of flagella expression in enterohaemorrhagic Escherichia coli O157: H7 by intestinal short-chain fatty acid mixes. Microbiology (United Kingdom) 2016;162:1761–1772. doi: 10.1099/mic.0.000357. [DOI] [PubMed] [Google Scholar]
- 46.Morgan JK, Vendura KW, Stevens SM, Riordan JT. RcsB determines the locus of enterocyte effacement (LEE) expression and adherence phenotype of Escherichia coli O157:H7 spinach outbreak strain TW14359 and coordinates bicarbonate-dependent LEE activation with repression of motility. Microbiology (United Kingdom) 2013;159:2342–2353. doi: 10.1099/mic.0.070201-0. [DOI] [PubMed] [Google Scholar]
- 47.Morgan JK, Carroll RK, Harro CM, Vendura KW, Shaw LN, Riordan JT. Global regulator of virulence A (GrvA) coordinates expression of discrete pathogenic mechanisms in enterohemorrhagic Escherichia coli through interactions with GadW-GadE. Journal of Bacteriology. 2015;198:394–409. doi: 10.1128/JB.00556-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abe H, Tatsuno I, Tobe T, Okutani A, Sasakawa C. Bicarbonate Ion Stimulates the Expression of Locus of Enterocyte Effacement-Encoded Genes in Enterohemorrhagic Escherichia coli O157:H7. Infection and Immunity. 2002;70:3500–3509. doi: 10.1128/IAI.70.7.3500-3509.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De la Cruz MA, Morgan JK, Ares MA, Yáñez-Santos JA, Riordan JT, Girón JA. The Two-Component System CpxRA Negatively Regulates the Locus of Enterocyte Effacement of Enterohemorrhagic Escherichia coli Involving σ32 and Lon protease. Frontiers in Cellular and Infection Microbiology. 2016;6 doi: 10.3389/fcimb.2016.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Connolly JPR, Gabrielsen M, Goldstone RJ, Grinter R, Wang D, Cogdell RJ, Walker D, Smith DGE, Roe AJ. A Highly Conserved Bacterial D-Serine Uptake System Links Host Metabolism and Virulence. PLoS Pathogens. 2016;12 doi: 10.1371/journal.ppat.1005359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Connolly JP, Goldstone RJ, Burgess K, Cogdell RJ, Beatson SA, Vollmer W, Smith DG, Roe AJ. The host metabolite D-serine contributes to bacterial niche specificity through gene selection. The ISME Journal. 2014;9:1039–1051. doi: 10.1038/ismej.2014.242. [DOI] [PMC free article] [PubMed] [Google Scholar]


