Abstract
Streptococcus (S.) suis is a frequent early colonizer of the upper respiratory tract of pigs. In fact, it is difficult to find S. suis-free animals under natural conditions, showing the successful adaptation of this pathogen to its porcine reservoir host. On the other hand, S. suis can cause life-threatening diseases and represents the most important bacterial cause of meningitis in pigs worldwide. Notably, S. suis can also cause zoonotic infections, such as meningitis, septicemia, endocarditis, and other diseases in humans. In Asia, it is classified as an emerging zoonotic pathogen and currently considered as one of the most important causes of bacterial meningitis in adults. The “two faces” of S. suis, one of a colonizing microbe and the other of a highly invasive pathogen, have raised many questions concerning the interpretation of diagnostic detection and the definition of virulence. Thus, one major research challenge is the identification of virulence-markers which allow differentiation of commensal and virulent strains. This is complicated by the high phenotypic and genotypic diversity of S. suis, as reflected by the occurrence of (at least) 33 capsular serotypes. In this review, we present current knowledge in the context of S. suis as a highly diverse pathobiont in the porcine respiratory tract that can exploit disrupted host homeostasis to flourish and promote inflammatory processes and invasive diseases in pigs and humans.
Keywords: Streptococcus suis, pathobiont, porcine respiratory tract, co-infections, respiratory infections
Introduction
Streptococcus (S.) suis is a commensal part of the respiratory microbiota of pigs, in particular of the tonsils and nasal cavities, but it can also cause highly invasive infections, such as meningitis, arthritis, endocarditis, bronchopneumonia, as well as septicemia and sudden death (Arends et al., 1984; Feng et al., 2014; Segura et al., 2016). Notably, though the colonization rate is up to 100%, clinical cases of S. suis infections, associated with meningitis, septicemia, or pneumonia, are by far less frequently reported (Goyette-Desjardins et al., 2014). S. suis is also considered an emerging zoonotic agent which can cause meningitis and sepsis in humans (Gottschalk et al., 2010). In contrast to swine, humans seem to be rarely colonized by S. suis. However, this remains to be studied in more detail and, therefore, human carrier rates (reported to be approximately 5% on average worldwide with respect to people in contact with pigs or pig products) may be underestimated (Strangmann et al., 2002; Goyette-Desjardins et al., 2014).
Streptococcus suis infections are known to be multi-factorial, unfavorable environmental conditions facilitate the development of disease. The nasopharynx is a reservoir niche for S. suis and various other (potentially) pathogenic microorganisms and commensals (Opriessnig et al., 2011). In this niche commensals can act as innocent bystander microbes, which inherently colonize the respiratory mucosa and can support other facultative pathogens to induce clinical disease. Those facultative pathogenic organisms are known as pathobionts. If pathogens play a dominant role in population changes of the microbiota and additionally manipulate the host response they are so-called keystone pathogens which can enhance the virulence of pathobionts leading to dysbiosis and inflammatory disease (Hajishengallis and Lamont, 2016). For S. suis, synergistic activities with other bacterial agents, such as Pasteurella multocida or Mycoplasma hyopneumoniae, as well as respiratory viruses like porcine reproductive and respiratory syndrome virus (PRRSV), porcine circovirus type 2, and swine influenza virus (SIV) (Fablet et al., 2011, 2012) may increase the risk of invasive infections (Meng et al., 2015). SIV and PRRVS are well-known keystone pathogens, since they pave the way for S. suis infections leading to severe respiratory symptoms and serious pneumonia (Thanawongnuwech et al., 2000; Lin et al., 2015; Meng et al., 2015).
Nevertheless, interactions of S. suis with the mucosal immune system and evasion of innate immune defense mechanisms are crucial for induction of disease. S. suis has several immune evasion strategies, for example, expression of polysaccharide capsule (CPS) to prevent phagocytosis-dependent killing mechanisms (Segura et al., 2004; Chabot-Roy et al., 2006), or biofilm formation which may protect S. suis from antimicrobials (Grenier et al., 2009; Bojarska et al., 2016). Such features seem to play a role in virulence but may also be important for survival as a pathobiont. In this review, we focus on the role of S. suis as a typical respiratory pathobiont in swine, which possesses a highly invasive potential and causes severe infectious diseases in pigs and humans. In particular, we address the epidemiology of S. suis in pigs and humans, its diversity, and its two “faces” as a commensal and invasive pathogen (illustrated in Figure 1). Finally, we also include possible models to study host–pathobiont interactions in the respiratory tract. Since a number of excellent reviews on virulence mechanisms and virulence-associated factors have been published in recent years, we will include such mechanisms and factors only with respect to their potential role for the lifestyle of S. suis as a pathobiont.
FIGURE 1.
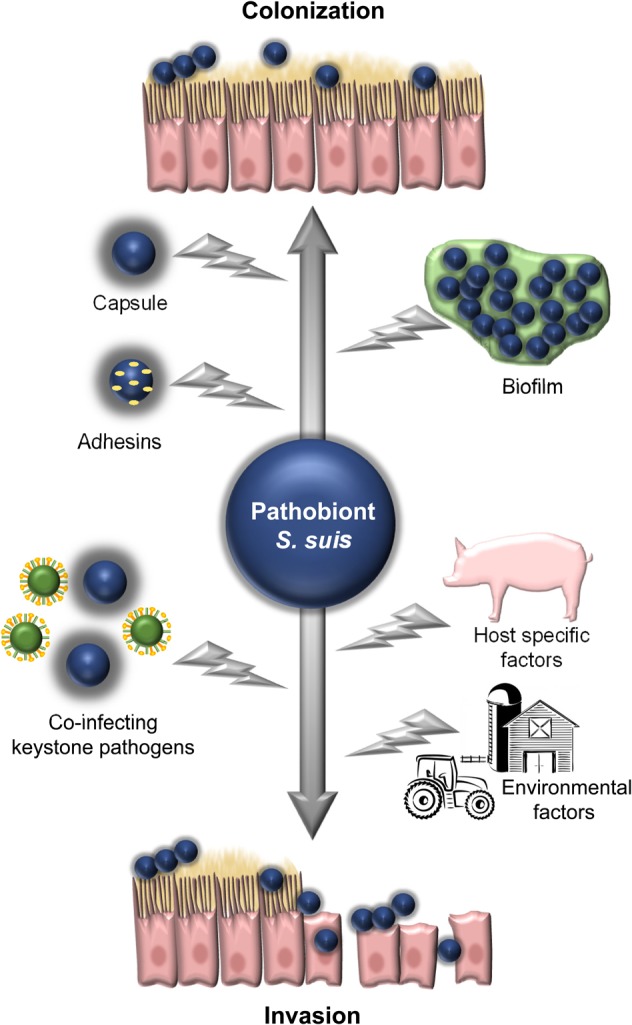
Illustration of the two phases (“faces”) of S. suis as a pathobiont in the respiratory tract. These are, firstly, as a colonization commensal bacterium and, secondly, as an invasive pathogen breaching different defense barriers. Some bacterial, host, and environmental factors, which contribute to the switch between both phases are depicted.
Epidemiology of S. suis in Pigs and Humans
Streptococcus suis is a widely distributed pathobiont and an emerging zoonotic pathogen. Its natural reservoir hosts are pigs (Lowe et al., 2011) and wild boars (Baums et al., 2007; Sanchez del Rey et al., 2014). Subclinical infected pigs play an important role in the epidemiology of S. suis as the main source of infection for other pigs and humans (Clifton-Hadley and Alexander, 1980). Susceptible pigs (especially weaning piglets) can suffer from meningitis, septicemia, pneumonia, endocarditis, or polyarthritis (Sanford and Tilker, 1982). Horizontal transmission via the respiratory tract due to nose-to-nose contact is the predominant route (Dekker et al., 2013), but vertical transmission from an infected sow to piglets via the genital tract during farrowing can also occur (Amass et al., 1997).
Humans can get infected with S. suis by eating raw or undercooked pork products (Fongcom et al., 2001; Gottschalk et al., 2010; Huong et al., 2014) or via cutaneous lesions when they get in contact with infected pigs or contaminated pork products (Wertheim et al., 2009). People in Asia are particularly affected because high-risk dishes, e.g., raw blood pudding, “tiet canh” (Huong et al., 2014), are common and the pig industry is more and more increasing while people are not aware of the risks of infection (Fulde and Valentin-Weigand, 2013). In Western countries only sporadic cases of human infections occur due to contact to infected pigs or raw pork meat (Goyette-Desjardins et al., 2014). In humans, S. suis causes mainly meningitis (Arends and Zanen, 1988) and septicemia [including streptococcal toxic shock-like syndrome/STSLS (Tang et al., 2006)], but cases of pneumonia, endocarditis, or peritonitis have also been reported (Huang et al., 2005). Some studies suggest that humans can also be healthy carriers of S. suis (Robertson and Blackmore, 1989; Elbers et al., 1999; Smith et al., 2008). Until end of December 2013 a total of 1642 human cases have been reported worldwide (Goyette-Desjardins et al., 2014), but this number has increased since then due to a high number of recent case reports and the high likelihood of misdiagnosis. Most of the human cases occurred in Asia (>90%), especially in Vietnam, Thailand, and China (Goyette-Desjardins et al., 2014). During the two outbreaks in China in 1998 and 2005, a total of 240 human cases were described (Tang et al., 2006; Yu et al., 2006).
Furthermore, S. suis can be found in the environment of slaughterhouses and wet markets (Ip et al., 2007; Ma et al., 2008; Nghia et al., 2011), particularly in Southeast Asia, which constitutes another source of infection for humans. In addition, S. suis was isolated from other animal species, such as rabbits, lambs, and dogs (Muckle et al., 2010; Sanchez del Rey et al., 2013; Muckle et al., 2014), though some of those had no close contact to pigs. These sources of infection should also be considered as a potential risk, especially since a few cases of human infections have been reported without any prior contact to pigs or pork products (Kerdsin et al., 2016).
Diversity of S. suis
Streptococcus suis is, genetically and phenotypically, a heterogeneous bacterial species. Strains belonging to different capsular serotypes or even to the same serotype differ from each other genetically (Blume et al., 2009; Gottschalk et al., 2013). For a comprehensive recent overview on the distribution of S. suis serotypes and genotypes, the reader is referred to a recent review by Goyette-Desjardins et al. (2014).
Multilocus sequence typing (MLST) is a method for genetic characterization, which allows to evaluate the epidemiology, the relation between different strains and virulent properties in more detail (Urwin and Maiden, 2003). King et al. (2002) have established a model of MLST for S. suis using seven different house-keeping genes (cpn60, dpr, recA, aroA, thrA, gki, and mutS). The nucleotide sequences of several alleles can be associated with each gene and the combination of those alleles of each isolate defines the sequence type (ST). Isolates with the same ST belong to the same clone. Occasionally, the same ST can comprise isolates with different serotypes and isolates with the same serotype could belong to different ST (King et al., 2002). In general, ST1 is mostly associated with clinical cases in both, pigs and humans, in Europe (Schultsz et al., 2012), Asia (King et al., 2002; Mai et al., 2008; Takamatsu et al., 2009), and Argentina (Callejo et al., 2016). ST7, which is responsible for the two large outbreaks in China in 1998 and 2005, seems to be endemic in China and Hong Kong (Ye et al., 2006; Li et al., 2010; Zhu et al., 2013). In North America, most of the isolated and analyzed serotype 2 strains from pigs and humans belong to ST25 and ST28 (Fittipaldi et al., 2011), the latter one has also been reported in Japan (Chang et al., 2006; Onishi et al., 2012) and other countries (Segura et al., 2017). ST101 to 104 is endemic in Thailand (Takamatsu et al., 2008, 2009; Kerdsin et al., 2011a), whereas ST20 was found only in humans in Europe [The Netherlands and France (Schultsz et al., 2012)].
So far, 35 serotypes based on the antigenicity of the capsular polysaccharides are known, but some of them have been suggested to belong to different bacterial species (Okura et al., 2016). Okura et al. (2013) developed a PCR which is able to detect all 35 serotypes but cannot distinguish serotypes 2 and 14 from serotypes 1/2 and 1, respectively. Especially these serotypes are commonly isolated from pigs (Wertheim et al., 2009). Additionally, there are many nontypable isolates, which do not agglutinate with any of the typing antisera (Messier et al., 2008) and which either belong to unknown encapsulated serotypes or non-encapsulated strains (Goyette-Desjardins et al., 2014). As reviewed recently by Goyette-Desjardins et al. (2014) the worldwide predominant serotype from diseased pigs is serotype 2 (27.9%), followed by serotypes 9 (19.4%) and 3 (15.9%). A total of 15.5% of the isolates were nontypable by serotyping (Goyette-Desjardins et al., 2014). The serotype distribution is different in healthy pigs, from which serotype 2 is less frequently isolated (Wisselink et al., 2000; Marois et al., 2007; Luque et al., 2010; Wang et al., 2013a). Unfortunately, there exist only few epidemiological data on S. suis in diseased pigs from those countries where most human cases are reported (Goyette-Desjardins et al., 2014). In some European countries, serotype 9 was more frequently found in diseased pigs and wild boars than serotype 2 (Vela et al., 2003; Tarradas et al., 2004; Luque et al., 2010; Schultsz et al., 2012; Sanchez del Rey et al., 2014), but until now no human serotype 9 cases have been reported. A recent study from Thailand revealed serotype 23 being the most prevalent in healthy pigs in Phayao Province, followed by serotypes 9, 7, and 2. Thirty-seven percent of the isolates was nontypable by multiplex PCR (Thongkamkoon et al., 2017). Another study from Northern Thailand (Chiang Mai Province) found mostly serotype 3 isolates in submaxillary glands of pig carcasses sold in wet markets (Wongsawan et al., 2015). A further study investigated samples from asymptomatic pigs from central Thailand and found mainly serotype 16 strains (Meekhanon et al., 2017). Taken together, S. suis serotype distribution differs worldwide, within a country, and even within the same region. Moreover, it was shown that pigs can be colonized by different serotypes at the same time (Flores et al., 1993). This raises the question whether the disease of a given animal is caused by a strain of a certain serotype or by interactions of several strains of different serotypes. Furthermore, it is unclear why some pigs in a herd get infected by a certain serotype while others do not (Higgins et al., 1990).
Worldwide, human cases are reported to be mainly due to serotype 2 (74.7%) and 14 (2.0%), both serotypes are equally involved in cases of meningitis (50–70%) and septicemia (20–25%) (Goyette-Desjardins et al., 2014). Only occasional cases were reported to be caused by serotypes 4, 5, 16, 21, 24, and 31 (Arends and Zanen, 1988; Nghia et al., 2008; Kerdsin et al., 2011b, 2016; Callejo et al., 2014; Gustavsson and Rasmussen, 2014; Hatrongjit et al., 2015; Taniyama et al., 2016). Most of the persons, who were infected by serotypes other than serotype 2, were suffering from a pre-existing liver cirrhosis (Kerdsin et al., 2011b; Taniyama et al., 2016) or other immunocompromising illnesses (Callejo et al., 2014). This suggests that those serotypes may be less virulent than serotype 2 strains. Callejo et al. (2016) reported that S. suis isolated from human cases in Argentina between 1995 and 2016 from cases of meningitis were caused by serotype 2 strains, except for one case caused by a serotype 5 isolate (Callejo et al., 2016). This is interesting because only few human cases have been reported from South America so far. One case of meningitis due to S. suis serotype 2 from Togo was reported in 2016 (Prince-David et al., 2016), which illustrates the emergence of this pathogen in a country, where it was previously unrecognized.
Although human cases occur only sporadically in Europe, the reported cases count for 8.5% of all human cases worldwide (Goyette-Desjardins et al., 2014). This may be explained by the fact that most of the European countries have a highly developed pig industry and the virulent serotype 2 can be found frequently in diseased pigs (Wisselink et al., 2000). In North America, only a few cases of human infections have been reported, although this country has the highest number of reports from diseased pigs (Goyette-Desjardins et al., 2014). One possible explanation may be that serotype 2 strains from North America are less virulent than Eurasian strains (Lachance et al., 2013).
The Commensal “Face” of S. suis
Although the survival mechanism of S. suis as a pathobiont remains to be elucidated, it seems clear that the S. suis genome of approximately 2 Mbp encodes for a variety of enzymes, putative adhesins, and other factors, which enable it to colonize the host with other commensals (and pathogens). Here, we focus on bacterial factors and mechanisms, which most likely are important for the commensal life of S. suis in the respiratory tract, though these factors may also contribute to virulence (Figure 1, upper part).
The innate and adaptive immune mechanisms in the respiratory tract play a major role in pathogen recognition, processing, and elimination thereby maintaining tissue homeostasis (Whitsett and Alenghat, 2015). Mucociliary activity of ciliated epithelial cells is a major defense barrier encountered by microbes entering the host via the respiratory tract. However, some pathogens, including S. suis, have adapted to colonize the respiratory cilia. Thus, the initial step in colonization, bacterial adherence, is crucial for development of a carrier state (Brassard et al., 2004). First studies on adhesins of S. suis were published in the early 1990s (Haataja et al., 1993, 1994, 1996; Tikkanen et al., 1995, 1996). In recent years, we learnt much more about mechanisms of adherence and tissue tropisms of S. suis, though we still do not know precisely what adhesins are essential for infection.
Salivary glycoproteins in humans have terminal sialic acids, which are reported to serve as glycan receptor motifs. These motifs are commonly recognized by commensal streptococcal bacteria such as, e.g., Streptococcus gordonii (Takahashi et al., 1997; Takahashi et al., 2002; Loimaranta et al., 2005; Deng et al., 2014). A recent study by Chuzeville et al. (2017) revealed that S. suis serotype 2 and 9 strains express genes coding for multimodal adhesion proteins known as antigen I/II (AgI/II). In the presence of salivary glycoproteins, AgI/II leads to the aggregation of S. suis, adherence, and colonization of the upper respiratory tract of pigs. Especially in serotype 9, the AgI/II is reported to facilitate aggregation and biofilm formation, and these aggregated bacteria could be swallowed, but are protected from the low pH in the stomach, which may enhance colonization of the intestine (Chuzeville et al., 2017). S. suis also has an adhesin known as factor H-binding protein, Fhb (Pian et al., 2012; Roy et al., 2016; Zhang et al., 2016). Factor H is an abundant host protein in the plasma, which is responsible in protecting the host from excessive complement effects and maintains complement homeostasis [reviewed in de Cordoba and de Jorge (2008)]. Binding of S. suis to factor H by Fhb results in enhanced adherence of the bacteria to epithelial and endothelial cells. Fhb also protects S. suis from phagocytosis and complement mediated killing (Pian et al., 2012; Roy et al., 2016). Zhang et al. (2016) reported the structural domains involved in binding of Fhb to the host cell receptor glycolipid GbO3, which is abundantly expressed on endothelial cells and certain epithelial cells. For further details on S. suis adhesins involved in adhesion to epithelial cells, the reader is referred to a recent comprehensive review on initial steps of S. suis pathogenesis (Segura et al., 2016).
The most prominent structure of S. suis is the polysaccharide capsule, of which several different antigen types exist, as described above. Most likely, the capsule covers adherence-mediating surface components, but it does not completely inhibit adherence to host cells. Accordingly, some studies showed that the absence (or downregulation) of the capsule increases the exposure of adhesins and subsequent bacterial adherence (Salasia et al., 1995; Lalonde et al., 2000; Benga et al., 2004; Esgleas et al., 2005). The thickness of the capsule depends on the bacterial environment in its host niche. It has been reported that the capsule is thinner during colonization and invasion of the respiratory epithelium, possibly to expose adhesins for better attachment to the epithelial cells (Gottschalk and Segura, 2000). Tanabe et al. (2010) also reported that the capsule hinders adhesins and hydrophobic components of S. suis, which are responsible for biofilm formation. However, in the bloodstream, the thickness of the capsule is higher and this enables S. suis to escape phagocytosis (Smith et al., 1999a,b; Gottschalk and Segura, 2000; Segura et al., 2004; Roy et al., 2016). This underlines that the expression of the capsule needs to be controlled during colonization (and subsequent infection).
The expression of genes responsible for S. suis capsule synthesis is regulated by transcriptional regulators such as catabolite control protein A (CcpA) (Willenborg et al., 2011) and small RNAs (sRNAs) like sRNA rss04. According to Willenborg et al. (2011) depletion of ccpA gene in S. suis resulted in a phenotype similar to a non-encapsulated strain, whereas the ccpA mutant showed reduced capsule thickness and higher susceptibility to phagocytosis compared to the wild-type (WT) parental strain (Willenborg et al., 2011). In contrast, the sRNA rss04 has an opposite effect. Transmission electron microscopic analysis revealed that S. suis Δrss04 had a thicker capsule compared to the WT and complemented strains and, therefore, its presence appears to repress CPS production by downregulating the expression of ccpA (Xiao et al., 2017). Thus, most likely capsule synthesis and its coordinated regulation are very important for the colonization and survival of S. suis as a pathobiont.
Some microorganisms escape hostile environments by aggregation in the form of biofilms that enable them to persist and colonize tissues, resist clearance from host defense mechanisms and antimicrobials, and facilitate exchange of genetic information (Donlan and Costerton, 2002). S. suis is able to form biofilms which is controlled by quorum sensing. This is a signaling network regulated by luxS gene (coding for the enzyme S-ribosylhomocysteinase, LuxS), which has been found in virulent S. suis serotype 2 strains. It has been reported that LuxS plays an important role by its ability to enhance the biosynthesis of auto-inducer 2 (AI-2), adherence, biofilm formation, cell metabolism, and resistance to host immune responses and antimicrobial therapy (Zhu et al., 2002; Vendeville et al., 2005; Han and Lu, 2009; Wang et al., 2011b, 2013c, 2015). Biofilm production by S. suis is induced via the activity of fibrinogen-mediated cross bridging of S. suis. The presence of fibrinogen could stimulate the expression of adhesins thereby facilitating adherence of the bacteria to each other (Bonifait et al., 2008). Moreover, mucin, produced by goblet cells, may enhance biofilm formation and promote survival in nutrient-limited condition as reported for Streptococcus mutans (Mothey et al., 2014). Bacteria forming biofilms resist antimicrobials better than planktonic cells (Olson et al., 2002; Grenier et al., 2009). It has been reported that virulent strains of S. suis have a higher ability to produce biofilms than avirulent strains (Wang et al., 2011a). The same authors reported that the adherence of S. suis forming a biofilm to human pharyngeal epithelial (HEp-2) cells was lower than that of planktonic cells suggesting a reduced virulence of S. suis in the former stage. On the one hand, in biofilms bacterial metabolism and expression of virulence-associated genes is reduced; on the other hand, secreted toxins may be trapped in the polysaccharide matrix resulting in less tissue damage to the host (Wang et al., 2011a). This may explain why virulent strains of S. suis can also be harmless components of the respiratory microbiome.
The Pathogenic “Face” of S. suis
As a facultative pathogenic bacterium, S. suis causes infectious diseases that are considered to be multifactorial, i.e., whether an initial infection remains subclinical or leads to clinical infection depends on several factors. It is long known that unfavorable environmental conditions such as overcrowding, poor ventilation and climatic conditions, poor hygiene status, high air pollution load, and other stressors correlate with an increasing clinical disease rate in pigs (Power, 1978; Sanford and Tilker, 1982; Chanter et al., 1993; Staats et al., 1997). Furthermore, host-specific factors, such as age, genetic background, and immunosuppression, influence disease development. Weaning piglets are most susceptible since protective maternal antibodies decline (Cloutier et al., 2003). Besides, pigs suffering from other bacterial and/or viral infections of the upper respiratory tract are more susceptible. In humans, especially advanced age and presence of pre-existing medical conditions that suppress the immune system are common predisposing factors for clinical S. suis infections (Arends and Zanen, 1988; Ma et al., 2008).
When (colonizing) S. suis encounters conditions that favor its replication, invasion, and evasion of immune control mechanisms, the opportunistic pathogen becomes pathogenic. This transition seems to depend on the individual strain and its equipment with virulence-(associated) factors, since only certain strains, geno-, and serotypes are isolated from diseased animals. In addition to the presence of virulence-related genes, their coordinated expression during infection is crucial for pathogenicity. Thus, host-, environment-, and pathogen-dependent factors are drivers of pathogenicity (Figure 1).
The respiratory tract can easily be colonized by environmental microorganisms which get access via direct contact or by aerosols. Thus, the upper airway tract harbors a complex and dynamic population of bacterial species including, e.g., Haemophilus parasuis, M. hyopneumoniae, Actinobacillus pleuropneumoniae, Actinobacillus suis, P. multocida, Bordetella bronchiseptica, and S. suis, as well as viruses like PRRSV, porcine circovirus type 2, and SIV (MacInnes et al., 2008; Opriessnig et al., 2011). Accordingly, porcine respiratory disease is often referred to as porcine respiratory disease complex due to its polymicrobial nature (Opriessnig et al., 2011).
The members of the respiratory microbiota differ in their intrinsic pathogenic potential and their role in shaping the population structure thereby building a mixture of non-pathogenic (accessory) commensal bacteria, which act as innocent bystander microbes or support pathogenic bacteria, and facultative pathogens known as pathobionts, such as S. suis. Moreover, keystone pathogens, sometimes also named master manipulators, play a dominant role in population changes, which may lead to subversion of the host immune system. This can affect the composition of the microbiota resulting in dysbiosis and increase of virulence of pathobionts, which then exploit the disrupted homeostasis for their invasion into deeper tissues (Hajishengallis and Lamont, 2016). For S. suis, PRRSV is considered to act as a keystone pathogen since PRRSV and S. suis coinfections in pig herds are frequently found (Schmitt et al., 2001) and co-infection of S. suis with PRRSV have been reported to enhance morbidity of S. suis infections (Thanawongnuwech et al., 2000; Feng et al., 2001; Auray et al., 2016). Although the precise mechanism by which PRRSV predisposes pigs to S. suis infection is unknown, recent studies showed that an altered innate immune system and exacerbating inflammatory responses are responsible for increasing the risk of S. suis infection in PRRSV-co-infected pigs. In vitro studies support the assumption that a decreased phagocytic activity by PRRSV-infected dendritic cells and porcine pulmonary macrophages may lead to a higher susceptibility to a subsequent S. suis infection (Auray et al., 2016). Furthermore, an epidemiological association between PRRSV in pigs and S. suis infections in pigs and humans was described (Hoa et al., 2013; Huong et al., 2016). To the best of our knowledge, there are no reports on associations of human viral respiratory infections with human S. suis infections.
Likewise, S. suis seems to be a pathobiont for infection by SIV. Experimental co-infections of pigs with SIV-S. suis revealed more severe clinical symptoms as well as more serious pathological changes and apoptosis of lungs compared to pigs mono-infected with either S. suis or SIV (Lin et al., 2015). Although very little is known about interactions between viral and bacterial pathogens and their role in the co-pathogenesis of respective diseases, in general virus-induced damage of the mucociliary barrier and a decreased immune response are considered to predispose pigs to secondary infections and pneumonia by opportunistic bacterial pathobionts (Fablet et al., 2011). Meng et al. (2015) found that SIV-facilitated adherence, colonization, and invasion of S. suis in a porcine precision-cut lung slices (PCLS) co-infection model was mediated by virus-induced impairment of the ciliary activity (Meng et al., 2015). Similarly, enhanced adherence of S. suis and direct binding in a capsule-dependent manner of S. suis to SIV- or SIV-infected cells were also found in co-infected newborn pig tracheal cells (Wang et al., 2013b; Wu et al., 2015). Thus, binding of S. suis to SIV-pre-infected cells appears to enable the bacterium to switch to an invasive pathogen. A further feature of SIV-S. suis co-infections seems to be an increased inflammatory response due to upregulation of inflammatory mediators like chemokines, interleukins, cell adhesion molecules, and eicosanoids, as it has been found in in vitro and in vivo experiments (Wang et al., 2013b; Dang et al., 2014; Lin et al., 2015). On the other hand, S. suis can also affect SIV infection. Lin et al. (2015) found increased viral loads in nasal swabs and lungs in co-infected pigs (Lin et al., 2015). Likewise, infection ability of SIV was enhanced after treatment with S. suis culture supernatants in vitro, most likely due to the proteolytic activity of a S. suis protease (Wang and Lu, 2008). In contrast, Wu et al. (2015) observed a negative effect on the growth capacity of SIV in S. suis co-infected cells (Wu et al., 2015).
Nevertheless, interactions of different microorganisms in the respiratory tract and their contribution to co-pathogenesis remain unclear. As suggested by Siqueira et al. (2017) the modes of actions of pathogens could be either additive or synergistic. Metatranscriptomic and metabolomic studies and appropriate infection models will surely help for better understanding of bacterial interactions and their roles in causing diseases or carrier states (Siqueira et al., 2017).
In Vitro Models to Study Host–Pathobiont Interactions in the Respiratory Tract
As outlined above, we are just beginning to understand the interplay of commensals, pathobionts, and keystone pathogens in the respiratory tract. Studies to dissect these complex processes should be carried out in respective animal models, e.g., in pigs, or under conditions which most closely mimic in vivo conditions. Since animal experiments have limitations for several reasons, ex vivo/in vitro tissue cell culture models receive more and more attention. Two of such models based on primary porcine respiratory epithelial cells are air–liquid-interface (ALI) cultures and PCLS. Both have been shown to be suitable to study host–pathobiont interactions of S. suis in the porcine respiratory tract (Meng et al., 2015, 2016).
For ALI cultures, primary porcine tracheal and bronchial epithelial cells (PTEC and PBEC) are isolated from swine lungs and cultured in a transwell system at ALI conditions for 4–5 weeks until the cells are well differentiated. Those well-differentiated respiratory epithelial cells build a pseudostratified epithelium, containing ciliated and mucin-producing cells (Figure 2A), and, therefore, represent the in vivo situation in the porcine respiratory tract. The expression of tight junction proteins and the development of a high trans-epithelial resistance indicate an epithelial barrier function (Prytherch et al., 2011). Human ALI culture systems have been proved suitable for modeling the respiratory tract by transcriptome analyses (Dvorak et al., 2011) and showing physiological responses to different pathogens (Krunkosky et al., 2007; Palermo et al., 2009). This cell culture model is particularly suitable to study the adherence and invasion of bacteria such as S. suis to well-differentiated respiratory epithelial cells and its effect on the epithelial barrier in vitro (Meng et al., 2016).
FIGURE 2.
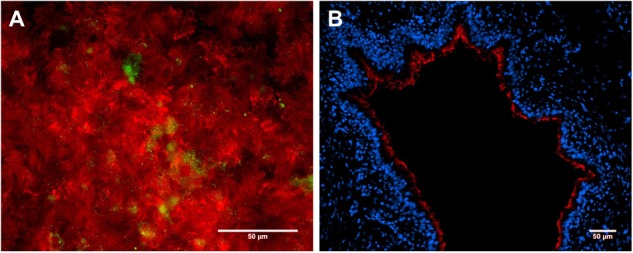
Primary porcine respiratory epithelial cell models to study host–pathobiont interactions in the respiratory tract. Immunofluorescence microscopy analysis of (A) primary porcine bronchial epithelial cells under air–liquid-interface (ALI) conditions after 3 weeks of differentiation and (B) a precision-cut lung slice (PCLS). Ciliated cells were stained by β-tubulin antibody (shown in red, A + B), mucin-producing cells were visualized by mucin 5-AC antibody (shown in green, A), and nuclei were stained by DAPI (shown in blue, B). Bars represent 50 μm.
The advantage of the ex vivo PCLS model is that it preserves the structural and functional integrity of the lung, including the ciliary activity at the bronchiolar surface (Figure 2B), since those slices are pieces of lung tissue which can be kept in cell culture medium for several days. PCLS have been proved to be convincing alternatives to in vivo experiments for physiological, pharmacological, and toxicological investigations (Morin et al., 2013). This method allows to investigate the adherence, colonization, and invasion of bacteria like S. suis and to study microbial effects on bronchial epithelial cells, e.g., the ciliary motility and bronchus-constriction, by light microscopy. A limitation of PCLS is the restricted time of viability of the cells, making it less suitable for studying long-term effects.
Air–liquid-interface cultures and PCLS as well as further models, such as organoids from the respiratory tract, will have to be further improved, e.g., by including immune cells. Such models and respective imaging techniques will enable researchers in the future to dissect the complex interactions of microbes on mucosal surfaces with each other and the host, which will contribute to a better understanding of the role that pathobionts play in infection processes in the airway system.
Author Contributions
PV-W developed the concept of the manuscript. DV, MW, YW, and PV-W wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This research was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany, to PV-W (Va23917-1).
References
- Amass S. F., SanMiguel P., Clark L. K. (1997). Demonstration of vertical transmission of Streptococcus suis in swine by genomic fingerprinting. J. Clin. Microbiol. 35 1595–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends J. P., Hartwig N., Rudolphy M., Zanen H. C. (1984). Carrier rate of Streptococcus suis capsular type 2 in palatine tonsils of slaughtered pigs. J. Clin. Microbiol. 20 945–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends J. P., Zanen H. C. (1988). Meningitis caused by Streptococcus suis in humans. Rev. Infect. Dis. 10 131–137. 10.1093/clinids/10.1.131 [DOI] [PubMed] [Google Scholar]
- Auray G., Lachance C., Wang Y., Gagnon C. A., Segura M., Gottschalk M. (2016). Transcriptional analysis of PRRSV-infected porcine dendritic cell response to Streptococcus suis infection reveals up-regulation of inflammatory-related genes expression. PLoS One 11:e0156019. 10.1371/journal.pone.0156019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baums C. G., Verkuhlen G. J., Rehm T., Silva L. M., Beyerbach M., Pohlmeyer K., et al. (2007). Prevalence of Streptococcus suis genotypes in wild boars of Northwestern Germany. Appl. Environ. Microbiol. 73 711–717. 10.1128/AEM.01800-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benga L., Goethe R., Rohde M., Valentin-Weigand P. (2004). Non-encapsulated strains reveal novel insights in invasion and survival of Streptococcus suis in epithelial cells. Cell Microbiol. 6 867–881. 10.1111/j.1462-5822.2004.00409.x [DOI] [PubMed] [Google Scholar]
- Blume V., Luque I., Vela A. I., Borge C., Maldonado A., Dominguez L., et al. (2009). Genetic and virulence-phenotype characterization of serotypes 2 and 9 of Streptococcus suis swine isolates. Int. Microbiol. 12 161–166. [PubMed] [Google Scholar]
- Bojarska A., Molska E., Janas K., Skoczynska A., Stefaniuk E., Hryniewicz W., et al. (2016). Streptococcus suis in invasive human infections in Poland: clonality and determinants of virulence and antimicrobial resistance. Eur. J. Clin. Microbiol. Infect. Dis. 35 917–925. 10.1007/s10096-016-2616-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifait L., Grignon L., Grenier D. (2008). Fibrinogen induces biofilm formation by Streptococcus suis and enhances its antibiotic resistance. Appl. Environ. Microbiol. 74 4969–4972. 10.1128/AEM.00558-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brassard J., Gottschalk M., Quessy S. (2004). Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet. Microbiol. 102 87–94. 10.1016/j.vetmic.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Callejo R., Prieto M., Salamone F., Auger J. P., Goyette-Desjardins G., Gottschalk M. (2014). Atypical Streptococcus suis in man, Argentina, 2013. Emerg. Infect. Dis. 20 500–502. 10.3201/eid2003.131148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callejo R., Zheng H., Du P., Prieto M., Xu J., Zielinski G., et al. (2016). Streptococcus suis serotype 2 strains isolated in Argentina (South America) are different from those recovered in North America and present a higher risk for humans. JMM Case Rep. 3:e005066. 10.1099/jmmcr.0.005066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot-Roy G., Willson P., Segura M., Lacouture S., Gottschalk M. (2006). Phagocytosis and killing of Streptococcus suis by porcine neutrophils. Microb. Pathog. 41 21–32. 10.1016/j.micpath.2006.04.001 [DOI] [PubMed] [Google Scholar]
- Chang B., Wada A., Ikebe T., Ohnishi M., Mita K., Endo M., et al. (2006). Characteristics of Streptococcus suis isolated from patients in Japan. Jpn. J. Infect. Dis. 59 397–399. [PubMed] [Google Scholar]
- Chanter N., Jones P. W., Alexander T. J. (1993). Meningitis in pigs caused by Streptococcus suis–a speculative review. Vet. Microbiol. 36 39–55. 10.1016/0378-1135(93)90127-S [DOI] [PubMed] [Google Scholar]
- Chuzeville S., Auger J. P., Dumesnil A., Roy D., Lacouture S., Fittipaldi N., et al. (2017). Serotype-specific role of antigen I/II in the initial steps of the pathogenesis of the infection caused by Streptococcus suis. Vet. Res. 48:39. 10.1186/s13567-017-0443-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifton-Hadley F. A., Alexander T. J. (1980). The carrier site and carrier rate of Streptococcus suis type II in pigs. Vet. Rec. 107 40–41. 10.1136/vr.107.2.40 [DOI] [PubMed] [Google Scholar]
- Cloutier G., D’Allaire S., Martinez G., Surprenant C., Lacouture S., Gottschalk M. (2003). Epidemiology of Streptococcus suis serotype 5 infection in a pig herd with and without clinical disease. Vet. Microbiol. 97 135–151. 10.1016/j.vetmic.2003.09.018 [DOI] [PubMed] [Google Scholar]
- Dang Y., Lachance C., Wang Y., Gagnon C. A., Savard C., Segura M., et al. (2014). Transcriptional approach to study porcine tracheal epithelial cells individually or dually infected with swine influenza virus and Streptococcus suis. BMC Vet. Res. 10:86. 10.1186/1746-6148-10-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cordoba S. R., de Jorge E. G. (2008). Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin. Exp. Immunol. 151 1–13. 10.1111/j.1365-2249.2007.03552.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker N., Bouma A., Daemen I., Klinkenberg D., van Leengoed L., Wagenaar J. A., et al. (2013). Effect of spatial separation of pigs on spread of Streptococcus suis serotype 9. PLoS One 8:e61339. 10.1371/journal.pone.0061339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Bensing B. A., Thamadilok S., Yu H., Lau K., Chen X., et al. (2014). Oral streptococci utilize a Siglec-like domain of serine-rich repeat adhesins to preferentially target platelet sialoglycans in human blood. PLoS Pathog. 10:e1004540. 10.1371/journal.ppat.1004540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan R. M., Costerton J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15 167–193. 10.1128/CMR.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak A., Tilley A. E., Shaykhiev R., Wang R., Crystal R. G. (2011). Do airway epithelium air-liquid cultures represent the in vivo airway epithelium transcriptome? Am. J. Respir. Cell Mol. Biol. 44 465–473. 10.1165/rcmb.2009-0453OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbers A. R., Vecht U., Osterhaus A. D., Groen J., Wisselink H. J., Diepersloot R. J., et al. (1999). Low prevalence of antibodies against the zoonotic agents Brucella abortus, Leptospira spp., Streptococcus suis serotype II, hantavirus, and lymphocytic choriomeningitis virus among veterinarians and pig farmers in the southern part of The Netherlands. Vet. Q. 21 50–54. 10.1080/01652176.1999.9694991 [DOI] [PubMed] [Google Scholar]
- Esgleas M., Lacouture S., Gottschalk M. (2005). Streptococcus suis serotype 2 binding to extracellular matrix proteins. FEMS Microbiol. Lett. 244 33–40. 10.1016/j.femsle.2005.01.017 [DOI] [PubMed] [Google Scholar]
- Fablet C., Marois C., Kuntz-Simon G., Rose N., Dorenlor V., Eono F., et al. (2011). Longitudinal study of respiratory infection patterns of breeding sows in five farrow-to-finish herds. Vet. Microbiol. 147 329–339. 10.1016/j.vetmic.2010.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fablet C., Marois-Crehan C., Simon G., Grasland B., Jestin A., Kobisch M., et al. (2012). Infectious agents associated with respiratory diseases in 125 farrow-to-finish pig herds: a cross-sectional study. Vet. Microbiol. 157 152–163. 10.1016/j.vetmic.2011.12.015 [DOI] [PubMed] [Google Scholar]
- Feng W., Laster S. M., Tompkins M., Brown T., Xu J. S., Altier C., et al. (2001). In utero infection by porcine reproductive and respiratory syndrome virus is sufficient to increase susceptibility of piglets to challenge by Streptococcus suis type II. J. Virol. 75 4889–4895. 10.1128/JVI.75.10.4889-4895.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Zhang H., Wu Z., Wang S., Cao M., Hu D., et al. (2014). Streptococcus suis infection: an emerging/reemerging challenge of bacterial infectious diseases? Virulence 5 477–497. 10.4161/viru.28595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittipaldi N., Xu J., Lacouture S., Tharavichitkul P., Osaki M., Sekizaki T., et al. (2011). Lineage and virulence of Streptococcus suis serotype 2 isolates from North America. Emerg. Infect. Dis. 17 2239–2244. 10.3201/eid1712.110609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores J. L., Higgins R., D’Allaire S., Charette R., Boudreau M., Gottschalk M. (1993). Distribution of the different capsular types of Streptococcus suis in nineteen swine nurseries. Can. Vet. J. 34 170–171. [PMC free article] [PubMed] [Google Scholar]
- Fongcom A., Pruksakorn S., Mongkol R., Tharavichitkul P., Yoonim N. (2001). Streptococcus suis infection in northern Thailand. J. Med. Assoc. Thai. 84 1502–1508. [PubMed] [Google Scholar]
- Fulde M., Valentin-Weigand P. (2013). Epidemiology and pathogenicity of zoonotic streptococci. Curr. Top. Microbiol. Immunol. 368 49–81. 10.1007/82_2012_277 [DOI] [PubMed] [Google Scholar]
- Gottschalk M., Lacouture S., Bonifait L., Roy D., Fittipaldi N., Grenier D. (2013). Characterization of Streptococcus suis isolates recovered between 2008 and 2011 from diseased pigs in Quebec, Canada. Vet. Microbiol. 162 819–825. 10.1016/j.vetmic.2012.10.028 [DOI] [PubMed] [Google Scholar]
- Gottschalk M., Segura M. (2000). The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet. Microbiol. 76 259–272. 10.1016/S0378-1135(00)00250-9 [DOI] [PubMed] [Google Scholar]
- Gottschalk M., Xu J., Calzas C., Segura M. (2010). Streptococcus suis: a new emerging or an old neglected zoonotic pathogen? Future Microbiol. 5 371–391. 10.2217/fmb.10.2 [DOI] [PubMed] [Google Scholar]
- Goyette-Desjardins G., Auger J. P., Xu J., Segura M., Gottschalk M. (2014). Streptococcus suis, an important pig pathogen and emerging zoonotic agent-an update on the worldwide distribution based on serotyping and sequence typing. Emerg. Microbes Infect. 3:e45. 10.1038/emi.2014.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenier D., Grignon L., Gottschalk M. (2009). Characterisation of biofilm formation by a Streptococcus suis meningitis isolate. Vet. J. 179 292–295. 10.1016/j.tvjl.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Gustavsson C., Rasmussen M. (2014). Septic arthritis caused by Streptococcus suis serotype 5 in pig farmer. Emerg. Infect. Dis. 20 489–490. 10.3201/eid2003.130535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haataja S., Tikkanen K., Hytonen J., Finne J. (1996). The Gal alpha 1-4 Gal-binding adhesin of Streptococcus suis, a gram-positive meningitis-associated bacterium. Adv. Exp. Med. Biol. 408 25–34. 10.1007/978-1-4613-0415-9_3 [DOI] [PubMed] [Google Scholar]
- Haataja S., Tikkanen K., Liukkonen J., Francois-Gerard C., Finne J. (1993). Characterization of a novel bacterial adhesion specificity of Streptococcus suis recognizing blood group P receptor oligosaccharides. J. Biol. Chem. 268 4311–4317. [PubMed] [Google Scholar]
- Haataja S., Tikkanen K., Nilsson U., Magnusson G., Karlsson K. A., Finne J. (1994). Oligosaccharide-receptor interaction of the Gal alpha 1-4Gal binding adhesin of Streptococcus suis. Combining site architecture and characterization of two variant adhesin specificities. J. Biol. Chem. 269 27466–27472. [PubMed] [Google Scholar]
- Hajishengallis G., Lamont R. J. (2016). Dancing with the Stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 24 477–489. 10.1016/j.tim.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X. G., Lu C. P. (2009). Detection of autoinducer-2 and analysis of the profile of luxS and pfs transcription in Streptococcus suis serotype 2. Curr. Microbiol. 58 146–152. 10.1007/s00284-008-9291-9 [DOI] [PubMed] [Google Scholar]
- Hatrongjit R., Kerdsin A., Gottschalk M., Takeuchi D., Hamada S., Oishi K., et al. (2015). First human case report of sepsis due to infection with Streptococcus suis serotype 31 in Thailand. BMC Infect. Dis. 15:392. 10.1186/s12879-015-1136-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins R., Gottschalk M., Mittal K. R., Beaudoin M. (1990). Streptococcus suis infection in swine. A sixteen month study. Can. J. Vet. Res. 54 170–173. [PMC free article] [PubMed] [Google Scholar]
- Hoa N. T., Chieu T. T., Do Dung S., Long N. T., Hieu T. Q., Luc N. T., et al. (2013). Streptococcus suis and porcine reproductive and respiratory syndrome, Vietnam. Emerg. Infect. Dis. 19 331–333. 10.3201/eid1902.120470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. T., Teng L. J., Ho S. W., Hsueh P. R. (2005). Streptococcus suis infection. J. Microbiol. Immunol. Infect. 38 306–313. [PubMed] [Google Scholar]
- Huong V. T., Hoa N. T., Horby P., Bryant J. E., Van Kinh N., Toan T. K., et al. (2014). Raw pig blood consumption and potential risk for Streptococcus suis infection, Vietnam. Emerg. Infect. Dis. 20 1895–1898. 10.3201/eid2011.140915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huong V. T., Thanh L. V., Phu V. D., Trinh D. T., Inui K., Tung N., et al. (2016). Temporal and spatial association of Streptococcus suis infection in humans and porcine reproductive and respiratory syndrome outbreaks in pigs in northern Vietnam. Epidemiol. Infect. 144 35–44. 10.1017/S0950268815000990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip M., Fung K. S., Chi F., Cheuk E. S., Chau S. S., Wong B. W., et al. (2007). Streptococcus suis in Hong Kong. Diagn. Microbiol. Infect. Dis. 57 15–20. 10.1016/j.diagmicrobio.2006.05.011 [DOI] [PubMed] [Google Scholar]
- Kerdsin A., Dejsirilert S., Puangpatra P., Sripakdee S., Chumla K., Boonkerd N., et al. (2011a). Genotypic profile of Streptococcus suis serotype 2 and clinical features of infection in humans, Thailand. Emerg. Infect. Dis. 17 835–842. 10.3201/eid1705.100754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerdsin A., Dejsirilert S., Sawanpanyalert P., Boonnark A., Noithachang W., Sriyakum D., et al. (2011b). Sepsis and spontaneous bacterial peritonitis in Thailand. Lancet 378:960 10.1016/S0140-6736(11)60923-9 [DOI] [PubMed] [Google Scholar]
- Kerdsin A., Gottschalk M., Hatrongjit R., Hamada S., Akeda Y., Oishi K. (2016). Fatal septic meningitis in child caused by Streptococcus suis serotype 24. Emerg. Infect. Dis. 22 1519–1520. 10.3201/eid2208.160452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. J., Leigh J. A., Heath P. J., Luque I., Tarradas C., Dowson C. G., et al. (2002). Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40 3671–3680. 10.1128/JCM.40.10.3671-3680.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krunkosky T. M., Jordan J. L., Chambers E., Krause D. C. (2007). Mycoplasma pneumoniae host-pathogen studies in an air-liquid culture of differentiated human airway epithelial cells. Microb. Pathog. 42 98–103. 10.1016/j.micpath.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Lachance C., Gottschalk M., Gerber P. P., Lemire P., Xu J., Segura M. (2013). Exacerbated type II interferon response drives hypervirulence and toxic shock by an emergent epidemic strain of Streptococcus suis. Infect. Immun. 81 1928–1939. 10.1128/IAI.01317-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde M., Segura M., Lacouture S., Gottschalk M. (2000). Interactions between Streptococcus suis serotype 2 and different epithelial cell lines. Microbiology 146 1913–1921. 10.1099/00221287-146-8-1913 [DOI] [PubMed] [Google Scholar]
- Li W., Ye C., Jing H., Cui Z., Bai X., Jin D., et al. (2010). Streptococcus suis outbreak investigation using multiple-locus variable tandem repeat number analysis. Microbiol. Immunol. 54 380–388. 10.1111/j.1348-0421.2010.00228.x [DOI] [PubMed] [Google Scholar]
- Lin X., Huang C., Shi J., Wang R., Sun X., Liu X., et al. (2015). Investigation of pathogenesis of H1N1 influenza virus and swine Streptococcus suis serotype 2 co-infection in pigs by microarray analysis. PLoS One 10:e0124086. 10.1371/journal.pone.0124086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loimaranta V., Jakubovics N. S., Hytönen J., Finne J., Jenkinson H. F., Strömberg N. (2005). Fluid- or surface-phase human salivary scavenger protein gp340 exposes different bacterial recognition properties. Infect. Immun. 73 2245–2252. 10.1128/IAI.73.4.2245-2252.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe B. A., Marsh T. L., Isaacs-Cosgrove N., Kirkwood R. N., Kiupel M., Mulks M. H. (2011). Microbial communities in the tonsils of healthy pigs. Vet. Microbiol. 147 346–357. 10.1016/j.vetmic.2010.06.025 [DOI] [PubMed] [Google Scholar]
- Luque I., Blume V., Borge C., Vela A. I., Perea J. A., Marquez J. M., et al. (2010). Genetic analysis of Streptococcus suis isolates recovered from diseased and healthy carrier pigs at different stages of production on a pig farm. Vet. J. 186 396–398. 10.1016/j.tvjl.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Ma E., Chung P. H., So T., Wong L., Choi K. M., Cheung D. T., et al. (2008). Streptococcus suis infection in Hong Kong: an emerging infectious disease? Epidemiol. Infect. 136 1691–1697. 10.1017/S0950268808000332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacInnes J. I., Gottschalk M., Lone A. G., Metcalf D. S., Ojha S., Rosendal T., et al. (2008). Prevalence of Actinobacillus pleuropneumoniae, Actinobacillus suis, Haemophilus parasuis, Pasteurella multocida, and Streptococcus suis in representative Ontario swine herds. Can. J. Vet. Res. 72 242–248. [PMC free article] [PubMed] [Google Scholar]
- Mai N. T., Hoa N. T., Nga T. V., Linh le D., Chau T. T., Sinh D. X., et al. (2008). Streptococcus suis meningitis in adults in Vietnam. Clin. Infect. Dis. 46 659–667. 10.1086/527385 [DOI] [PubMed] [Google Scholar]
- Marois C., Le Devendec L., Gottschalk M., Kobisch M. (2007). Detection and molecular typing of Streptococcus suis in tonsils from live pigs in France. Can. J. Vet. Res. 71 14–22. [PMC free article] [PubMed] [Google Scholar]
- Meekhanon N., Kaewmongkol S., Phimpraphai W., Okura M., Osaki M., Sekizaki T., et al. (2017). Potentially hazardous Streptococcus suis strains latent in asymptomatic pigs in a major swine production area of Thailand. J. Med. Microbiol. 66 662–669. 10.1099/jmm.0.000483 [DOI] [PubMed] [Google Scholar]
- Meng F., Wu N. H., Nerlich A., Herrler G., Valentin-Weigand P., Seitz M. (2015). Dynamic Virus-bacterium interactions in a porcine precision-cut lung slice coinfection model: swine influenza virus paves the way for Streptococcus suis infection in a two-step process. Infect. Immun. 83 2806–2815. 10.1128/IAI.00171-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F., Wu N. H., Seitz M., Herrler G., Valentin-Weigand P. (2016). Efficient suilysin-mediated invasion and apoptosis in porcine respiratory epithelial cells after streptococcal infection under air-liquid interface conditions. Sci. Rep. 6:26748. 10.1038/srep26748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messier S., Lacouture S., Gottschalk M. Groupe de Recherche sur les Maladies Infectieusesdu Porc Centre de Recherche en Infectiologie Porcine (2008). Distribution of Streptococcus suis capsular types from 2001 to 2007. Can. Vet. J. 49 461–462. [PMC free article] [PubMed] [Google Scholar]
- Morin J. P., Baste J. M., Gay A., Crochemore C., Corbiere C., Monteil C. (2013). Precision cut lung slices as an efficient tool for in vitro lung physio-pharmacotoxicology studies. Xenobiotica 43 63–72. 10.3109/00498254.2012.727043 [DOI] [PubMed] [Google Scholar]
- Mothey D., Buttaro B. A., Piggot P. J. (2014). Mucin can enhance growth, biofilm formation, and survival of Streptococcus mutans. FEMS Microbiol. Lett. 350 161–167. 10.1111/1574-6968.12336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckle A., Giles J., Lund L., Stewart T., Gottschalk M. (2010). Isolation of Streptococcus suis from the urine of a clinically ill dog. Can. Vet. J. 51 773–774. [PMC free article] [PubMed] [Google Scholar]
- Muckle A., Lopez A., Gottschalk M., Lopez-Mendez C., Giles J., Lund L., et al. (2014). Isolation of Streptococcus suis from 2 lambs with a history of lameness. Can. Vet. J. 55 946–949. [PMC free article] [PubMed] [Google Scholar]
- Nghia H. D., Hoa N. T., Linh le D., Campbell J., Diep T. S., Chau N. V., et al. (2008). Human case of Streptococcus suis serotype 16 infection. Emerg. Infect. Dis. 14 155–157. 10.3201/eid1401.070534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nghia H. D., Tu le T. P., Wolbers M., Thai C. Q., Hoang N. V., Nga T. V., et al. (2011). Risk factors of Streptococcus suis infection in Vietnam. A case-control study. PLoS One 6:e17604. 10.1371/journal.pone.0017604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M., Osaki M., Nomoto R., Arai S., Osawa R., Sekizaki T., et al. (2016). Current taxonomical situation of Streptococcus suis. Pathogens 5:45. 10.3390/pathogens5030045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okura M., Takamatsu D., Maruyama F., Nozawa T., Nakagawa I., Osaki M., et al. (2013). Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl. Environ. Microbiol. 79 2796–2806. 10.1128/AEM.03742-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M. E., Ceri H., Morck D. W., Buret A. G., Read R. R. (2002). Biofilm bacteria: formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 66 86–92. [PMC free article] [PubMed] [Google Scholar]
- Onishi H., Sugawara M., Okura M., Osaki M., Takamatsu D. (2012). Prevalence of Streptococcus suis genotypes in isolates from porcine endocarditis in East Japan. J. Vet. Med. Sci. 74 1681–1684. 10.1292/jvms.12-0301 [DOI] [PubMed] [Google Scholar]
- Opriessnig T., Gimenez-Lirola L. G., Halbur P. G. (2011). Polymicrobial respiratory disease in pigs. Anim. Health Res. Rev. 12 133–148. 10.1017/S1466252311000120 [DOI] [PubMed] [Google Scholar]
- Palermo L. M., Porotto M., Yokoyama C. C., Palmer S. G., Mungall B. A., Greengard O., et al. (2009). Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J. Virol. 83 6900–6908. 10.1128/JVI.00475-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pian Y., Gan S., Wang S., Guo J., Wang P., Zheng Y., et al. (2012). Fhb, a novel factor H-binding surface protein, contributes to the antiphagocytic ability and virulence of Streptococcus suis. Infect. Immun. 80 2402–2413. 10.1128/IAI.06294-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power S. B. (1978). Streptococcus suis type 2 infection in pigs. Vet. Rec. 102 215–216. 10.1136/vr.102.10.215 [DOI] [PubMed] [Google Scholar]
- Prince-David M., Salou M., Marois-Crehan C., Assogba K., Plainvert C., Balogou K. A., et al. (2016). Human meningitis due to Streptococcus suis in Lome, Togo: a case report. BMC Infect. Dis. 16:651. 10.1186/s12879-016-2006-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prytherch Z., Job C., Marshall H., Oreffo V., Foster M., BeruBe K. (2011). Tissue-Specific stem cell differentiation in an in vitro airway model. Macromol. Biosci. 11 1467–1477. 10.1002/mabi.201100181 [DOI] [PubMed] [Google Scholar]
- Robertson I. D., Blackmore D. K. (1989). Occupational exposure to Streptococcus suis type 2. Epidemiol. Infect. 103 157–164. 10.1017/S0950268800030454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy D., Grenier D., Segura M., Mathieu-Denoncourt A., Gottschalk M. (2016). Recruitment of factor H to the Streptococcus suis cell surface is multifactorial. Pathogens 5:E47. 10.3390/pathogens5030047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salasia S. I., Lammler C., Herrmann G. (1995). Properties of a Streptococcus suis isolate of serotype 2 and two capsular mutants. Vet. Microbiol. 45 151–156. 10.1016/0378-1135(95)00036-A [DOI] [PubMed] [Google Scholar]
- Sanchez del Rey V., Fernandez-Garayzabal J. F., Briones V., Iriso A., Dominguez L., Gottschalk M., et al. (2013). Genetic analysis of Streptococcus suis isolates from wild rabbits. Vet. Microbiol. 165 483–486. 10.1016/j.vetmic.2013.04.025 [DOI] [PubMed] [Google Scholar]
- Sanchez del Rey V., Fernandez-Garayzabal J. F., Mentaberre G., Briones V., Lavin S., Dominguez L., et al. (2014). Characterisation of Streptococcus suis isolates from wild boars (Sus scrofa). Vet. J. 200 464–467. 10.1016/j.tvjl.2014.03.013 [DOI] [PubMed] [Google Scholar]
- Sanford S. E., Tilker M. E. (1982). Streptococcus suis type II-associated diseases in swine: observations of a one-year study. J. Am. Vet. Med. Assoc. 181 673–676. [PubMed] [Google Scholar]
- Schmitt C. S., Halbur P. G., Roth J. A., Kinyon J. M., Kasorndorkbua C., Thacker B. (2001). Influence of ampicillin, ceftiofur, attenuated live PRRSV vaccine, and reduced dose Streptococcus suis exposure on disease associated with PRRSV and S. suis coinfection. Vet. Microbiol. 78 29–37. 10.1016/S0378-1135(00)00289-3 [DOI] [PubMed] [Google Scholar]
- Schultsz C., Jansen E., Keijzers W., Rothkamp A., Duim B., Wagenaar J. A., et al. (2012). Differences in the population structure of invasive Streptococcus suis strains isolated from pigs and from humans in The Netherlands. PLoS One 7:e33854. 10.1371/journal.pone.0033854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura M., Calzas C., Grenier D., Gottschalk M. (2016). Initial steps of the pathogenesis of the infection caused by Streptococcus suis: fighting against nonspecific defenses. FEBS Lett. 590 3772–3799. 10.1002/1873-3468.12364 [DOI] [PubMed] [Google Scholar]
- Segura M., Fittipaldi N., Calzas C., Gottschalk M. (2017). Critical Streptococcus suis virulence factors: are they all really critical? Trends Microbiol. 25 585–599. 10.1016/j.tim.2017.02.005 [DOI] [PubMed] [Google Scholar]
- Segura M., Gottschalk M., Olivier M. (2004). Encapsulated Streptococcus suis inhibits activation of signaling pathways involved in phagocytosis. Infect. Immun. 72 5322–5330. 10.1128/IAI.72.9.5322-5330.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira F. M., Perez-Wohlfeil E., Carvalho F. M., Trelles O., Schrank I. S., Vasconcelos A. T. R., et al. (2017). Microbiome overview in swine lungs. PLoS One 12:e0181503. 10.1371/journal.pone.0181503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. E., Damman M., van der Velde J., Wagenaar F., Wisselink H. J., Stockhofe-Zurwieden N., et al. (1999a). Identification and characterization of the cps locus of Streptococcus suis serotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect. Immun. 67 1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. E., Veenbergen V., van der Velde J., Damman M., Wisselink H. J., Smits M. A. (1999b). The cps genes of Streptococcus suis serotypes 1,2, and 9: development of rapid serotype-specific PCR assays. J. Clin. Microbiol. 37 3146–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. C., Capuano A. W., Boese B., Myers K. P., Gray G. C. (2008). Exposure to Streptococcus suis among US swine workers. Emerg. Infect. Dis. 14 1925–1927. 10.3201/eid1412.080162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staats J. J., Feder I., Okwumabua O., Chengappa M. M. (1997). Streptococcus suis: past and present. Vet. Res. Commun. 21 381–407. 10.1023/A:1005870317757 [DOI] [PubMed] [Google Scholar]
- Strangmann E., Froleke H., Kohse K. P. (2002). Septic shock caused by Streptococcus suis: case report and investigation of a risk group. Int. J. Hyg. Environ. Health 205 385–392. 10.1078/1438-4639-00165 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Konishi K., Cisar J. O., Yoshikawa M. (2002). Identification and characterization of hsa, the gene encoding the sialic acid-binding adhesin of Streptococcus gordonii DL1. Infect. Immun. 70 1209–1218. 10.1128/IAI.70.3.1209-1218.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Sandberg A. L., Ruhl S., Muller J., Cisar J. O. (1997). A specific cell surface antigen of Streptococcus gordonii is associated with bacterial hemagglutination and adhesion to alpha2-3-linked sialic acid-containing receptors. Infect. Immun. 65 5042–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D., Nishino H., Ishiji T., Ishii J., Osaki M., Fittipaldi N., et al. (2009). Genetic organization and preferential distribution of putative pilus gene clusters in Streptococcus suis. Vet. Microbiol. 138 132–139. 10.1016/j.vetmic.2009.02.013 [DOI] [PubMed] [Google Scholar]
- Takamatsu D., Wongsawan K., Osaki M., Nishino H., Ishiji T., Tharavichitkul P., et al. (2008). Streptococcus suis in humans, Thailand. Emerg. Infect. Dis. 14 181–183. 10.3201/eid1401.070568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe S., Bonifait L., Fittipaldi N., Grignon L., Gottschalk M., Grenier D. (2010). Pleiotropic effects of polysaccharide capsule loss on selected biological properties of Streptococcus suis. Can. J. Vet. Res. 74 65–70. [PMC free article] [PubMed] [Google Scholar]
- Tang J., Wang C., Feng Y., Yang W., Song H., Chen Z., et al. (2006). Streptococcal toxic shock syndrome caused by Streptococcus suis serotype 2. PLoS Med. 3:e151. 10.1371/journal.pmed.0030151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniyama D., Sakurai M., Sakai T., Kikuchi T., Takahashi T. (2016). Human case of bacteremia due to Streptococcus suis serotype 5 in Japan: the first report and literature review. IDCases 6 36–38. 10.1016/j.idcr.2016.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarradas C., Perea A., Vela A. I., Goyache J., Dominguez L., Fernandez-Garaizabal J. F., et al. (2004). Distribution of serotypes of Streptococcus suis isolated from diseased pigs in Spain. Vet. Rec. 154 665–666. 10.1136/vr.154.21.665 [DOI] [PubMed] [Google Scholar]
- Thanawongnuwech R., Brown G. B., Halbur P. G., Roth J. A., Royer R. L., Thacker B. J. (2000). Pathogenesis of porcine reproductive and respiratory syndrome virus-induced increase in susceptibility to Streptococcus suis infection. Vet. Pathol. 37 143–152. 10.1354/vp.37-2-143 [DOI] [PubMed] [Google Scholar]
- Thongkamkoon P., Kiatyingangsulee T., Gottschalk M. (2017). Serotypes of Streptococcus suis isolated from healthy pigs in Phayao Province, Thailand. BMC Res Notes 10:53. 10.1186/s13104-016-2354-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen K., Haataja S., Finne J. (1996). The galactosyl-(alpha 1-4)-galactose-binding adhesin of Streptococcus suis: occurrence in strains of different hemagglutination activities and induction of opsonic antibodies. Infect. Immun. 64 3659–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen K., Haataja S., Francois-Gerard C., Finne J. (1995). Purification of a galactosyl-alpha 1-4-galactose-binding adhesin from the gram-positive meningitis-associated bacterium Streptococcus suis. J. Biol. Chem. 270 28874–28878. 10.1074/jbc.270.48.28874 [DOI] [PubMed] [Google Scholar]
- Urwin R., Maiden M. C. (2003). Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11 479–487. 10.1016/j.tim.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Vela A. I., Goyache J., Tarradas C., Luque I., Mateos A., Moreno M. A., et al. (2003). Analysis of genetic diversity of Streptococcus suis clinical isolates from pigs in Spain by pulsed-field gel electrophoresis. J. Clin. Microbiol. 41 2498–2502. 10.1128/JCM.41.6.2498-2502.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendeville A., Winzer K., Heurlier K., Tang C. M., Hardie K. R. (2005). Making ’sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3 383–396. 10.1038/nrmicro1146 [DOI] [PubMed] [Google Scholar]
- Wang K., Lu C. (2008). Streptococcus suis type 2 culture supernatant enhances the infection ability of the Swine influenza virus H3 subtype in MDCK cells. Berl. Munch. Tierarztl. Wochenschr. 121 198–202. [PubMed] [Google Scholar]
- Wang K., Zhang W., Li X., Lu C., Chen J., Fan W., et al. (2013a). Characterization of Streptococcus suis isolates from slaughter swine. Curr. Microbiol. 66 344–349. 10.1007/s00284-012-0275-4 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yi L., Zhang Z., Fan H., Cheng X., Lu C. (2013c). Overexpression of luxS cannot increase autoinducer-2 production, only affect the growth and biofilm formation in Streptococcus suis. Sci. World J. 2013:924276. 10.1155/2013/924276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gagnon C. A., Savard C., Music N., Srednik M., Segura M., et al. (2013b). Capsular sialic acid of Streptococcus suis serotype 2 binds to swine influenza virus and enhances bacterial interactions with virus-infected tracheal epithelial cells. Infect. Immun. 81 4498–4508. 10.1128/IAI.00818-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yi L., Wang S., Fan H., Ding C., Mao X., et al. (2015). Crystal structure and identification of two key amino acids involved in AI-2 production and biofilm formation in Streptococcus suis LuxS. PLoS One 10:e0138826. 10.1371/journal.pone.0138826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Wu Z., Lu C. (2011a). Reduced virulence is an important characteristic of biofilm infection of Streptococcus suis. FEMS Microbiol. Lett. 316 36–43. 10.1111/j.1574-6968.2010.02189.x [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang W., Wu Z., Zhu X., Lu C. (2011b). Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet. Microbiol. 152 151–160. 10.1016/j.vetmic.2011.04.029 [DOI] [PubMed] [Google Scholar]
- Wertheim H. F., Nghia H. D., Taylor W., Schultsz C. (2009). Streptococcus suis: an emerging human pathogen. Clin. Infect. Dis. 48 617–625. 10.1086/596763 [DOI] [PubMed] [Google Scholar]
- Whitsett J. A., Alenghat T. (2015). Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat. Immunol. 16 27–35. 10.1038/ni.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenborg J., Fulde M., de Greeff A., Rohde M., Smith H. E., Valentin-Weigand P., et al. (2011). Role of glucose and CcpA in capsule expression and virulence of Streptococcus suis. Microbiology 157 1823–1833. 10.1099/mic.0.046417-0 [DOI] [PubMed] [Google Scholar]
- Wisselink H. J., Smith H. E., Stockhofe-Zurwieden N., Peperkamp K., Vecht U. (2000). Distribution of capsular types and production of muramidase-released protein (MRP) and extracellular factor (EF) of Streptococcus suis strains isolated from diseased pigs in seven European countries. Vet. Microbiol. 74 237–248. 10.1016/S0378-1135(00)00188-7 [DOI] [PubMed] [Google Scholar]
- Wongsawan K., Gottschalk M., Tharavichitkul P. (2015). Serotype- and virulence-associated gene profile of Streptococcus suis isolates from pig carcasses in Chiang Mai Province, Northern Thailand. J. Vet. Med. Sci. 77 233–236. 10.1292/jvms.14-0380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N. H., Meng F., Seitz M., Valentin-Weigand P., Herrler G. (2015). Sialic acid-dependent interactions between influenza viruses and Streptococcus suis affect the infection of porcine tracheal cells. J. Gen. Virol. 96 2557–2568. 10.1099/jgv.0.000223 [DOI] [PubMed] [Google Scholar]
- Xiao G., Tang H., Zhang S., Ren H., Dai J., Lai L., et al. (2017). Streptococcus suis small RNA rss04 contributes to the induction of meningitis by regulating capsule synthesis and by inducing biofilm formation in a mouse infection model. Vet. Microbiol. 199 111–119. 10.1016/j.vetmic.2016.12.034 [DOI] [PubMed] [Google Scholar]
- Ye C., Zhu X., Jing H., Du H., Segura M., Zheng H., et al. (2006). Streptococcus suis sequence type 7 outbreak, Sichuan, China. Emerg. Infect. Dis. 12 1203–1208. 10.3201/eid1708.060232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Jing H., Chen Z., Zheng H., Zhu X., Wang H., et al. (2006). Human Streptococcus suis outbreak, Sichuan, China. Emerg. Infect. Dis. 12 914–920. 10.3201/eid1206.051194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Hao H., Yu Y., Kong D., Chen S., Jiang H., et al. (2016). Structural basis of the interaction between the meningitis pathogen Streptococcus suis adhesin Fhb and its human receptor. FEBS Lett. 590 1384–1392. 10.1002/1873-3468.12174 [DOI] [PubMed] [Google Scholar]
- Zhu J., Miller M. B., Vance R. E., Dziejman M., Bassler B. L., Mekalanos J. J. (2002). Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 99 3129–3134. 10.1073/pnas.052694299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Wu C., Sun X., Zhang A., Zhu J., Hua Y., et al. (2013). Characterization of Streptococcus suis serotype 2 isolates from China. Vet. Microbiol. 166 527–534. 10.1016/j.vetmic.2013.06.009 [DOI] [PubMed] [Google Scholar]


