Abstract
Cicer arietinum L. (chickpea) is the world's fourth most widely grown pulse. Chickpea seeds are a primary source of dietary protein for humans, and chickpea cultivation contributes to biological nitrogen fixation in the soil, given its symbiotic relationship with rhizobia. Therefore, chickpea cultivation plays a pivotal role in innovative sustainable models of agro-ecosystems inserted in crop rotation in arid and semi-arid environments for soil improvement and the reduction of chemical inputs. Indeed, the arid and semi-arid tropical zones of Africa and Asia have been primary areas of cultivation and diversification. Yet, nowadays, chickpea is gaining prominence in Canada, Australia, and South America where it constitutes a main ingredient in vegetarian and vegan diets. Viruses and plant parasitic nematodes (PPNs) have been considered to be of minor and local impact in primary areas of cultivation. However, the introduction of chickpea in new environments exposes the crop to these biotic stresses, compromising its yields. The adoption of high-throughput genomic technologies, including genome and transcriptome sequencing projects by the chickpea research community, has provided major insights into genome evolution as well as genomic architecture and domestication. This review summarizes the major viruses and PPNs that affect chickpea cultivation worldwide. We also present an overview of the current state of chickpea genomics. Accordingly, we explore the opportunities that genomics, post-genomics and novel editing biotechnologies are offering in order to understand chickpea diseases and stress tolerance and to design innovative control strategies.
Keywords: Cicer arietinum L., plant viruses, plant parasitic nematodes, RNA silencing, viral metagenomics, plant transformation, genome editing
Cicer arietinum L.: uses, origin, and distribution
In many developing countries, grain legumes have gained much importance in view of acute shortages in the production of animal proteins and the prevalence of protein malnutrition. Conversely, they are a valid alternative as a source of protein for specific (vegetarian or vegan) or balanced diets worldwide, particularly in developed countries.
Legumes are able to fix atmospheric nitrogen, in association with bacteria, and play a central role in low-input and sustainable agricultural systems (Graham and Vance, 2003). With a global production of ca. 77 × 106 tons, grain legumes (also known as “pulses”) rank third after cereals and oilseeds (FAO, 2014). The world production of chickpea in 2014 was more than 13 × 106 tons (FAO, 2014), making chickpea rank fourth among the pulses after soybean, peanut, and common bean. However, chickpea can be considered the most important crop at regional level, especially in semi-arid areas of the world and in Mediterranean regions (FAO, 2014). The genus Cicer L. includes 44 taxa, 9 annuals, and 35 perennials, and has a narrow genetic base, probably as a consequence of it being a monophyletic descendent from its wild progenitor Cicer reticulatum, grown in the Fertile Crescent region (the center of chickpea domestication and diversification) (Abbo et al., 2003). The most popularly known species is the cultivated Cicer arietinum L., with 2n = 2x = 16 chromosomes and a genome size of ~738 Mb (Varshney et al., 2013). Commercially, the cultivated chickpea varieties are grouped according to the plant's flowers pigmentation as well as size and color of seeds; i.e., desi-type (small-seeded) and kabuli-type (large-seeded). Desi-type accounts for about 85% of the world's production and is mainly grown in India, Pakistan, Iran, Afghanistan, and Ethiopia. kabuli-type, instead, is grown in the Middle East, India, Mexico, North and South America, Australia, Spain, and Italy. A third type is characterized by a medium-to-small size and cream-colored seed, and it is designated as “pea-shaped” (Upadhyaya et al., 2008). Seed color (black, red, or white, and their variations) is a key commercial characteristic, which is also associated with the content of phenylpropanoid pathway-derived bioactive secondary metabolites such as flavonoids, lignans, and isoflavones. In addition to seed coat color determination, these secondary products have potential medicinal properties (Sirtori, 2001), and varied and important functions in processes, such as UV protection, disease resistance, and nodulation (N2 fixation) (Reinprecht et al., 2013).
The ex-situ collections of chickpea landraces and wild relatives are stored in 44 genebanks worldwide (Smýkal, 2015) and hold a combination of 98,313 accessions. The largest collections are conserved at the International Crops Research Institute for the Semi-Arid Tropics (ICRISAT) in India (20,140 accessions) and International Center for Agricultural Research in the Dry Areas (ICARDA) in Syria (13,818 accessions) (Table 1). Chickpea underwent a drastic loss of genetic diversity due to a series of bottlenecks unique to this crop, i.e., (i) reluctant cross-compatibility with wild species, (ii) difficulty in domestication, and (iii) winter-spring annual phenology (Abbo et al., 2003). Consequently, C. arietinum displays a lack of adaptive diversity for a range of biotic and abiotic stress. Susceptibility to viruses, pathogens and pests, sensitivity to environmental stress and poor cross-pollination are the main reasons for the limited diffusion and low production of chickpea.
Table 1.
Major “depositor institutes” conserving chickpea accessions.
| Genebank and link | Acronym | Country | Accessions |
|---|---|---|---|
| International Crop Research Institute for the Semi-Arid Tropics http://www.icrisat.org |
ICRISAT | India | 20,140 |
| International Center for Agricultural Research in the Dry Areas http://www.icarda.org |
ICARDA | Syria | 13,818 |
| United States Department of Agriculture https://www.ars-grin.gov |
USDA | USA | 6,789 |
| Aegean Agricultural Research Institute http://www.gfar.net |
AARI | Turkey | 2,075 |
| Australian Temperate Field Crops Collection http://elibrary.grdc.com.au |
GRDC | Australia | 8,655 |
| National Plant Gene Bank http://medomed.org |
NPGB | Iran | 5,700 |
| Vavilov Institute of Plant Genetic Resources https://www.gbif.org |
VIR | Russia | 2,091 |
| Institute for Agrobotany Tapi' oszele https://www.nebih.gov.hu |
nèbih | Hungary | 1,170 |
| Mediterranean Germplasm Database http://ibbr.cnr.it/mgd/ |
MGR_IBBR_CNR | Italy | 358 |
Viruses and virus-like entities hosted by C. arietinum L.
Several viruses have been isolated from naturally infected chickpea worldwide, but only a few cause diseases, which under specific environmental conditions can lead to significant economic loss (Bos et al., 1988; Kumar et al., 2008). The most relevant viruses reported to infect and induce disease in chickpea are: Alfalfa mosaic virus (AMV, Alfamovirus, Bromoviridae), Cucumber mosaic virus (CMV, Cucumovirus, Bromoviridae), Bean leafroll virus (BLRV) and Beet western yellows virus (BWYV) (both Luteovirus, Luteoviridae), Pea enation mosaic virus complex (PEMV-1, Enamovirus, Luteoviridae) and (PEMV-2, Umbravirus), Chickpea stunt disease-associated virus (CpSDaV, genus unassigned, Luteoviridae), and a number of geminiviruses of the genus Mastrevirus, the most important being Chickpea chlorotic dwarf virus (CpCDV). Faba bean necrotic yellows virus (FBNYV, Nanovirus, Nanoviridae) has also been reported (Makkouk et al., 2012). Table 2 contains a list of all the viruses to date associated with chickpea. Figure 1 contains a schematic representations of life cycles and spread of two groups of plant viruses included in Table 2 (i.e., with RNA or DNA genome).
Table 2.
Viruses reported to infect chickpea.
| Family | Virus | Type of transmission | Countries | Genetic resistance | Some References |
|---|---|---|---|---|---|
| Bromoviridae | Alfalfa mosaic virus (AMV, genus Alfamovirus) | Aphids (non-pers.), seeds, sap* | Iran | – | Kaiser and Danesh, 1971; Makkouk et al., 2003 |
| Cucumber mosaic virus (CMV, genus Cucumovirus) | Aphids (non-pers.), seeds, sap* | Iran, Morocco | – | Kaiser and Danesh, 1971; Ouizbouben and Fortass, 1997; Makkouk et al., 2003 | |
| Luteoviridae | Bean leafroll virus (BLRV, genus Luteovirus) | Aphid (pers.) | Azerbaijan, Iran, India, Turkey | – | Kaiser and Danesh, 1971 |
| Beet western yellows virus (BWYV, genus Polerovirus) | Aphids (pers.) | Azerbaijan, Iran | – | Makkouk et al., 2003; Mustafayev et al., 2011 | |
| Soybean dwarf virus (SbDV, genus Lutetovirus) | Aphids | Iran, Syria | – | Makkouk et al., 2003; Kumari and Makkouk, 2007 | |
| Chickpea chlorotic stunt virus (CpCSV, genus Polerovirus) | Aphids (pers.) | Ethiopia, Syria, Egypt, Eritrea, Iran, Morocco, Sudan | – | Abraham et al., 2006, 2009; Asaad et al., 2009; Banane et al., 2010 | |
| Pea enation mosaic virus-1 (PEMV-1, genus Enamovirus) | Aphids (pers.), seeds, sap* | Canada, USA, Iran, Syria | yes | Makkouk et al., 2003 | |
| Potyviridae | Bean yellow mosaic virus (BYMV, genus Potyvirus) | Aphids (non-pers.), seeds, sap* | Iran, Algeria, Morocco | – | Makkouk et al., 2003 |
| Turnip mosaic virus (TuMV, genus Potyvirus) | Aphids (non-pers.), seeds, sap* | Australia | – | Schwinghamer et al., 2007 | |
| Pea seed-borne mosaic virus (PSbMV, genus Potyvirus) | Aphid, seeds, sap* | Iran, Australia, Morocco, Algeria | yes | Ouizbouben and Fortass, 1997 | |
| Nanoviridae | Faba bean necrotic yellows virus (FBNYV, genus Nanovirus) | Aphids (pers.) | Jordan, Syria, Turkey, Lebanon, Iran, Egypt, Algeria | – | Yahia et al., 1997 |
| Geminiviridae | Chickpea chlorotic dwarf virus (CpCDV, genus Mastrevirus) | Leafhoppers | India, Iran, Syria, Turkey | – | Horn et al., 1993; Makkouk et al., 2003 |
| Tobacco yellow dwarf virus (TYDV, genus Geminiviridae) | Leafhoppers | Australia | – | Thomas et al., 2010 | |
| Virgaviridae | Tomato mottle mosaic Virus (ToMMV, genus Tobamovirus) | Seeds, sap* | Italy | – | Pirovano et al., 2014 |
| Tombusviridae | Turnip crinkle virus (TCV, genus Carmovirus) | Coleoptera, sap* | Italy | – | Ghasemzadeh et al., 2018 |
| Pospiviroidae | Hop stunt viroid (HSVd, genus Hostuviroid) | Sap | Italy | – | Pirovano et al., 2014 |
pers., persistent;
Mechanical infection by wounding using infectious sap as inoculum.
Figure 1.
Virus cycles in agro-ecosystems. (A) Schematic representation of replication cycles of Turnip crinkle virus (TCV) family Tombusviridae, genus Carmovirus), a virus that has been found associated to chickpea in open field. TCV has a positive (+) sense RNA genome that replicates (blue line). The viral RNA-dependent RNA polymerase (RdRp) amplifies the viral genome in the cytoplasm via negative (−) sense RNA template synthesis (red line). (+) RNA enters into the cellular translation machinery and codes for the RdRp. Moreover, movement proteins (MPs) and coat protein (CP) are the products of translation on viral sub-genomic RNAs. TCV genomic RNA can be encapsidated by the CP to form an icosahedral virion that can be then acquired by insects such as coleoptera. (B) Schematic representation of replication cycles of single stranded (ss) DNA viruses of the family Geminiviridae. Circular viral genomic ssDNA (1) functions as template for the synthesis of antisense ssDNA (orange line) due to the activity of host DNA-dependent DNA polymerase (yellow element) (2) to form a viral double stranded (ds)DNA intermediate (3). Viral dsDNA can be transcribed in the nucleus by the host DNA-dependent RNA polymerase PolII. Viral RNA transcripts are transferred to the cytoplasm, and enter into the translational machinery to release viral replicase (rep, blue element), MP and CP. One strand of the viral dsDNA can undergo cleavage by viral rep (4), thus allowing the access to the host DNA polymerase that extends the viral ssDNA and generates several copies of the circular genome (5). The ssDNA can be encapsidated and acquired by leafhopper vectors. (C) Chickpea is a permissive, non-symptomatic host for several viruses and it is often used in rotation with and/or in proximity to other crops for a sustainable agriculture. It therefore functions as a reservoir of virus inoculum that can be spread through insect vectors to other permissive crops that can show viral symptoms such as leaf yellowing, curling deformation and a general impact on the crop production. Metagenomics of nucleic acids of viral origin can be applied on either symptomatic or non-symptomatic plant tissues, as well as to other environmental samples (soil, insects) in order to explore viral entities associated to agro-ecosystems.
In recent years, the most invasive chickpea virus has been CpCDV. This virus, first reported in India in 1993 (Horn et al., 1993), has recently spread in many countries and among several crops, including other leguminous species (faba bean, lentil, bean), some solanaceous (tomato, pepper) and cucurbits (squash, cucumber), as well as other unrelated species such as cotton, sugar beet, okra, and papaya (Manzoor et al., 2014; Fahmy et al., 2015; Kraberger et al., 2015; Ouattara et al., 2017). In a newly discovered disease of watermelon in Tunisia, causing fruit hardness, CpCDV has been found as the causal agent (Zaagueri et al., 2017a,b). CpCDV is known to be transmitted by leafhopper species of the genus Orosius in a persistent manner (Horn et al., 1994). Today, CpCDV has attained a very wide distribution, including the Indian subcontinent, the Middle East and North Africa. Being so polyphagous and having a very widespread vector, CpCDV is certainly an emerging pest that will most likely colonize new areas (and possibly hosts) in forthcoming years.
In the last two decades, chickpea cultivation has been exposed to viral infections in novel areas of cultivation, such as Australia, where a high incidence of disease due to outbreak of viruses has been detected. The Australian food and agriculture stakeholders are closely observing chickpea cultivations and claiming the need to develop strategies that can assist in avoiding future viral epidemics in chickpea and other pulse crops. The Australian Grains Research and Development Corporation (GRDC) (Table 1) is supporting surveys of chickpea viruses in Central and West Asia (Kumari et al., 2011). As a result, other geminiviruses similar to CpCDV (but not CpCDV) have been found (Thomas et al., 2010; Hadfield et al., 2012), though currently limited to Australia.
Some chickpea viruses have a recognized quarantine significance, as tested by the Germplasm Health Laboratory of ICRISAT and ICARDA (see the Crop Genebank Knowledge Base website: https://cropgenebank.sgrp.cgiar.org/index.php/management-mainmenu-433/stogs-mainmenu-238/chickpea/guidelines/viruses). They are: Pea seed-borne mosaic virus, Bean yellow mosaic virus (PSbMV and BYMV, respectively; Potyvirus, Potyviridae), AMV and CMV (Table 2). Although belonging to different families, these viruses are transmitted by aphids and are also seed-transmitted to variable degrees. This last feature is of paramount importance for international trade, because viruses can reach and invade new habitats by the long distance human transport of seeds. Table 2 highlights the commercial sources of resistance against viruses, which to date are only two: PEMV-1 and PSbMV. For other viruses, no resistance has been described in the literature.
Recently, next-generation sequencing (NGS) approaches have opened the door to reconstructing viral populations in a high-throughput and cost-effective manner. Nowadays, NGS can be employed in environmental studies in the agro-ecosystem to either analyze known plant viruses by means of a reference-guided approach or to discover novel plant viruses using a de novo-based strategy (Massart et al., 2014).
Viral surveys using metagenomics in C. arietinum L. based on short (s)RNA analysis have been carried out in Apulia, Southern Italy, during the 2013–2017 time period. The surveys revealed that a large number of known viral species co-infect chickpea plants without causing any symptoms. Surprisingly, among the viruses found were Tomato mottle mosaic virus (ToMMV, Tobamovirus, Virgaviridae), which had not yet been observed in chickpea or reported in Europe, and one viroid referring to Hop stunt viroid (HSVd, Hostuviroid, Pospiviroidae) (Pirovano et al., 2014). In the same surveys, but in different plant samples, a high level of Turnip crinkle virus (TCV, Carmovirus, Tombusviridae) was found, though never reported before in chickpea (Ghasemzadeh et al., 2018). Worthy of note, viral metagenomics is showing that chickpea in open field is a highly permissive host for viruses and mixed infections are not uncommon. This means that most of the symptomatology that in the literature was ascribed to specific infections deserves further studies. In Figure 2 some viral symptoms that could be unequivocally ascribed to infection by a single virus.
Figure 2.
Selected photos showing symptoms induced by viruses on chickpea plants. (A) Tip wilting induced by mechanical inoculation with TuMV (from Schwinghamer et al., 2007). (B) Symptoms of chlorotic stunt disease caused by CpCDV on chickpea (from Kanakala et al., 2013).
To date, other virus-like infectious agents, such as phytoplasmas, have been reported only in sporadic cases, i.e., Australia, Ethiopia, Oman, and Pakistan. In most cases, phytoplasmas were associated with yellowing, phyllody and little leaves. Generally, infectious phytoplasmas are recognized as members of the 16SrII peanut witches' broom group (Ghanekar et al., 1988; Saqib et al., 2005; Al-Saady et al., 2006; Akhtar et al., 2008).
PPNs associated with C. arietinum L.
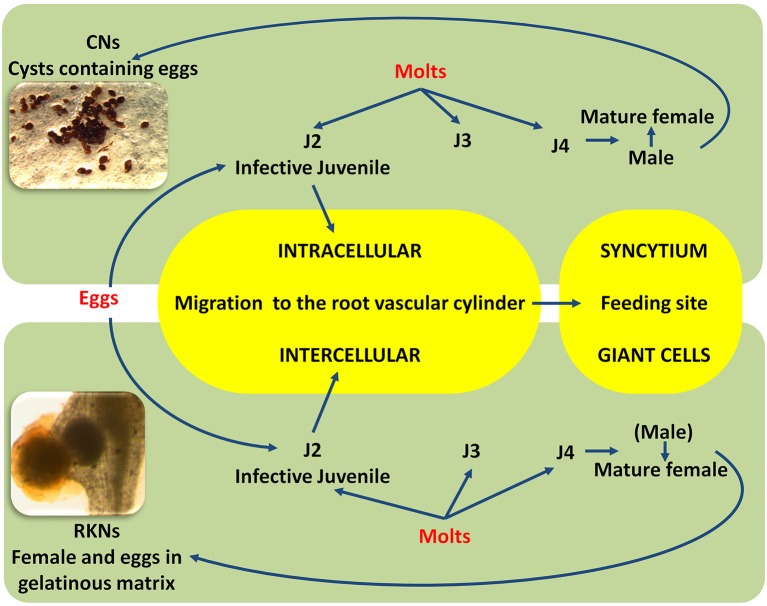
Diseases caused by soil-borne PPNs can generate significant yield losses in economically relevant crops (De Coninck et al., 2015). The estimation of plant parasitic nematodes (PPN) constrains to chickpea production was estimated in 14% (Castillo et al., 2008). PPNs are biotrophic (i.e., obligate parasites that are completely dependent on the host as the only source of nutrients) and polyphagous, because they can infect many different hosts among monocots and dicots. In the most representative PPNs families, root-lesion nematodes (Pratylenchus spp.), reniform nematodes (Rotylenchus reniformis), cyst-forming nematodes (CNs) (Heterodera spp.) and root-knot nematodes (RKNs) (Meloidogyne spp.) have been found pathogenic for chickpea and studies were carried out to characterized chickpea-nematode interactions, to describe geographical distributions, general symptoms even at histopathological levels (Figures 3, 4 and Table 3). The two PPNs largest groups most represented in the world's agro-ecosystem (Hussey, 1989) with interesting infections trategies and life cycles (Figure 5) are CNs and RKNs (Figure 5). Although three CN species of Heterodera have been found associated with chickpea worldwide (Castillo et al., 2008), Heterodera ciceri (Figure 4) is the only nematode that can lead to significant economic loss. In semi-arid areas of cultivation, the eggs do not undergo dormancy but hatch in the presence of chickpea root diffusates (exudates), where there are suitable soil moisture and temperature conditions of at least 10°C. Chickpea is highly susceptible to damage by H. ciceri and, therefore, efforts have been dedicated to search for potential sources of resistance to transfer them into genotypes of commercial varieties. However, resistant accessions have been identified only in C. bijugum, C. pinnatifidum, and C. reticulatum, and were deposited in the ICARDA genebank (Table 1) (Malhotra et al., 2002). Meloidogyne arenaria, Meloidogyne incognita (Figure 3), and Meloidogyne javanica are the RKN species that cause damage to chickpea. All three are typically found in areas with warm climatic conditions, and attack chickpea especially in the Indian subcontinent. On the other hand, Meloidogyne artiellia (Figure 4), being well-adapted to cool and wet conditions, is widely distributed in the Mediterranean region, including Italy (Castillo et al., 2008). Particularly M. arenaria, M. incognita, and M. javanica induce large galls in chickpea roots, whereas M. artiellia gives rise to very small galls surrounding the feeding sites (Vovlas et al., 2005) or no galls in the infected roots (Table 3). Ansari and co-authors (Ansari et al., 2004) screened more than 7,000 accessions of chickpea germplasm for resistance to M. javanica (Treub) Chitwood; four promising nematode-tolerant genotypes were found and conserved in the chickpea ICRISAT genebank (Table 1).
Figure 3.
Symptoms of nematode infection on chickpea plants in field and greenhouse. (A) Symptoms of infection visible in the field on C. arietinum: plant reduced in crop yield with chlorotic, pale, and yellow leaves. (B) Greenhouse pot test: control plant (left) and M. incognita infected plant (right). (C) Root system of control (c) and M. incognita infected plants (+n). (D) Egg masses (em) generated by M. incognita mature female, in root galling tissue. (E) Newly formed cysts (cy) of H. goettingiana.
Figure 4.
Three important PPNs associated to chickpea roots. Meloidogyne artiellia: (A) Longitudinal root section showing anatomical alterations; (B) Scanning electron microscopy (SEM) photo of a female on the root. Heterodera ciceri: (C) The tissues disruption caused by the cyst nematode is shown in longitudinal root section; (D) SEM image of a mature female. Pratylenchus thornei: (E) Longitudinal section of the root showing lesions caused by the nematode; (F) Fuchsin-stained root cortex section, showing the migratory endoparasite. n, nematode; e, eggs; gc, giant cell; ne, necrotic tissues; s, syncytium. Scale bars: (A,C,E) = 500 μm; (B,D,F) = 200 μm (Source: Nicola Vovlas, CNR).
Table 3.
Selection of PPNs, associated with chickpea (font: https://www.cabi.org).
| Family | Species | Generic and characteristic symptoms | Countries | References |
|---|---|---|---|---|
| Meloidogynidae |
Meloidogyne incognita, Meloidogyne arenaria, Meloidogyne javanica |
Whole plant: early senescence; Leaves: abnormally colored and wilted Roots: galls, swollen and reduced root system |
Indian Subcontinent | Ali and Sharma, 2003; Vovlas et al., 2005 |
| Meloidogyne artiellia | Root with small or absent galls and protruded adult female | Mediterranean Basin | Vovlas et al., 2005 | |
| Heteroderidae | Heterodera goettingiana | Whole plant: stunted Leaves: pale green at an early stage, later chlorotic. Reduced number of flowers and pods, small or no seeds Roots: poorly developed, lacking nitrogen-fixation nodules. |
North Africa | Di Vito et al., 1994 |
| Heterodera ciceri | Soil infestation in small circular area that should extend to entire field. Eggs don't undergo dormancy. | Turkey, Syria | Greco et al., 1988; Castillo et al., 2008 |
|
| Pratylenchidae | Pratylenchus thornei | Whole plant: dwarfing distributed in patches Leaves: chlorosis and reduction shoot weight Roots: necrotic streaks or lesions, soft rot of cortex |
Australia, India, Mexico, North Africa, Spain | Castillo et al., 1996 |
| Pratylenchus penetrans | Whole plant: reduced crop yield Leaves: chlorotic (pale, yellowing) Roots: may be thin, and with a reduced number of lateral roots. |
North Africa, Spain, Turkey | Di Vito et al., 1994 | |
| Hoplolaimidae | Rotylenchus reniformis | Whole plant: distorted Leaves: abnormal colors Stems: stunting or rosetting Roots: external feeding and reduced root system |
India, Egypt, Ghana | Mahapatra and Pahdi, 1996 |
Figure 5.
Simplified life cycles of cyst nematodes (CNs) and root-knot (RKNs) nematodes. Larvae hatch from cysts or from egg masses; the first-stage juvenile molts inside the eggshell become an invasive second-stage juvenile (J2) adapted to penetrate the root using an intra, inter-cellular migration and to the establishment of the feeding site (Syncytium and Giant cell). The nematode has to change molts (J3, J4) to become a fully mature (male or female) adult. Parthenogenetic and amphimictic reproduction modalities are different between CNs and RKNs.
Genomics of C. arietinum L.: host response to pathogens and new applications of biotechnology
The chickpea genome has recently been released by two research groups (Jain et al., 2013; Varshney et al., 2013) and further analyzed (Parween et al., 2015; Thudi et al., 2016). The availability of a rich genomic platform of chickpea and its relatives, such as C. reticulatum (a source of interesting characteristics) (Gupta et al., 2017), provides insight into both genome diversity and domestication and therefore should be considered as a resource to improve chickpea resistance against biotic and abiotic stress.
One of the most recurrent themes in plant pathology research is the highly adaptable nature of pathogens, including viruses and nematodes. These organisms possess the ability to harness and modify cellular resources in order to coexist with the plant host. Current genomics in legumes make it possible to study specific layers of plant-pathogen interactions directly using crop plants, including chickpea. A phylogenetic analysis of legume species constructed with genome-wide, single-copy orthologous genes shows that the closest relative to chickpea is Medicago truncatula, and secondarily Glycine max (Zheng et al., 2016) (Figure 6). M. truncatula and G. max are widely considered as model legumes and, therefore, studies in chickpea could benefit from those carried out in the model relative species. The availability of a genomic platform of the chickpea, together with recent advances in understanding the mechanisms of immune responses to plant pathogens, presents interesting perspectives for attenuating the damage caused in chickpea by biotic stress. Below we highlight the promising main topics.
Figure 6.
Phylogenetic tree of seven legume species with Vitis vinifera as the out-group. The phylogenetic tree was constructed with a genome-wide single-copy orthologous genes of legume species i.e., Glycine max, (cultivated soybean), Glycine soja (wild soybean), Medicago truncatula (barrel clover), Lotus japonicus (bird's-foot trefoil), Cajanus cajan (Pigeonpea) Cicer arietinum (chickpea), Phaseolus vulgaris (common bean). Modified from Zheng et al. (2016).
Plant immune defense response, effector targets, and RNA silencing in pathogen attack
Plants recognize pathogens and microbes through pathogen/microbe-associated molecular patterns (P/MAMPs). PAMPs are evolutionarily conserved molecules across kingdoms; in plants they carry out critical functions against several microbial attacks (Boutrot and Zipfel, 2017), including invasion of viruses, bacteria, fungi, and nematodes. For instance, it is widely accepted that the early stages of pathogen attack could be considered the key target step in plant defense strategies; this idea has also been recently extended to nematode parasitism (Holbein et al., 2016). PAMPs activate host defense responses (PAMP-triggered immunity or PTI) through a complex signaling cascade. Effectors should interfere with PTI responses, thereby leading to effector-triggered susceptibility (ETS). Manosalva and coauthors (Manosalva et al., 2015) showed that PPNs secrete conserved pheromones named “ascarosides,” eliciting MAMP response in various plants, and are exclusively identified in the phylum Nematoda. In turn, microbial virulent pathogens are able to overcome plant defense mechanisms by secreting effectors into the host. An effector protein can also be the elicitor of effector-triggered immunity (ETI) (Mandadi and Scholthof, 2013). If this first defense system is defeated, then plant resistance initiates a second mechanism which is more amplified and faster than PTI and usually develops in a form of programmed cell death known as the hypersensitive response (HR), leading the infected host cell to apoptosis. In this second detection system level, plants are able to recognize pathogenic effectors through nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins and are characterized by leucine-rich repeats (LRR) that give them binding specificity. In fact, among the largest gene families in plants deputed to play roles in response to a broad range of pests and pathogens is the R-gene family, which mainly includes NBS-LRR genes (Zheng et al., 2016). Chickpea contains at least 153 NBS-LRR homolog genes in eight chromosomes (Varshney et al., 2013). This number is considerably lower than the number of orthologs in other legume species (Jain et al., 2013). Once discovered in M. truncatula and G. max, the cascade regulation of NBS-LRRs triggered by micro (mi)RNAs of the miR2118/482 superfamily members has been associated with nodulation events (plant-rhizobium interactions) and not to better specified plant pathogen defense strategies (Zhai et al., 2011). The interaction between miRNA and R-genes might have long-term evolutionary benefits by buffering NBS-LRR levels to reduce the fitness cost of these genes (Zheng et al., 2016). More recently, NBS-LRR secondary siRNA cascade mechanisms have been revealed to spawn valuable layers of non-race-specific resistance against viral and bacterial pathogens (Shivaprasad et al., 2012) (Figure 7). This recently discovered plant strategy seems to be independent from either the NBS-LRR protein additive effect of expression or from the R-gene-to-pathogen gene interaction. In chickpea leaf/shoot/floral tissues, 22 nt-long miR2118 is fairly present and targets NBS-LRRs (Srivastava et al., 2015), and the secondary siRNA mechanism involved in cascade regulation of NBS-LRR is present as well. Importantly, NBS-LRR regulation can be subverted by plant viruses. RNA silencing in plants and insects can function as a defense mechanism against invading viruses, and viruses have evolved viral suppressors of RNA silencing (VSRs) to overcome the host defense (reviewed by Csorba et al., 2015). VSRs can act on various steps of the different silencing pathways and, thus, can have a profound impact on host endogenous RNA-silencing regulatory pathways, including the generation and function of plant endogenous siRNA, such as miRNAs and secondary siRNAs (Figure 7).
Figure 7.
NBS-LRR silencing cascade mechanism. Schematic representation of NBS-LRR silencing cascade mechanism triggered by miR2118 (a legume specific miRNAs discovered in soybean), highly conserved in C. arietinum. In red circles, viral silencing suppressors (Csorba et al., 2015) that can impair the cascade mechanism.
Chickpea seems to be a permissive host for many plant viruses that are considered capable of inducing pathogenesis in many plant species. All of the plant viruses families infecting chickpea (Table 2) are known to express VSRs, which, in turn, have been shown to subvert RNA silencing machinery. For instance, PEMV-1 expresses the P0 protein, which has been shown to destabilize AGO1 protein (Fusaro et al., 2012) and, therefore, could hinder the miR2118-triggered, NBS-LRR–mediated cascade mechanism of R-gene silencing at several stages (Figure 7). Similarly, but with a different mechanism, TCV P38 can alter AGO1 activity (Azevedo et al., 2010) (Figure 7). Tobamovirus replicase (i.e., P122/P126) is known to bind miRNA and siRNA, preventing their stabilization and incorporation into the RNA-induced silencing complex (Csorba et al., 2007; Vogler et al., 2007) (Figure 7). All these VSRs from viruses infecting chickpea can block downregulation of NBS-LRR and the downstream cascade mechanism, inducing overexpression of R-genes with a wider coverage against viral and other pathogens, despite the low number of R-genes in the chickpea genome.
Recently, it has become clear that silencing pathways also play an important role in other plant pathosystems, including the onset of nematode parasitism. Indeed, through a transgenic approach, it has been shown that VSRs can subvert host RNA silencing machinery and increase the susceptibility to nematode parasitism (Walsh et al., 2017).
RNA silencing approaches have also been exploited in plants to control PPNs, given that RNA silencing mechanisms are also conserved in nematodes (Fire et al., 1998). Double-stranded RNA (dsRNA) can be produced through engineered plants that have the ability to silence target genes in nematode body. The delivery of dsRNAs from plant to nematode occurs by the ingestion process and can trigger RNA interference (RNAi), resulting in the inactivation of targeted genes (Gheysen and Vanholme, 2007). Availability of a genomic platform of PPNs is a prerequisite to identify the nematode genes responsible for the interactions and run loss-of-function (Abad et al., 2008; Denver et al., 2016). This could lead, for example, to adopt strategies based on the manipulation of nematode-derived protein elicitor(s), molecules able to induce a PTI-like response (Mendy et al., 2017). The ETI defense response in plant-nematode interaction is relatively better investigated than PTI, and often involves an HR reaction due to the initiation of the two characteristic “feeding structures” induced in the root by sedentary endoparasitic nematodes (Goverse and Smant, 2014) (Figure 5). A noteworthy case is the HR that takes place in the induction of several individual “giant cells” in Mi-1-resistant tomato plants infected by RKN-infective second-stage juveniles (J2) (Figure 5). By contrast, the deterioration of the “syncitium” (composed of hundreds of fused root cells, induced by H. glycines in soybean genotypes harboring a natural resistance gene at the Rgh1 locus, is not characterized by typical cell death. Rgh1-mediated resistance seems to involve the collapse of the feeding site by nuclear and cytoplasmic fragmentation.
Recently, a novel and unique mechanism of plant resistance has been discovered through mutation analysis, gene silencing and transgenic complementation in soybean–H. glycines interactions. Wu et al. (2016) have demonstrated that the single dominant Rgh4 locus, which is a major quantitative trait locus encoding serine hydroxymethyltransferase (SHMT), confers resistance to CNs (Wu et al., 2016). SHMT is an enzyme that is ubiquitous in nature and structurally conserved across kingdoms. The resistant allele possesses two functional single nucleotide polymorphisms (SNPs, denoted as P130R and N358Y) compared to that of the sensitive allele, rhg4. These mutations affect the kinetic activity of SHMT, which could result in folate deficiency inside syncytia and a nutritional deficiency that starves the nematode. This is a novel plant defense strategy against roundworm that could readily be extended to other important crops. Preliminary exploration within the chickpea genome has confirmed the presence of at least two shmt loci (Figure 8). These findings will likely boost research to extend the use of SHMT resistance to chickpea by identifying the source of positive functional SNPs in ancient local varieties or, alternatively, to apply novel technologies such as genome editing of functional SNPs.
Figure 8.
Serine hydroxymethyltransferase gene model. Shmt model and polymorphism in resistant (Forrest) and susceptible (Essex) soybean cultivars and alignment of predicted chickpea shmt 1-like mRNA sequences (NCBI reference XM_004504310.1 and XM_004502186.1) showing the two functional SNPs positions.
Biotechnology applications: current status on the genetic transformation of Cicer arietinum L.
The enhancement of tolerance to biotic and abiotic stress in chickpea can significantly increase its yield potential. However, due to the limited genetic pool, cross compatibility and lack of resistance sources to biotic constraints in the available germplasm the improvement of chickpea by means of conventional breeding faces serious limitations. Modern plant biotechnology tools offer several possibilities to finally overcome these limitations. The main objectives are (i) to enhance chickpea resistance to pests and other biotic and abiotic stress, and (ii) to achieve more sustainable food production in developing countries, such as in the semi-arid tropics where agrochemicals are inaccessible to low-income farmers (Sharma and Ortiz, 2000; Sharma et al., 2001). Most biotechnology approaches require skills and tools for manipulating the genome of a plant, either through transgenics or other means, and the process always includes plant transformation and regeneration steps. Chickpea, like other large-seed grain legumes such as faba bean, pigeonpea, and common bean, is considered to be “reluctant” to in vitro transformation and regeneration (Somers et al., 2003). Therefore, one critical point of chickpea productivity improvement remains the development of reliable genetic transformation techniques.
Plant genetic transformation is defined as the method for the delivery, integration, and expression of foreign genes into plant genomes (Atif et al., 2013). There are two main methods that enable delivery of the gene into the plant genome: (i) direct gene transfer (mediated by physical or chemical forces) and (ii) Agrobacterium-mediated genetic transformation, where Agrobacterium tumefaciens is used as a vehicle to introduce foreign genes into the plant genome.
In the case of the chickpea, many research efforts were undertaken to improve resistance against major biotic stress, such as pod borers (Heliothis armigera [Hub.]), aphids (Aphis craccivora), bruchids, fungal diseases (Fusarium oxysporium/F. udum), and abiotic (drought and salinity) stress, as well as the nutritional quality by increasing the sulfur-containing amino acid content. The first transformation studies with chickpea were performed by Srinivasan et al. (1991) and Islam et al. (1994) using callus culture; shoot regeneration was not possible. Although these studies were unsuccessful due to poor regeneration, they showed the susceptibility of chickpea to infection with A. tumefaciens and proved its potential as a transformation vector for chickpea. Afterwards, generation of transgenic chickpea was reported with varying degrees of success. To our knowledge, however, the number of reports describing the successful production of transgenic chickpea using either Agrobacterium-mediated or particle bombardment transformation is still very limited (Mishra et al., 2012; Atif et al., 2013; Tripathi et al., 2013). Table 4 summarizes chickpea transformation studies. Most of the first attempts on genetic transformation used the Agrobacterium-mediated method with few exceptions, where particle bombardment was employed (Tewari-Singh et al., 2004; Ganguly et al., 2014). Indurker et al. (2007) reported a successful transformation protocol (16% transformation frequency) using particle bombardment with gold particles as micro-carrier, in combination with helium pressure of 900 psi on epicotyl explants of the cultivars ICCC37 and PG-12. The construct was a pHS102 plasmid harboring the reporter gene uidA, neomycin phosphotransferase II (nptII) and insecticidal cry1Ac. Fontana and colleagues (Fontana et al., 1993) reported the first successful chickpea transformation protocol after transformation of embryonic axes with A. tumefaciens. The transferred genes were successfully inherited into subsequent generations. Molecular evidence for the transgenic nature was confirmed by studying the integration and expression of β-D-glucuronidase and nptII genes as well as the integration and expression of the transferred genes. Later, other reports described new protocols (Krishnamurthy et al., 2000; Polowick et al., 2004; Sarmah et al., 2004; Senthil et al., 2004; Sanyal et al., 2005) improved for their simplicity and relatively short time required to produce transgenic plants (T0) without the callus phase (4–6 months). From surveying the literature on chickpea transformation, it can be concluded that the average frequency of Agrobacterium-mediated transformation is 0.1–5.1%, which is very low compared to model plants such as G. max and M. truncatula (96 and 80%, respectively) (Iantcheva et al., 2001; Li et al., 2017). However, with the ICC10943 cultivar and using sonication-assisted, Agrobacterium-mediated transformation (SAAT) cases of transformation efficiency higher that 25% have been reported (Bhattacharjee et al., 2010; Table 4). Therefore a wider utilization of SAAT for chickpea transformation can be foreseen, which should be nonetheless tested on several other varieties.
Table 4.
Genetic transformation studies in chickpea.
| Transformation method | Cultivar | Explant type | Transferred genes | Transformation frequency % | References |
|---|---|---|---|---|---|
| Agrobacterium-mediated transformation | Local ecotype | EAx | uidA, nptII | 4* | Fontana et al., 1993 |
| ICCV1, ICCV6 and desi (local) variety | EAx | uidA, nptlI | e.g., ICCV-6: 1.96 | Kar et al., 1996 | |
| PG1, PG12, Chafa and Turkey | EAx | uidA, nptII | e.g., Turkey < 1.5 | Krishnamurthy et al., 2000 | |
| H-208, ICCL87322, K-850, Annigiri, and ICCV5 | EAx | uidA, bar | 5.1§ | Senthil et al., 2004 | |
| Semsen | Halved EAx attached to cotyledon | nptII, bean αAI1 | 0.72¢ | Sarmah et al., 2004 | |
| CDC Yuma | EAx | uidA, nptII | 3.1∧ | Polowick et al., 2004 | |
| C-235, BG-256, Pusa-362 and Pusa-372 | Pre-conditioned CNs | cry1Ac, nptII | e.g., BG 256: 1.12 | Sanyal et al., 2005 | |
| K-850 | EAx | α-ai, uidA, nptII | 0.3 | Ignacimuthu and Prakash, 2006 | |
| Sonication-assisted Agrobacterium mediated transformation (SAAT) | ICC10943 and ICC10386 | Decapitated embryo | uidA, hpt II | ICC 10943: 26 ± 2, ICC 10386: 24 ± 3Δ | Pathak and Hamzah, 2008 |
| ICCV89314 | Single cotyledon with half embryo | ASAL, nptII, gusA | 0.066 ± 0.003 (mean ± SE) | Chakraborti et al., 2009 | |
| C-235, Annigiri and K-850 | Wounded apical dome of shoot apex | uidA, bar | 2.43 | Singh et al., 2009 | |
| C-235 | EAx with half portions of both cotyledons | pmi | 3 | Patil et al., 2009 | |
| C-235 | AMEs | P5CSF129A, npt II, uidA | Not mentioned | Bhatnagar-Mathur et al., 2009 | |
| Semsen, ICCV89314 | Cotyledon with half EAx | cry2Aa, nptII | 0.3 | Acharjee et al., 2010 | |
| Pusa-256, KWR-108, Pusa-1003 and local line (from market) | Cotyledon- and cotyledonary-node-derived-callus and EAx | uidA, hpt | e.g., KWR 108: 23.45 | Bhattacharjee et al., 2010 | |
| Annigeri | CNs | P5CS, hpt, uidA | Not mentioned | Ghanti et al., 2011 | |
| P-362 | CNs | cry1Ab, cry1Ac, nptII | 2.77 | Mehrotra et al., 2010 | |
| C-235, BG-256, P-362 and P-372 | Immature cotyledons, EAx | uidA, nptII | e.g., P 362: 2.08 | Tripathi et al., 2013 | |
| C-235 | AMEs | DREB1A, nptII | Not mentioned | Anbazhagan et al., 2015 | |
| DCP-923 | EAx | fused cry1Ab/Ac, hpt | Not mentioned | Ganguly et al., 2014 | |
| P-362 | CNs explants | nptII, uidA, modified human α1-PI, cry1Ab, cry1Ac | Not mentioned | Yadav et al., 2017 | |
| C-235 | AMEs explants | uidA, nptII | 1.2 | Srivastava et al., 2017 | |
| Particle bombardment | ICCV1, ICCV6 | EAx | nptII, cry1Ac | Not mentioned | Kar et al., 1997 |
| P-362, P-1042 and P-1043 | Decapitated embryo | pat, nptII, uidA, AK | Not mentioned | Tewari-Singh et al., 2004 | |
| Chaffa, PG12, ICCC37 and ICCC32 | EAx, epicotyl and stem | nptII, uidA, cry1Ac | 16 ± 0.33£ | Indurker et al., 2007 |
EAx, embryonic axis; AMEs, Axillary meristem explants; CNs, Cotyledonary Nodes; uidA, β-Glucuronidase, commonly referred to as the gus gene; nptII, neomycin phospho transferase II; bar, Basta (bialaphos) resistance; αAI1, bean-α amylase inhibitor 1; pat, phosphinothricin-acetyltransferase; AK aspartate kinase; cry1Ac, insecticidal crystal toxins 1Ac; α-ai, α-amylase inhibitor; ASAL, Allium sativum leaf agglutinin; gusA, β-Glucuronidase; pmi, phosphomannose isomerase; P5CSF129A, D1- pyrroline-5-carboxylate synthetase F129A; cry2Aa, insecticidal crystal toxins 2Aa; hpt, hygromycin phosphotransferase; P5CS, pyrroline-5-carboxylate; DREB1A, dehydration response element B1A; α1-PI, modified human Alpha-1-proteinase inhibitor.
Number of whole plants transformed/initial number of embryos;
Number of confirmed independent lines/number of initial seeds;
18 independently derived transgenic plants obtained from a total of 2,500 explants (explant that consisted of one cotyledon attached to half embryonic axis);
7 separate experiments with the use of shoot elongation media (MS);
Obtained by dividing [100 times the number of confirmed transformed plants of independent lines (both PCR and Southern blot positive)] by the number of treated explants;
Epicotyl, average of three experiments with 150 explants each.
Perspectives
The strong potential of genetic transformation techniques for crop improvement is unquestionable. The clustered regularly interspaced short palindromic repeat (CRISPR)-associated protein 9 (Cas9) DNA editing system has recently been developed as a new method for genome engineering. It is based on the type II CRISPR-associated immune system that protects bacteria against invading DNA viruses and/or plasmids (Jinek et al., 2012). Genome editing by CRISPR/Cas9 as well as other techniques, including zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), have been applied to edit the genome in several plant species (Kim and Kim, 2014). The successful utilization of CRISPR/Cas9-directed genome editing in plant species has been reported and also includes the two relatives, i.e., the legume models G. max and M. truncatula (Li et al., 2015; Meng et al., 2017). CRISPR/Cas9 gene editing technology is currently revolutionizing genetic studies and crop improvement because it can be applied with high-throughput and at genome-scale (Yang et al., 2017). To our knowledge, no research effort has been made to implement this system in chickpea. The application of CRISP/Cas9 in chickpea genome editing will not only provide answers to basic biological questions but will also reduce public concern about transgenic plants, owing to its non-GMC nature. In most cases, Cas9 and guide (g)RNAs are delivered into plant cells by Agrobacterium-mediated T-DNA transformation or by physical means, such as PEG-mediated transformation of protoplasts or biolistic transformation of calluses. In the case of chickpea and other legumes, this approach could face limitations due to the difficulties of transformation and regeneration from callus. An alternative approach that could help to overcome these limitations is the identification and engineering of plant viruses as a tool for systemic gene editing in plants. Some successful examples are already available. Cabbage leaf curl virus, a geminivirus, is able to deliver gRNA and induce systemic gene mutations in plants (Yin et al., 2015). An RNA viral vector based on Tobacco rattle virus (TRV) has been demonstrated to serve as a vehicle to deliver genome-engineering reagents to all plant parts, including meristems. This provides a general method for easily recovering seeds with the desired modifications, obviating the need for transformation and/or tissue culture (Ali et al., 2015). More recently a legume virus, the Pea early-browning virus, has been demonstrated to be more efficient than TRV for these applications (Ali et al., 2017). An additional challenge would be the identification of the best DNA or RNA viruses able to fully infect chickpea to be engineered and used as viral vectors dedicated to genome editing.
Genome editing of chickpea may help improving specific characteristics of a crop with limited genetic pool and lack of resistance sources. An emblematic case would be the modification of functional SNPs in the SHMT gene (Figure 8) in order to confer resistance to nematodes or to modify miRNA target sites in NBS-LRR genes (Figure 7), ensuring the up-regulation of certain functional R-genes.
Perspectives for the improvement of chickpea should also take into account the genomic selection approach. It facilitates the rapid selection of superior genotypes and accelerates the breeding cycle and it has been applied with a large success in many other crops (Crossa et al., 2017).
Chickpea cultivations may constitute a reservoir of viral entities. Indeed, chickpea seems to be a permissive host for many viruses and viroids, thus ensuring their maintenance in agro-ecosystems: most of the hosted viruses are symptomless in chickpea, but pathogenic for other plant species. Viral metagenomics is currently the tool most indicated for surveys of virus-infected plants. In addition, metagenomics approaches can help to discover novel infectious entities and microbes either hosted by or associated to chickpea. This could help scientists better identify and describe the multi-trophic interactions that may influence nematode reproduction or plant-rhizobia interactions.
To modulate plant PPNs, several transgenic strategies have been used, such as (i) cloning of resistance genes from natural resources and transfer to other plant species; (ii) overexpression of different protease inhibitors; and (iii) suppression of nematode effectors in plants using RNAi (Ali et al., 2017). RNAi (Rosso et al., 2009; Banerjee et al., 2017) is worth exploring more in depth, particularly RNAi-based technology combined with peptide expression which disrupt nematode sensory activities (Fosu-Nyarko and Jones, 2015). Moreover, a number of CRISPR/Cas9 genome-editing protocols have been established in Caenorhabditis elegans (Friedland et al., 2013; Zamanian and Andersen, 2016). Genome manipulation with novel developments in this model organism, research, and advances in parasite genomics could open new doors to the biology of closely related nematode parasites during their interaction with legumes. A more in-depth understanding of the potentiality in biotechnologies for legume pest management will at least modernize chickpea breeding programs, targeting a greater impact on food and nutrition security, climate change adaptation and worldwide diffusion.
Author contributions
All the authors have contributed to the review with their proper specific expertise in plant science, plant pathology, and plant biotechnology.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank CNR-IPSP (Italy)-NRC (Egypt) 2016-2017 Bilateral Project Cicer arietinum ancient varieties in Egypt: exploiting associated pathogens and modern defense strategies for supporting all the authors and SaveGraINPuglia project (PSR 2007-2013, Misura 214, 4a and PSR 2014-2020, Misura10.2.1) for supporting PL and VP. The authors are particularly grateful to Dr. Nicola Vovlas for the photos A and E in Figure 3 and the photos in Figure 4.
References
- Abad P., Gouzy J., Aury J. M., Castagnone-Sereno P., Danchin E. G., Deleury E., et al. (2008). Genome sequence of the metazoan plant-parasitic nematode Meloidogyne incognita. Nat. Biotechnol. 26, 909–915. 10.1038/nbt.1482 [DOI] [PubMed] [Google Scholar]
- Abbo S., Shtienberg D., Lichtenzveig J., Lev-Yadun S., Gopher A. (2003). The chickpea, summer cropping, and a new model for pulse domestication in the ancient near east. Q. Rev. Biol. 78, 435–448. 10.1086/378927 [DOI] [PubMed] [Google Scholar]
- Abraham A. D., Menzel W., Lesemann D. E., Varrelmann M., Vetten H. J. (2006). Chickpea chlorotic stunt virus: a new polerovirus infecting cool-season food legumes in Ethiopia. Phytopathology 96, 437–446. 10.1094/PHYTO-96-0437 [DOI] [PubMed] [Google Scholar]
- Abraham A. D., Menzel W., Varrelmann M., Vetten H. J. (2009). Molecular, serological and biological variation among chickpea chlorotic stunt virus isolates from five countries of North Africa and West Asia. Arch. Virol. 154, 791–799. 10.1007/s00705-009-0374-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharjee S., Sarmah B. K., Kumar P. A., Olsen K., Mahon R., Moar W. J., et al. (2010). Transgenic chickpeas (Cicer arietinum L.) expressing a sequence-modified cry2Aa gene. Plant Sci. 178, 333–339. 10.1016/j.plantsci.2010.02.001 [DOI] [Google Scholar]
- Akhtar K. P., Shah T. M., Atta B. M., Dickinson M., Jamil F. F., Haq M. A., et al. (2008). Natural occurrence of phytoplasma associated with chickpea phyllody disease in Pakistan – a new record. Plant Pathol. 57, 771 10.1111/j.1365-3059.2007.01800.x [DOI] [Google Scholar]
- Ali S. S., Sharma S. B. (2003). Nematode survey of chickpea production areas in Rajasthan, India. Nematol. Mediterr. 31, 147–149. Available online at: http://journals.fcla.edu/nemamedi/article/view/86740/83656 [Google Scholar]
- Ali Z., Abul-Faraj A., Li L., Ghosh N., Piatek M., Mahjoub A., et al. (2015). Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 8, 1288–1291. 10.1016/j.molp.2015.02.011 [DOI] [PubMed] [Google Scholar]
- Ali Z., Eid A., Ali S., Mahfouz M. M. (2017). Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res. 244, 333–337. 10.1016/j.virusres.2017.10.009 [DOI] [PubMed] [Google Scholar]
- Al-Saady N. A., Al-Subhi A. M., Al-Nabhan I. A., Khan A. (2006). First record of a Group 16SrII phytoplasma infecting chickpea in Oman. Plant Dis. 90, 273 10.1094/PD-90-0973C [DOI] [PubMed] [Google Scholar]
- Anbazhagan K., Bhatnagar-Mathur P., Vadez V., Dumbala S. R., Kishor P. B., Sharma K. K. (2015). DREB1A overexpression in transgenic chickpea alters key traits influencing plant water budget across water regimes. Plant Cell Rep. 34, 199–210. 10.1007/s00299-014-1699-z [DOI] [PubMed] [Google Scholar]
- Ansari M. A., Patel B. A., Mhase N. L., Patel D. J., Douaik A., Sharma S. B. (2004). Tolerance of chickpea (Cicer arietinum L.) lines to root-knot nematode, Meloidogyne javanica (Treub) Chitwood. Genet. Resour. Crop Evol. 51, 1449–1453. 10.1023/B:GRES.0000023460.26690.23 [DOI] [Google Scholar]
- Asaad N. Y., Kumari S. G., Haj-Kassem A. A., Shalaby A. B. A., Al-Shaabi S., Malhotra R. S. (2009). Detection and characterization of chickpea chlorotic stunt virus in Syria. J. Phytopathol. 157, 756–761. 10.1111/j.1439-0434.2009.01574.x [DOI] [Google Scholar]
- Atif R. M., Patat-Ochatt R. M., Svabova L., Ondrej V., Klenoticova H., Jacas L., et al. (2013). Gene transfer in legumes, in Progress in Botany, eds Lüttge U., Beyschlag W., Francis D., Cushman J. (Berlin; Heidelberg: Springer; ), 37–100. [Google Scholar]
- Azevedo J., Garcia D., Pontier D., Ohnesorge S., Yu A., Garcia S., et al. (2010). Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24, 904–915. 10.1101/gad.1908710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banane J. K., Vahdat A., Menzel W., Vetten H. J. (2010). Serological and molecular identification of Chickpea chlorotic stunt virus from chickpea in iran. Plant Dis. 94, 788 10.1094/PDIS-94-6-0788C [DOI] [PubMed] [Google Scholar]
- Banerjee S., Banerjee A., Gill S. S., Gupta O. P., Dahuja A., Jain P. K., et al. (2017). RNA interference: a novel source of resistance to combat plant parasitic nematodes. Front. Plant Sci. 8:834. 10.3389/fpls.2017.00834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatnagar-Mathur P., Vadez V., Devi M. J., Lavanya M., Vani G., Sharma K. K. (2009). Genetic engineering of chickpea (Cicer arietinum L.) with the P5CSF129A gene for osmoregulation with implications on drought tolerance. Mol. Breed. 23, 591–606. 10.1007/s11032-009-9258-y [DOI] [Google Scholar]
- Bhattacharjee B., Mohan M., Nair S. (2010). Transformation of chickpea: effect of genotype, explant, Agrobacterium-strain and composition of culture medium. Biol. Plant. 54, 21–32. 10.1007/s10535-010-0004-4 [DOI] [Google Scholar]
- Bos L., Hampton R. O., Makkouk K. M. (1988). Viruses and virus diseases of pea, lentil, faba bean and chickpea, in World Crops: Cool Season Food Legumes, ed Summerfield R. J. (Dordrecht: Springer; ), 591–615. [Google Scholar]
- Boutrot F., Zipfel C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286. 10.1146/annurev-phyto-080614-120106 [DOI] [PubMed] [Google Scholar]
- Castillo P., Gomez-Barcina A., Jimenez-Diaz R. M. (1996). Plant parasitic nematodes associated with chickpea in Southern Spain and effect of soil temperature on reproduction of Pratylenchus thornei. Nematologica 42, 211–219. 10.1163/004325996X00057 [DOI] [Google Scholar]
- Castillo P., Navas-Cortés J. A., Landa B. B., Jiménez-Díaz R. M., Vovlas N. (2008). Plant-parasitic nematodes attacking chickpea andtheir in planta interactions with rhizobia and phytopathogenic fungi. Plant Dis. 92, 840–853. 10.1094/PDIS-92-6-0840 [DOI] [PubMed] [Google Scholar]
- Chakraborti D., Sarkar A., Mondal H. A., Das S. (2009). Tissue specific expression of potent insecticidal, Allium sativum leaf agglutinin (ASAL) in important pulse crop, chickpea (Cicer arietinum L.) to resist the phloem feeding Aphis craccivora. Transgenic Res. 18, 529–544. 10.1007/s11248-009-9242-7 [DOI] [PubMed] [Google Scholar]
- Crossa J., Perez-Rodriguez P., Cuevas J., Montesinos-Lopez O., Jarquin D., De Los Campos G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. 10.1016/j.tplants.2017.08.011 [DOI] [PubMed] [Google Scholar]
- Csorba T., Bovi A., Dalmay T., Burgyan J. (2007). The p122 subunit of Tobacco Mosaic Virus replicase is a potent silencing suppressor and compromises both small interfering RNA- and microRNA-mediated pathways. J. Virol. 81, 11768–11780. 10.1128/JVI.01230-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T., Kontra L., Burgyan J. (2015). Viral silencing suppressors: tools forged to fine-tune host-pathogen coexistence. Virology 479–480, 85–103. 10.1016/j.virol.2015.02.028 [DOI] [PubMed] [Google Scholar]
- De Coninck B., Timmermans P., Vos C., Cammue B. P., Kazan K. (2015). What lies beneath: belowground defense strategies in plants. Trends Plant Sci. 20, 91–101. 10.1016/j.tplants.2014.09.007 [DOI] [PubMed] [Google Scholar]
- Denver D. R., Brown A. M., Howe D. K., Peetz A. B., Zasada I. A. (2016). Genome skimming: a rapid approach to gaining diverse biological insights into multicellular pathogens. PLoS Pathog. 12:e1005713. 10.1371/journal.ppat.1005713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Vito M., Greco N., Oreste G., Saxena M. C., Singh K. B., Kusmenoglu I. (1994). Plant parasitic nematodes of legumes in Turkey. Nematol. Mediterr. 22, 245–251. [Google Scholar]
- Fahmy I. F., Taha O., El-Ashry A. N. (2015). First genome analysis and molecular characterization of Chickpea chlorotic dwarf virus Egyptian isolate infecting squash. VirusDisease 26, 33–41. 10.1007/s13337-014-0246-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO (2014). Pulse Quantity Production. Available online at: http://www.fao.org/faostat/en/
- Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., Mello C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. 10.1038/35888 [DOI] [PubMed] [Google Scholar]
- Fosu-Nyarko J., Jones M. J. J. (2015). Application of biotechnology for nematode control in crop plants. Adv. Bot. Res. 73, 339–376. 10.1016/bs.abr.2014.12.012 [DOI] [Google Scholar]
- Fontana G. S., Santini L., Caretto S., Frugis G., Mariotti D. (1993). Genetic transformation in the grain legume Cicer arietinum L. (chickpea). Plant Cell Rep. 12, 194–198. 10.1007/BF00237052 [DOI] [PubMed] [Google Scholar]
- Friedland A. E., Tzur Y. B., Esvelt K. M., Colaiacovo M. P., Church G. M., Calarco J. A. (2013). Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 10, 741–743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusaro A. F., Correa R. L., Nakasugi K., Jackson C., Kawchuk L., Vaslin M. F., et al. (2012). The Enamovirus P0 protein is a silencing suppressor which inhibits local and systemic RNA silencing through AGO1 degradation. Virology 426, 178–187. 10.1016/j.virol.2012.01.026 [DOI] [PubMed] [Google Scholar]
- Ganguly M., Molla K. A., Karmakar S., Datta K., Datta S. K. (2014). Development of pod borer-resistant transgenic chickpea using a pod-specific and a constitutive promoter-driven fused cry1Ab/Ac gene. Theor. Appl. Genet. 127, 2555–2565. 10.1007/s00122-014-2397-5 [DOI] [PubMed] [Google Scholar]
- Ghanekar A. M., Manohar S. K., Reddy S. V., Nene Y. L. (1988). Association of a mycoplasma-like organism with chickpea phyllody. Indian Phytopathol. 41, 462–464. [Google Scholar]
- Ghanti K. K. S., Sujata K. G., Vijay Kumar B. M., Nataraja Karba N., Reddy K. J., Srinath Rao M., et al. (2011). Heterologous expression of P5CS gene in chickpea enhances salt tolerance without affecting yield. Biol. Plant. 55, 634 10.1007/s10535-011-0161-0 [DOI] [Google Scholar]
- Ghasemzadeh A., Małgorzata Ter Haar M., Shams-Bakhsh M., Pirovano W., Pantaleo V. (2018). Shannon entropy to evaluate substitution rate variation among viral nucleotide positions in datasets of viral siRNAs. Methods Mol. Biol. 1746. 10.1007/978-1-4939-7683-6_15 [DOI] [PubMed] [Google Scholar]
- Gheysen G., Vanholme B. (2007). RNAi from plants to nematodes. Trends Biotechnol. 25, 89–92. 10.1016/j.tibtech.2007.01.007 [DOI] [PubMed] [Google Scholar]
- Goverse A., Smant G. (2014). The activation and suppression of plant innate immunity by parasitic nematodes. Annu. Rev. Phytopathol. 52, 243–265. 10.1146/annurev-phyto-102313-050118 [DOI] [PubMed] [Google Scholar]
- Graham P. H., Vance C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. 10.1104/pp.017004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco N., Di Vito M., Saxena M. C., Reddy M. V. (1988). Effect of Heterodera cicero on yeld of chickpea and lentil and development of this nematode on chickpea in Syria. Nematologica 34, 98–114. 10.1163/002825988X00099 [DOI] [Google Scholar]
- Gupta S., Nawaz K., Parween S., Roy R., Sahu K., Kumar Pole A., et al. (2017). Draft genome sequence of Cicer reticulatum L., the wild progenitor of chickpea provides a resource for agronomic trait improvement. DNA Res. 24, 1–10. 10.1093/dnares/dsw042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J., Thomas J. E., Schwinghamer M. W., Kraberger S., Stainton D., Dayaram A., et al. (2012). Molecular characterisation of dicot-infecting mastreviruses from Australia. Virus Res. 166, 13–22. 10.1016/j.virusres.2012.02.024 [DOI] [PubMed] [Google Scholar]
- Holbein J., Grundler F. M., Siddique S. (2016). Plant basal resistance to nematodes: an update. J. Exp. Bot. 67, 2049–2061. 10.1093/jxb/erw005 [DOI] [PubMed] [Google Scholar]
- Horn N. M., Reddy S. V., Reddy R. D. V. (1994). Virus-vector relationships of chickpea chlorotic dwarf geminivirus and the leafhopper Orosius orientalis (Hemiptera: Cicadellidae). Ann. Appl. Biol. 124, 441–450. 10.1111/j.1744-7348.1994.tb04149.x [DOI] [Google Scholar]
- Horn N. M., Reddy S. V., Roberts I. M., Reddy R. D. V. (1993). Chickpea chlorotic dwarf virus, a new leafhopper-transmitted geminivirus of chickpea in India. Ann. Appl. Biol. 122, 467–479. 10.1111/j.1744-7348.1993.tb04050.x [DOI] [Google Scholar]
- Hussey R. S. (1989). Disease-inducing secretions of plant-parasitic nematodes. Annu. Rev. Phytopathol. 27, 127–141. 10.1146/annurev.py.27.090189.001011 [DOI] [Google Scholar]
- Iantcheva A., Vlahova M., Trinh T. H., Brown S. C., Slater A., Elliott M. C., et al. (2001). Assessment of polysomaty, embryo formation and regeneration in liquid media for various species of diploid annual Medicago. Plant Sci. 160, 621–627. 10.1016/S0168-9452(00)00432-5 [DOI] [PubMed] [Google Scholar]
- Ignacimuthu S., Prakash S. (2006). Agrobacterium-mediated transformation of chickpea with alpha-amylase inhibitor gene for insect resistance. J. Biosci. 31, 339–345. 10.1007/BF02704106 [DOI] [PubMed] [Google Scholar]
- Indurker S., Misra H. S., Eapen S. (2007). Genetic transformation of chickpea (Cicer arietinum L.) with insecticidal crystal protein gene using particle gun bombardment. Plant Cell Rep. 26, 755–763. 10.1007/s00299-006-0283-6 [DOI] [PubMed] [Google Scholar]
- Islam R., Malik T., Husnain T., Riazuddin S. (1994). Strain and cultivar specificity in the Agrobacterium-chickpea interaction. Plant Cell Rep. 13, 561–563. 10.1007/BF00234511 [DOI] [PubMed] [Google Scholar]
- Jain M., Misra G., Patel R. K., Priya P., Jhanwar S., Khan A. W., et al. (2013). A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 74, 715–729. 10.1111/tpj.12173 [DOI] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser W. J., Danesh D. (1971). Biology of Four Viruses affecting Cicer arietinum in Iran. Phytopathology 61, 372–375. 10.1094/Phyto-61-372 [DOI] [Google Scholar]
- Kanakala S., Verma H. N., Vijay P., Saxena D. R., Malathi V. G. (2013). Response of chickpea genotypes to Agrobacterium-mediated delivery of Chickpea chlorotic dwarf virus (CpCDV) genome and identification of resistance source. Appl. Microbiol. Biotechnol. 97, 9491–9501. 10.1007/s00253-013-5162-9 [DOI] [PubMed] [Google Scholar]
- Kar S., Basu D., Das S., Ramkrishnan N. A., Mukherjee P. P. N., Sen S. K. (1997). Expression of cryIA(c) gene of Bacillus thuringiensis in transgenic chickpea plants inhibits development of pod- borer (Heliothis armigera) larvae. Transgenic Res. 6, 177–185. 10.1023/A:1018433922766 [DOI] [Google Scholar]
- Kar S., Johnson T. M., Nayak P., Sen S. K. (1996). Efficient transgenic plant regeneration through Agrobacterium-mediated transformation of Chickpea (Cicer arietinum L.). Plant Cell Rep. 16, 32–37. 10.1007/BF01275444 [DOI] [PubMed] [Google Scholar]
- Kim H., Kim J. S. (2014). A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 15, 321–334. 10.1038/nrg3686 [DOI] [PubMed] [Google Scholar]
- Kraberger S., Kumari S. G., Hamed A. A., Gronenborn B., Thomas J. E., Sharman M., et al. (2015). Molecular diversity of Chickpea chlorotic dwarf virus in Sudan: high rates of intra-species recombination - a driving force in the emergence of new strains. Infect. Genet. Evol. 29, 203–215. 10.1016/j.meegid.2014.11.024 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy K. V., Suhasini K., Sagare A. P., Meixner M., De Kathen A., Pickardt T., et al. (2000). Agrobacterium mediated transformation of chickpea (Cicer arietinum L.) embryo axes. Plant Cell Rep. 19, 235–240. 10.1007/s002990050005 [DOI] [PubMed] [Google Scholar]
- Kumar P. L., Kumari S. M. G., Waliyar F. (2008). Virus diseases of Chickpea, in Characterization, Diagnosis & Management of Plant Viruses, Vol. 3: Vegetable and Pulse Crops, eds Rao G. P., Kumar P. L., Holguín-Peña R. J. (Houston, TX: Studium Press LLC; ), 213–234. [Google Scholar]
- Kumari S. G., Makkouk K. M. (2007). Virus diseases of faba bean (Vicia faba L.) in Asia and Africa, in Plant Viruses (Aleppo: Global Science Book; ), 93–95. [Google Scholar]
- Kumari S. G., Assad N., Alnaasan Y., Attar N., Van Leur J. A. G. (2011). Chickpea chlorotic stunt virus: host range, purification, serology and transmission characteristics and occurrence in Central and West Asia and North Africa (CWANA), in Fourth Conference of the International Working Group On Legume & Vegetable Viruses (IWGLVV), ed. Horticulture Australia. (Antequera: ). [Google Scholar]
- Li S., Cong Y., Liu Y., Wang T., Shuai Q., Chen N., et al. (2017). Optimization of Agrobacterium-mediated transformation in soybean. Front. Plant Sci. 8:246. 10.3389/fpls.2017.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu Z. B., Xing A., Moon B. P., Koellhoffer J. P., Huang L., et al. (2015). Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 169, 960–970. 10.1104/pp.15.00783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahapatra B. C., Pahdi N. N. (1996). Pathogenicity and control of Rotylenchulus reniformis on Cicer arietinum. Nematol. Mediterr. 14, 287–290. [Google Scholar]
- Makkouk K. M., Kumari S. G., Shahraeen N., Fazlali Y., Farzadfar S., Ghotbi T., et al. (2003). Identification and seasonal variation of viral diseases of chickpea and lentil in Iran. J. Plant Dis. Protect. 110, 157–169. Available online at: https://www.cabdirect.org/cabdirect/abstract/20033060880 [Google Scholar]
- Makkouk K., Pappu H., Kumari S. G. (2012). Virus diseases of peas, beans, and faba bean in the Mediterranean region. Adv. Virus Res. 84, 367–402. 10.1016/B978-0-12-394314-9.00011-7 [DOI] [PubMed] [Google Scholar]
- Malhotra R. S., Singh K. B., Di Vito M., Greco N., Saxena M. C. (2002). Registration of ILC10765 and ILC 10766 chickpea germplasm lines resistant to cyst nematodes. Crop Sci. 42:1756 10.2135/cropsci2002.1756 [DOI] [Google Scholar]
- Mandadi K. K., Scholthof K. B. (2013). Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25, 1489–1505. 10.1105/tpc.113.111658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manosalva P., Manohar M., Von Reuss S. H., Chen S., Koch A., Kaplan F., et al. (2015). Conserved nematode signalling molecules elicit plant defenses and pathogen resistance. Nat. Commun. 6, 7795. 10.1038/ncomms8795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzoor M. T., Ilyas M., Shafiq M., Haider M. S., Shahid A. A., Briddon R. W. (2014). A distinct strain of chickpea chlorotic dwarf virus (genus Mastrevirus, family Geminiviridae) identified in cotton plants affected by leaf curl disease. Arch. Virol. 159, 1217–1221. 10.1007/s00705-013-1911-4 [DOI] [PubMed] [Google Scholar]
- Massart S., Olmos A., Jijakli H., Candresse T. (2014). Current impact and future directions of high throughput sequencing in plant virus diagnostics. Virus Res. 188, 90–96. 10.1016/j.virusres.2014.03.029 [DOI] [PubMed] [Google Scholar]
- Mehrotra M., Singh A. K., Sanyal I., Altosaar I., Amla D. V. (2010). Pyramiding of modified cry1Ab and cry1Ac genes of Bacillus thuringiensis in transgenic chickpea (Cicer arietinum L.) for improved resistance to pod borer insect Helicoverpa armigera. Euphytica 182, 87–102. 10.1007/s10681-011-0501-3 [DOI] [Google Scholar]
- Mendy B., Wang'ombe M. W., Radakovic Z. S., Holbein J., Ilyas M., Chopra D., et al. (2017). Arabidopsis leucine-rich repeat receptor-like kinase NILR1 is required for induction of innate immunity to parasitic nematodes. PLoS Pathog. 13:e1006284. 10.1371/journal.ppat.1006284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Hou Y., Wang H., Ji R., Liu B., Wen J., et al. (2017). Targeted mutagenesis by CRISPR/Cas9 system in the model legume Medicago truncatula. Plant Cell Rep. 36, 371–374. 10.1007/s00299-016-2069-9 [DOI] [PubMed] [Google Scholar]
- Mishra S., Sanyal I., Amla D. V. (2012). Changes in protein pattern during different developmental stages of somatic embryos in chickpea. Biol. Plant. 56, 613–619. 10.1007/s10535-012-0124-0 [DOI] [Google Scholar]
- Mustafayev E., Kumari S. G., Attar N., Zeynal A. (2011). Viruses infecting chickpea and lentil crops in Azerbaijan. Austral. Plant Pathol. 40, 611–620. 10.1007/s13313-011-0094-2 [DOI] [Google Scholar]
- Ouattara A., Tiendrebeogo F., Lefeuvre P., Hoareau M., Claverie S., Traore E. V., et al. (2017). New strains of chickpea chlorotic dwarf virus discovered on diseased papaya and tomato plants in Burkina Faso. Arch. Virol. 162, 1791–1794. 10.1007/s00705-017-3262-z [DOI] [PubMed] [Google Scholar]
- Ouizbouben A., Fortass M. (1997). Survey of chickpea for viruses in Morocco. EPPO Bull. 27, 249–254. 10.1111/j.1365-2338.1997.tb00642.x [DOI] [Google Scholar]
- Parween S., Nawaz K., Roy R., Pole A. K., Venkata Suresh B., Misra G., et al. (2015). An advanced draft genome assembly of a desi type chickpea (Cicer arietinum L.). Sci. Rep. 5:12806. 10.1038/srep12806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak M. R., Hamzah R. Y. (2008). An effective method of sonication-assisted Agrobacterium-mediated transformation of chickpeas. Plant Cell Tissue Organ Cult. 93, 65–71. 10.1007/s11240-008-9344-6 [DOI] [Google Scholar]
- Patil G., Deokar A., Jain P. K., Thengane R. J., Srinivasan R. (2009). Development of a phosphomannose isomerase-based Agrobacterium-mediated transformation system for chickpea (Cicer arietinum L.). Plant Cell Rep. 28, 1669–1676. 10.1007/s00299-009-0766-3 [DOI] [PubMed] [Google Scholar]
- Pirovano W., Miozzi L., Boetzer M., Pantaleo V. (2014). Bioinformatics approaches for viral metagenomics in plants using short RNAs: model case of study and application to a Cicer arietinum population. Front. Microbiol. 5:790. 10.3389/fmicb.2014.00790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polowick P. L., Baliski D. S., Mahon J. D. (2004). Agrobacterium tumefaciens-mediated transformation of chickpea (Cicer arietinum L.): gene integration, expression and inheritance. Plant Cell Rep. 23, 485–491. 10.1007/s00299-004-0857-0 [DOI] [PubMed] [Google Scholar]
- Reinprecht Y., Yadegari Z., Perry G. E., Siddiqua M., Wright L. C., McClean P. E., et al. (2013). In silico comparison of genomic regions containing genes coding for enzymes and transcription factors for the phenylpropanoid pathway in Phaseolus vulgaris L. and Glycine max L. Merr. Front. Plant Sci. 4:317. 10.3389/fpls.2013.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso M. N., Jones J. T., Abad P. (2009). RNAi and functional genomics in plant parasitic nematodes. Annu. Rev. Phytopathol. 47, 207–232. 10.1146/annurev.phyto.112408.132605 [DOI] [PubMed] [Google Scholar]
- Sanyal I., Singh A. K., Kaushik M., Amla D. V. (2005). Agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) with Bacillus thuringiensis cry1Ac gene for resistance against pod borer insect Helicoverpa armigera. Plant Sci. 168, 1135–1146. 10.1016/j.plantsci.2004.12.015 [DOI] [Google Scholar]
- Saqib M., Bayliss K. L., Dell B., Hardy Gest J., Jones M. G. K. (2005). First record of a phytoplasma-associated disease of chickpea (Cicer arietinum) in Australia. Austral. Plant Pathol. 34, 425–426. 10.1071/AP05047 [DOI] [Google Scholar]
- Sarmah B. K., Moore A., Tate W., Molvig L., Morton R. L., Rees D. P., et al. (2004). Transgenic chickpea seeds expressing high levels of a bean α-amylase inhibitor. Mol. Breed. 24, 73–82. 10.1023/B:MOLB.0000037996.01494.12 [DOI] [Google Scholar]
- Schwinghamer M. W., Thomas J. E., Parry J. N., Schilg M. A., Dann E. K. (2007). First record of natural infection of chickpea by Turnip mosaic virus. Australas. Plant Dis. Notes 2, 41–43. 10.1071/DN07020 [DOI] [Google Scholar]
- Senthil G., Williamson B., Dinkins R. D., Ramsay G. (2004). An efficient transformation system for chickpea (Cicer arietinum L.). Plant Cell Rep. 23, 297–303. 10.1007/s00299-004-0854-3 [DOI] [PubMed] [Google Scholar]
- Sharma H. C., Sharma K. K., Seetharama N., Ortiz R. (2001). Genetic transformation of crop plants: risks and opportunities for the rural poor. Curr. Sci. 80, 1495–1508. Available online at: http://www.jstor.org/stable/24106285 [Google Scholar]
- Sharma K. K., Ortiz R. (2000). Program for the application of genetic transformation for crop improvement in the semi-arid tropics. In Vitro Cell. Dev. Biol. Plant 36, 83–92. 10.1007/s11627-000-0019-1 [DOI] [Google Scholar]
- Shivaprasad P. V., Chen H. M., Patel K., Bond D. M., Santos B. A., Baulcombe D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. 10.1105/tpc.111.095380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R. R., Cho-Vega J. H., Davuluri Y., Ma S., Kasbidi F., Milito C., et al. (2009). Sonic hedgehog signaling pathway is activated in ALK-positive anaplastic large cell lymphoma. Cancer Res. 69, 2550–2558. 10.1158/0008-5472.CAN-08-1808 [DOI] [PubMed] [Google Scholar]
- Sirtori C. R. (2001). Risks and benefits of soy phytoestrogens in cardiovascular diseases, cancer, climacteric symptoms and osteoporosis. Drug Saf. 24, 665–682. 10.2165/00002018-200124090-00003 [DOI] [PubMed] [Google Scholar]
- Smýkal P., Coyne C. J., Ambrose M. J., l Maxted N., Schaefer H., Blair M. W., et al. (2015). Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 34, 43–104. 10.1080/07352689.2014.897904 [DOI] [Google Scholar]
- Somers D. A., Samac D. A., Olhoft P. M. (2003). Recent advances in legume transformation. Plant Physiol. 131, 892–899. 10.1104/pp.102.017681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan R., Mohapatra T., Sharma R. P. (1991). Agrobacterium mediated genetic transformation of chickpea, Cicer arietinum L. Indian J. Exp. Biol. 29, 758–761. [PubMed] [Google Scholar]
- Srivastava J., Datta S., Mishra S. P. (2017). Development of an efficient Agrobacterium mediated transformation system for chickpea (Cicer arietinum). Biol. Plant. 72, 153–170. 10.1515/biolog-2017-0015 [DOI] [Google Scholar]
- Srivastava S., Zheng Y., Kudapa H., Jagadeeswaran G., Hivrale V., Varshney R. K., et al. (2015). High throughput sequencing of small RNA component of leaves and inflorescence revealed conserved and novel miRNAs as well as phasiRNA loci in chickpea. Plant Sci. 235, 46–57. 10.1016/j.plantsci.2015.03.002 [DOI] [PubMed] [Google Scholar]
- Tewari-Singh N., Sen J., Kiesecker H., Reddy V. S., Jacobsen H. J., Guha-Mukherjee S. (2004). Use of a herbicide or lysine plus threonine for non-antibiotic selection of transgenic chickpea. Plant Cell Rep. 22, 576–583. 10.1007/s00299-003-0730-6 [DOI] [PubMed] [Google Scholar]
- Thomas J. E., Parry J. N., Schwinghamer M. W., Dann E. K. (2010). Two novel mastreviruses from chickpea (Cicer arietinum) in Australia. Arch. Virol. 155, 1777–1788. 10.1007/s00705-010-0763-4 [DOI] [PubMed] [Google Scholar]
- Thudi M., Chitikineni A., Liu X., He W., Roorkiwal M., Yang W., et al. (2016). Recent breeding programs enhanced genetic diversity in both desi and kabuli varieties of chickpea (Cicer arietinum L.). Sci. Rep. 6:38636. 10.1038/srep38636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi L., Singh A. K., Singh S., Singh R., Chaudhary S., Sanya I., et al. (2013). Optimization of regeneration and Agrobacterium-mediated transformation of immature cotyledons of chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult. 113, 513–527. 10.1007/s11240-013-0293-3 [DOI] [Google Scholar]
- Upadhyaya H. D., Dwivedi S. L., Baum M., Varshney R. K., Udupa S. M., Gowda C. L., et al. (2008). Genetic structure, diversity, and allelic richness in composite collection and reference set in chickpea (Cicer arietinum L.). BMC Plant Biol. 8:106. 10.1186/1471-2229-8-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R. K., Song C., Saxena R. K., Azam S., Yu S., Sharpe A. G., et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246. 10.1038/nbt.2491 [DOI] [PubMed] [Google Scholar]
- Vogler H., Akbergenov R., Shivaprasad P. V., Dang V., Fasler M., Kwon M. O., et al. (2007). sRNA modification associated with suppression of RNA silencing by tobamovirus replicase protein. J. Virol. 81, 10379–10388. 10.1128/JVI.00727-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vovlas N., Rapoport H. F., Jimenez Diaz R. M., Castillo P. (2005). Differences in feeding sites induced by root-knot nematodes, Meloidogyne spp., in chickpea. Phytopathology 95, 368–375. 10.1094/PHYTO-95-0368 [DOI] [PubMed] [Google Scholar]
- Walsh E., Elmore J. M., Taylor C. G. (2017). Root-knot nematode parasitism suppresses host RNA silencing. Mol. Plant Microbe Interact. 30, 295–300. 10.1094/MPMI-08-16-0160-R [DOI] [PubMed] [Google Scholar]
- Wu X. Y., Zhou G. C., Chen Y. X., Wu P., Liu L. W., Ma F. F., et al. (2016). Soybean cyst nematode resistance emerged via artificial selection of duplicated serine hydroxymethyltransferase genes. Front. Plant Sci. 7:998. 10.3389/fpls.2016.00998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav R., Mehrotra M., Singh A. K., Niranjan A., Singh R., Sanyal I., et al. (2017). Improvement in Agrobacterium-mediated transformation of chickpea (Cicer arietinum L.) by the inhibition of polyphenolics released during wounding of cotyledonary node explants. Protoplasma 254, 253–269. 10.1007/s00709-015-0940-0 [DOI] [PubMed] [Google Scholar]
- Yahia A. A., Ouada M. A., Illoul H., Tair M. I. (1997). First occurrence of bean yellow mosaic potyvirus on chickpea in Algeria. EPPO Bull. 27, 261–263. 10.1111/j.1365-2338.1997.tb00645.x [DOI] [Google Scholar]
- Yang N., Wang R., Zhao Y. (2017). Revolutionize genetic studies and crop improvement with high-throughput and genome-scale CRISPR/Cas9 gene editing technology. Mol. Plant 10, 1141–1143. 10.1016/j.molp.2017.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K., Han T., Liu G., Chen T., Wang Y., Yu A. Y., et al. (2015). A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 5:14926. 10.1038/srep14926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaagueri T., Miozzi L., Mnari-Hattab M., Noris E., Accotto G. P., Vaira A. M. (2017a). Deep sequencing data and infectivity assays indicate that chickpea chlorotic dwarf virus is the etiological agent of the “hard fruit syndrome” of watermelon. Viruses 9:E311 10.3390/v9110311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaagueri T., Mnari-Hattab M., Zammouri S., Hajlaoui M. R., Accotto G. P., Vaira A. M. (2017b). First report of chickpea chlorotic dwarf virus in watermelon (Citrullus lanatus) in Tunisia. Plant Dis. 101, 392 10.1094/PDIS-07-16-1028-PDN [DOI] [Google Scholar]
- Zamanian M., Andersen E. C. (2016). Prospects and challenges of CRISPR/Cas genome editing for the study and control of neglected vector-borne nematode diseases. FEBS J. 283, 3204–3221. 10.1111/febs.13781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J., Jeong D. H., De Paoli E., Park S., Rosen B. D., Li Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. 10.1101/gad.177527.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng F., Wu H., Zhang R., Li S., He W., Wong F. L., et al. (2016). Molecular phylogeny and dynamic evolution of disease resistance genes in the legume family. BMC Genomics 17:402. 10.1186/s12864-016-2736-9 [DOI] [PMC free article] [PubMed] [Google Scholar]