Abstract
Many bacterial pathogens coordinately regulate genes encoding important metabolic pathways during disease progression, including the phosphoenolpyruvate (PEP)-phosphotransferase system (PTS) for uptake of carbohydrates. The Gram-positive Group A Streptococcus (GAS) is a pathogen that infects multiple tissues in the human host. The virulence regulator Mga in GAS can be phosphorylated by the PTS, affecting Mga activity based on carbohydrate availability. Here, we explored the effects of glucose availability on the Mga regulon. RNA-seq was used to identify transcriptomic differences between the Mga regulon grown to late log phase in the presence of glucose (THY) or after glucose has been expended (C media). Our results revealed a correlation between the genes activated in C media with those known to be repressed by CcpA, indicating that C media mimics a non-preferred sugar environment. Interestingly, we found very little overlap in the Mga regulon from GAS grown in THY versus C media beyond the core virulence genes. We also observed an alteration in the phosphorylation status of Mga, indicating that the observed media differences in the Mga regulon may be directly attributed to glucose levels. Thus, these results support an in vivo link between glucose availability and virulence regulation in GAS.
Introduction
Coordinated expression of virulence factors is essential for bacterial pathogens to successfully colonize and elicit an infection in the host. Since expression of virulence genes is often linked to the availability of essential nutrients such as carbohydrates, pathogenic bacteria often utilize carbon catabolism regulatory pathways to sense the presence of preferred carbohydrates and repress the genes involved in alternative sugar utilization1,2. In fact, many pathogens have been shown to coordinately regulate the expression of carbohydrate metabolism genes with disease progression during in vivo studies3–9.
The phosphoenolpyruvate (PEP)-phosphotransferase system (PTS) allows bacteria to uptake carbohydrates and monitor carbon utilization1 via two cytoplasmic enzymes, EI (ptsI) and Hpr (ptsH), and several sugar-specific EII components. Each EII comprises two cytosolic subunits (EIIAB), an integral membrane transporter (EIIC), and sometimes a second membrane component (EIID). Phosphotransfer begins when the PEP generated from glycolysis transfers its phosphate to EI, which then is transferred to Hpr on His-15, then a a sugar-specific EIIA, then EIIB and finally to the transported sugar via its EIIC1. When Gram-positive bacteria are in nutrient rich conditions, Hpr is phosphorylated at Ser-46 by the kinase, HprK, allowing HprSer~P to dimerize with the carbon catabolite protein, CcpA, and elicit carbon catabolite repression (CCR) by binding to catabolite response elements (cre) found in promoter sequences10. In the absence of a preferred carbon source, Hpr-His15~P and EIIB~P are capable of phosphorylating histidines within PTS regulatory domains (PRDs) of transcriptional regulators (LicT, MtlR, LevR, etc.), regulating their activity and the expression of non-preferred sugar operons1. Thus, the PTS represents a signal transduction network through which Gram-positive pathogens alter gene regulation in response to carbohydrate utilization.
The Group A Streptococcus (GAS, Streptococcus pyogenes) is a Gram-positive pathogen that can colonize a variety of tissues in the human host, resulting in both life-threatening invasive as well as benign diseases. Each year around the world, GAS elicits over 700 million self-limiting infections and results in more than 500,000 deaths due to invasive infections and nonsuppurative sequelae11. Both ex vivo and in vivo studies have established that GAS exhibits significant changes in its transcriptome during infection4,12–15. As a fastidious fermentative organism that relies heavily on carbohydrates, metabolic genes under CCR are induced in vivo and are often required for full virulence in GAS16–21, directly linking GAS carbohydrate metabolism and virulence. Here we use the M1T1 strain 5448, a representative of one of the most prevalent serotypes of GAS isolated from invasive forms of infections worldwide.
GAS utilizes global transcriptional regulators such as the ubiquitous stand-alone regulator Mga to coordinate transcriptome changes impacting virulence22. Mga is critical for multiple in vivo phenotypes, including biofilm formation, growth in whole human blood and soft tissue, resistance to phagocytosis, and attachment to keratinocytes4,12,23–26. Mga regulates approximately 10% of the GAS genome during exponential phase of growth in rich medium (THY), including transcription of several sugar transport and utilization operons27. The ‘core’ Mga regulon consists of virulence genes critical for attachment and immune evasion such as M protein (emm), the emm-superfamily (mrp, enn, arp, etc.), C5a peptidase (scpA), and fibronectin-binding protein (fba)28. The Mga regulon was found to be expressed during the acute phase of GAS-mediated pharyngitis in macaques concurrently with the PTS and sugar metabolism operons15. Taken together, these data underscore that Mga and its virulence regulon are linked to carbohydrate utilization.
Mga belongs to a novel family of PTS regulatory domain (PRD)-containing virulence regulators (PCVR) that allows the PTS to directly phosphorylate and affect the activity of Mga based on carbohydrate availability3,29. Genetic, biochemical, and structural studies on the homologous Bacillus anthracis toxin regulator, AtxA, indicate that it is also a PCVR30–33. Two reiterative PRD domains (PRD-1 and PRD-2) of Mga allow PTS phosphorylation to impact its function likely through controlling dimerization of the carboxy-terminal EIIBGat-like domain comparable to AtxA and other Gram-positive sugar-specific PRD-containing activators3,24,31,32,34. Although it is known that Mga activity is modulated by the PTS, we still do not know whether carbohydrate availability affects Mga-dependent gene regulation. In this study, we explored the effects of glucose availability on Mga and its impact on the Mga regulon. RNA-Seq was used to identify transcriptomic differences between the Mga regulon grown to late exponential phase either in the presence (THY) or absence (C media) of glucose. We observed that Mga was differentially phosphorylated in THY in comparison to C media, which led to a high degree of plasticity of the regulon that is correlated to glucose availability.
Results
Transcriptome of M1T1 5448 growing in presence or absence of glucose
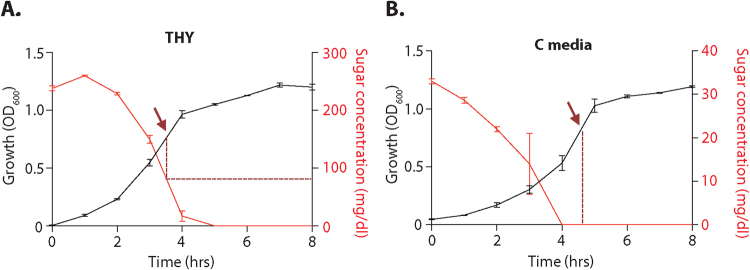
When GAS invades into deep tissue niches in the host, it can encounter an environment where the preferred carbohydrate, glucose, is not as readily available. Previous work found that C media accurately mimics this environment due to its initial low glucose, high peptide composition35. Here, we wanted to assess whether the presence or absence of glucose in the environment has an impact on the Mga virulence regulon of the M1T1 GAS 5448. The rich Todd-Hewitt Yeast (THY) media has an initial level of 0.5% glucose (w/v) and was used as a representative of a glucose-rich environment, while C media has a much lower initial level of 0.05% glucose (w/v) and was used to mimic a low- or no-glucose deep-tissue environment35. We directly assayed the change in the concentration of carbohydrates (primarily glucose) in THY (initially 240 mg/dl) and C media (initially 32 mg/dl) over an 8-h time course during GAS 5448 growth (Fig. 1). As expected, C media initially contained 8-fold less glucose than THY and it was expended earlier in log phase growth (Fig. 1). Total RNA was then isolated from four biological replicates from the WT 5448 grown in either THY or C media to late logarithmic phase (Fig. 1, arrows), the point of maximum Mga regulon expression36, and then processed for RNA-seq as described in Methods. At this point in growth, THY contained ca. 80 mg/dl glucose whereas all of the glucose had been consumed by GAS in C media for over 30 minutes (Fig. 1, dashed lines). RNA-Seq data generated from cells grown in THY was compared to C media-grown cells (THY/C media), where a 1.8-fold change in gene expression (log2 ≤ −0.90 or ≥0.90) and a p value of ≤0.05 were considered significant (Supplemental Table S1 and Fig. S1). Principal component analysis (PCA) of the RNA-seq data indicated reproducibility of the datasets from the same media, as well as a significant difference between WT 5448 grown in THY and C media (Supplemental Fig. S2).
Figure 1.
Comparison of glucose concentration over growth of GAS in THY and C media. M1T1 GAS 5448 was grown in either THY (A) or C media (B), with growth (black, OD600) and total sugar (glucose) concentration (red, mg/dl measured every hour as described in Methods). Data represent the average of three replicates, shown with the standard error of the mean. The right Y axis (red) represent different levels of glucose concentrations in (A) and (B). Arrows indicate sampling point for RNA isolation and dashed lines show the glucose level at that time point.
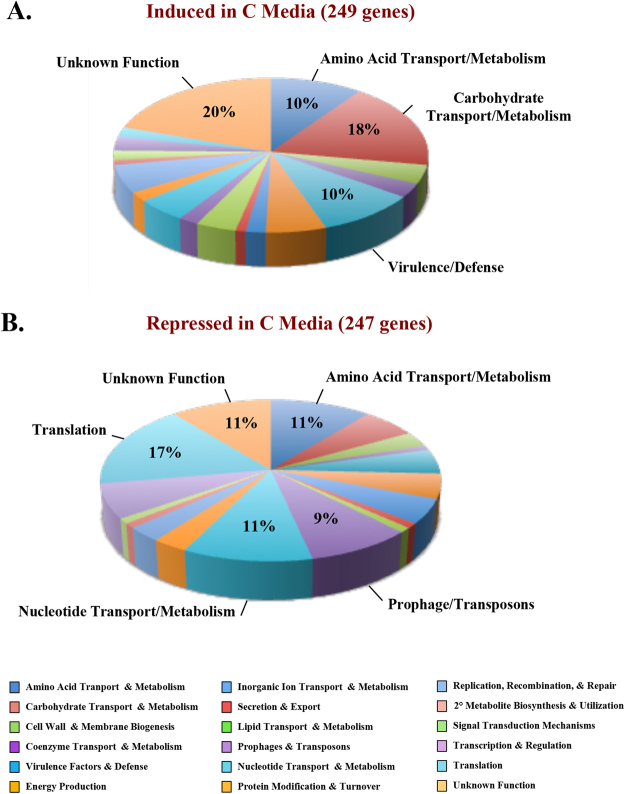
A total of 496 genes (26.9% of the non-tRNA genome) were found to be differentially expressed (249 induced, 247 repressed) in C media compared to THY (Fig. 2, Supplemental Table S1). Analysis of gene ontology using Cluster of Orthologous Groups (COGs) found that C media primarily induced the expression of genes encoding proteins involved in carbohydrate transport and metabolism (44 genes; 18% of induced genes), amino acid transport and metabolism (25 genes; 10% of induced genes), and proteins of Unknown Function (50 genes; 20% of induced genes) (Fig. 2A). In contrast, C media led to the down regulation of genes related to protein synthesis (43 genes; 17% of total repressed genes), amino acid transport and metabolism (26 genes; 11% of repressed genes), nucleotide transport and metabolism (26 genes; 11% of repressed genes), and proteins of Unknown Function (26 genes; 11% of repressed genes) (Fig. 2B). Induction of non-glucose carbohydrate and amino acid metabolism pathways in C media correlates with the composition of C media, which is high in peptides and low in glucose.
Figure 2.
Functional categories of M1T1 GAS 5448 genes differentially expressed in C media versus THY. RNA-seq on M1T1 GAS 5448 grown in THY (high glucose) and C media (low glucose) to late logarithmic phase was analyzed for differential gene expression. GAS genes either induced (A) or repressed (B) in C media compared to THY were assessed based on their Cluster of Orthologous Group (COG) designation and displayed in pie graph format. The most prevalent COG designations are indicated with their overall percentage. Color scheme for COG categories are indicated at the bottom.
CcpA represses the expression of alternative carbohydrate utilization genes (i.e., CCR) through directly binding to cre sites under favorable glucose-replete conditions. Since C media contains no glucose at the time point taken, we predicted that CCR would be relieved. 119 (~24%) of the genes differentially expressed in C media (Supplemental Table S1) were also found to be regulated by CcpA in published transcriptome studies17,18,37. Of these, 59 had cre or cre2 sites37 located upstream of genes that were either induced or repressed in C media (Supplemental Table S1), supporting that they could be directly regulated via CcpA. Thus, growth of WT 5448 in C media at late logarithmic phase partially resembles expression of a CcpA regulon in the absence of glucose (non-repressing condition)17,18,37.
In general, we found that most metabolic genes involved in utilization and transport of alternative (non-glucose) sugars were induced during late logarithmic phase growth in C media. These included many alternative PTS carbohydrate EII transporter genes such as those for 3-keto-L-gulonate (ptxABC), mannitol (spy_1664), galactose (spy_1399), both cellobiose operons (spy_1079–1083 and celB), and trehalose (treCB). The ABC transporters for sialic acid (spy_0213–0215) and cyclomaltodextrin (malADC) were also upregulated. Interestingly, the β-glucoside-specific EIIABC and both mannose operons ptsABCD and manLMN were repressed in C media, suggesting they may represent sugar uptake systems for more preferred sugars (Supplemental Table S1).
Several virulence-related genes were upregulated during growth in C media, including the has (capsule synthesis) and sag (Streptolysin S) operons; which are also under CCR17,18. Although there is a considerable overlap between the C media and CcpA regulons, there were also media-specific regulatory phenotypes. An additional 25 genes found in this study have been previously shown to be regulated by CcpA17,18, but were regulated in a different direction. Interestingly, we observed a significant increase in the expression of the lacD.1 operon and rgg/ropB, both of which encode for regulators of the secreted cysteine protease virulence factor gene speB. C media is an inducer of speB expression in M14 GAS35; however, we did not see speB being differentially expressed in M1T1 GAS 5448. We also found several genes of the core Mga virulence regulon to be down regulated in C media (Table 1) in comparison to THY, with the exception of emm and mga. Using real-time qPCR, we validated that mga and emm transcript levels were unaffected by growth in C media whereas the Mga-regulated genes encoding the fibronectin binding protein (fba) and the C5A peptidase (scpA) were down regulated.
Table 1.
Expression of Mga core virulence regulon in THY compared to C media (THY/C).
| Spy # | Annotation | Gene | RNA-Seq Log2 FC |
qPCR Log2 FC |
|---|---|---|---|---|
| M5005_Spy1714 | cell surface protein; Mga-regulated | fba | 2.51 | 1.32 ± 0.17 |
| M5005_Spy1715 | C5A peptidase precursor; Mga-regulated | scpA | 2.20 | 1.26 ± 0.33 |
| M5005_Spy1716 | Transposase | −1.66 | — | |
| M5005_Spy1717 | Transposase | −2.47 | — | |
| M5005_Spy1718 | inhibitor of complement; Mga-regulated | sic1.0 | 2.32 | 0.49 ± 0.14 |
| M5005_Spy1719 | M protein; Mga-regulated | emm1 | 0.74 | 0.84 ± 0.21 |
| M5005_Spy1720 | multi-virulence gene regulator Mga | mga | 0.67 | 0.32 ± 0.11 |
| M5005_Spy1721 | hypothetical protein | −2.86 | — |
Taken together, the transcriptome of M1T1 GAS 5448 in C media exhibits similarity to the release of CcpA-mediated repression likely due to the absence of glucose in C media at late logarithmic phase. Additionally, expression of some of the Mga regulon, but not mga itself, is also reduced in C media compared to a glucose-rich environment.
Comparison of the M1T1 GAS Mga regulon in THY to C media
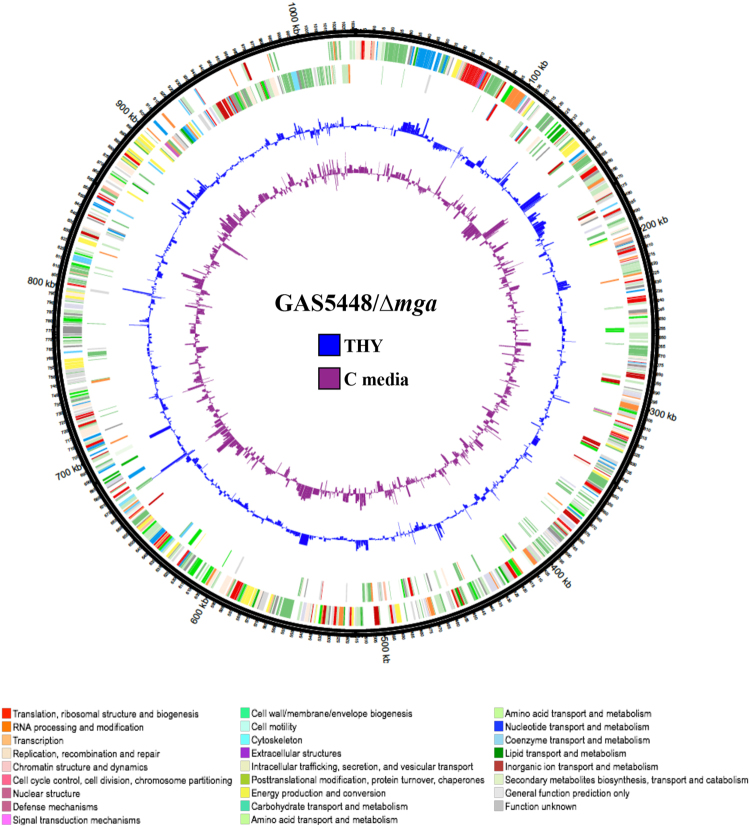
Since we observed differential expression of Mga-regulated genes in different glucose concentrations without changes in mga itself, we hypothesized that the Mga regulon in these two medias would vary. An insertional inactivation of mga in M1T1 5448 was constructed using a temperature-sensitive plasmid to generate the mga mutant, 5448.930 (see Methods). Growth kinetics of 5448.930 were found to be comparable to WT 5448 in both THY and C media (data not shown). Total RNA was isolated in two biological replicates from the WT 5448 and the ∆mga mutant grown in either THY (+glucose) or C media (low glucose) to late logarithmic phase and processed for RNA-seq as described in Methods. Data obtained from the WT 5448 was compared to the ∆mga grown cells in either media (WT/∆mga) with changes in gene expression over 1.8-fold (log2 ≤ −0.90 or ≥0.90) and a p value of ≤0.05 considered significant (Fig. 3 and Supplemental Fig. S3).
Figure 3.
Transcriptomic landscape of M1T1 5448 ∆mga mutant during growth in low and high glucose. Circos plot of genes differentially expressed (DE) between GAS 5448 compared to an isogenic ∆mga 5448.930 grown in either C media (low glucose) or THY (high glucose) at late logarithmic growth. The outer most ring represents a size ruler and the next two rings represent the GAS open reading frames (ORFs) on the (+) and (−) strand of the genome, respectively. Color of ORFs reflect their functional Cluster of Orthologous Group (COG) as indicated at bottom. The inner rings show transcript levels (inside is down regulation, outside is up regulation) with the height of the bars representing the log2 fold-change in DE in THY (blue) vs. C media (purple).
The most activated genes in both media conditions were those of the core Mga regulon (e.g., emm, sic, scpA), showing between 7-fold to 18-fold reduction in transcript levels in the ∆mga mutant (Table 2; Supplemental Figs S3 and S4). Interestingly, we did not observe a Mga-specific phenotype for sclA in our RNA-Seq datasets, a gene considered to be part of the core Mga regulon27,29,38,39. The mga mutant 5448.930 was passaged at the permissive temperature to generate the 5448.930R rescue strain as a complement control (Supplemental Fig. S3B). Real-time qPCR using the same RNA that was assayed by RNA-seq found that sclA was still regulated by Mga and it could be complemented in a strain 5448.930 R rescued for the mga mutation (Supplemental Figs S3AB and S4AB). This may result from a read alignment issue due to two scl alleles (sclA/scl1 and scl2) encoded in the GAS genome, the second of which is not regulated by Mga.
Table 2.
Genes regulated by Mga in both THY and C Media (WT 5448/∆mga).
| Spy Number | Annotation | Gene | THY Log2 FC |
C Media Log2 FC |
|---|---|---|---|---|
| M5005_Spy0113 | transposase | 1.47 | 1.32 | |
| M5005_Spy0143 | hypothetical protein | 2.54 | 3.35 | |
| M5005_Spy0522 | unsaturated glucuronyl hydrolase | 1.45 | 0.98 | |
| M5005_Spy0668 | IgG-degrading protease | mac | 1.62 | 1.37 |
| M5005_Spy0824 | tetrahydropholate synth., pyrophosphokinase | folK | −1.00 | −0.87 |
| M5005_Spy1076 | putative bicarbonate transporter | glnH | −1.31 | −0.85 |
| M5005_Spy1077 | putative bicarbonate transporter | glnQ.2 | −1.04 | −1.04 |
| M5005_Spy1714 | fibronectin-binding surface protein | fba | 3.20 | 4.79 |
| M5005_Spy1715 | C5A peptidase precursor; Mga-regulated | scpA | 3.43 | 5.04 |
| M5005_Spy1718 | secreted inhibitor of complement | sic1.0 | 4.63 | 5.53 |
| M5005_Spy1719 | M protein | emm1 | 9.08 | 10.06 |
| M5005_Spy1720 | multi-virulence gene regulator Mga | mga | 4.60 | 5.84 |
| M5005_Spy1721 | hypothetical protein | −2.78 | −5.00 | |
| M5005_Spy1733 | hypothetical protein | −2.54 | 1.45 | |
| M5005_Spy1738 | secreted DNase-Streptodornase-chromosomal | spd | 0.98 | 1.03 |
| M5005_Spy1798 | suppressor of clpP/X, SpxA homolog, allele 2 | spxA2 | −1.86 | −1.07 |
*Bold, shaded grey denotes the core Mga virulence regulon.
M1T1 Mga regulon in THY (+glucose)
Published transcriptome studies of the Mga regulon from M1 SF370, M6 JRS4, M4 GA40634, and M59 MGAS15249 GAS strains at log phase in THY have indicated significant inter-serotype variation27,29. For RNA-seq of M1T1 5448 in THY, a total of 148 genes (excluding tRNA genes) were significantly regulated by Mga with approximately equal numbers of repressed (73) and activated (75) genes (Supplemental Table S2; Fig. 3, blue ring). Overall, 7.9% of the genome (excluding tRNA genes) was regulated by Mga in the M1T1 GAS 5448 background grown in THY to late logarithmic phase. To confirm the RNA-seq results generated from THY, 8 genes were chosen for analysis via qPCR, representing 6 activated and 2 repressed genes (Supplemental Fig. S3A). Real-time qPCR results validated the RNA-seq data with a correlation coefficient of 0.87 (Supplemental Fig. S3C).
Interestingly, the chromosomal spd gene, encoding the DNase Spd (Mitogenic Factor/MF, DNase B), exhibited 4-fold lower transcript in the mutant compared to WT 5448 (Supplemental Table S2), representing a new virulence gene activated by Mga. Mga repressed expression of the cysteine protease SpeB (speB) and its inhibitor (spi) in the M1T1 5448 background in THY (Supplemental Table S2, Fig. S3). This is in contrast to M1 SF370, where Mga activated speB27. Mga did regulate operons involved in sugar metabolism in M1T1 5448 comparable to M1 SF37027 (Supplemental Table S2). However, the majority of sugar-related operons were activated by Mga in 5448, the opposite of what was previously observed for SF37027. Therefore, despite similarities between many genes regulated by Mga in M1 SF370 and M1T1 5448, there were also clear differences in genes and/or directionality of regulation by Mga (Table 3). This further supported published evidence that the Mga regulon can vary across or even within serotypes grown under identical conditions.
Table 3.
Comparison of the M1 SF370 and M1T1 5448 Mga regulons in THY.
| MGAS5005 | Gene Name |
Annotation | Log2FC 5448 RNA-seq |
SF370 | Log2FC SF370 Microarray* |
|---|---|---|---|---|---|
| M5005_spy0143 | hypothetical protein | 2.54 | SPy0169 | 1.95 | |
| M5005_spy0340 | lctO | L-lactate oxidase | 2.47 | SPy0414 | 1.30 |
| M5005_spy1632 | lacG | 6-phospho-beta-galactosidase | 2.06 | SPy1916 | 1.59 |
| M5005_spy1714 | fba | fibronectin-binding surface protein | 3.20 | SPy2009 | 5.59 |
| M5005_spy1715 | scpA | C5A peptidase | 3.43 | SPy2010 | 5.67 |
| M5005_spy1718 | sic1.0 | secreted inhibitor of complement | 4.63 | SPy2016 | 6.13 |
| M5005_spy1719 | emm1 | M protein | 9.08 | SPy2018 | 6.13 |
| M5005_spy1720 | mga | Multi-virulence gene regulator Mga | 4.60 | SPy2019 | 1.96 |
*Reference27.
M1T1 Mga regulon in C media (low glucose)
Until now, the Mga regulon has only been described for GAS grown in THY27,29. RNA-seq analysis of the Mga regulon in C media at a point where glucose was consumed revealed a total of 135 genes (excluding tRNA genes) that were regulated by Mga (44 repressed, 91 activated) in C media, representing 7.2% of the GAS genome (Supplemental Table S3, Fig. 3; purple ring). Confirmation of 9 genes was assessed via real-time qPCR using RNA isolated from WT 5448, the mga mutant 5448.930, and its rescue strain (Supplemental Fig. S4), showing a correlation coefficient of 0.95.
In the absence of glucose, Mga activated expression of all three M1T1 DNases (streptodornases). As with THY, the chromosomally-encoded spd (MF, DNaseB) was also activated by Mga. In addition, the phage-encoded streptodornases sdaD2 (Sda1) and spd3 (Spd-3) were also regulated by Mga in C media, with both exhibiting ca. 5-fold reduction in a ∆mga mutant (Supplemental Table S3). Mga regulation of sda2 in C media was confirmed by qPCR, showing an ca. 8-fold activation (Supplemental Fig. S4; green bar) that could be complemented upon glucose supplementation, indicating that Mga regulation of this streptodornase occurs in a glucose-dependent manner. In support of this finding, E64-treated (cysteine protease inhibitor) supernatants from ∆mga mutants grown to late logarithmic phase in C media exhibited a decrease in steady-state Sda1 levels compared to WT 5448 (Supplemental Fig. S5) based on Western blots using polyclonal α-Sda1 antisera (gift from M. Walker).
Of note, we found a large number of virulence and metabolic genes to be Mga-regulated in C media that were not seen in the THY regulon. For example, genes encoding for Streptolysin S (sagA-H), Streptolysin O (slo), Streptokinase (ska), Streptococcal secreted esterase (sse), the heme utilization operon (hupYZ), and the osmotic stress operon (opuAA/opuABC) were regulated by Mga only in the glucose-depleted C media (Supplemental Table S3). Interestingly, Mga did not regulate speB in C media (Supplemental Fig. S4A, light blue bar), indicating that speB is potentially Mga-regulated in a glucose-specific manner in M1T1 GAS. Similarly, the known category B M1 Mga-regulated gene grm, encoding a 78-amino acid hypothetical cytosolic protein21,27, was found to be a part of the C media-specific regulon (Supplemental Table S3). Overall, we observed a broader induction of known virulence factors in a ∆mga background in C media, which appears to indicate that the presence or absence of glucose may impact the M1T1 GAS Mga regulon.
Glucose levels alter the Mga regulon
Comparison of the M1T1 5448 Mga regulon from THY and C media revealed significant differences in the genes that are Mga-regulated in the presence or absence of glucose (Table 2). Only 16 genes were found to be common between the two Mga regulons (Fig. 4; Table 2) comprising the core regulon virulence genes (Table 2; bold), as well as glnH and glnQ.2 (bicarbonate transporter), mac (IgG-degrading protease), spd (chromosomally-encoded streptodornase), M5005_spy0522 (sugar hydrolase), M5005_ spy0143 (hypothetical protein), and spxA.2 (a transcriptional regulator). Despite the stark difference in genes that are regulated by Mga in a glucose versus a no glucose condition, the broad COG categories represented were not different. In THY, the top three categories of Mga-regulated genes were unknown function (23 total), carbohydrate utilization (32 total), and defense mechanism/virulence (18 total) (Fig. 5). In C media, it was unknown function (32 total), defense mechanism/virulence (19 total), and energy production & conservation (17 total) (Fig. 5). This further supports that the PRD-containing Virulence Regulator (PCVR) Mga is a master regulator for genes involved in GAS pathogenesis and carbohydrate metabolism regardless of glucose levels.
Figure 4.
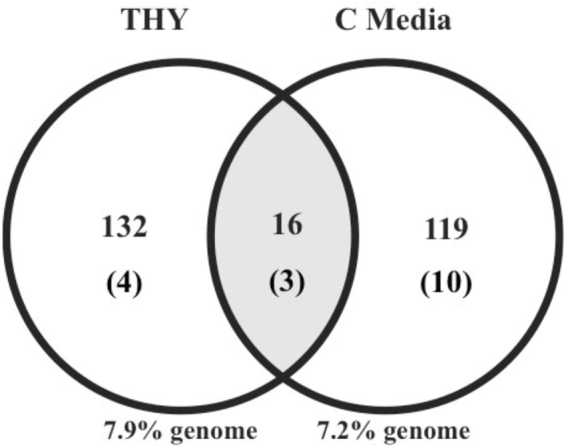
Comparison Mga regulons of GAS in high glucose THY and low glucose C Media. Venn diagram represents the numbers of Mga-regulated genes in either high glucose THY (left) or low glucose C media (right), with 16 genes found in common between the two data sets in the center (shaded). The number of tRNA genes found in each dataset (parentheses) are indicated, although percentage of regulation was calculated by excluding tRNA genes.
Figure 5.
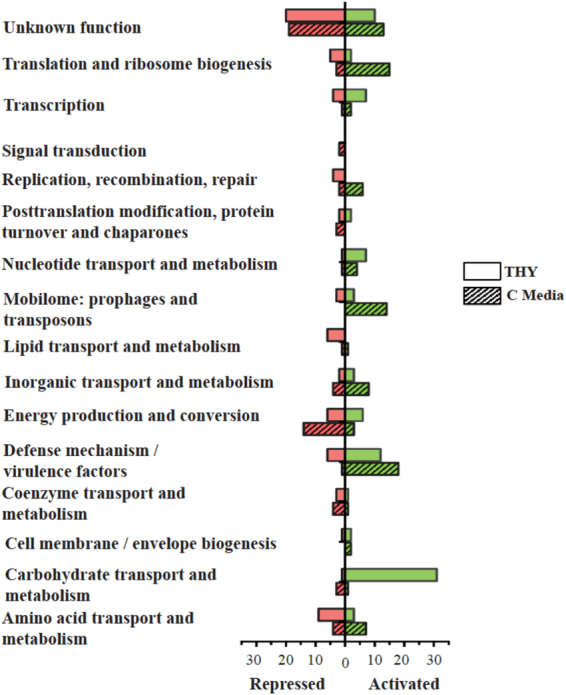
COG categories for Mga-regulated genes observed in THY and C media. A comparison of differentially expressed (DE) genes in various cluster of orthologous genes (COG) categories from THY (open bars) versus C media (crossed bars) of the GAS 5448 ∆mga 5448.930 mutant at late logarithmic growth when compared to the WT 5448. Bar length depicts the number of DE genes either down regulated (red) or up regulated (green) at p value of ≤0.05 with the total number of down and up regulated genes shown.
Importantly, media-dependent Mga regulation was recapitulated using an independent Krmit transposon mga mutant KM16.1. Real-time qPCR was performed on selected targets from RNA isolated from WT 5448 and the KM16.1 mga mutant grown in either C media or THY. Four genes previously chosen for validation in THY (Supplemental Fig. S6A) and nine genes from C media (Supplemental Fig. S6B) were tested and identical media-dependent phenotypes in Mga-regulated gene regulation were seen for KM16.1 as in the 5448.930 mutant (Supplemental Fig. S3, S4 and S6).
Supplementation of glucose in C media mimics a THY regulon
Mga has an altered regulon in C media (low glucose) in comparison to THY (+glucose) (Fig. 3). We were interested in testing whether supplementing C media (32 mg/dl glucose) to a starting concentration comparable to THY (240 mg/dl) would mimic the THY regulon results. RNA was isolated for real-time qPCR from ∆mga 5448.930 and the parental 5448 grown in C media supplemented to ca. 240 mg/dl glucose. A total of 5 differentially-regulated genes were chosen that were exclusive to the C media RNA-seq dataset (Supplemental Table S3; Fig. 6), as well as speB, which was only found to be regulated in THY (Fig. S6) and emm, which was observed in both datasets. We saw that 4 out of the 5 differentially regulated genes specific to C Media were no longer regulated by Mga when glucose was supplemented back to levels present in THY, including the phage-encoded DNase sdaD2 (Fig. 6; green bar). Interestingly, opuAA (Fig. 6; pink bar) was the only gene that was not influenced by glucose.
Figure 6.
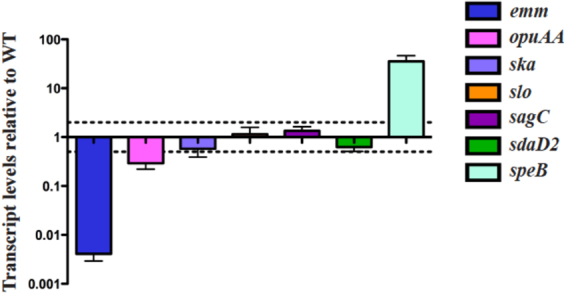
Supplementation of C media with glucose mimics some phenotypes observed for a ∆mga strain in THY. Transcript levels were determined using qPCR on RNA taken from WT GAS 5448 grown in C media supplemented to 0.5% glucose at late-log compared to ∆mga 5448.930 grown under the same conditions. Error bars represent the standard error from three biological replicates. Differences greater than 2-fold in expression for WT compared to mutant cells (dashed line) were considered significant.
Mga was shown to repress the transcription of speB in THY at late log phase (Supplemental Fig. S3A; light blue bar) and this phenotype was absent in C media (Supplemental Fig. S4; light blue bar). When glucose levels in C media were supplemented to levels found in THY, Mga was observed to repress speB again (Fig. 6; light blue bar). Therefore, while the mechanism by which Mga regulates speB is most likely indirect, there is a correlation between glucose availability and Mga regulation of speB. Overall, there appears to be two causes to the changes observed between THY and C media: (1) the majority of the changes appear to be specific to glucose availability, and (2) some changes in the regulon dynamics are due to C media-specific differences.
Mga is differentially phosphorylated in C Media
We observed differential expression of core Mga regulon genes of GAS 5448 grown in THY (+glucose) compared to C media (low glucose), yet mga itself was not affected (Table 1). Since Mga is a PRD-containing virulence regulator (PCVR) whose activity can be influenced by direct phosphorylation via the PTS in GAS, we asked whether the phosphorylation state of Mga was altered between these two environments. The Phos-tag gel matrix retards the migration of phosphorylated proteins and allows the identification of phosphorylation events via band shift compared to the non-phosphorylated protein of interest (see Methods). To study PTS-Mga interactions in GAS, we previously generated3 an M4 ∆mga (79% identity to M1 Mga) GAS strain (KSM547) that expressed a His-tagged version of either WT Mga (pKSM808) or a phosphoablative A/A/A Mga (pKSM871) that substitutes all three PTS-phosphorylatable histidines with alanines from a replicating plasmid (see Methods). The two strains were grown in either THY or C media to late log growth, whole cell lysates were isolated, and separated by Phos-tag SDS-PAGE for Western blot using α-Mga4 antibodies. The blot revealed two bands for WT 5448 lysates grown in THY, a slow-migrating more diffuse upper band arising from phosphorylated Mga at one or several of 3 potential histidines (Fig. 7A, Mga4~P-His6) and a larger fast-migrating band corresponding to non-phosphorylated Mga (Fig. 7A, Mga4-His6). In C media at a point where glucose is depleted, the slow-migrating Mga4~P-His6 is reduced, indicating less PTS phosphorylation (Fig. 7A). As would be expected, the Mga4~P-His6 band is absent in the phosphoablative A/A/A M4 Mga control mutant regardless of the media (Fig. 7A). The ratio of Mga4~P-His6 compared to total Mga4-His6 in the two WT lanes from two biological replicates was determined by densitometry analysis and normalized to the same ratio in the respective A/A/A Mga lanes for each (Fig. 7C). This showed a 3-fold increase in PTS-dependent phospho-Mga in THY compared to C media. As a control for loading, the same cell lysates were subjected to Western blot after SDS-PAGE using α-Mga4 antibodies (Fig. 7B). While a comparable amount of protein was detected for all strains in the Western blot, we consistently observed less protein for the A/A/A M4 Mga mutant grown in C media and assessed by Phos-tag and this phenomenon is currently being explored further. Together, these data provide the first evidence that Mga can be alternatively phosphorylated in the presence or absence of glucose.
Figure 7.
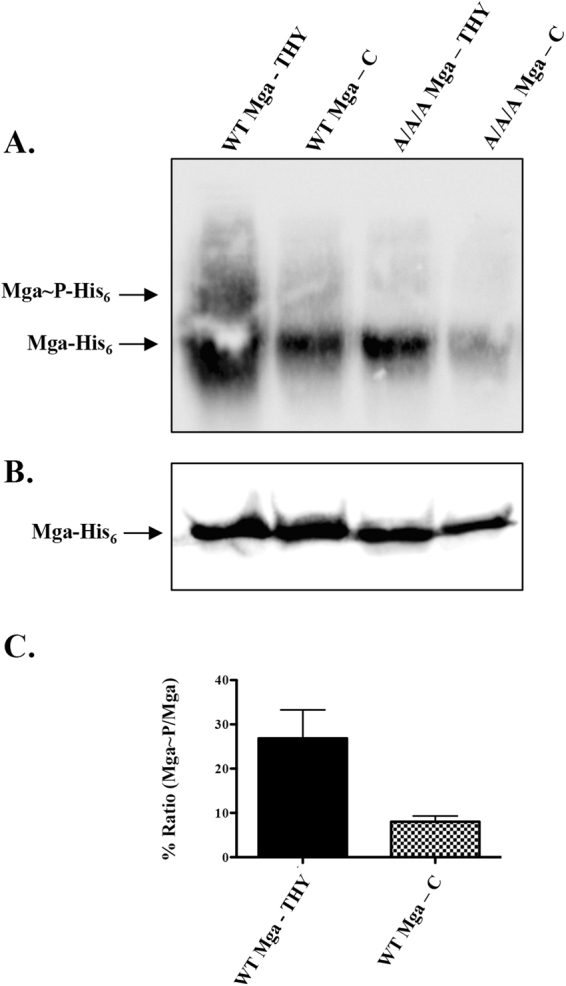
Mga is differentially phosphorylated based on glucose availability. The isogenic M4 mga mutant strain KSM547 complemented in trans with the His6- tagged WT strain (pKSM808) or phosphoablative A/A/A mga mutant (pKSM871) was grown in THY or C medium to the late exponential growth phase. Shown is a representative image of multiple replicates. (A) Equal concentrations of cell lysates of WT and A/A/A expressing strains grown in each media were resolved on a Phos-tag gel and immunoblotted using polyclonal αMga4 antibody. Bands corresponding to non-phosphorylated Mga and phosphorylated Mga are indicated by arrows. (B) Mga levels in same cell lysates were determined by 10% SDS-PAGE and immunoblotting using polyclonal αMga4 antibody. (B) Densitometric ratio of WT Mga~P-His6 bands to total Mga-His6 from THY and (C) Media lysates after separation on Phos-tag and Western blotting with a-Mga. Results were normalized to the same ratio in the respective A/A/A Mga-His6 control lanes from 2 biological replicates. Standard Error of the Mean (SEM) is indicated.
Discussion
The current study was undertaken to assess how glucose affects global transcriptional changes in the GAS cell through Mga. Our RNA-seq results revealed that in the WT M1T1 5448 some of the Mga regulon is up regulated in THY (+glucose), but not emm and mga itself. There was a strong correlation between the genes activated in C media (low glucose) with those known to be repressed by CcpA, indicating that C media relieves CCR. Interestingly, when comparing the Mga regulon in both medias, we found very little overlap beyond the core virulence genes, despite regulating similar functional categories (COGs). Many of these differences in Mga-regulated genes between the two medias could be directly attributed to glucose levels, which led to an alteration in the PRD-mediated phosphorylation patterns of Mga. Thus, Mga and its regulon are likely influenced by glucose and further support an in vivo link between glucose availability and virulence regulation in GAS.
The C media transcriptome of M1T1 GAS
For low G + C Gram-positive bacteria, carbon catabolite repression (CCR) is controlled by the master regulator, CcpA, through direct binding to catabolite response elements (cre) sites. Metabolic genes that enable the utilization of alternative (non-glucose) sugars are classically induced in the absence of the preferred carbohydrate source, glucose. For GAS, two studies have determined the genome-wide, CcpA-induced regulons in M1T1 serotype17,18, which found that ~8% of the GAS genome was regulated by CcpA in THY. Here, we observed that 16% of the GAS genome was differentially expressed when grown in the absence of glucose (C media at late log phase), which overlapped with the published CcpA regulons17,18. Thus, it appears that the lack of glucose encountered by GAS in C media at this point in growth is capable of relieving CCR and stimulating the transcription of alternative carbohydrate uptake systems. Although there are clear commonalities, our data also showed that GAS undergoes a much broader transcriptomic change during growth in C media (476 genes differentially expressed, 25.5% of genome) when compared to growth in THY (Supplemental Table S1). Therefore, growth in C media mimics a low- to no-glucose environment, but it also creates other changes in the cell that are likely attributed to high levels of peptides present in C media.
The C media-induced regulon was established for the serotype M14 HSC5 using altered buffered and osmotic conditions at early stationary growth phase35. The authors found C media to be a representative of conditions encountered by GAS during deep tissue infection due to up regulation of the virulence factor and cysteine protease gene, speB, to levels comparable to those observed during in vivo infection35. Thus, C media has been used in several studies for optimal expression of speB. Surprisingly, we did not observe a significant induction of speB in C media. However, the current study was conducted in late logarithmic phase, and not stationary phase, where speB is known to be most highly expressed. We did observe an increase in expression of both lacD.1 and rgg/ropB, which have been shown to interact with and stimulate speB expression40. A previous study found that in vivo/environmental signals suppress the expression of speB, and induce the expression of speA41. However, we saw no observable change in speA in our RNA-Seq dataset of 5448 grown in THY compared to C media. Therefore, the conditions tested here may not be optimal for the induction of speB expression in M1T1 5448.
Plasticity of the M1T1 Mga virulence regulon under varying glucose levels
The Mga regulon was initially determined by our group based on microarray analysis of 3 diverse serotypes representing both mga-1 and mga-2 alleles (M1, M6 and M4) grown to late-logarithmic growth in THY, involving ~10% of the GAS genome that encompassed mostly virulence and carbohydrate metabolism genes27. Recently, RNA-seq of Mga-regulated genes in an M59 (mga-2) serotype, also at late-logarithmic growth in THY, revealed ~7% of the genome was involved, primarily related to virulence, metabolism, and unknown functions29. Our RNA-seq results revealed that the Mga regulon in an M1T1 GAS strain under similar growth conditions regulated ~8% of the genome, in line with the recent M59 regulon. Although the M1 (SF370) and M1T1 (5448) strains analyzed differ by only 7% in their genomes (mostly prophage content), comparison of the Mga (mga-1 allele) regulon datasets revealed very little overlap. A similar trend was observed when regulons from strains expressing two mga-2 alleles (M59 and M4) were compared29. We saw that SF370 and 5448 shared only 8 Mga-regulated genes that were regulated in the same direction, with 5 genes being members of the core Mga-regulated virulence genes (Table 3). Thus, a common theme amongst all Mga regulons is the activation of core virulence genes when present (e.g., emm, sic, fba, scpA, sclA, sof), although the degree of activation appears to vary27,29.
The other three Mga-regulated genes shared between M1 and M1T1 GAS were lacG (6-phospho-beta-galactosidase), lctO (lactate oxidase), and M5005_spy0143 (hypothetical protein). Both lacG and lctO are involved in lactose metabolism. LctO converts lactate into pyruvate and leads to production of hydrogen peroxide (H2O2) in a growth phase- and glucose-dependent manner through direct repression by CcpA19. In S. pneumoniae, increase of H2O2 production acts as a virulence trait that aids in colonization by slowing the clearance of bacteria through the inhibition of ciliary beating in the upper respiratory tract and by promoting bactericidal activity to outcompete other microbial species from the microflora42–44. Therefore, it’s possible that Mga activates the transcription of lctO in order to up regulate hydrogen peroxide production just enough to aid in colonization, which is the stage of infection that correlates with the highest level of Mga activation15. Interestingly, M5005_spy0143 encoding a small hypothetical protein was also found to be regulated in C media (Table 2 and Supplemental Table S3), and was the only gene other than the core regulon found in common across the different serotypes and medias.
The THY Mga regulon of M1T1 5448 showed activation of several sugar-specific operons, in contrast to the repression previously observed in the published regulons27,29. M1T1 Mga appears to regulate carbon metabolism genes comparable to what was observed for MafR, a recently characterized Mga-homolog and PCVR implicated in the pathogenesis of E. faecalis45. Further, the cysteine protease, speB, and the SpeB inhibitor (speI) were both observed as being repressed by Mga in M1T1 5448 grown in THY (Supplemental Table S2 and Fig. S3, light blue bar) whereas speB was previously shown to be activated by Mga in a serotype-specific manner27. Interestingly, no change in the transcript of ropB/rgg or lacD.1 was observed in the 5448 ∆mga mutant, both of which are known to regulate speB expression16,40. Given that regulation of speB by Mga was restored when glucose was added back to C media (Fig. 6), there appears to be a glucose-dependent Mga regulation of speB in the M1T1 background. Regardless of the differences, it is apparent that Mga regulates a variety of genes important for colonization and, albeit indirect, metabolic operons in the presence of glucose.
In both THY and C media, Mga regulates the same approximate number (155 versus 148 genes, respectively) of the non-prophage GAS genome and representing comparable COG categories (Figs 4 and 5). Yet, there was a stark contrast in the genes represented between the datasets from the two medias. A total of only 16 genes (~10%) overlapped between THY and C media, half of which are established members of the core Mga virulence regulon (Table 2) and the remaining 8 genes being unique to the M1T1 5448 Mga regulon. Of these, glnH and glnQ encode a putative bicarbonate transporter. Mga is activated under elevated CO2 conditions46, and this environmental signal may act independently of other activation signals, as is seen for the B. anthracis PCVR AtxA and the group G Streptococcus (GBS) DmgB, both of which are homologs of Mga24. Therefore, a potential link between bicarbonate transport and Mga gene regulation may exist. Since the number of genes that overlap between the THY and C media Mga regulon are so few, we propose that this set of nine novel genes may also be a part of the core M1T1 Mga regulon.
Streptodornases represent novel Mga-regulated virulence genes
Both M1T1 phage-encoded DNases (sda1and spd3) were activated by Mga in C media (Supplemental Table S3) whereas the chromosomally-encoded spd was activated in both the presence and absence of glucose (Table 2). In M49 NZ131, the stand-alone regulator Rgg was found to bind directly to the upstream region of spd3, and directly repress transcription of this phage-encoded nuclease47,48. We also demonstrated that Mga regulates expression of sdaD2 in a glucose-dependent manner (Figs 6 and 7). Interestingly, sdaD2 is one of two horizontally transferred genes that distinguishes the invasive M1T1 serotype49 and has been suggested to provide selective pressure for the covS switch in the M1T1 5448 (covS+) in vivo50,51. It has been suggested that in the covS+ M1T1 MGAS2221, the Mga regulon, not Sda1, plays a much more important role in the covS switch in vivo52. It may be that during invasive infection with covS+ M1T1 strains, a glucose-deplete environment leads to Mga-dependent activation of sdaD2 (and other streptodornases) that impacts the covS switch.
Glucose levels affect Mga phosphorylation and activation of its regulon
We observed a 4- to 5-fold repression of some core Mga regulon genes (fba, scpA, sic, etc.) in C Media (Table 1) in the WT 5448; however, a significant change in transcription of emm was not observed. Two possibilities for the reduction of Mga regulon expression in C media compared to THY are: (1) CcpA has been shown to bind to a cre site located upstream of the P1 promoter region of mga and stimulates transcription53; therefore, in the low-glucose environment of C media, CCR is relieved, causing a decrease in expression of the Mga regulon under a low glucose condition. Alternatively, (2) PTS-mediated phosphorylation of Mga activity may impact regulation of the core genes3,29. The latter scenario is the most probable, as the transcript levels of mga were not significantly different between THY and C media, indicating possible posttranslational modification. The lack of differential emm expression is not clear at this time and we are exploring this mechanism further.
The differential phosphorylation patterns of Mga observed using Phos-tag supports that PTS involvement can affect the transcription of the regulon in one of three ways: (1) no phosphorylation produces an active Mga, (2) phosphorylation on only PRD2 can cause an increase in activity, and (3) phosphorylation on PRD1 inhibits Mga activity, and is dominant over PRD2 phosphorylation3. Therefore, we hypothesize that the low- to no-glucose environment encountered in C media would likely cause phosphorylation on PRD1, causing a reduction in Mga activity and regulon expression, whereas growth in THY (high glucose) would result in an active non-phosphorylated Mga and remain active. Hence this would help explain the activation of the regulon in THY over C media (Table 1). Furthermore, a phosphoablative Mga mutant would eliminate the PTS phosphorylation phenotype regardless of media.
Surprisingly, Mga from GAS grown in THY showed two bands on a Phos-tag gel representing a phosphorylated and unphosphorylated species. In C media, far less of the phosphorylated form of Mga was observed, while the phosphoablative (A/A/A) construct was unphosphorylated under both media conditions (Fig. 7). Therefore, we now hypothesize that in C media, when glucose is depleted, Mga still may be in its “active” form (condition 1), and THY represents a “hyper-active” form of Mga (condition (2). This would be in line with the model that was previously proposed by Hondorp et al.3, since glucose is initially present in both THY and C media. Further, since RNA was isolated from strains grown to late log, there still may be a small but significant amount of glucose present in C media to maintain the phosphorylation status of Mga. Regardless, we now believe it may not only be the level of glucose that is important for Mga activity, but the mere presence of it during growth which could affect the phosphorylation status of Mga. The unphosphorylated band for the A/A/A Mga in both C media and THY indicates that phosphorylation by the PTS occurs at the phosphohistidines on the PRD domains of Mga.
Concluding Remarks
This work shows that glucose directly affects Mga activity and the composition of the Mga regulon. Furthermore, the differences between the regulons in the presence or absence of glucose is most likely attributed to a phosphorylation event that occurs on the phosphohistidines located on the PRD domains of Mga. While it is unclear which phosphohistdine(s) are affected and the mechanism by which Mga is able to regulate genes alternatively, it is clear that in the presence of glucose, the PTS plays an important role in activating this global transcriptional regulator and virulence genes downstream.
Methods
Bacterial Strains and Media
Streptococcus pyogenes (GAS) strain M1T1 544854 is isolated from an invasive infection. The reference genome used in this study was generated from the M1T1 strain MGAS500555. GA40634 is a clinical isolate of the GAS M4 serotype and SF370 is a sequenced M1 strain. KSM547 and KSM165-L are isogenic strains of GA40634 and SF370, respectively, with an insertional inactivation of the mga gene27,38. GAS bacteria were either cultured in Todd-Hewitt medium supplemented with 0.2% yeast extract (THY) or in C Media56 buffered to pH 7.5 with NaOH. A 25% (w/v) stock solution of glucose was used to supplement C media to the levels indicated in select experiments. GAS growth was assayed via absorbance using a Klett-Summerson colorimeter (A filter) and expressed in Klett Units.
Escherichia coli (E. coli) strain DH5α (hsdR17 recA1 gyrA endA1 relA1) and C43[DE]57 were used as the host for plasmid constructions, and C41[DE3]57 was used for protein expression, as indicated in Table 4. E. coli strains were grown in Luria-Bertani (LB) broth or ZYP-505258 for protein expression. Antibiotics were added to media, as needed, at the following concentrations: spectinomycin at 100 µg/ml for both E. coli and GAS, kanamycin at 50 µg/ml for E. coli and 300 µg/ml for GAS, and 100 µg/ml of ampicillin for E. coli.
Table 4.
Bacterial strains and plasmids used in this study.
| Strain/Plasmid | Relevant genotype | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | hsdR17 recA1 gyrA endA1 relA1 | 72 |
| C41[DE3] | F- ompT gal dcm hsdSB (rB−mB−)(DE3) | 57 |
| C43[DE3] | F- ompT gal dcm hsdSB (rB−mB−)(DE3) | 57 |
| S. pyogenes | ||
| 5448 | M1T1 | 54 |
| 5448.930 | ∆mga mutant in 5448 (insertional inactivation) | This study |
| 5448.930 R | mga + 5448, cured strain | This study |
| KM16.1 | ∆mga mutant in 5448 (transposon) | This study |
| GA40634 | M4 GAS, clinical isolate | 73 |
| KSM547 | ∆mga mutant in GA40634 | 38 |
| SF370 | M1 GAS | 74 |
| KSM165L | ∆mga mutant in SF370 | 27 |
| Plasmids | ||
| pCRS | Temperature-sensitive conditional vector; SpR | 23 |
| pJRS525 | Replicating vector for GAS with SpR | 36 |
| pKSM807 | WT mga1 under native Pmga1 | 34 |
| pKSM808 | WT mga4-his6 (GA40634) under native Pmga4 | 34 |
| pKSM809 | WT mga1-his6 under native Pmga1 | 34 |
| pKSM871 | H204A/H270A/H324A mga4-his6 under native Pmga4 | 3 |
| pKSM930 | pCRS construct; insertional inactivation of mga | This study |
| pLZ12-Spec | Broad host range cloning vector, SpR | 75 |
| pMga1-His | WT mga1 (SF370) with C-terminal His6 | 34 |
| pUC19 | ColE1 ori ApR lacZα | 76 |
DNA manipulations
Plasmid DNA was extracted from E. coli using the Wizard Plus SV miniprep system (Promega). DNA fragments were agarose gel purified using the Wizard SV gel and PCR cleanup system (Promega). PCR for generating probes and cloning was conducted using Accuprime Pfx (Life Technologies) according to the manufacturer’s recommended protocol. PCR for diagnostic assays was performed using Taq DNA polymerase (NEB). Genewiz, Inc performed all DNA sequencing. The Master-Pure complete DNA and RNA purification kit for Gram-positive bacteria (Epicentre, Illumina) was used to extract genomic DNA.
Insertional inactivation and rescue of GAS 5448 mga mutants
An insertional inactivation construct was generated using a 300 bp PCR internal fragment of mga from 5448 gDNA, which was amplified using the primers Mga InIn F and Mga InIn R (Supplemental Table S4). The fragment was cloned via BamHI sites into pCRS, generating pKSM930 (Table 4). GAS 5448 was transformed with 20 µg of plasmid (pKSM930) and grown on THY agar containing spectinomycin at 30 °C. Potential integration mga mutants (GAS 5448.930) were identified following selection with spectinomycin and growth at the non-permissive temperature (37 °C). PCR on isolated gDNA was used to confirm plasmid insertion for each GAS mutant using primers SPR1 and SPR2 (Supplemental Table S4).
The strain 5448.930 (Table 4) was cured of the plasmid inactivating mga by passage in liquid culture four times at 30 °C with no drug selection. Cultures were then plated, and patched on THY supplemented with spectinomycin and THY alone. Patches that showed spectinomycin sensitivity were then screened for loss of the spectinomycin gene by PCR using the primer SPR1 and SPR 2 (Supplemental Table S4).
A transposon insertion in mga was identified from a pool of Krmit (for kanamycin-resistant transposon for massive identification of transposants) transposon mutants generated in 544859. Library #16 was screened using AP-PCR60 followed by sequencing and alignment of sequence to the 5448 genome. Transposant #1 (16.1) was mapped to the 5′-end of mga in the sense direction.
RNA-Seq and data analysis
RNA sequencing (RNA-Seq) was performed as previously described21. Briefly, Direct-zol RNA MiniPrep kit (Zymo Research) was used to isolate total RNA using a modified procedure to improve GAS cell disruption. Frozen cell pellets were resuspended in 700 µl of Trizol plus 300 mg of acid-washed glass beads (Sigma Life Science) and disrupted by vortexing for 5 min. RNA samples were DNase-treated using the Turbo DNase-free kit (Life Technologies). A total of 5 µg of this RNA was treated for ribosomal RNA (rRNA) removal using the Ribo-Zero Magnetic kit for Gram-positive bacteria (Epicentre). Quality and quantity were assessed using a 2100 Bioanalyzer (Agilent) NanoDrop 8000 spectrophotometer (Thermo Scientific), respectively. Directional RNA-Seq libraries were created using the ScriptSeq v2 RNA-Seq Library Preparation kit (Illumina) according to the manufacturer’s recommendations.
A rapid-run 100 bp single-read DNA sequencing was then performed at the Institute for Bioscience and Biotechnology Research (IBBR) Sequencing Facility at the University of Maryland, College Park, using the Illumina HiSeq 1500 platform. Data were generated in the standard Sanger FastQ format and raw reads were deposited with the Sequence Read Archive (SRA) at the National Center for Biotechnology Institute (accession PRJN412519).
Read quality was measured using FastQC61, filtered and trimmed using trimmomatic62 and mapped against the MGAS5005 genome (accession CP000017) using alignment software, as previously described. Differential expression analyses were performed following size-factor and quantile normalization of read counts. Limma63 and DESeq64 statistical models were used to account for batch effects. The resulting metrics of expression were visualized using Circos65, and tested for ontology enrichment using a variety of ontology software packages (KEGG66, goseq67, clusterProfiler68, GOstats69, and topGO70).
qRT-PCR
The quantitative (real-time) reverse transcription-PCR (qRT-PCR) experiments were performed as previously described21. Briefly, DNase-treated total RNA from strains were added to SYBR green master mix (Applied Biosystems) with 6.5 µl of each gene-specific real-time primer from a 20 nM stock (Table 2) using the one-step protocol on a Light Cycler 480 (Roche). Real-time primers were designed using the interactive tool Primer3 (http://biotools.umassmed.edu/bioapps/primer3_www.cgi).
RNA-Seq data validation was performed as previously described21, using cDNA generated separately in a two-step protocol in order to take into account the strand-specificity of the RNA-Seq results.
All qPCR results are depicted as ratios of the experimental/wild type levels relative to gyrA transcripts, which acts as an internal control. Standard error was calculated from three biological replicates, and differences over 2-fold in expression were considered significant. Correlation coefficients were determined by graphing the log value of the RNA-seq result on the X-axis to the log value of the RT-qPCR on the Y-axis. The R2 value was calculated using a linear regression model, which represented the fitness of the data (Supplemental Fig. S1).
Glucose utilization monitored by Glucose Test Strips
Glucose utilization was monitored using a blood glucometer (AimStrip® Plus). Although typically used to monitor blood sugar, we used this apparatus to monitor glucose concentration of THY, CDM, and whole human blood during GAS growth. Values are given in mg/dl. Readings were taken by first inserting a test strip (AimStrip® Plus) into the monitor, placing the test strip into the culture to draw up liquid droplets, and waiting for 10 seconds for the final reading.
Mga-1 purification and expression
GAS Mga1-His6 was purified as previously described34. Briefly, E. coli containing the plasmid pMga1-His was grown in ZYP auto-induction media for ~50 h at 37 °C. Cells were harvested by centrifugation at 4 °C. Pellets were then resuspended in Lysis Buffer A (20 mM NaPi, 500 mM NaCl, 20 mM imidazole, pH 7.4) and 1× EDTA-free cOmplete protease inhibitor (Roche), and incubated on ice with lysozyme for 30 min followed by sonication using a Branson Sonifier 450 with a tapered microtip (setting 6, 50% duty cycle) pulsing 3 × 1 min with 3 min breaks on ice between cycles. The lysate was spun for clarification at 20,000 × g for 30 min at 4 °C 3–4 times. The lysate was loaded on a 5 ml NiNTA agarose resin (Qiagen) and rotated at 4 °C for 1 h. The column was then washed twice with NiNTA Wash Buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0). Protein was eluted with 10 ml of NiNTA elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 8.0) and chosen fractions were stored at −20 °C. Protein concentration was analyzed using the Bio-Rad protein assay kit, reading the absorbance at 595 nm on a spectrophotometer.
GAS whole cell lysates
Soluble GAS protein extractions were performed using the bacteriophage lysine, PlyC (kindly provided by D. Nelson) as previously described34. Briefly, GAS cells were inoculated 1:20 into 12 ml of THY or C media56, grown to late-log, and pelleted by centrifugation for 15 min at 6,000 × g. Pellets were resuspended in 200 µl of Buffer D (30 mM Tris pH 7.5, 0.1 mM dithiothreitol [DTT], 40% (v/v) glycerol), 1 × cOmplete EDTA-free protease inhibitor (Roche), 10 µl (20 U) of Turbo DNase (Life Technologies), and 5 µl (250 U) of PlyC71. The cells were mixed by flicking and incubated on ice for 20 min, followed by centrifugation at 13,000 × g for 10 min at 4 °C. Clarified supernatants containing soluble proteins were extracted and boiled with 3 × cracking buffer (0.15% [w/v] Bromophenol Blue, 0.6% [w/v] SDS, 3 ml glycerol, 3.9 ml Tris-HCl [500 mM, pH 6.8], 1.5 ml β-mercaptoethanol, and 4 ml dH2O) at 95 °C for 5 min, and stored at −20 °C. Protein concentration was assayed using the Bio-Rad protein assay kit, reading the absorbance at 595 nm on a spectrophotometer.
Western Blots
Selected protein samples were run on a 10% SDS-PAGE, with 6% stacking gel, for 1 h at 180 V. The gels were then transferred to nitrocellulose membranes using the Mini-Protean apparatus (Bio-Rad) in 1X transfer buffer (25 mM Tris base, 0.2 M glycine, 20% methanol). Membranes were blocked overnight at 4 °C in blocking solution (5% (w/v) dried milk in PBS-tween). Primary antibodies were incubated at a 1:1,000 dilution, unless otherwise stated. The following primary antibodies were used at a 1:1,000 dilution: α-His antibody (Roche), α-Mga4, α-Mga1-HRP, and α-Sda1 (kindly provided by M. Walker). Blots were then washed three times for 5 min in PBS-tween. The primary antibody Hsp60 (Enzo Life Sciences) was used at a 1:2,500 dilution. Blots were then incubated with secondary antibodies: α-rabbit-HRP (1:5,000) or α-mouse-HRP (1:20,000) for 1 h. The blots were then washed with PBS-tween three times for 5 min, and visualized using SuperSignal West Femto Substrate (Thermo Scientific) and a LAS-3000 CCD camera (FUJIFILM).
Phos-tag phosphate affinity gel
The phos-tag gel system was used to visualize the phosphorylation state of Mga. Briefly, GAS whole cell lysates were run on a Mn2+-Phos-tag 10% SDS-PAGE gel (WAKO), with 6% stacking gel. The resolving gel was made as follows: 1.75 ml of 1.5 M Tris-HCl, 1.75 ml of 40% bis-acrylamide, 140 µl of 10 mM MnCl, 140 µl of phos-tag solution, 3.1 ml of dH2O, 100 µl of 10% ammonium persulfate, and 20 µl of TEMED (Bio-Rad). Protein samples were run for 2 h at room temperature and 2 h on ice, for a total of 4 h at 180 V, prior to western blot as described above.
Data Availability Statement
The authors have deposited all RNA-seq raw sequencing reads with the Sequence Read Archive (SRA) at the National Center for Biotechnology Institute (accession PRJN412519) for public availability.
Electronic supplementary material
Acknowledgements
We thank Rezia Era Braza for critical comments of this manuscript. This work was directly supported by grants from the NIH National Institute of Allergy and Infectious Diseases (K.S.M., AI047928; N.M.E.S., AI094773) and in part by a NIH F31 pre-doctoral fellowship (K.M.V., AI100576).
Author Contributions
K.M.V., Y.L.B. and K.S.M. conceived and designed the research plan, supervised the project, analyzed the data and interpreted the results. K.M.V. performed most of the experiments with technical contributions from G.S.S. and E.I. A.T.B. and N.M.E. performed the bioinformatics analyses. K.M.V., Y.L.B., A.T.B., N.M.E. and K.S.M. wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23366-7.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yoann Le Breton, Email: lebreton@umd.edu.
Kevin S. McIver, Email: kmciver@umd.edu
References
- 1.Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gorke B, Stulke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nature Reviews Microbiology. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 3.Hondorp ER, et al. PTS phosphorylation of Mga modulates regulon expression and virulence in the group A streptococcus. Mol. Microbiol. 2013;88:1176–1193. doi: 10.1111/mmi.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho KH, Caparon MG. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 2005;57:1545–1556. doi: 10.1111/j.1365-2958.2005.04786.x. [DOI] [PubMed] [Google Scholar]
- 5.Munoz-Elias EJ, McKinney JD. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer R, Camilli A. Sucrose metabolism contributes to in vivo fitness of Streptococcus pneumoniae. Mol. Microbiol. 2007;66:1–13. doi: 10.1111/j.1365-2958.2007.05878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Son MS, Matthews WJ, Jr., Kang Y, Nguyen DT, Hoang TT. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect. Immun. 2007;75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rollenhagen C, Bumann D. Salmonella enterica highly expressed genes are disease specific. Infect. Immun. 2006;74:1649–1660. doi: 10.1128/IAI.74.3.1649-1660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Mol. Microbiol. 2007;64:771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- 10.Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. P-Ser-HPr–a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochimica et biophysica acta. 2005;1754:118–125. doi: 10.1016/j.bbapap.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 11.Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 12.Graham MR, et al. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. American Journal of Pathology. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graham MR, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. American Journal of Pathology. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of Group A Streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Path. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Virtaneva K, et al. Longitudinal analysis of the Group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc. Natl. Acad. Sci. USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loughman JA, Caparon MG. A novel adaptation of aldolase regulates virulence in Streptococcus pyogenes. EMBO J. 2006;25:5414–5422. doi: 10.1038/sj.emboj.7601393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinkel TL, McIver KS. CcpA-mediated repression of streptolysin S expression and virulence in the Group A Streptococcus. Infect. Immun. 2008;76:3451–3463. doi: 10.1128/IAI.00343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelburne SA, 3rd, et al. A direct link between carbohydrate utilization and virulence in the major human pathogen Group A Streptococcus. Proc. Natl. Acad. Sci. USA. 2008;105:1698–1703. doi: 10.1073/pnas.0711767105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kietzman CC, Caparon MG. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 2010;78:241–252. doi: 10.1128/IAI.00746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gera K, Le T, Jamin R, Eichenbaum Z, McIver KS. The Phosphoenolpyruvate Phosphotransferase System in Group A Streptococcus Acts To Reduce Streptolysin S Activity and Lesion Severity during Soft Tissue Infection. Infect. Immun. 2014;82:1192–1204. doi: 10.1128/IAI.01271-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valdes, K. M. et al. The fruRBA operon is necessary for Group A Streptococcal growth in fructose and for resistance to neutrophil killing during growth in whole human blood. Infect. Immun (2016). [DOI] [PMC free article] [PubMed]
- 22.Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes and their impact on pathogen-host interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/S0966-842X(03)00098-2. [DOI] [PubMed] [Google Scholar]
- 23.Le Breton Y, et al. Genome-wide identification of genes required for fitness of Group A Streptococcus in human blood. Infect. Immun. 2013;81:862–875. doi: 10.1128/IAI.00837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hondorp ER, McIver KS. The Mga virulence regulon: infection where the grass is greener. Mol. Microbiol. 2007;66:1056–1065. doi: 10.1111/j.1365-2958.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- 25.Kihlberg BM, Cooney J, Caparon MG, Olsén A, Björck L. Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 1995;19:299–315. doi: 10.1016/S0882-4010(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 26.Le Breton Y, et al. Genome-wide discovery of novel M1T1 group A streptococcal determinants important for fitness and virulence during soft-tissue infection. PLoS Path. 2017;13:e1006584. doi: 10.1371/journal.ppat.1006584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ribardo DA, McIver KS. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 2006;62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 28.McIver KS. Stand-alone response regulators controlling global virulence networks in streptococcus pyogenes. Contributions to Microbiology. 2009;16:103–119. doi: 10.1159/000219375. [DOI] [PubMed] [Google Scholar]
- 29.Sanson M, et al. Phosphorylation events in the multiple gene regulator of group A Streptococcus significantly influence global gene expression and virulence. Infect. Immun. 2015;83:2382–2395. doi: 10.1128/IAI.03023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dale JL, Raynor MJ, Dwivedi P, Koehler T. M. cis-Acting elements that control expression of the master virulence regulatory gene atxA in Bacillus anthracis. J. Bacteriol. 2012;194:4069–4079. doi: 10.1128/JB.00776-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammerstrom TG, et al. Crystal structure of Bacillus anthracis virulence regulator AtxA and effects of phosphorylated histidines on multimerization and activity. Mol. Microbiol. 2015;95:426–441. doi: 10.1111/mmi.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammerstrom TG, Roh JH, Nikonowicz EP, Koehler TM. Bacillus anthracis virulence regulator AtxA: oligomeric state, function and CO(2) -signalling. Mol. Microbiol. 2011;82:634–647. doi: 10.1111/j.1365-2958.2011.07843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsvetanova B, et al. Opposing effects of histidine phosphorylation regulate the AtxA virulence transcription factor in Bacillus anthracis. Mol. Microbiol. 2007;63:644–655. doi: 10.1111/j.1365-2958.2006.05543.x. [DOI] [PubMed] [Google Scholar]
- 34.Hondorp ER, et al. Characterization of the Group A Streptococcus Mga virulence regulator reveals a role for the C-terminal region in oligomerization and transcriptional activation. Mol. Microbiol. 2012;83:953–967. doi: 10.1111/j.1365-2958.2012.07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loughman JA, Caparon M. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: a model for in vivo gene expression. J. Bacteriol. 2006;188:399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McIver KS, Scott JR. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 1997;179:5178–5187. doi: 10.1128/jb.179.16.5178-5187.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DebRoy S, et al. A Multi-Serotype Approach Clarifies the Catabolite Control Protein A Regulon in the Major Human Pathogen Group A Streptococcus. Sci. Rep. 2016;6:32442. doi: 10.1038/srep32442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almengor AC, McIver KS. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J. Bacteriol. 2004;186:7847–7857. doi: 10.1128/JB.186.23.7847-7857.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen R, Eden A, Björck L. SclA, a novel collagen-like surface protein of Streptococcus pyogenes. Infect. Immun. 2000;68:6370–6377. doi: 10.1128/IAI.68.11.6370-6377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loughman JA, Caparon MG. Comparative functional analysis of the lac operons in Streptococcus pyogenes. Mol. Microbiol. 2007;64:269–280. doi: 10.1111/j.1365-2958.2007.05663.x. [DOI] [PubMed] [Google Scholar]
- 41.Kazmi SU, et al. Reciprocal, temporal expression of SpeA and SpeB by invasive M1T1 group a streptococcal isolates in vivo. Infect. Immun. 2001;69:4988–4995. doi: 10.1128/IAI.69.8.4988-4995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirst RA, et al. Relative roles of pneumolysin and hydrogen peroxide from Streptococcus pneumoniae in inhibition of ependymal ciliary beat frequency. Infect. Immun. 2000;68:1557–1562. doi: 10.1128/IAI.68.3.1557-1562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 2000;68:3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. Interference between Streptococcus pneumoniae and Staphylococcus aureus: In vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J. Bacteriol. 2006;188:4996–5001. doi: 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruiz-Cruz S, Espinosa M, Goldmann O, Bravo A. Global Regulation of Gene Expression by the MafR Protein of Enterococcus faecalis. Frontiers in Microbiology. 2015;6:1521. doi: 10.3389/fmicb.2015.01521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McIver KS, Heath AS, Scott JR. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 1995;63:4540–4542. doi: 10.1128/iai.63.11.4540-4542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anbalagan S, Chaussee MS. Transcriptional regulation of a bacteriophage encoded extracellular DNase (Spd-3) by Rgg in Streptococcus pyogenes. PLoS One. 2013;8:e61312. doi: 10.1371/journal.pone.0061312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anbalagan S, McShan WM, Dunman PM, Chaussee MS. Identification of Rgg binding sites in the Streptococcus pyogenes chromosome. J. Bacteriol. 2011;193:4933–4942. doi: 10.1128/JB.00429-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maamary PG, et al. Tracing the evolutionary history of the pandemic group A streptococcal M1T1 clone. FASEB J. 2012;26:4675–4684. doi: 10.1096/fj.12-212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aziz RK, et al. Invasive M1T1 Group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol. Microbiol. 2004;51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 51.Walker MJ, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat. Med. 2007;13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 52.Horstmann N, et al. Characterization of the effect of the histidine kinase CovS on response regulator phosphorylation in group A Streptococcus. Infect. Immun. 2015;83:1068–1077. doi: 10.1128/IAI.02659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Almengor AC, Kinkel TL, Day SJ, McIver KS. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the group A streptococcus. J. Bacteriol. 2007;189:8405–8416. doi: 10.1128/JB.01038-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatellier S, et al. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 2000;68:3523–3534. doi: 10.1128/IAI.68.6.3523-3534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sumby P, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 56.Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 58.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 59.Le Breton Y, et al. Essential Genes in the Core Genome of the Human Pathogen Streptococcus pyogenes. Sci. Rep. 2015;5:9838. doi: 10.1038/srep09838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Le Breton, Y. & McIver, K. S. Genetic Manipulation of Streptococcus pyogenes (The Group A Streptococcus, GAS). Curr. Prot. Microbiol. Oct 2; 30: Unit 9D.3 (2013). [DOI] [PMC free article] [PubMed]
- 61.Andrews, S. FASTQC. A quality control tool for high throughput sequence data, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 62.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq 2. bioRxiv (2014). [DOI] [PMC free article] [PubMed]
- 65.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ogata H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu G, Wang LG, Han Y, He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- 70.Alexa A, Rahnenfuhrer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 71.Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: a multimeric bacteriophage lysin. Proc. Natl. Acad. Sci. USA. 2006;103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanahan D, Meselson M. Plasmid screening at high colony density. Methods Enzymol. 1983;100:333–342. doi: 10.1016/0076-6879(83)00066-X. [DOI] [PubMed] [Google Scholar]
- 73.Schuchat A, et al. Active bacterial core surveillance of the emerging infections program network. Emerging Infect. Dis. 2001;7:92–99. doi: 10.3201/eid0701.010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferretti JJ, et al. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Husmann LK, Scott JR, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect. Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors have deposited all RNA-seq raw sequencing reads with the Sequence Read Archive (SRA) at the National Center for Biotechnology Institute (accession PRJN412519) for public availability.