Abstract
By applying the culturomics concept and using culture conditions containing a high salt concentration, we herein isolated the first known halophilic archaeon colonizing the human gut. Here we described its phenotypic and biochemical characterization as well as its genome annotation. Strain Arc-HrT (= CSUR P0974 = CECT 9307) was mesophile and grew optimally at 37 °C and pH 7. Strain Arc-HrT was also extremely halophilic with an optimal growth observed at 15% NaCl. It showed gram-negative cocci, was strictly aerobic, non-motile and non-spore-forming, and exhibited catalase and oxidase activities. The 4,015,175 bp long genome exhibits a G + C% content of 65.36% and contains 3911 protein-coding and 64 predicted RNA genes. PCR-amplified 16S rRNA gene of strain Arc-HrT yielded a 99.2% sequence similarity with Haloferax prahovense, the phylogenetically closest validly published species in the Haloferax genus. The DDH was of 50.70 ± 5.2% with H. prahovense, 53.70 ± 2.69% with H. volcanii, 50.90 ± 2.64% with H. alexandrinus, 52.90 ± 2.67% with H. gibbonsii and 54.30 ± 2.70% with H. lucentense. The data herein represented confirm strain Arc-HrT as a unique species and consequently we propose its classification as representative of a novel species belonging to the genus Haloferax, as Haloferax massiliense sp. nov.
Keywords: Culturomics, Taxono-genomics, Halophilic archaea, Haloferax massiliense
Introduction
The human intestinal microbiota is a complex ecosystem consisting of a wide diversity including bacteria (Lagier et al. 2012), archaea (Khelaifia et al. 2013), and unicellular eukaryotes (Nam et al. 2008). The culturomics concept, recently introduced in our laboratory to study the prokaryotes diversity in the human gut (Lagier et al. 2012), allowed the isolation of a huge halophilic bacteria diversity including several new species (Lagier et al. 2016). Among the diverse culture conditions and several culture media used by culturomics to isolate new prokaryotes, some conditions targeting specifically extremophile organisms were also used (Lagier et al. 2016). Indeed, culture media containing high salt concentration are essentially used to select halophilic bacteria and archaea.
Currently, the determination of the affiliation of a new prokaryote is based on the 16S rDNA sequence, G + C content % and DNA–DNA hybridization (DDH). This approach is limited because of the very low cutoff between species and genera (Welker and Moore 2011). In some cases, 16S rRNA gene sequence comparison has been proved to poorly discriminate some species belonging to a same genus and remain ineffective (Stackebrandt and Ebers 2006). Recently, we proposed a polyphasic approach based on phenotypic and biochemical characterization, MALDI-TOF MS spectrum and total genome sequencing and annotation to better define and classify new taxa (Ramasamy et al. 2014).
Using culturomics techniques to isolate halophilic prokaryotes colonizing the human gut (Lagier et al. 2016), strain Arc-HrT was isolated from a stool specimen of a 22-year-old Amazonian obese female patient (Khelaifia and Raoult 2016). This strain presented different characteristics enabling its classification as a new species of the Haloferax genus. The Haloferax genus was first described by Torreblanca et al. (1986) and actually includes 12 species with validly published names. Members of the Haloferax genus are essentially extremely halophilic archaea that inhabit hypersaline environments such as the Dead Sea and the Great Salt Lake. They are classified in the family Haloferacaceae within the Euryarchaeota phylum and the various species constitute 57 recognized genera (Arahal et al. 2017).
In this study, we present a classification and a set of characteristics for Haloferax massiliense sp. nov., strain Arc-HrT (= CSUR P0974 = CECT 9307) with its complete genome sequencing and annotation.
Materials and methods
Ethics and samples collection
The stool specimens were collected from a 22-year-old Amazonian obese female patient after defecation in sterile plastic containers, sampled and stored at – 80 °C until use. Informed and signed consent was obtained from the patient. The study and the assent procedure were approved by the Ethics Committees of the IHU Méditerranée Infection (Faculty of Medicine, Marseille, France), under agreement number 09-022. Salt concentration of the stool specimen was measured by digital refractometer (Fisher scientific, Illkirch, France) and the pH was measured using a pH-meter.
Isolation of the strain
Strain Arc-HrT was isolated in December 2013 by aerobic culture of the stool specimen in a home-made culture medium consisting of a Columbia broth (Sigma-Aldrich, Saint-Quentin Fallavier, France) modified by the addition of (per liter): MgCl2·6H2O, 15 g; MgSO4·7H2O, 20 g; KCl, 4 g; CaCl2·2H2O, 2 g; NaBr, 0.5 g; NaHCO3, 0.5 g, glucose, 2 g and 150 g of NaCl. pH was adjusted to 7.5 with 10 M NaOH before autoclaving. Approximately, 1 g of stool specimen was inoculated into 100 mL of this liquid medium in a flask incubated aerobically at 37 °C with stirring at 150 rpm. Subcultures were realized after 10, 15, 20 and 30 days of incubation. Then, serial dilutions of 10−1–10−10 were performed in the home-made liquid culture medium and then plated onto agar plates consisting of the previously detailed liquid medium with 1.5% agar.
Strain identification by MALDI-TOF MS and 16S rRNA gene sequencing
MALDI-TOF MS protein analysis was carried out as previously described (Seng et al. 2013). The resulting 12 spectra of strain Arc-HrT were imported into the MALDI BioTyper software (version 2.0, Bruker) and analyzed by standard pattern matching (with default parameter settings) against the main spectra of halophilic and methanogenic archaea including the spectra from Haloferax alexandrinus, Methanobrevibacter smithii, Methanobrevibacter oralis, Methanobrevibacter arboriphilus, and Methanomassilicoccus massiliensis. The 16S rRNA gene amplification by PCR and sequencing were performed as previously described (Lepp et al. 2004). The phylogenetic tree was reconstructed according to the method described by Elsawi et al. (2017).
Growth conditions
The optimum growth temperature of strain Arc-HrT was tested on the solid medium by inoculating 105 CFU/mL of an exponentially growing culture incubated aerobically at 28, 37, 45***, and 55 °C. Growth atmosphere was tested under aerobic atmosphere, in the presence of 5% CO2, and also in microaerophilic and anaerobic atmospheres created using GENbag microaer and GENbag anaer (BioMérieux, Marcy l’Etoile, France) respectively. The optimum NaCl concentration required for growth was tested on solid media at 0, 1, 5, 7.5, 10, 15, 20, 25** and 30% of NaCl. The optimum pH was determined by growth testing at pH 5, 6, 7, 7.5, 8 and 9.
Biochemical, sporulation and motility assays
To characterize the biochemical properties of strain Arc-HrT, we used the commercially available Api ZYM, Api 20 NE, Api 50 CH strips (bioMérieux), supplemented by 15% NaCl (w/v) and 30 g/L of MgSO4. The sporulation test was done by thermic-shock at 80 °C for 20 min and subculturing on the solid medium. The motility of strain Arc-HrT was assessed by observing a fresh culture under DM1000 photonic microscope (Leica Microsystems, Nanterre, France) with a 100X oil-immersion objective lens. The colonies’ surface was observed on the agar culture medium after 3 days of incubation under aerobic conditions at 37 °C.
Antibiotic susceptibility testing
Susceptibility of strain Arc-HrT to antibiotics was tested using antibiotic disks (B. Braun Medical SAS, Boulogne, France) containing the following antibiotics: fosfomycin 50 µg, doxycycline 30UI, rifampicin 30 µg, vancomycin 30 µg, amoxicillin 20 µg, erythromycin 15UI, ampicillin 25 µg, cefoxitin 30 µg, colistin 50 µg, tobramycin 10 µg, gentamicin 500 µg, penicillin G 10UI, trimethoprim 1.25 µg/sulfamethoxazole 23.75 µg, oxacillin 5 µg, imipenem 10 µg, metronidazole 4 µg and anisomycin 10 µg.
Microscopy and gram test
Cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer for at least 1 h at 4 °C. A drop of cell suspension was deposited for approximately 5 min on glow-discharged formvar carbon film on 400 mesh nickel grids (FCF400-Ni, EMS). The grids were dried on blotting paper and cells were negatively stained for 10 s with 1% ammonium molybdate solution in filtered water at RT. Electron micrographs were acquired with a Morgagni 268D (Philips) transmission electron microscope operated at 80 keV. The gram stain was performed using the color GRAM 2 kit (Biomerieux) and observed using a DM1000 photonic microscope (Leica Microsystems).
Analysis of fatty acid methyl ester and membrane polar lipids
Polar lipids were extracted and identified by one-dimensional TLC as described by Cui and Zhang (2014). Cellular fatty acid methyl ester (FAME) analysis was performed by GC/MS. Three samples were prepared with approximately 80 mg of bacterial biomass per tube harvested from several culture plates. Fatty acid methyl esters were prepared as described by Sasser (2006). GC/MS analyses were carried out as described before (Dione et al. 2016). Briefly, fatty acid methyl esters were separated using an Elite 5-MS column and monitored by mass spectrometry (Clarus 500—SQ 8 S, Perkin Elmer, Courtaboeuf, France). Spectral database search was performed using MS Search 2.0 operated with the Standard Reference Database 1A (NIST, Gaithersburg, USA) and the FAMEs mass spectral database (Wiley, Chichester, UK).
DNA extraction and genome sequencing
After scraping 5 Petri dishes in 1 mL TE buffer, the genomic DNA (gDNA) of strain Arc-HrT was extracted from 200 µL of the bacterial suspension after a classical lysis treatment with a final concentration of lysozyme at 40 mg/mL for 2 h at 37 °C followed by an incubation time of 1 h at 37 °C in SDS 1% final and 30µL RNAse. Proteinase K treatment was realized with at 37 °C. After three phenol extractions and alcohol precipitation, the sample was eluted in the minimal volume of 50 µL in EB buffer. DNA was quantified by a Qubit assay with the high sensitivity kit (Life technologies, Carlsbad, CA, USA) to 14 ng/µL.
GDNA was sequenced on the MiSeq Technology (Illumina Inc, San Diego, CA, USA) with the mate pair strategy as previously described (Dione et al. 2016). Total information of 10.6 Gb was obtained from a 1326 K/mm2 cluster density with a cluster passing quality control filters of 99.1% (20,978,044 pass filter clusters). Within this run, the index representation for strain Arc-HrT was determined to be of 6.22%. The 1,303,974 paired reads were filtered according to the read qualities, trimmed and then assembled.
Genome assembly
Illumina reads were trimmed using Trimmomatic (Lohse et al. 2012), then assembled thought Spades software (Nurk et al. 2013; Bankevich et al. 2012). Contigs obtained were combined together by SSpace (Boetzer et al. 2011) and Opera software (Gao et al. 2011) helped by GapFiller (Boetzer and Pirovano 2012) to reduce the set. Some manual refinements using CLC Genomics v7 software (CLC bio, Aarhus, Denmark) and homemade tools in Python improved the genome. Finally, the draft genome of strain Arc-HrT consisted of 8 contigs.
Genome annotation and comparison
Non-coding genes and miscellaneous features were predicted using RNAmmer (Lagesen et al. 2007), ARAGORN (Laslett and Canback 2004), Rfam (Griffiths-Jones et al. 2003), PFAM (Punta et al. 2012), and Infernal (Nawrocki et al. 2009). Coding DNA sequences (CDSs) were predicted using Prodigal (Hyatt et al. 2010) and functional annotation was achieved using BLAST + (Camacho et al. 2009) and HMMER3 (Eddy 2011) against the UniProtKB database (The UniProt Consortium 2011). A brief genomic comparison was also made between strain Arc-HrT (CSTE00000000), Haloferax alexandrinus strain Arc-Hr (CCDK00000000), Haloferax gibbonsii strain ARA6 (CP011947), Haloferax lucentense strain DSM 14919 (AOLH00000000), Haloferax volcanii strain DS2 (CP001956) and Haloferax prahovense strain DSM 18310 (AOLG00000000). To estimate the mean level of nucleotide sequence similarity at the genome level between strain Arc-HrT and the four closest species with an available genome, we used the Average Genomic Identity of Orthologous gene Sequences (AGIOS), in a laboratory’s pipeline. Briefly, this pipeline combines the Proteinortho (Lechner et al. 2011) software (with the following parameters: e value 1e−5, 30% identity, 50% coverage and algebraic connectivity of 50%) for the detection of orthologous proteins between genomes compared pairwise, retrieves the corresponding genes and determines the mean percentage of nucleotide sequence identity between orthologous ORFs using the Needleman–Wunsch global alignment algorithm (Ramasamy et al. 2014). Strain Arc-HrT genome was locally aligned 2-by-2 using BLAT algorithm (Kent 2002; Auch et al. 2010) against each selected genomes previously cited and DNA–DNA hybridization (DDH) values were estimated by using the genome-to genome sequence comparison (Auch et al. 2010).
Results
Strain identification and phylogenetic analysis
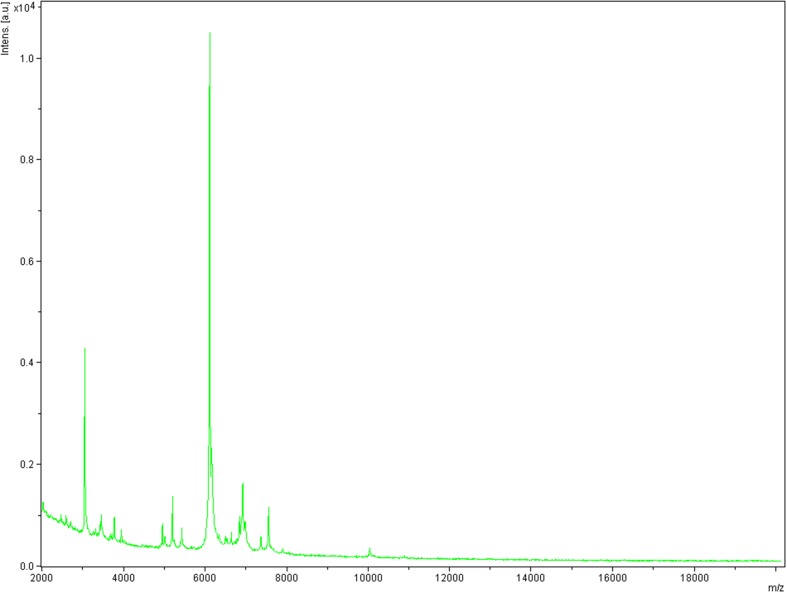
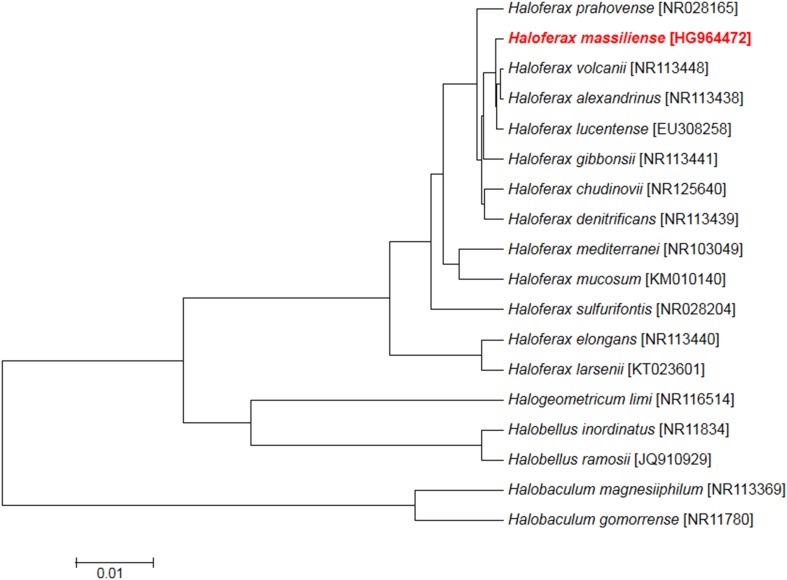
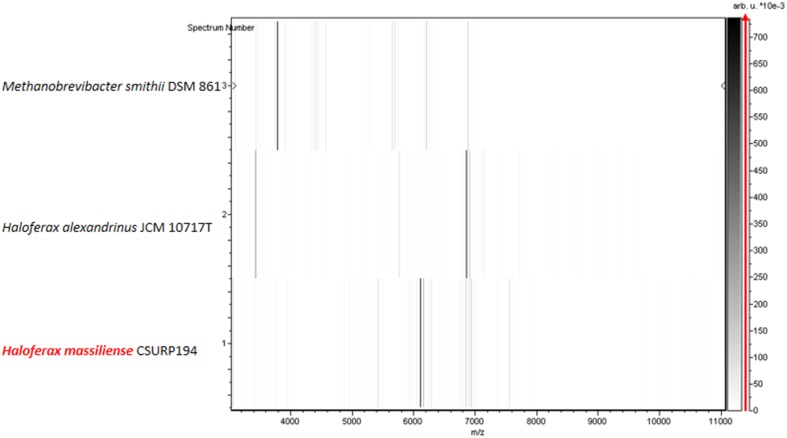
Using MALDI-TOF MS identification, no significant score allowing a correct identification was obtained for strain Arc-HrT against our database (the Bruker database is constantly incremented with URMITE data), suggesting that our isolate did not belong to any known species; and consequently, spectra from strain Arc-HrT was added to our database (http://www.mediterranee-infection.com/article.php?laref=256&titre=urms) (Fig. 1). PCR-amplified 16S rRNA gene of strain Arc-HrT (HG964472) exhibited a 99.2% sequence similarity with Haloferax prahovense JCM 13924 (NR113446), the phylogenetically closest validly published species with standing in nomenclature (Fig. 2). As 16S rRNA gene sequence comparison has been proven to poorly discriminate Haloferax species, we sequenced the complete genome of strain Arc-HrT and a digital DNA–DNA hybridization (dDDH) was made with four of the closest Haloferax species (see the part on genome comparison). These data confirmed strain Arc-HrT as a unique species. Finally, the gel view showed the protein spectral differences with other members of the genus Haloferax (Fig. 3).
Fig. 1.
Reference mass spectrum from Haloferax massiliense strain Arc-HrT. Spectra from 12 individual colonies were compared and a reference spectrum was generated
Fig. 2.
Phylogenetic tree highlighting the position of Haloferax massiliense strain Arc-HrT relative to other type strains within Haloferax, Halogeometricum, Halobellus and Halobaculum genus. The respective GenBank accession numbers for 16S rRNA genes are indicated in parenthesis. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using the maximum-likelihood method within the MEGA software. The scale bar represents 0.005% nucleotide sequence divergence
Fig. 3.
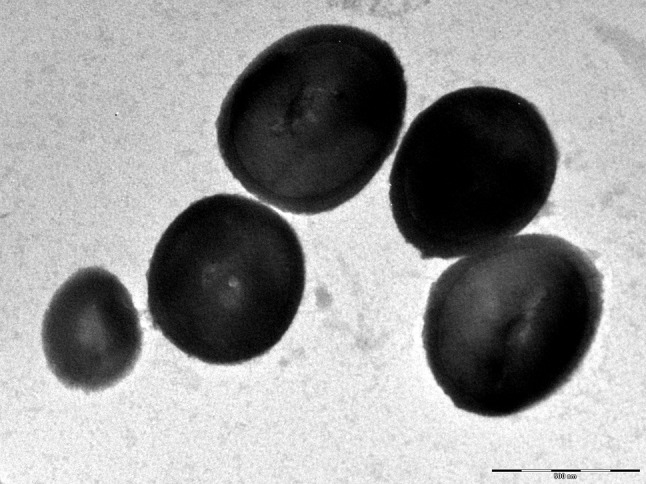
Transmission electron microscopy of Haloferax massiliense strain Arc-HrT, using a Morgani 268D (Philips) at an operating voltage of 80 keV. The scale bar represents 500 nm
Phenotypic and biochemical characteristics
Salt concentration of the stool specimen measured by digital refractometer was about 2.5% and the pH was 7.2. Strain Arc-HrT colonies were circular, red, shiny and smooth with a diameter of 0.5–1 mm. Cells were gram-negative, non-motile and non-spore-forming. Cells were very pleomorphic (irregular cocci, short and long rods, triangles and ovals) and had a diameter between 1 to 4 µm (Fig. 4). Strain Arc-HrT was mesophilic and grew at temperatures ranging from 25 to 45 °C, with an optimum at 37 °C. NaCl was required for growth and the strain grew at a salinity ranging from 10 to 25% of NaCl with an optimum at 15%; cells underwent lysis below 100 g/L NaCl. The optimum pH for growth was 7 (range between pH 6.5 and 8). The strain was strictly aerobic and grew in the presence of 5% CO2; no growth was observed in microaerophilic or anaerobic condition by using alternative electron acceptors such as nitrate or DMSO, or by fermenting l-arginine. Principal features are presented in Table 1.
Fig. 4.
Gel view comparing Haloferax massiliense strain Arc-HrT to other species within the genus Haloferax. The gel view displays the raw spectra of loaded spectrum files arranged in a pseudo-gel like look. The x-axis records the m/z value. The left y-axis displays the running spectrum number originating from subsequent spectra loading. The peak intensity is expressed by a Gray scale scheme code. The color bar and the right y-axis indicate the relation between the color of a peak and the peak intensity, in arbitrary units. Displayed species are indicated on the left
Table 1.
Classification and general features of Haloferax massiliense strain Arc-HrT according to the MIGS recommendations (Field et al. 2008)
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain: Archaea | TAS (Woese et al. 1990) | |
| Phylum: Euryarchaeota | TAS (Cavalier-Smith 2002; Garrity and Holt 2001) | ||
| Class: Halobacteria | TAS (Grant et al. 2001a, b; Gupta et al. 2015) | ||
| Order: Haloferacales | TAS (Grant and Larsen 1989; Gupta et al. 2015) | ||
| Family: Haloferacaceae | TAS (Grant and Larsen 1989; Gupta et al. 2015) | ||
| Genus: Haloferax | TSA (Torreblanca et al. 1986) | ||
| Species: Haloferax massiliense | IDA | ||
| Type strain: Arc-HrT | IDA | ||
| Gram stain | negative | IDA | |
| Cell shape | Cocci | IDA | |
| Motility | Non motile | IDA | |
| Sporulation | Non spore-forming | IDA | |
| Temperature range | Mesophile | IDA | |
| Optimum temperature | 37 °C | IDA | |
| pH | pH 6.5–8 | ||
| Optimum pH | 7 | ||
| MIGS-6.3 | Salinity | 10–25% | IDA |
| Optimum salinity | 15% NaCl | IDA | |
| MIGS-22 | Oxygen requirement | Strictly aerobic | IDA |
| Carbon source | Unknown | IDA | |
| Energy source | Unknown | IDA | |
| MIGS-6 | Habitat | Human gut | IDA |
| MIGS-15 | Biotic relationship | Free living | IDA |
| Pathogenicity | Unknown | NAS | |
| Biosafety level | 2 | IDA | |
| MIGS-14 | Isolation | Human feces | IDA |
| MIGS-4 | Geographic location | France | IDA |
| MIGS-5 | Sample collection time | December 2013 | IDA |
| MIGS-4.3 | Depth | surface | IDA |
| MIGS-4.4 | Altitude | 0 m above sea level | IDA |
aEvidence codes—IDA, Inferred from Direct Assay; TAS, Traceable Author Statement (i.e., a direct report exists in the literature); NAS, non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from http://www.geneontology.org/GO.evidence.shtml of the Gene Ontology project (Ashburner et al. 2000). If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements
Strain Arc-HrT exhibited positive catalase and oxidase activities. Using an API ZYM strip, positive reactions were observed for alkaline phosphatase, acid phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, β-glucuronidase, and negative reactions were observed for lipase (C14), valine arylamidase, trypsin, α-chymotrypsin, β-galactosidase, N-acetyl- β-glucosaminidase, α-galactosidase, α-glucosidase, β-glucosidase, α-fucosidase, α-mannosidase. An API 50CH strip showed positive reaction for glycerol, d-fructose, l-rhamnose, potassium 2-ketogluconate and potassium 5-ketogluconate, and negative reactions for arbutin, salicin, d-maltose, d-sucrose, d-raffinose,, erythritol, d-ribose, d-xylose, l-xylose, d-adonitol, methyl-βd-xylopyranoside, d-glucose, d-galactose, d-lactose, l-sorbose, dulcitol, inositol, d-mannitol, d-sorbitol, methyl-αd-mannopyranoside, methyl-αd-glucopyranoside, d-cellobiose, d-melibiose, d-trehalose, d-melezitose, starch, glycogen, xylitol, gentiobiose, d-turanose, d-lyxose, d-tagatose, d-fucose, d-fucose, d-arabitol, d-arabitol***, and potassium gluconate. The phenotypic characteristics of strain Arc-HrT were compared with the most closely related species (Table 2).
Table 2.
Differential characteristics of Haloferax massiliense strain Arc-HrT, Haloferax prahovense (Enache et al. 2007), Haloferax volcanii (Torreblanca et al. 1986), Haloferax denitrificans (Tindall et al. 1989); 4, Haloferax mediterranei (Torreblanca et al. 1986), Haloferax gibbonsii (Juez et al. 1986), Haloferax alexandrinus (Asker and Ohta 2002) and Haloferax lucentense (Gutierrez et al. 2002). na: No available data
| Properties | H. massiliense | H. prahovense | H. volcanii | H. denitrificans | H. gibbonsii | H. mediterranei | H. alexandrinus | H. lucentense |
|---|---|---|---|---|---|---|---|---|
| Oxygen requirement | + | + | + | + | + | + | + | + |
| Gram stain | – | – | – | – | – | – | – | – |
| Salt requirement | + | + | + | + | + | + | + | + |
| Motility | – | – | – | – | – | + | – | + |
| Endospore formation | – | – | – | – | – | – | – | – |
| Indole | – | – | + | + | – | + | + | + |
| Tween 80 hydrolysis | + | + | + | – | – | + | + | + |
| Production of | ||||||||
| Alkaline phosphatase | + | na | na | na | na | na | + | na |
| Catalase | + | + | + | + | + | + | + | + |
| Oxidase | + | + | + | + | + | + | + | + |
| Nitrate reductase | + | – | + | + | – | + | + | – |
| Urease | – | na | na | – | na | na | – | na |
| β-galactosidase | – | na | + | + | + | na | + | – |
| N-acetyl-glucosamine | – | Na | na | na | na | na | + | na |
| Acid from | ||||||||
| l-Arabinose | – | – | + | na | + | + | + | + |
| Ribose | – | + | – | – | na | na | + | na |
| Mannose | – | – | – | – | + | + | – | na |
| Mannitol | – | – | na | na | na | + | na | – |
| Sucrose | – | – | + | + | + | + | + | – |
| d-Glucose | – | – | + | + | + | + | + | + |
| d-Fructose | + | + | + | + | + | + | + | + |
| d-Maltose | – | + | + | + | + | + | + | + |
| d-Lactose | – | + | – | – | – | + | – | – |
| Gelatin hydrolysis | – | – | – | + | + | + | + | – |
| Starch hydrolysis | – | + | – | – | – | + | – | – |
| Casein hydrolysis | + | – | – | – | + | + | – | – |
| Habitat | Human Gut | Salt lake | Bottom sedimen | solar saltern | Solar salterns | Solar salt pond | solar saltern | Water of a saltern |
Antimicrobial susceptibility testing demonstrated that strain Arc-HrT was susceptible to rifampicin, trimethoprim/sulfamethoxazole and anisomycin, and resistant to fosfomycin, doxycycline, vancomycin, amoxicillin, erythromycin, ampicillin, cefoxitin, colistin, tobramycin, gentamicin, penicillin G, oxacillin, imipenem and metronidazole.
The fatty acid was 3-methyl-butanoic acid (5:0 iso), a branched short chain fatty acid. Phenylacetic acid, known as an antifungal agent (Ryan et al. 2009), was also detected. Membrane polar lipids were diglycosyl diether analogs of phosphatidylglycerol, phosphatidylglycerol, diglycosyl diether,**** and sulfated diglycosyl diether (S-DGD-1), the glycolipid marker of Haloferax spp. (Cui and Zhang. 2014). This fatty acid’s profile is completely different from that of bacteria described by (Dione et al. 2016).
Genome sequencing information and annotation
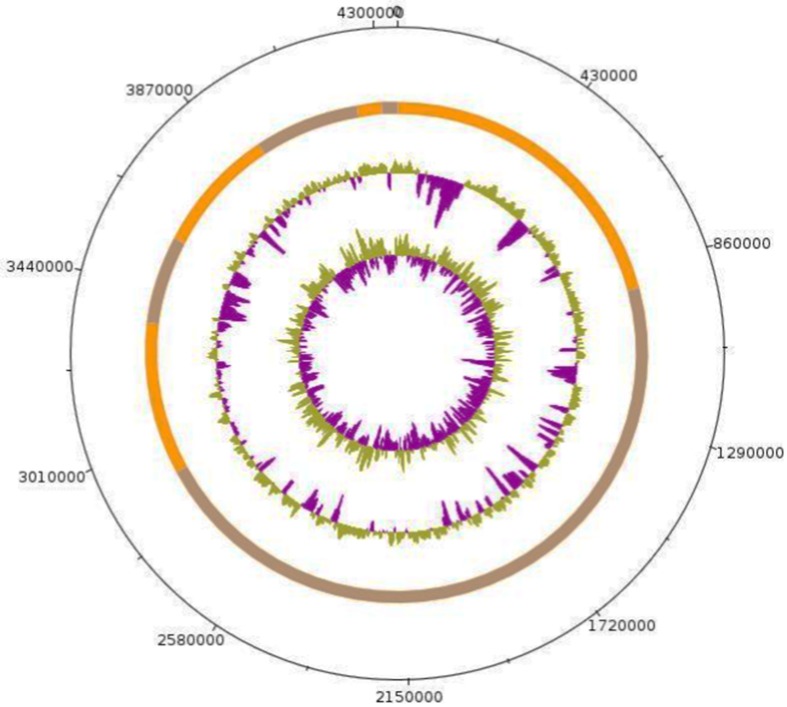
Strain Arc-HrT’s genome was sequenced as part of a culturomic study aiming at isolating all prokaryotes species colonizing the human gut (Lagier et al. 2016) and because of its phylogenetic affiliation to the Haloferax genus. Strain Arc-HrT represents the 13th genome sequenced in the Haloferax genus. The draft genome of strain Arc-HrT contains 4,015,175 bp with a G + C content of 65.36% and consists of 8 contigs without gaps (Fig. 5). The genome was shown to encode at least 64 predicted RNA including 3 rRNA, 57 tRNA, 4 miscellaneous RNA and 3911 protein-coding genes. Among these genes, 490 (13%) were found to be putative proteins and 291 (8%) were assigned as hypothetical proteins. Moreover, 2335 genes matched at least one sequence in Clusters of Orthologous Groups (COGs) database (Tatusov et al. 1997, 2000) with BLASTP default parameters. Table 3 shows the detailed project information and its association with MIGS version 2.0 compliance. The properties and the statistics of the genome are summarized in Table 4. The distribution of genes into COGs functional categories is presented in Table 5.
Fig. 5.
Circular representation of the Haloferax massiliense Arc-HrT genome. Circles from the center to the outside: GC screw (green/purple), GC content (green/purple) and contigs (orange/brown)
Table 3.
Project information
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | 1 mate-paired |
| MIGS-29 | Sequencing platforms | MiSeq Illumina |
| MIGS-31.2 | Sequencing coverage | 620 |
| MIGS-30 | Assemblers | Spades |
| MIGS-32 | Gene calling method | Prodigal |
| Genbank ID | CSTE01000001–CSTE01000008 | |
| Genbank date of release | Apr, 2014 | |
| MIGS-13 | Source material identifier | Arc-HrT |
| Project relevance | Mar, 2014 |
Table 4.
Nucleotide content and gene count levels of the genome
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 4,015,175 | 100 |
| DNA coding region (bp) | 3,414,159 | 78.50 |
| DNA G + C content (bp) | 2,624,318 | 65.36 |
| Total protein-coding genes | 3911 | 100 |
| rRNA | 3 | 0.08 |
| tRNA | 57 | 1.46 |
| tmRNA | 0 | 0 |
| miscRNA | 4 | 0.11 |
| Genes with function prediction | 2825 | 72.23 |
| Genes assigned to COGs | 3116 | 79.68 |
aThe total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome
Table 5.
Number of genes associated with the 25 general COG functional categories
| Code | Description | Value | % of total |
|---|---|---|---|
| J | Translation, ribosomal structure and biogenesis | 165 | 4.22 |
| A | RNA processing and modification | 1 | 0.03 |
| K | Transcription | 172 | 4.40 |
| L | Replication, recombination and repair | 133 | 3.41 |
| B | Chromatin structure and dynamics | 6 | 0.16 |
| D | Cell cycle control, cell division, chromosome partitioning | 27 | 0.69 |
| Y | Nuclear structure | 0 | 0.0 |
| V | Defense mechanisms | 37 | 0.95 |
| T | Signal transduction mechanisms | 157 | 4.02 |
| M | Cell wall/membrane biogenesis | 119 | 3.05 |
| N | Cell motility | 38 | 0.98 |
| Z | Cytoskeleton | 0 | 0.0 |
| W | Extracellular structures | 0 | 0.0 |
| U | Intracellular trafficking and secretion, and vesicular transport | 35 | 0.90 |
| O | Posttranslational modification, protein turnover, chaperones | 113 | 2.89 |
| C | Energy production and conversion | 208 | 5.32 |
| G | Carbohydrate transport and metabolism | 219 | 5.6 |
| E | Amino acid transport and metabolism | 341 | 8.72 |
| F | Nucleotide transport and metabolism | 75 | 1.92 |
| H | Coenzyme transport and metabolism | 154 | 3.94 |
| I | Lipid transport and metabolism | 79 | 2.02 |
| P | Inorganic ion transport and metabolism | 202 | 5.17 |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 54 | 1.39 |
| R | General function prediction only | 490 | 12.53 |
| S | Function unknown | 291 | 7.45 |
Genome comparison
The draft genome of strain Arc-HrT is larger than that of H. prahovense, H. alexandrinus, H. gibbonsii, H. lucentense and H. volcanii (4.35, 4, 3.9, 3.62, 2.95 and 2.85 Mb respectively). The G + C content of strain Arc-HrT is smaller than that of H. alexandrinus, H. lucentense, H. volcanii and H. gibbonsii (65.36, 66, 66.4, 66.6 and 67.1%, respectively) but smaller than that of H. prahovense (65.7%). The gene content of strain Arc-HrT is larger than that of H. alexandrinus, H. prahovense, H. lucentense, H. gibbonsii and H. volcanii (3.911, 3.770, 3.766, 3.593, 2.997 and 2.917).
The distribution of genes into COG categories was identical (Fig. 6) in all compared genomes.
Fig. 6.
Distribution of functional classes of predicted genes according to cluster of orthologous groups of proteins from Haloferax massiliense strain Arc-HrT
The Average Genomic Identity of Orthologous gene Sequences (AGIOS) shows that strain Arc-HrT shared 2.690, 2.353, 2.958, 2.975 and 2.459 orthologous genes with H. lucentense, H. volcanii, H. prahovense, H. alexandrinus and H. gibbonsii, respectively (Table 6). Among compared species, except for strain Arc-HrT, AGIOS values ranged from 92.08 to 98.83%. AGIOS values between strain Arc-HrT and compared species were in the same range (from 92.24% with H. alexandrinus to 93.29% with H. volcanii). The DDH values ranged from 50.70 to 82.20%, among compared species, except for strain Arc-HrT. Among compared species and strain Arc-HrT, the DDH values ranged from 50.70% with H. prahovense, to 54.30% with H. lucentense, these values were lower than the 70% cutoff (Meier-Kolthoff et al. 2013) (Table 7).
Table 6.
Number of orthologous proteins shared between genomes (upper right), average percentage similarity of nucleotides corresponding to orthologous protein shared between genomes (lower left) and number of proteins per genome (bold)
| H. lucentense | H. volcanii | H. prahovense | H. alexandrinus | H. massiliense | H. gibbonsii | |
|---|---|---|---|---|---|---|
| Haloferax lucentense | 4086 | 2348 | 2754 | 2761 | 2690 | 2355 |
| Haloferax volcanii | 97.07 | 2995 | 2409 | 2400 | 2353 | 2412 |
| Haloferax prahovense | 92.1 | 93.21 | 4180 | 3238 | 2958 | 2696 |
| Haloferax alexandrinus | 92.08 | 92.84 | 98.83 | 4109 | 2973 | 2685 |
| Haloferax massiliense | 93.07 | 93.29 | 92.33 | 92.24 | 4259 | 2459 |
| Haloferax gibbonsii | 92.44 | 93.67 | 96.68 | 96.3 | 92,82 | 3053 |
Table 7.
Pairwise comparison of Haloferax massiliense with other species using GGDC, formula 2 (DDH estimates based on identities/HSP length)
| H. massiliense | H. alexandrinus | H. gibbonsii | H. lucentense | H. volcanii | H. prahovense | |
|---|---|---|---|---|---|---|
| H. massiliense | 100 ± 00% | 50.90 ± 2.64% | 52.90 ± 2.67% | 54.30 ± 2.70% | 53.70 ± 2.69% | 50.70 ± 5.2% |
| H. alexandrinus | 100 ± 00% | 73.50 ± 2.90% | 50. 80 ± 2.63% | 53.50 ± 2.68% | 96 ± 2.4% | |
| H. gibbonsii | 100 ± 00% | 51.90 ± 2.65% | 53.80 ± 2.69% | 72 ± 5.8% | ||
| H. lucentense | 100 ± 00% | 82.20 ± 2.69% | 50.70 ± 5.3% | |||
| H. volcanii | 100 ± 00% | 52.80 ± 5.3% | ||||
| H. prahovense | 100 ± 00% |
The confidence intervals indicate the inherent uncertainty in estimating DDH values from intergenomic distances based on models derived from empirical test data sets (which are always limited in size). These results are in accordance with the 16S rRNA (Fig. 4) and phylogenomic analyses as well as the GGDC results
Discussion
Here, we describe the genome sequence and most of the biochemical characteristics of the first isolate of Haloferax massiliense sp. nov., an extremely halophilic archaea isolated from the human gut. Halophilic organisms are generally known to colonize hypersaline environments where the salt concentration is close to saturation, such as salt lakes and salt marshes (Oren 1994). Here, using a culture medium containing high salt concentration, we successfully isolated strain Arc-HrT belonging to the Haloferax genus within the Haloferacaceae family. This strain presents the first halophilic archaea isolated from the human gut. Recently, DNA sequences belonging to some halophilic archaea frequently present or abundant in extreme environments were detected by PCR in the human gastro-intestinal tract as well as some members of the Halobacteriaceae family (Oxley et al. 2010). Bacterial halophilism has become a subject of considerable interest for microbiologists and molecular biologists during the past 20 years, because of their development on salty foods (Fukushima et al. 2007). Indeed, these organizations have also been detected in refined salt (Diop et al. 2016) as well as food products where salt is used in large quantities in the process of their conservation such as salted fish, pork ham, sausages and fish sauces (Tanasupawat et al. 2009; Kim et al. 2010). Additionally, the limitation of these organisms to extreme environments has been recently contested after their detection in habitats with relatively low salinity, suggesting an ability of adaptation to survive in more moderate environments (Purdy et al. 2004).
This work does not intend to demonstrate a medical or biotechnological interest regarding strain Arch-HrT; its only aim is to expand knowledge about the human microbiota and isolating all the prokaryotes that colonize the human digestive tract (Lagier et al. 2016).
Conclusion
Based on the characteristics reported here and the phylogenetic affiliation of strain Arc-HrT, we proposed the creation of Haloferax massiliense sp. nov., as a new species belonging to the Haloferax genus with strain Arc-HrT as its type strain. Haloferax massiliense sp. nov., (= CSURP0974 = CECT 9307), described here, was isolated from the human gut as part of a culturomic study aiming at expanding the repertoire of microorganisms colonizing the human gut.
Description of Haloferax massiliense sp. nov
Haloferax massiliense (mas.si.li.en’se, N.L. neut. adj., massiliense of Massilia, the Roman name of Marseille, France, where the type strain was isolated).
Haloferax massiliense strain Arc-HrT is a strictly aerobic gram negative, non-motile and non-spore-forming. Cells were very pleomorphic (irregular cocci, short and long rods, triangles and ovals) and had a diameter between 1 and 4 µm. An optimal growth was observed at 37 °C, pH 7 and 15% of NaCl. Colonies are red, smooth, shiny and measure 0.5–1 mm. Strain Arc-HrT has exhibited positive catalase and oxidase activities.
Using API strips, positive reactions were observed for alkaline phosphatase, acid phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, naphthol-AS-BI-phosphohydrolase, β-glucuronidase, glycerol, d-fructose, l-rhamnose, potassium 2-ketogluconate and potassium 5-ketogluconate. Strain Arc-HrT was susceptible to rifampicin and trimethoprim/sulfamethoxazole. The genome of Haloferax massiliense is 4,349,774 bp long and exhibits a G + C% content of 65.36%. The 16S rRNA and genome sequences are deposited in EMBL-EBI under accession numbers HG964472 and CSTE00000000, respectively. The type strain Arc-HrT (= CSUR P0974 = CECT 9307) was isolated from a stool specimen of 22-year-old Amazonian obese female patient as part of a culturomics study.
Acknowledgements
The authors thank Magdalen Lardière for English reviewing. This study was funded by the “Fondation Méditerranée Infection”.
Abbreviations
- FAME
Fatty acid methyl ester
- GC/MS
Gas chromatography/mass spectrometry
- CSUR
Collection de Souches de l’Unité des Rickettsies
- CECT
Colección Española de Cultivos Tipo
- MALDI-TOF MS
Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry
- URMITE
Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes
- IU
International unit
Compliance with ethical standards
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Arahal DR, Oren A, Ventosa A. International committee on systematics of prokaryotes subcommittee on the taxonomy of halobacteria and subcommittee on the taxonomy of halomonadaceae. Minutes of the joint open meeting, 11 July 2017, Valencia, Spain. Int J Syst Evol Microbiol. 2017;67:4279–4283. doi: 10.1099/ijsem.0.002296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asker D, Ohta Y. Haloferax alexandrinus sp. nov., an extremely halophilic canthaxanthin-producing archaeon from a solar saltern in Alexandria (Egypt) Int J Syst Evol Microbiol. 2002;52(P3):729–738. doi: 10.1099/00207713-52-3-729. [DOI] [PubMed] [Google Scholar]
- Auch AF, von Jan M, Klenk HP, Göker M. Digital DNA–DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genom Sci. 2010;2(1):117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Pirovano W. Toward almost closed genomes with GapFiller. Genome Biol. 2012;13:R56. doi: 10.1186/gb-2012-13-6-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int J Syst Evol Microbiol. 2002;52:7–76. doi: 10.1099/00207713-52-1-7. [DOI] [PubMed] [Google Scholar]
- Cui HL, Zhang WJ. Salinigranum rubrum gen. nov., sp. nov., a member of the family Halobacteriaceae isolated from a marine solar saltern. Int J Syst Evol Microbiol. 2014;64(6):2029–2033. doi: 10.1099/ijs.0.061606-0. [DOI] [PubMed] [Google Scholar]
- Dione N, Sankar SA, Lagier JC, Khelaifia S, Michele C, Armstrong N, Richez M, Abrahão J, Raoult D, Fournier PE. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diop A, Khelaifia S, Armstrong N, Labas N, Fournier PE, Raoult D, Million M. Microbial culturomics unravels the halophilic microbiota repertoire of table salt: description of Gracilibacillus massiliensis sp. nov. Microb Ecol Health Dis. 2016;18(27):32049. doi: 10.3402/mehd.v27.32049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated profile HMM searches. PLoS Comp Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsawi Z, Togo AH, Beye M, Dubourg G, Andrieu C, Armsrtong N, Richez M, di Pinto F, Bittar F, Labas N, Fournier PE, Raoult D, Khelaifia S. Hugonella massiliensis gen. nov., sp. nov., genome sequence, and description of a new strictly anaerobic bacterium isolated from the human gut. Microbiologyopen. 2017 doi: 10.1002/mbo3.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enache M, Itoh T, Kamekura M, Teodosiu G, Dumitru L. Haloferax prahovense sp. nov., an extremely halophilic archaeon isolated from a Romanian salt lake. Int J Syst Evol Microbiol. 2007;57(2):393–397. doi: 10.1099/ijs.0.64674-0. [DOI] [PubMed] [Google Scholar]
- Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, Ashburner M, Axelrod N, Baldauf S, Ballard S, Boore J, Cochrane G, Cole J, Dawyndt P, De Vos P, DePamphilis C, Edwards R, Faruque N, Feldman R, Gilbert J, Gilna P, Glöckner FO, Goldstein P, Guralnick R, Haft D, Hancock D, Hermjakob H, Hertz-Fowler C, Hugenholtz P, Joint I, Kagan L, Kane M, Kennedy J, Kowalchuk G, Kottmann R, Kolker E, Kravitz S, Kyrpides N, Leebens-Mack J, Lewis SE, Li K, Lister AL, Lord P, Maltsev N, Markowitz V, Martiny J, Methe B, Mizrachi I, Moxon R, Nelson K, Parkhill J, Proctor L, White O, Sansone SA, Spiers A, Stevens R, Swift P, Taylor C, Tateno Y, Tett A, Turner S, Ussery D, Vaughan B, Ward N, Whetzel T, San Gil I, Wilson G, Wipat A. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima T, Usami R, Kamekura M. A traditional Japanese-style salt field is a niche for haloarchaeal strains that can survive in 0.5% salt solution. Saline Syst. 2007;3(1):1. doi: 10.1186/1746-1448-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Sung WK, Nagarajan N. Opera: reconstructing optimal genomic scaffolds with high-throughput paired-end sequences. J Comput Biol. 2011;18(11):1681–1691. doi: 10.1089/cmb.2011.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Holt JG. Phylum AII. Euryarchaeota phy. nov. In: Boone DR, Castenholz RW, editors. Bergey’s manual of systematic bacteriology, 2nd edn, vol 1. The archaea and the deeply branching and phototrophic bacteria. New York: Springer; 2001. p. 211. [Google Scholar]
- Grant WD, Larsen H. Extremely halophilic archaeobacteria. Order Halobacteriales ord. nov. In: Pfennig N, editor. Bergey’s manual of systematic bacteriology. Baltimore: Williams & Wilkins; 1989. pp. 2216–2233. [Google Scholar]
- Grant WD, Kamekura M, McGenity TJ, Ventosa A. Class III. Halobacteria class. nov. In: Boone DR, Castenholz RW, editors. Bergey’s Manual of systematic bacteriology, 2nd edn, vol 1. The archaea and the deeply branching and phototrophic bacteria. New York: Springer; 2001. p. 294. [Google Scholar]
- Grant WD, Kamekura M, McGenity TJ, Ventosa A. The order halobacteriales. In: Boone DR, Castenholz RW, editors. Bergey’s manual of systematic bacteriology. 2. New York: Springer; 2001. pp. 294–334. [Google Scholar]
- Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS, Naushad S, Baker S. Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int J Syst Evol Microbiol. 2015;65:1050–1069. doi: 10.1099/ijs.0.070136-0. [DOI] [PubMed] [Google Scholar]
- Gutierrez MC, Kamekura M, Holmes ML, Dyall-Smith ML, Ventosa A. Taxonomic characterization of Haloferax sp. (H. alicantei) strain Aa 2.2: description of Haloferax lucentensis sp. nov. Extremophiles. 2002;6:479–483. doi: 10.1007/s00792-002-0282-7. [DOI] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juez G, Rodriguez-Valera F, Ventosa A, Kushner DJ. Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extremely halophilic archaebacteria. Syst Appl Microbiol. 1986;8:75–79. doi: 10.1016/S0723-2020(86)80152-7. [DOI] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelaifia S, Raoult D. Haloferax massiliensis sp. nov., the first human-associated halophilic archaea. New Microbes New Infect. 2016;12:96–98. doi: 10.1016/j.nmni.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelaifia S, Raoult D, Drancourt M. A versatile medium for cultivating methanogenic archaea. PLoS One. 2013;8(4):e61563. doi: 10.1371/journal.pone.0061563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Roh SW, Bae JW. Halomonas jeotgali sp. nov., a new moderate halophilic bacterium isolated from a traditional fermented seafood. J Microbiol. 2010;48(3):404–410. doi: 10.1007/s12275-010-0032-y. [DOI] [PubMed] [Google Scholar]
- Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012;18(12):1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, Dubourg G, Durand G, Mourembou G, Guilhot E, Togo A, Bellali S, Bachar D, Cassir N, Bittar F, Delerce J, Mailhe M, Ricaboni D, Bilen M, Dangui Nieko NP, Dia Badiane NM, Valles C, Mouelhi D, Diop K, Million M, Musso D, Abrahão J, Azhar EI, Bibi F, Yasir M, Diallo A, Sokhna C, Djossou F, Vitton V, Robert C, Rolain JM, La Scola B, Fournier PE, Levasseur A, Raoult D. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;7(1):16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinform. 2011;28(12):124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA. Methanogenic archaea and human periodontal disease. Proc Natl Acad Sci USA. 2004;101(16):6176–6181. doi: 10.1073/pnas.0308766101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 2012;40(Web Server issue):W622–W627. doi: 10.1093/nar/gks540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013;21(14):60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam YD, Chang HW, Kim KH, Roh SW, Kim MS, Jung MJ, Lee SW, Kim JY, Yoon JH, Bae JW. Bacterial, archaeal, and eukaryal diversity in the intestines of Korean people. J Microbiol. 2008;46(5):491–501. doi: 10.1007/s12275-008-0199-7. [DOI] [PubMed] [Google Scholar]
- Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25(10):1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20(10):714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. The ecology of the extremely halophilic archaea. FEMS Microbiol Rev. 1994;13(4):415. doi: 10.1111/j.1574-6976.1994.tb00060.x. [DOI] [PubMed] [Google Scholar]
- Oxley AP, Lanfranconi MP, Würdemann D, Ott S, Schreiber S, McGenity TJ, Timmis KN, Nogales B. Halophilic archaea in the human intestinal mucosa. Environ Microbiol. 2010;12(9):2398–2410. doi: 10.1111/j.1462-2920.2010.02212.x. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdy KJ, Cresswell-Maynard TD, Nedwell DB, McGenity TJ, Grant WD, Timmis KN, Embley TM. Isolation of haloarchaea that grow at low salinities. Environ Microbiol. 2004;6(6):591–595. doi: 10.1111/j.1462-2920.2004.00592.x. [DOI] [PubMed] [Google Scholar]
- Ramasamy D, Mishra AK, Lagier JC, Padhmanabhan R, Rossi M, Sentausa E, Raoult D, Fournier PE. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int J Syst Evol Microbiol. 2014;64:384–391. doi: 10.1099/ijs.0.057091-0. [DOI] [PubMed] [Google Scholar]
- Ryan LA, Dal Bello F, Czerny M, Koehler P, Arendt EK. Quantification of phenyllactic acid in wheat sourdough using high resolution gas chromatography–mass spectrometry. J Agric Food Chem. 2009;57(3):1060–1064. doi: 10.1021/jf802578e. [DOI] [PubMed] [Google Scholar]
- Sasser M (2006) Bacterial identification by gas chromatographic analysis of Fatty Acids Methyl Esters (GC-FAME)”, MIDI. Technical Note #101
- Seng P, Abat C, Rolain JM, Colson P, Lagier J-C, Gouriet F, Fournier Pierre Edouard, Drancourt Michel, La Scola Bernard, Raoult Didier. Identification of rare pathogenic bacteria in a clinical microbiology laboratory: impact of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(7):2182–2194. doi: 10.1128/JCM.00492-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152. [Google Scholar]
- Tanasupawat S, Namwong S, Kudo T, Itoh T. Identification of halophilic bacteria from fish sauce (nam-pla) in Thailand. J Cult Collect. 2009;6:69–75. [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genomoe-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall BJ, Tomlinson GA, Hochstein LI. Transfer of Halobacterium denitrificans (Tomlinson, Jahnke, and Hochstein) to the genus Haloferax as Haloferax denitrificans comb. nov. Int J Syst Bacteriol. 1989;39(3):359–360. doi: 10.1099/00207713-39-3-359. [DOI] [PubMed] [Google Scholar]
- Torreblanca M, Rodriquez-Valera F, Juez G, Ventosa A, Kamekura M, Kates M. Classification of non-alkaliphilic halobacteria based on numerical taxonomy and polar lipid composition, and description of Haloarcula gen. nov. and Haloferax gen. nov. Syst Appl Microbiol. 1986;8:89–99. doi: 10.1016/S0723-2020(86)80155-2. [DOI] [Google Scholar]
- Welker M, Moore ER. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol. 2011;34(1):2–11. doi: 10.1016/j.syapm.2010.11.013. [DOI] [PubMed] [Google Scholar]
- Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]