Abstract
There are more than 2300 genes that are predominantly expressed in mouse testes. The role of hundreds of these genes has been studied in mouse spermatogenesis but still there are many genes whose function is unknown. Gene knockout (KO) strategy in mice is widely used for in vivo study of gene function. The present study was designed to explore the function of the four genes: Tex37, Ccdc73, Prss55 and Nxt2, which were evolutionarily conserved in eutherians. We found that these genes had a testis-enriched expression pattern in mice except Nxt2. We knocked out these genes by CRISPR/Cas9 individually and found that all the KO mice had normal fertility with no detectable difference in testis/body weight ratios, epididymal sperm counts, as well as testicular and epididymal histology from wild type mice. Although these genes are evolutionarily conserved in eutherians including human and mouse, they are not individually essential for spermatogenesis, testis development and male fertility in mice in laboratory conditions. Our report of these fertile KO data could avoid the repetition and duplication of efforts which will help in prioritizing efforts to focus on genes that are indispensable for male reproduction.
Introduction
The transmission of male heritance occurs through a highly-sophisticated process called spermatogenesis. During mammalian spermatogenesis, spermatogonia undergo mitosis to maintain their population and produce primary spermatocytes (2n). It is followed by the meiotic phase in which the haploid spermatids (n) are produced by two consecutive divisions. The spermatids undergo the process of spermiogenesis forming mature spermatozoa1. All these stages of spermatogenesis are spatio-temporally well-regulated and thousands of genes are involved in its successful completion.
It has been indicated that over 2300 genes are predominantly expressed in testes2. Most of these genes are believed to be essential for spermatogenesis or sperm function but the roles of many of these are still unknown. To study the specific function of such testis-enriched mammalian genes, gene knockout (KO) strategies are the efficient tools which have been used extensively3–5. The functional roles of more than 400 genes have been elaborated, with most of them being indispensable for male fertility6–13. However, a small number of genes have been studied to be dispensable for spermatogenesis and male fertility even though they have predominant testicular expression14,15. Indeed, the infertile KO mice are always reported but fertile KO mice are only published when the gene is well known.
Recently, Miyata et al. used various genome editing techniques to knockout 54 evolutionarily conserved and testis-enriched mouse genes. They did not find any essential roles of these genes in male reproduction15. They elaborated the importance of disseminating the fertile KO mice data to scientific community and academia, which can save valuable resources by preventing the same KO recapitulation15. Hence, it is vital to publicize such important data which would help researchers in prioritizing energies to focus on genes essential for male reproduction.
Here we selected five human genes (TEX37, CCDC73, PRSS55, LYZL11 and NXT2) which have predominant testicular expression and are evolutionarily conserved in eutherians. We knocked out these genes using CRISPR/Cas9 system in mice. However, during the course of our work, Lyzl1 KO mice were reported by Miyata et al.15. As the deletion region of our Lyzl1−/− mice (c.[188_206del; 210_242del]/c.[188_206del; 210_242del]) was different from those reported by Miyata et al. (c.21_34del/c.21_34del)15, therefore, we still included the generation and analysis of Lyzl1−/− mice as independent confirmation of the Miyata et al. data in our study. We did not observe any obvious defects in spermatogenesis and fertility in all these KO mice. Thus, our study revealed that, these selected genes are not individually vital for mouse spermatogenesis and fertility.
Results
Identification and phylogeny of the testis-enriched genes
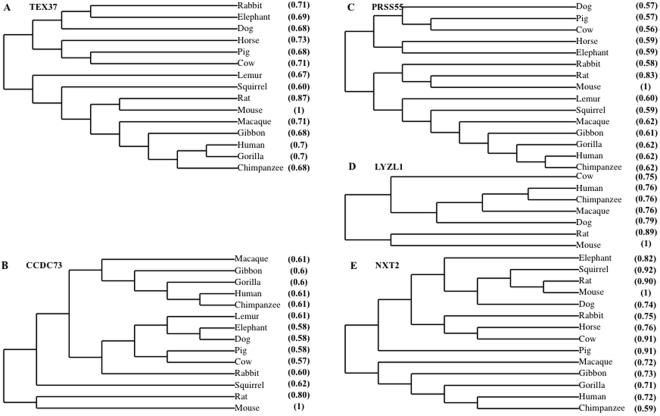
Human genes with predominant testicular expression and conserved open reading frames (ORFs) in mouse were identified by NCBI searches (Table S1). We selected 5 human genes: TEX37, CCDC73, PRSS55, LYZL11 and NXT2 which were shown to be highly expressed in testes (Table S2). Phylogenetic analysis of their orthologs showed a higher level of sequence resemblance implying that the proteins are conserved in many of the eutherians (Fig. 1). In mouse, these selected genes also had testis-enriched expression except Nxt2 which showed expression in other tissues based on NCBI searches (Table 1). We further carried out RT-PCR to check the temporal expression of these selected genes during postnatal testis development using testicular cDNAs from mice of different ages (Fig. 2). The findings revealed that Nxt2 expression began in 1-week-old testes, which suggested its possible role in early spermatogenesis. Whereas Tex37, Ccdc73, Prss55 and Lyzl1 were initially expressed in 4-week-old testes, implying their possible role in late spermatogenesis (spermiogenesis) or fertilization.
Figure 1.
Conservation of selected genes in eutherians. Multiple proteins sequence alignments were performed by T-coffee. Phylogenetic trees were constructed using online database TreeDyn from multiple protein sequence alignment36–40. Phylogeny of (A) Tex37, (B) Ccdc73, (C) Prss55, (D) Lyzl1, and (E) Nxt2. Parentheses show percent identity to reference sequence (mouse:1).
Table 1.
Expression of the selected genes in mouse tissues.
| Tissue | Tex37 (RPKM) | Ccdc73 (RPKM) | Prss55 (RPKM) | Lyzl1 (RPKM) | Nxt2 (RPKM) |
|---|---|---|---|---|---|
| Brain | 0.00 | 1.87 | 0.00 | 0.00 | 1.98 |
| Heart | 0.00 | 0.22 | 0.00 | 0.00 | 1.08 |
| Intestine large | 0.00 | 0.05 | 0.00 | 0.00 | 1.69 |
| Intestine small | 0.00 | 0.00 | 0.00 | 0.00 | 0.13 |
| Kidney | 0.00 | 0.12 | 0.00 | 0.00 | 4.49 |
| Liver | 0.00 | 0.05 | 0.00 | 0.00 | 1.65 |
| Lung | 0.00 | 0.09 | 0.00 | 0.00 | 1.25 |
| Mammary gland | 0.00 | 0.07 | 0.00 | 0.00 | 0.89 |
| Ovary | 0.00 | 0.06 | 0.00 | 0.00 | 0.20 |
| Placenta | 0.00 | 0.64 | 0.00 | 0.00 | 1.07 |
| Spleen | 0.00 | 0.06 | 0.00 | 0.00 | 0.27 |
| Stomach | 0.00 | 0.00 | 0.00 | 0.00 | 0.09 |
| Testis | 105.01 | 80.39 | 127.78 | 49.15 | 0.56 |
RPKM: Reads per kilo base per million base mapped reads.
Figure 2.
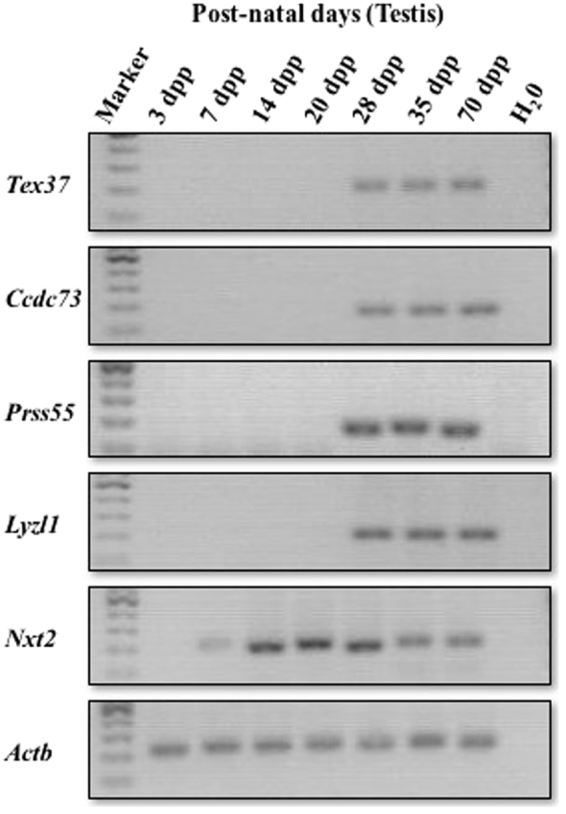
mRNA expression of the selected genes. Postnatal temporal expression of genes in testes of 3, 7, 14, 20, 28, 35 and 70-dpp-old mice was analyzed by RT-PCR. Actb was used as positive control. H20 was used as negative control. Cropped gels are shown here. Full-length gels are provided for review in the supplementary Figure S2a.
Generation of KO mice
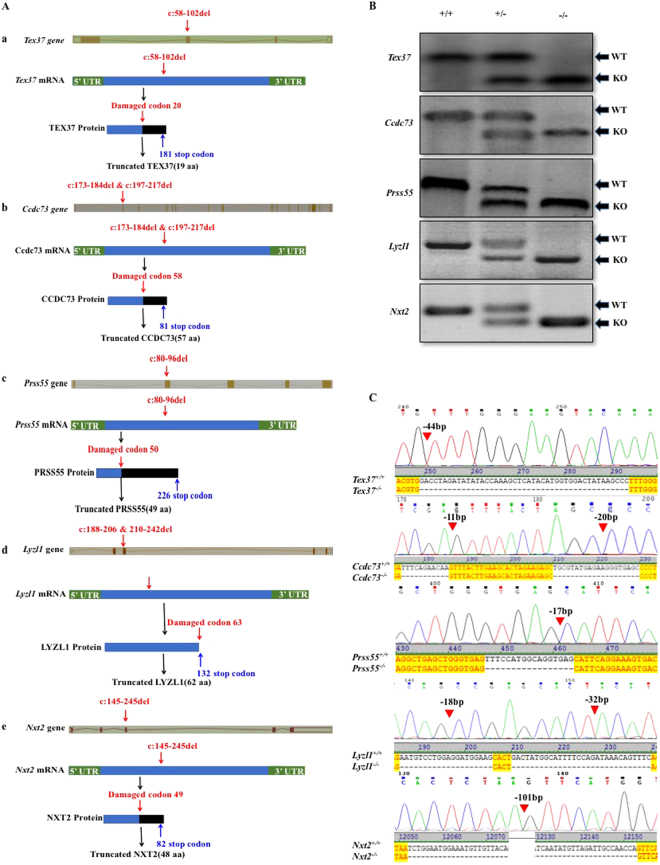
To investigate the function of these 5 genes in a short span of time, KO mice were produced by CRISPR/Cas9 technique. To be noted, Lyzl1 KO was previously reported by Miyata et al.15. Homozygous mice having large deletions forming frameshift mutations, were obtained by selective breeding (Fig. 3A). PCR and Sanger sequencing results confirmed the deletions of targeted regions (Fig. 3B,C). All the KO mouse lines exhibited normal development.
Figure 3.
Knockout strategy and genotyping of mutant mice. (A) Schematic strategies for the generation of KO mice using CRISPR/Cas9. 44 bp of Tex37 (a), 11 bp and 20 bp of Ccdc73 (b) and 17 bp of Prss55 (c) were deleted from Exon 2. While, 18 bp and 32 bp of Lyzl1 (d) and 101 bp of Nxt2 (e) were deleted from exon 3. (B) Genotype of each KO mouse was confirmed with PCR. (C) Representative Sanger sequence image for the verification of each KO mouse. Red arrow heads above the chromatograms and dashes in aligned cds sequences show the deletions.
Normal fertility and sperm count of the KO mice
After the confirmation of successful gene deletions, we analyzed the fertility of each KO male by breeding it with wild type (WT) females. The mating behavior of the KO mice was similar to that of WT with 100% fertility. The average number of litters per male/month and the average number of pups per litter produced by WT and each KO male were comparable (Table 2). Next, the testes from adult WT and each KO mouse were inspected for morphology and testis/body weight ratio. The examination revealed no significant difference between the WT and each KO group (Fig. 4A,B). Similarly, no significant difference was observed for sperm counts between WT and each KO group (Fig. 4C). Finally, histological sections from WT and each KO testis and epididymides were assessed. The examination illustrated intact seminiferous tubules with normal spermatogenesis having spermatogenic cells from spermatogonia to spermatozoa, and abundant spermatozoa in epididymides of WT and KO mice (Fig. 4D). Altogether these results reflected that complete knockout of Tex37, Ccdc73, Prss55, Lyzl1 and Nxt2, which are testis-expressed genes, have no detectable effects on spermatogenesis and fertility of mice under normal laboratory conditions.
Table 2.
Fertility assay.
| Genotype | Mating period (months) | No. of fertile males (%) | Average litters/male/month | Average pups/litter |
|---|---|---|---|---|
| WT | 4 | 5 (100) | 1.94 ± 0.24 | 7.52 ± 1.03 |
| Tex37 −/− | 2 | 2 (100) | 1.75 ± 0.36NS | 7.71 ± 2.36NS |
| Ccdc73 −/− | 2 | 3 (100) | 1.83 ± 0.29NS | 6.82 ± 1.54NS |
| Prss55 −/− | 4 | 4 (100) | 1.94 ± 0.13NS | 7.68 ± 1.01NS |
| Lyzl1 −/− | 3 | 3 (100) | 1.56 ± 0.19NS | 6.86 ± 2.32NS |
| Nxt2 −/− | 2 | 3 (100) | 1.83 ± 0.29NS | 7.45 ± 1.29NS |
Each male was bred with two females. WT, wild type. Student’s t test was performed for average litters/male/month and average pups per litter between WT and each knockout mouse group. NS, no significant difference. Data are presented as mean ± SD.
Figure 4.

Fertility and spermatogenesis of the KO mice. (A) Representative images of testes from 70-dpp-old WT and KO mice. Scale bars, 2 mm. (B) Testis/body weight ratio of 70-dpp-old WT and KO mice. Error bars represent SD. n, the number of animals. NS, no significant difference, Student’s t-test was performed between WT and each KO mouse group for testis/body weight ratio. (C) Sperm count of 70-dpp-old WT and KO mice. Error bars represent SD. n, the number of animals. NS, no significant difference, Student’s t-test was performed between WT and each KO mouse group for sperm count. (D) H&E staining of testes and epididymides caput and cauda from 70-dpp-old WT and KO mice. Scale bars, 100μm. The data shown is representative images from at least three mice.
Discussion
According to the report of Schultz et al., 2300 mouse genes have predominant testicular expression2. Extensive work has been done on genomic and transcriptomic levels to determine the function of these genes. Still, the role of many of these genes needs to be elaborated, we thus selected the four mouse genes expressed in testis (Tex37, Ccdc73, Prss55 and Nxt2) for the study of their functional roles in mouse spermatogenesis and fertility.
The five genes have conserved ORFs both in human and mouse (along with some other eutherians) with at least 75% identity in coding nucleotide and 60% identity in amino acid sequences (Table S1). These similarities suggested critical roles played by these genes during spermatogenesis and male fertility. From NCBI, we checked the expression of these genes in various mouse tissues which showed that all the genes had testis-enriched expression except Nxt2 which have expression in other mouse tissues (Table 1). It has been reported that deficiency of the genes with similar expression profile, such as Zmym3, Ku70, Fam46d, Pdha2, Tex101 and Spata19, etc. always resulted in spermatogenic problems leading to male infertility, thus validating the essential roles of these genes16–18.
Therefore, to figure out the functional roles of the selected genes, we generated homozygous knockout mice by CRISPR/Cas9. Using similar procedures by Miyata et al.15, we confirmed the disruption of targeted genes by PCR genotyping and Sanger sequencing (Fig. 3). The antibody was available only for TEX37, hence, we could only check TEX37 at protein level in the WT and Tex37 KO mice. The examination further verified the successful deletion of targeted gene (Supplementary figure S1). Indeed, the absence of any alternate translation initiation site in the ORFs of disrupted genes excluded the possibility of any functional proteins produced from alternate translation initiation sites19. Moreover, our deletions were large enough that truncated protein, if formed, would be non-functional (Fig. 3C).
To determine the roles of each gene in male fertility, we compared the number (average litters/male/month) and size (average pups per litter) of litters produced by each KO group with those in WT males and found no significant difference. The Lyzl1 KO mice have also been reported by Miyata et al. with their number and size of litters being comparable with the controls15, which is consistent with our observations. Although some KO mice with normal fertility but decreased testicular size and sperm count have been reported20–22, our KO mice were not only fertile but also exhibited normal testicular size and sperm count. The possible reasons for the lack of obvious phenotypes in these KO mice might be functional redundancy. In current scenario, deficiency of these five targeted genes would be compensated by other factors23. For example, there are several members in Ccdc family, thus the insufficiency of Ccdc73 could be counteracted by other members, such as Ccdc42, which is necessary for proper sperm development24. Likewise, we determined that Tex37 KO males are fertile, possibly its paralogs, Tex11 and Tex14, may compensate its deficiency25,26. Similarly, Nxt1 has testis enriched expression, presenting 75% of its amino acid sequence identity with NXT2, which implies that Nxt1 might compensate for Nxt2 deficiency. Thus, future studies on double/triple knockout animals for these genes with their paralogs may further enhance the understanding of their roles in testes and male fertility.
To be noted, we only analyzed the phenotype in normal laboratory conditions, we cannot exclude the possibility that these genes may be functional in some stimulated condition. Additionally, though all the KO mouse lines apparently exhibited normal development, some of these genes have expression at basal/low level in tissue(s) other than testis, therefore, the possible existence of unnoticed phenotype(s) cannot be excluded. Thus, we could only conclude that Tex37, Ccdc73, Prss55, Lyzl1 and Nxt2 are dispensable for fertility of male mice in the same genetic backgrounds under normal laboratory conditions.
In summary, we generated KO mice of the genes Tex37, Ccdc73, Prss55, Lyzl1 and Nxt2 which have testis-enriched expression in human and are conserved across most of the eutherian species. These KO mice exhibited normal fertility, and testicular development and function. As the fertile Lyzl1 KO mice have been reported previously by Miyata et al.15, our results further identified four new genes role in spermatogenesis. These results not only indicate that these genes do not play a prominent role in mouse fertility, but also help scientific community, reproductive biologists and academia to focus on genes indispensable for testis development and male fertility.
Materials and Methods
RNA extraction and RT-PCR
We performed RNA extraction and RT-PCR as explained previously27,28. TRIzol reagents (Takara, 9109) were used for the extraction of total RNAs. cDNAs were synthesized by using the PrimeScript RT kit (TaKaRa, RR047A) from total RNAs as described in manufacturer’s procedure. EasyTaq DNA Polymerase (TransGen Biotech, AP111) was used to perform RT-PCR with the following cycle conditions: 94 °C for 5 minutes, following 35 cycles; each at 98 °C for 15 seconds, 53 °C for 30 seconds, 72 °C for 20 seconds and finally, 72 °C for 2 minutes. RT-PCR was performed with total volume of 30 μl PCR mix by adding 15 μl of 10xEasy Taq DNA Polymerase buffer (Trans, AP111), 1.8 μl of 2.5 mM dNTPs (Trans, AD101), 0.8 μl of reverse and forward PCR primers (10 μM), 0.3 μl of Easy Taq DNA Polymerase (Trans, AP111) and 2 μl of template cDNA. RT-PCR for the selected genes and Actb (used as positive control) was performed simultaneously and gel electrophoresis was run in parallel.
Generation of KO mice
KO mice were produced by one-cell embryo injection using the CRISPR/Cas9 genome editing strategy as reported29. The second or third exon of genes were targeted by two guide RNAs (Table S3). We designed single guide RNAs (sgRNAs) targeting a specific exon as described previously and selected these sgRNAs on the basis of rare off-target hits using computational predictions by CRISPRdirect (https://crispr.dbcls.jp) or the Bowtie software30–32. After in vitro transcription of sgRNAs, they were co-injected with Cas9 mRNA into zygotes of B6D2F1 (C57BL/6 × DBA/2 J) mice and then embryos were transferred to pseudo pregnant ICR females30. After Sanger sequencing verification, the male and female founders carrying a heterozygous (+/−) deletion mutation were inbred to produce homozygous (−/−) mice. These mice were fed with food and ddH2O ad libitum and held in specified photoperiod (lights on 08:00–20:00) in the laboratory animal center of University of Science and Technology of China (USTC). All the procedures and analyses on laboratory animals were conducted according to the institutional guidelines which were approved by Institutional Animal Care Committee of USTC.
Fertility test
For fertility testing, each 10 to 12-week-old WT or KO male was separately housed with two sexually mature WT C57BL/6 females for indicated mating periods (Table 2). The number of litters and pups from each pregnant female was recorded at birth.
Hematoxylin and eosin (H&E) staining
The 70-day-old WT and KO mice were euthanized by cervical dislocation. After the removal, testes and epididymides were instantly fixed overnight in Bouin’s solution. The tissues embedding were carried out in paraffin for block formation. Tissue sections were prepared by microtome and subsequently H&E staining was performed as described previously33. The experiments were repeated thrice by using samples from different sets of mouse testes. To keep the inter-experimental deviations minimum, all the procedures on WT and knockout testes were carried out concurrently. Digital Nikon DS-Ri1 camera installed on a Nikon Eclipse 80i microscope was used to capture images.
Sperm counting
For sperm counting, 70-day-old mice were euthanized, their epididymides were isolated and followed by multiple incisions. Sperm were released in 1 ml buffer after incubation of these epididymides for 30 minutes at 37 °C in a CO2 (5%) incubator. This buffer comprised of NaCl (75 mM), EDTA (24 mM), and 0.4% bovine serum albumin (Sigma, A2058). Sperm were collected from this buffer solution by nylon-mesh filtration. Subsequently hemocytometer was used for sperm counting as reported previously33.
Western blot
Testicular lysates were prepared in lysis buffer containing NaCl (300 mM), EDTA (5 mM), Tris/HCl (50 mM) with pH of 7.4, Triton X-100 (1%) and protease inhibitors (Roche, 04693116001, Basel, Switzerland). Western blot was then performed followed by band quantification as outlined34,35. Primary antibodies for TEX37 (1:500; Protein tech, 25464-1-AP, USA) and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:1000; Millipore, MAB374, MA, USA) were used.
Electronic supplementary material
Acknowledgements
This work was supported by the National Key Research and Developmental Program of China (2016YFC1000600), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), the National Natural Science Foundation of China (31630050, 31371519 and 31501199) and CAS-TWAS President’s PhD Fellowship Program.
Author Contributions
Conceived and designed the experiments: Q.S., M.K., N.J. and X.J. Performed the experiments: M.K., L.J., H.M.J.H., R.K., T.L., Q.T., X.Z., H.Y. and C.Y. Analyzed the data: M.K., X.J., A.A. and N.J. Paper wrote up: Q.S. and M.K. Modification of the manuscript: X.J., T.K. and N.J.
Competing Interests
The authors declare no competing interests.
Footnotes
Manan Khan and Nazish Jabeen contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23176-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaohua Jiang, Email: biojxh@ustc.edu.cn.
Qinghua Shi, Email: qshi@ustc.edu.cn.
References
- 1.Chocu S, Calvel P, Rolland AD, Pineau C. Spermatogenesis in mammals: proteomic insights. Systems biology in reproductive medicine. 2012;58:179–190. doi: 10.3109/19396368.2012.691943. [DOI] [PubMed] [Google Scholar]
- 2.Schultz N, Hamra FK, Garbers DL. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proceedings of the National Academy of Sciences. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzuk, M. M. & Lamb, D. J. Genetic dissection of mammalian fertility pathways. Translocations 45, 46XY (2002). [DOI] [PubMed]
- 4.O’Bryan, M. K. & de Kretser, D. Mouse models for genes involved in impaired spermatogenesis. Int J Androl 29, 76–89; discussion 105–108, 10.1111/j.1365-2605.2005.00614.x (2006). [DOI] [PubMed]
- 5.Jiang X-H, et al. Blood-testis barrier and spermatogenesis: lessons from genetically-modified mice. Asian journal of andrology. 2014;16:572. doi: 10.4103/1008-682X.125401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenbaum MP, et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4982–4987. doi: 10.1073/pnas.0505123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kherraf ZE, et al. SPINK2 deficiency causes infertility by inducing sperm defects in heterozygotes and azoospermia in homozygotes. EMBO Mol Med. 2017;9:1132–1149. doi: 10.15252/emmm.201607461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khelifa MB, et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. The American Journal of Human Genetics. 2014;94:95–104. doi: 10.1016/j.ajhg.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takasaki N, et al. A heterozygous mutation of GALNTL5 affects male infertility with impairment of sperm motility. Proc Natl Acad Sci USA. 2014;111:1120–1125. doi: 10.1073/pnas.1310777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ayhan O, et al. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet. 2014;51:239–244. doi: 10.1136/jmedgenet-2013-102102. [DOI] [PubMed] [Google Scholar]
- 11.Öllinger R, et al. Deletion of the pluripotency-associated Tex19. 1 gene causes activation of endogenous retroviruses and defective spermatogenesis in mice. PLoS genetics. 2008;4:e1000199. doi: 10.1371/journal.pgen.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun R, et al. Lyzl4, a novel mouse sperm-related protein, is involved in fertilization. Acta Biochim Biophys Sin. 2011;43:346–353. doi: 10.1093/abbs/gmr017. [DOI] [PubMed] [Google Scholar]
- 13.Scarman AL, et al. Organization and chromosomal localization of the murine Testisin gene encoding a serine protease temporally expressed during spermatogenesis. The FEBS Journal. 2001;268:1250–1258. doi: 10.1046/j.1432-1327.2001.01986.x. [DOI] [PubMed] [Google Scholar]
- 14.Okabe M. Mechanism of fertilization: a modern view. Exp Anim. 2014;63:357–365. doi: 10.1538/expanim.14-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyata H, et al. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proceedings of the National Academy of Sciences. 2016;113:7704–7710. doi: 10.1073/pnas.1608458113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmed EA, Sfeir A, Takai H, Scherthan H. Ku70 and non-homologous end joining protect testicular cells from DNA damage. J Cell Sci. 2013;126:3095–3104. doi: 10.1242/jcs.122788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, et al. Comparative and functional analysis of testis-specific genes. Biol Pharm Bull. 2011;34:28–35. doi: 10.1248/bpb.34.28. [DOI] [PubMed] [Google Scholar]
- 18.Hu X, et al. Gene knockout of Zmym3 in mice arrests spermatogenesis at meiotic metaphase with defects in spindle assembly checkpoint. Cell Death & Disease. 2017;8:e2910. doi: 10.1038/cddis.2017.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Klerk E. & T. Hoen, P. A. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015;31:128–139. doi: 10.1016/j.tig.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Schürmann A, et al. Reduced sperm count and normal fertility in male mice with targeted disruption of the ADP-ribosylation factor-like 4 (Arl4) gene. Molecular and cellular biology. 2002;22:2761–2768. doi: 10.1128/MCB.22.8.2761-2768.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudson DF, et al. Centromere protein B null mice are mitotically and meiotically normal but have lower body and testis weights. The Journal of cell biology. 1998;141:309–319. doi: 10.1083/jcb.141.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar TR, Wang Y, Lu N, Matzuk MM. Follicle stimulating hormone is required for ovarian follicle maturation but not male fertility. Nature genetics. 1997;15:201–204. doi: 10.1038/ng0297-201. [DOI] [PubMed] [Google Scholar]
- 23.Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod. 2004;71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 24.Pasek RC, et al. Coiled-coil domain containing 42 (Ccdc42) is necessary for proper sperm development and male fertility in the mouse. Dev Biol. 2016;412:208–218. doi: 10.1016/j.ydbio.2016.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adelman CA, Petrini JH. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet. 2008;4:e1000042. doi: 10.1371/journal.pgen.1000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenbaum MP, et al. TEX14 is essential for intercellular bridges and fertility in male mice. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:4982–4987. doi: 10.1073/pnas.0505123103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin, S. et al. Histone acetyltransferase KAT8 is essential for mouse oocyte development by regulating ROS levels. Development (2017). [DOI] [PMC free article] [PubMed]
- 28.Jiang, X. et al. Specific deficiency of Plzf paralog, Zbtb20, in Sertoli cells does not affect spermatogenesis and fertility in mice. 4, 7062, 10.1038/srep07062, https://www.nature.com/articles/srep07062#supplementary-information (2014). [DOI] [PMC free article] [PubMed]
- 29.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen B, et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat Methods. 2014;11:399–402. doi: 10.1038/nmeth.2857. [DOI] [PubMed] [Google Scholar]
- 31.Mashiko D, et al. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep. 2013;3:3355. doi: 10.1038/srep03355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang X, et al. Specific deletion of Cdh2 in Sertoli cells leads to altered meiotic progression and subfertility of mice. Biol Reprod. 2015;92:79. doi: 10.1095/biolreprod.114.126334. [DOI] [PubMed] [Google Scholar]
- 34.Zhang H, et al. microRNA 376a regulates follicle assembly by targeting Pcna in fetal and neonatal mouse ovaries. Reproduction. 2014;148:43–54. doi: 10.1530/REP-13-0508. [DOI] [PubMed] [Google Scholar]
- 35.Yi Q, et al. p53 dependent centrosome clustering prevents multipolar mitosis in tetraploid cells. PLoS One. 2011;6:e27304. doi: 10.1371/journal.pone.0027304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dereeper A, et al. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic acids research. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chevenet F, Brun C, Bañuls A-L, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dereeper A, Audic S, Claverie JM, Blanc G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 2010;10:8. doi: 10.1186/1471-2148-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavrent’eva, I., Antipova, A., Semenov, A. & Bichurina, M. Genotyping of parvovirus B19 isolates circulating in Northwestern Federal District of Russia. Zhurnal mikrobiologii, epidemiologii, i immunobiologii, 36–43 (2013). [PubMed]
- 40.Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.