Summary
Heterochromatin plays a central role in the process of immune evasion, pathogenesis, and transmission of the malaria parasite Plasmodium falciparum during blood stage infection. Here, we use ChIP sequencing to demonstrate that sporozoites from mosquito salivary glands expand heterochromatin at subtelomeric regions to silence blood-stage-specific genes. Our data also revealed that heterochromatin enrichment is predictive of the transcription status of clonally variant genes members that mediate cytoadhesion in blood stage parasites. A specific member (here called NF54varsporo) of the var gene family remains euchromatic, and the resultant PfEMP1 (NF54_SpzPfEMP1) is expressed at the sporozoite surface. NF54_SpzPfEMP1-specific antibodies efficiently block hepatocyte infection in a strain-specific manner. Furthermore, human volunteers immunized with infective sporozoites developed antibodies against NF54_SpzPfEMP1. Overall, we show that the epigenetic signature of var genes is reset in mosquito stages. Moreover, the identification of a strain-specific sporozoite PfEMP1 is highly relevant for vaccine design based on sporozoites.
Keywords: malaria, P. falciparum, sporozoite, var genes, epigenetic, PfHP1, heterochromatin, PfEMP1, hepatocyte infection
Graphical Abstract
Highlights
-
•
Sporozoites expand subtelomeric heterochromatin to silence blood-stage-specific genes
-
•
A strain-specific PfEMP1 is expressed on the surface of sporozoites
-
•
NF54_SpzPfEMP1 is immunogenic in sporozoite-infected human volunteers
-
•
Antibodies against NF54_SpzPfEMP1 block sporozoite infection of hepatocytes
P. falciparum PfEMP1 surface adhesion proteins mediate immune evasion and pathogenesis during asexual blood stage development. Zanghì et al. find that a strain-specific member of PfEMP1 is expressed at the sporozoite surface. Antibodies against this protein inhibit sporozoite invasion of hepatocytes in a strain-specific manner.
Introduction
The most devastating form of human malaria is caused by the protozoan parasite P. falciparum, with more than 200 million people infected annually and an estimated 445,000 deaths in 2017 (World Health Organization, 2017). Malaria is transmitted by the bite of an infected Anopheles mosquito, which harbors sporozoites in its salivary glands. From the point of injection into the skin, sporozoites migrate via blood vessels to the liver, cross the sinusoidal cell layer separating the blood and the liver, and finally invade hepatocytes where asexual reproduction leads to the release of thousands of merozoites into the bloodstream (Prudêncio and Mota, 2007). Merozoites infect mature red blood cells and, through asexual reproduction, generate daughter merozoites to initiate a new infective cycle. The persistence and pathogenesis of P. falciparum during blood stage proliferation relies on the exclusive and successive expression of variant surface adhesion molecules, PfEMP1, expressed at the membranes of infected red blood cells (iRBCs) and mediate cytoadhesion in the microvasculature (Smith, 2014). This immune evasion mechanism, termed “antigenic variation,” depends on monoallelic expression of one of approximately 60 var genes that encode PfEMP1 proteins.
Different epigenetic factors lead to the default transcriptional silencing of all but one var gene via the establishment of facultative heterochromatin (Guizetti and Scherf, 2013). Heterochromatin protein 1 (PfHP1) is a key regulator of facultative heterochromatin in P. falciparum (Flueck et al., 2009, Pérez-Toledo et al., 2009), and conditional depletion of PfHP1 disrupts transcriptional repression of var genes as well as the master regulator of sexual commitment, PfAP2-G (Brancucci et al., 2014). In addition, transcription of a single var gene is associated with antisense transcription of a long non-coding RNA (lncRNA) originating from its intron, a conserved feature shared by all members of the var family (Ralph et al., 2005, Jiang et al., 2013, Amit-Avraham et al., 2015). Thus, the parasite uses multiple layers of epigenetic regulation to ensure monoallelic expression of variant gene families, which creates phenotypic plasticity in genetically identical parasites during blood stage development (Lopez-Rubio et al., 2009, Rovira-Graells et al., 2012).
Although variegated gene expression appears to have evolved as a survival strategy to promote prolonged blood stage infections in humans, it is unknown whether heterochromatin-mediated control of variant gene families is important in other parasite stages, such as the sporozoite stage. Sporozoites have been successfully used to provide immune protection to human volunteers and are a key stage to target for malaria vaccine development (Richie et al., 2015).
To study the organization of heterochromatin in sporozoites, which are relatively low in abundance in the mosquito salivary glands, we developed a robust, low-cell-input chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq) protocol. ChIP-seq of PfHP1 revealed a remarkable organization of heterochromatin in sporozoites that differs from that observed in asexual blood stage parasites. Furthermore, our epigenetic analysis predicted the expression of a specific PfEMP1 on the surface of sporozoites. Antibodies raised against this particular PfEMP1 efficiently blocked sporozoite infection of human hepatocytes in a strain-specific manner, demonstrating a previously unknown role of the var gene family in malaria parasite transmission.
Results
Heterochromatin Islands Form Nuclear Clusters in Sporozoites
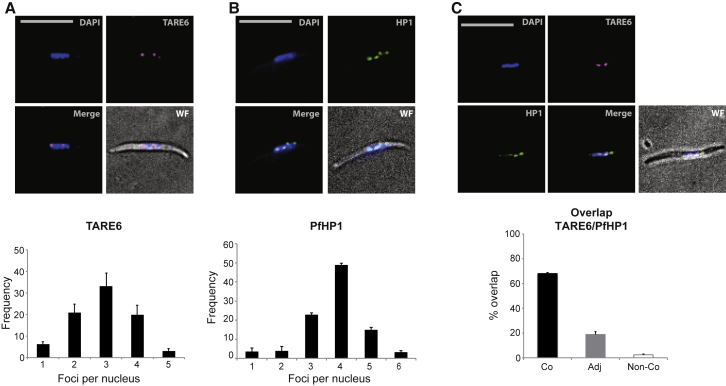
In P. falciparum blood stage parasites, the maintenance of heterochromatin islands is linked to the physical tethering of these genomic regions to the nuclear periphery, forming 4–7 perinuclear foci (Lopez-Rubio et al., 2009). PfHP1 is a major component of heterochromatin in perinuclear chromosome clusters. To determine whether a similar spatial chromosome arrangement exists in sporozoites, we performed DNA fluorescence in situ hybridization (FISH) using a probe corresponding to the conserved sub-telomeric repeat TARE6 (Telomere-Associated Repeat 6) (Figure 1A) and indirect immunofluorescene assays (IFAs) with antibodies against PfHP1 (Figure 1B). We observed an average of 2–4 TARE6-containing foci per nucleus, which localized to the nuclear periphery and overlapped with PfHP1-containing foci (Figure 1C). These data indicate a conserved spatial organization of heterochromatin in the nucleus between asexual blood stage parasites and salivary gland sporozoites.
Figure 1.
Chromosome Ends and Heterochromatin Islands Cluster at the Nuclear Periphery in P. falciparum Sporozoites
(A) DNA fluorescence in situ hybridization (FISH) of telomere distribution in sporozoites using a probe (magenta) against sub-telomeric repeat TARE6 (top) and corresponding quantification of the number of TARE6 foci per nucleus (bottom).
(B) Immunofluorescence of PfHP1 (green) in sporozoites using anti-PfHP1 antibodies (top) and corresponding quantification of the number of PfHP1 foci per nucleus (bottom).
For (A) and (B), error bars represent the 95% confidence intervals (±SEM) established for 50 sporozoites from three independent experiments.
(C) Combined DNA FISH of telomere distribution (using a TARE6 probe in magenta) and immunofluorescence of PfHP1 distribution (green) using anti-PfHP1 antibodies in sporozoites (top) and corresponding quantification of the percent overlap of TARE6 and PfHP1 foci per nucleus (bottom). Co, colocalized; Adj, adjacent; Non-Co, non-colocalized. Error bars represent SD from two independent experiments.
For (A)–(C), DNA was stained with DAPI (blue), and the wide-field (WF) image is merged with the fluorescent images. Scale bars, 5 μm.
PfHP1 Genome-wide Distribution in P. falciparum Sporozoites
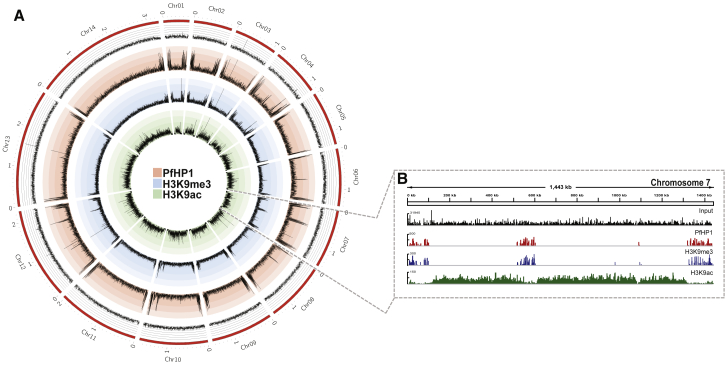
Previous studies have used ChIP to elucidate the role of histone post-translational modifications (PTMs), histone variants, and other chromatin factors in gene expression during the asexual stage of the human malaria parasite, P. falciparum (Lopez-Rubio et al., 2013). In asexual blood stages, PfHP1 and H3K9me3 regulate transcription of clonally variant virulence gene families such as var by establishing facultative heterochromatin at promoter regions and gene bodies (Lopez-Rubio et al., 2009, Flueck et al., 2009). These heterochromatic regions span approximately 100 kb, forming so-called heterochromatin islands at sub-telomeric regions and several internal chromosome clusters. Because the number of sporozoites that can be recovered from the infected mosquito salivary gland is low, we developed a robust low input ChIP-seq protocol, which allowed us to use approximately 600,000–800,000 sporozoites per immunoprecipitation (IP). We investigated the genome-wide distribution of heterochromatin by performing ChIP with highly specific antibodies against PfHP1 and the transcriptionally repressive histone H3 modification lysine 9 trimethylation (H3K9me3) (Tables S1 and S2). We observed an enrichment of PfHP1 and H3K9me3 in sub-telomeric regions of all 14 chromosomes, as well as in central chromosome regions of chromosomes 4, 6, 7, 8, and 12 (Figure 2A; Table S2). These regions contain variant gene families and show a similar heterochromatin enrichment in blood stage parasites (Lopez-Rubio et al., 2009). In contrast, ChIP-seq of the transcriptionally activating mark H3K9ac revealed its absence from these heterochromatic loci and enrichment within chromosome regions that primarily contain housekeeping genes (Figures 2A and 2B). Notably, ApiAP2-G, a member of the AP2 DNA-binding protein family that is involved in gametocyte formation, appears to be enriched in PfHP1 and H3K9me3 in sporozoites as it is in asexual blood stage parasites, suggesting a re-setting of the chromatin of this gene after gametogenesis is complete (Figure S1). We conclude that the heterochromatin organization is largely preserved in different life cycle stages.
Figure 2.
Genome-wide Distribution of Eu- and Heterochromatin in P. falciparum Sporozoites
(A) Circos plot of ChIP-seq data showing genome-wide enrichment of PfHP1 (red), H3K9me3 (blue), and H3K9ac (green) in sporozoites. Coverage plots are represented as average reads per million (RPM) over bins of 1,000 nt with a maximum y axis value of 400 for PfHP1, 500 for H3K9me3, and 400 for H3K9Ac. The 14 chromosomes are represented by the outer gray circle, with chromosome sizes given in megabases (Input, black). Statistical analysis of the biological replicates is presented in Table S1. MACS2-based peak calling analysis of PfHP1, H3K9me3, and H3K9ac enrichment in sporozoites is presented in Table S2.
(B) PfHP1, H3K9me3, and H3K9Ac enrichment across chromosome 7 in sporozoites, with genomic position indicated at the top in kilobases. Coverage plots are represented as average RPM over bins of 1,000 nt, with the maximum value of y axis indicated.
Data are representative of three or more independent experiments. See also Tables S1 and S2.
Heterochromatin Spreads into Genes Coding for Exported Blood Stage Proteins in Sporozoites
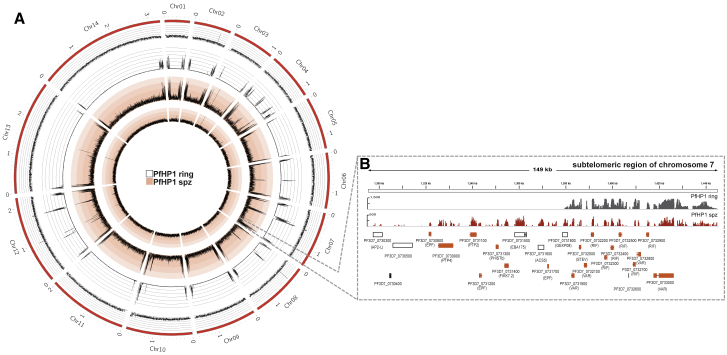
When comparing the genome-wide PfHP1 pattern in sporozoites to that in asexual blood stage parasites (Figure 3A), we observed an overall similar organization of PfHP1 islands along all 14 chromosomes. We noted, however, a significant sporozoite-specific spreading of sub-telomeric heterochromatin from nearly all chromosome ends toward central chromosomal regions (Figures 3A and 3B). These extended heterochromatic regions of approximately 50 kb are highly enriched in genes encoding parasite proteins that are typically exported to the host erythrocyte during blood stage development, remodeling the erythrocyte to accommodate the parasite’s needs (de Koning-Ward et al., 2016) (Figures 3B and S2). RNA sequencing in sporozoites showed that steady-state mRNA levels of these subtelomeric genes encoding blood-stage-exported proteins have an inverse correlation with H3K9me3/HP1 occupancy (Figure S3; Table S3), linking sporozoite heterochromatin extension to stage-specific gene silencing.
Figure 3.
Heterochromatin Islands at Chromosome Ends Are Extended in P. falciparum Sporozoites Relative to Blood Stages
(A) Circos plot of ChIP-seq data compares genome-wide PfHP1 enrichment in sporozoites (spz; red lines) and asexual blood stage parasites (ring; gray lines). Coverage plots are represented as average RPM over bins of 1,000 nt with a y-axis maximum value of 400 for PfHP1 in sporozoites and 1,500 for PfHP1 in blood stages. The 14 chromosomes are represented by the outer gray circle, with chromosome sizes given in megabases. Statistical analysis of the biological replicates is presented in Table S1. MACS2-based peak calling analysis of PfHP1, H3K9me3, and H3K9ac enrichment in sporozoites is presented in Table S2.
(B) An enlarged view of a 149-kb sub-telomeric region of the right arm of chromosome 7 shows PfHP1 enrichment in the genomes of sporozoites (spz; red) and blood stage parasites (ring; black). Genes encoding exported proteins of iRBCs are highlighted in orange below the plot. Coverage plots are represented as average RPM over bins of 1 nt, with the maximum value indicated on the y axis.
Data are representative of three or more independent experiments. See also Tables S1 and S2.
Heterochromatin Enrichment Predicts Expression of a var Gene in Sporozoites
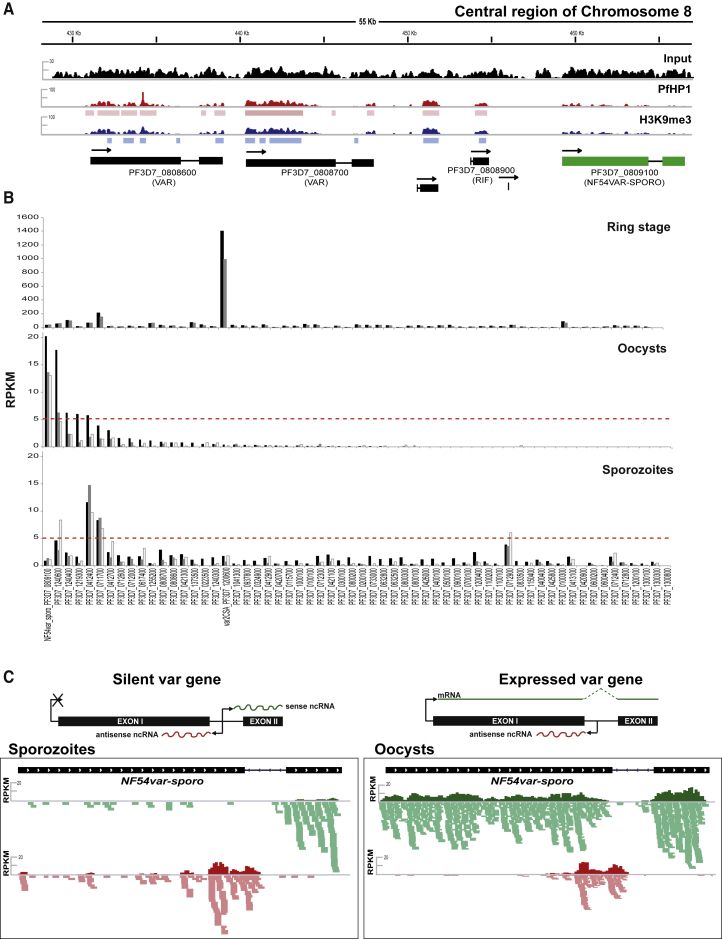
In asexual blood stage parasites, a single PfEMP1 is expressed at any given time on the erythrocyte surface, and this expression correlates with low levels of PfHP1 and H3K9me3 within the promoter and gene body of the corresponding var gene. In contrast, the silent var genes are enriched for these heterochromatic marks (Lopez-Rubio et al., 2009). We performed MACS2-based peak enrichment analysis of four independent PfHP1 (Tables S2A and S4A) and three independent H3K9me3 (Table S2B) ChIP-seq sporozoite experiments. All var genes present in the NF54 P. falciparum strain were enriched for PfHP1 and H3K9me3 (Table S4A, “Peaks”). Intriguingly, one var gene (PF3D7_0809100) consistently showed the lowest enrichment of PfHP1 within its promoter and gene body (Figure 4A; Table S4A), suggesting that the corresponding PfEMP1 may be expressed in sporozoites. PF3D7_0809100 (subsequently referred to as NF54varsporo) is located in the central region of chromosome 8, adjacent to two other var genes and two members of a second clonally variant gene family rif, all of which retain higher levels of PfHP1 and H3K9me3 (Figure 4A). In general, other clonally variant gene families, including rif, Pfmc2TM, and PfACS, which are located proximal to sub-telomeric var genes on 13 of the 14 chromosomes, appear to maintain blood-stage-like heterochromatin patterns: only two of 180 rif genes, one of 13 Pfmc2TM genes, and five of 13 PfACS genes are depleted for PfHP1 and H3K9me3 (Table S5).
Figure 4.
PfHP1 Occupancy Predicts Transcription of NF54varsporo in Sporozoites
(A) ChIP sequencing data show enrichment of PfHP1 (red) and H3K9me3 (blue) over a 55-kb genomic region of chromosome 8 (chromosome coordinates are shown at top) in P. falciparum sporozoites. The coverage of the ChIP input DNA is indicated in black, and the MACS2-based peaks (Table S2) are indicated as rectangular boxes underneath the coverage plots. Coverage plots are represented as average RPM over bins of 1,000 nt with the maximum value indicated on the y axis. Data are representative of three or more independent experiments. The var gene PF3D7_0809100, or NF54varsporo, shows low H3K9me3/PfHP1 enrichment and is highlighted in green.
(B) RNA-seq shows var gene transcription across different P. falciparum life-cycle stages: asexual-ring-stage parasites 12 hr post-invasion (two replicates at top), mosquito-derived oocysts (three replicates in the middle), and mosquito salivary gland sporozoites (three replicates at bottom). Transcripts corresponding to exon I of each var gene (shown on the x axis) are indicated as RPKM (reads per kilobase of exon per one million mapped reads), indicated on the y axis. var gene transcription was only considered when above the red dotted line. See Table S4 for more details.
(C) Each var gene has two exons that flank a conserved intron. Shown in the top panel are schematics representing transcription of var mRNA and intronic ncRNAs from silent and active var genes. The bottom panel shows stranded RNA-seq data that demonstrate NF54varsporo gene transcription in sporozoites (left) and oocysts (right). Coverage plots of steady-state RNA levels are indicated as RPKM (reads per kilobase of exon per one million mapped reads), with the mapped raw reads for sense (green) and antisense (red) transcription shown below the corresponding coverage plot.
See also Table S4.
Given the major role of PfEMP1 in parasite virulence and immune evasion (Wahlgren et al., 2017), we focused on the expression of var genes in sporozoites. We investigated NF54varsporo transcription by performing strand-specific RNA sequencing (RNA-seq) analysis of the sporozoite steady-state transcriptome (Figure S4; Table S3). As shown in Figure 4B (lower panel) and Figure 4C, we did not detect full-length mRNA transcripts for NF54varsporo in the sporozoite (Tables S4B and S4C), but only bidirectional intronic promoter activity (Figure 4C, left panel) similar to that observed for silent var genes in the late-stage asexual blood stage parasites (Ralph et al., 2005, Epp et al., 2009). Despite identifying low-level, full-length mRNAs for two other var genes, PF3D7_0711700 and PF3D7_0412400 (Figure 4B, bottom panel; Figure S5; Tables S4B and S4C), these lack antisense noncoding RNAs (ncRNAs) to exon I that has been associated with var gene transcriptional activation and monoellelic expression in asexual blood stage parasites (Figure S5, left panel; and Table S4D) (Ralph et al., 2005, Jiang et al., 2013, Amit-Avraham et al., 2015). For this reason, we consider that var genes PF3D7_0711700 and PF3D7_0412400 may not be expressed in the sporozoite stage.
Since var mRNA transcription and PfEMP1 expression show limited overlap during asexual blood stage development, it is possible that NF54varsporo is transcribed at the stage that precedes sporozoites: the oocysts in the mosquito midgut. Indeed, RNA-seq of oocysts (day 8 post-infection of the mosquito) revealed the presence of the full-length transcript of NF54varsporo, which showed the highest RPKM (reads per kilobase of exon model per million reads) value of any var gene in three biological replicates (Figures 4B, middle panel, and 4C, right panel; Tables S4B and S4C), indicating that NF54_SpzPfEMP1 could be synthesized already in late mosquito stages. We observed another full-length var transcript, PF3D7_1240600; however, in the absence of intron-derived exon I antisense ncRNA, we speculate that this prohibits protein expression (Figure S5, right panel; Tables S4C and S4D). Alternatively, the observed oocyst var transcripts are expressed in individual parasites. Sporozoites expressing NF54varsporo may have a selective advantage in the process of migration to the salivary gland, leading to, predominantly, sporozoites that express NF54_SpzPfEMP1.
In contrast, RNA-seq data from the parental ring asexual blood stage parasites detected almost no reads mapping to NF54varsporo compared to the predominantly transcribed var gene, PF3D7_1200600 (top panel of Figure 4B; Tables S4B and S4C), suggesting a reset of var gene transcription during the parasite transmission stage.
A Strain-Specific PfEMP1 Is Expressed on the Surface of Sporozoites
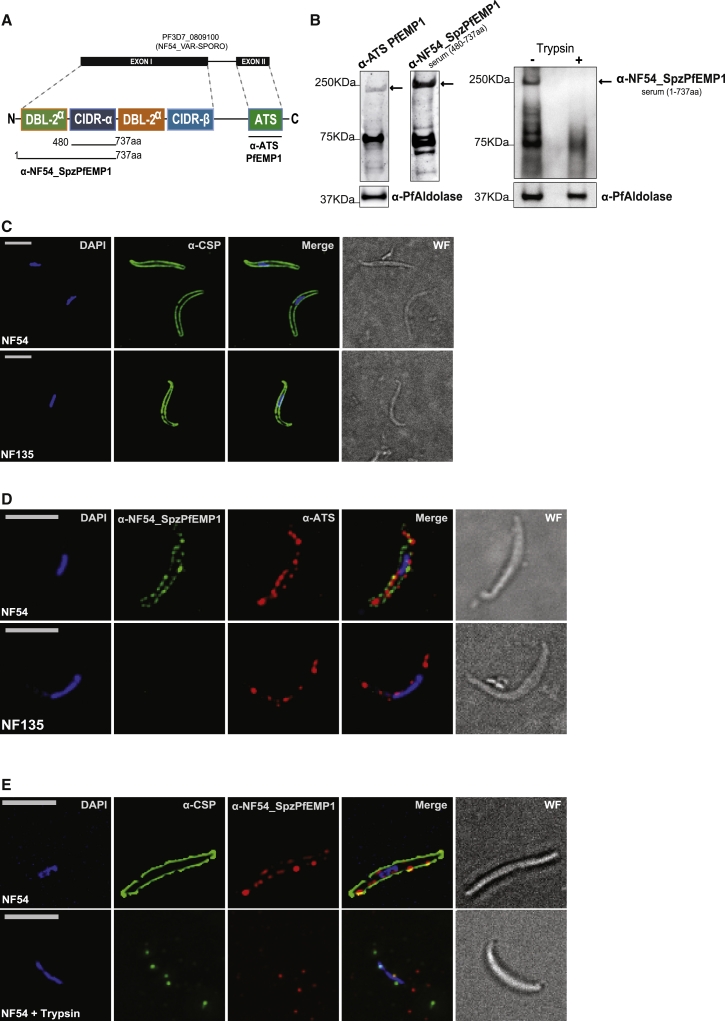
To determine whether the PfEMP1 protein encoded by NF54varsporo is synthesized in sporozoites, we prepared NF54 sporozoite extracts and performed immunoblotting assays using either an antibody against the conserved intracellular C-terminal ATS (Acid Terminal Segment) domain that reacts with all PfEMP1 proteins (Nacer et al., 2015) or specific antibodies against the extracellular variable domain of NF54_SpzPfEMP1 (Figure 5A). Anti-NF54_SpzPfEMP1 antibodies and the anti-ATS reacted with a band corresponding to the predicted molecular weight of NF54_SpzPfEMP1 (241 kDa in Figures 5B and S6A). These sera detect smaller bands of approximately 70 to 75 KDa that may represent degraded forms of PfEMP1. Furthermore, IFAs of fixed NF54 sporozoites using mouse anti-NF54_SpzPfEMP1 and ATS antibodies showed a surface membrane-like staining pattern (Figure 5D). Almost all sporozoites (>98%) react with the anti-NF54_SpzPfEMP1 antibodies. We observed a similar IFA staining pattern in six independent NF54 sporozoite preparations, supporting the selective expression of a member of the var gene family. No surface staining was observed with antibodies against the extracellular domain of the PfEMP1 encoded by var2CSA (Pf3D7_1200600) or the CIDR (cysteine-rich interdomain region) domain of an unrelated central var gene (Pf3D7_0412700), both of which retain H3K9me3/PfHP1 enrichment in sporozoites (Figure S6B; data not shown).
Figure 5.
NF54_SpzPfEMP1 Protein Is Strain Specific and Exported to the Sporozoite Surface
(A) Schematic representation of the predicted domains of the PfEMP1 encoded by PF3D7_0809100 and the corresponding antibodies against the semi-conserved ATS region and the polymorphic domains of the NF54_SpzPfEMP1.
(B) Western blot analysis of sporozoite extracts using anti-ATS or anti-NF54_SpzPfEMP1 antibodies. Anti-PfAldolase antibodies served as a positive control for protein extraction. Molecular weights are shown at the left of the blot, and PfEMP1 is indicated with an arrow.
(C) Immunofluorescence analysis of fixed sporozoites of two different strains (NF54 at top and NF135 on bottom) using anti-CSP antibodies (green).
(D) Immunofluorescence analysis of fixed and permeabilized sporozoites of two different strains (NF54 at top and NF135 on bottom) using anti-NF54_SpzPfEMP1 (green) and anti-ATS (red) antibodies.
(E) Surface immunofluorescence of live sporozoites (top) and live sporozoites after trypsin treatment (bottom) using anti-CSP (green) and anti-NF54_SpzPfEMP1 (red) antibodies.
For (C)–(E), DNA was stained with DAPI (blue), and the wide-field (WF) image is shown at the right. Scale bars, 5 μm.
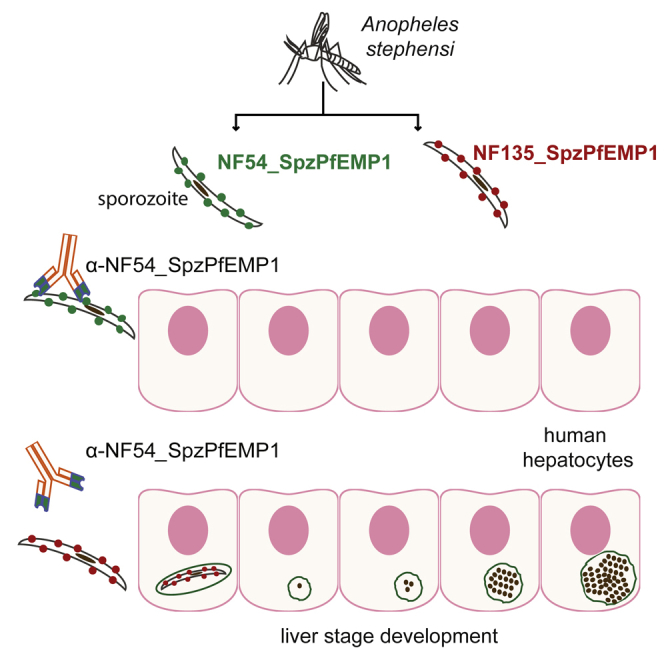
Since NF54varsporo is not conserved in other P. falciparum parasite strains (http://www.plasmodb.org) we performed IFA analysis on sporozoites of a genetically distinct strain, the Cambodian clone NF135 (Teirlinck et al., 2013). NF135 reacts with antibodies directed against the major surface antigen of sporozoites, Circumsporozoite protein (CSP) (Figure 5C). However, no cross-reactivity with anti-NF54_SpzPfEMP1 was observed. Expression of a PfEMP1 was confirmed by using anti-ATS (Figure 5D). These data suggest that an antigenically variant PfEMP1 antigen, possibly with a similar function as that of NF54_SpzPfEMP1, is expressed on the surface of NF135 P. falciparum sporozoites. Moreover, we performed blastp on the NF135 proteome (UniProt ID: UP000019114, based on the assembly deposited at GenBank [GEO: AOPS00000000], Broad Institute). No ortholog of NF54varsporo was detectable in NF135, supporting the hypothesis that anti- NF54_SpzPfEMP1 antibodies do not crossreact with NF135 sporozoites.
The IFAs on live sporozoites, with intact cell membranes, and the anti-NF54_SpzPfEMP1 antibody, but not the anti-ATS antibody, stained the surface of sporozoites (Figure 5E). Trypsin treatment of live sporozoites (50 μg/mL) abolished antibody surface reactivity in immunofluorescence and by western blot, confirming that the CIDR domain of NF54_SpzPfEMP1 is extracellular in sporozoites (Figures 5B and 5E). Together, these results provide evidence of expression of a clonally variant and strain-specific PfEMP1 protein at the surface of sporozoites with a similar orientation to that seen at the membrane of iRBCs.
To study further the specificity of the anti-NF54_SpzPfEMP1, we incubated NF54 asexual blood stage parasites, with magnetic beads coated with immunoglobulin G (IgG) of anti-NF54_SpzPfEMP1. After three rounds of selection, we obtained parasites that react (>30%) with anti-NF54_SpzPfEMP1 (Figure S6C). qRT-PCR analysis on RNA prepared from ring stage parasites before and after panning showed an enrichment for NF54varsporo transcripts of approximately 50-fold (Figure S6D). We demonstrate that the same PfEMP1 can be expressed in two distinct phases of the life cycle.
Antibodies against NF54_SpzPfEMP1 Block Sporozoite Infection of Hepatocytes
In intra-erythrocytic parasites, PfEMP1 proteins are exported across the parasite membrane and the parasitophorous vacuolar membrane to the surface of the erythrocyte, where they serve as adhesion molecules that can bind to a variety of endothelial receptors such as CD36, ICAM, etc., causing malaria pathogenesis by obstructing blood vessels in critical organs such as the brain (Wahlgren et al., 2017). Antibodies against the extracellular PfEMP1 region have been shown to block the rosetting phenotype and/or adhesion to various host endothelial receptors, such as CD36 or CSA.
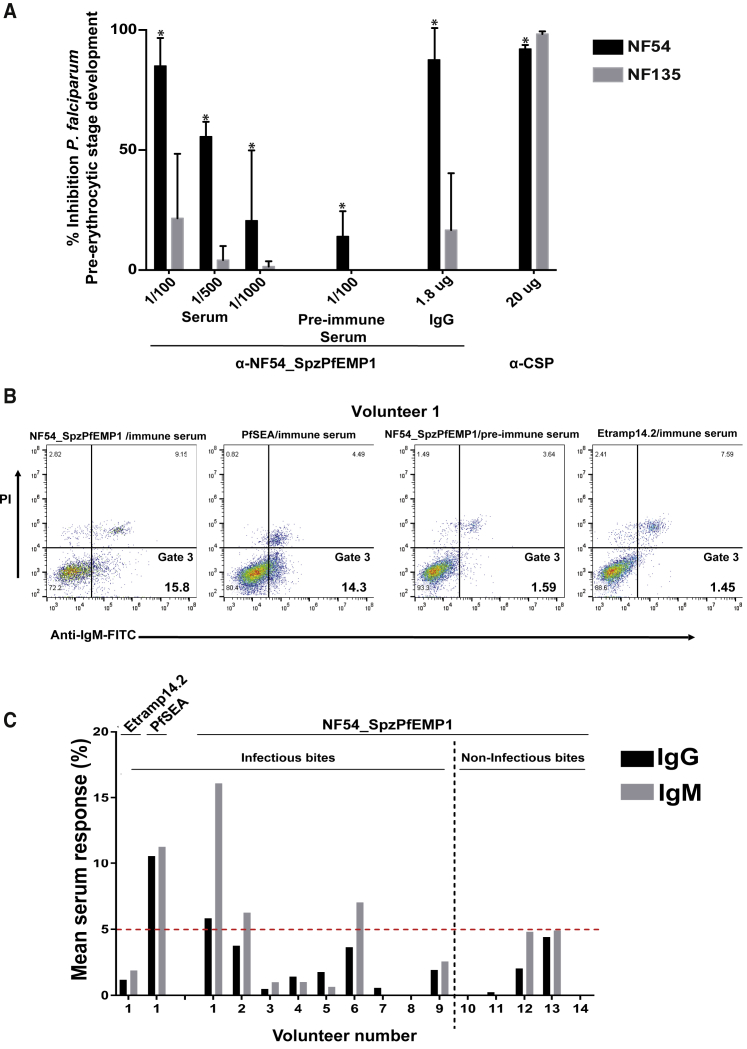
Because NF54_SpzPfEMP1 is expressed on the surface of mature sporozoites, we evaluated its potential to interact with hepatocytes, thus contributing to hepatocyte infection. We pre-incubated freshly isolated salivary gland sporozoites with either the anti-NF54_SpzPfEMP1 mouse antibody (both serum and the purified IgG) against the first 737 amino acids containing the DBL1 and CIDR1 domains (Figure 5A) or the corresponding pre-immune serum before adding the sporozoites to a primary human hepatocyte culture. After 3 days of infection, we observed that the anti-NF54_SpzPfEMP1 serum strongly reduced hepatocyte infection in a dilution-dependent manner, whereas the preimmune mouse serum showed only very low inhibition (Figure 6A). It is noteworthy that the purified anti-NF54_SpzPfEMP1 IgG (1.8 μg/mL) inhibits hepatocyte infection at a level similar to that of the anti-CSP IgG (20 μg/mL) (Figure 6A). Importantly, anti-NF54_SpzPfEMP1 did not block infection with clone NF135, which does not express NF54_SpzPfEMP1 (Figures 5D and 6A), whereas the anti-CSP antibody blocked both strains at levels >90% (Figure 6A).
Figure 6.
Antibodies against NF54_SpzPfEMP1 Inhibit Sporozoite Infection of Primary Human Hepatocytes
(A) NF54_SpzPfEMP1 antibody inhibition of hepatocyte infection of NF54 sporozoites expressing NF54_SpzPfEMP1 (black) and of NF135 sporozoites that did not express NF54_SpzPfEMP1 (gray). Data are presented as the percentage of inhibition over the untreated control after treatment with the following antibodies: anti-CSP, anti-NF54_SpzPfEMP1 immune serum, or the corresponding anti-NF54_SpzPfEMP1 pre-immune serum or purified IgG. Data represent the average of two independent biological experiments. ∗p < 0.0205, as measured using a two-way ANOVA. Error bars indicate SD.
(B and C) Detection of antibodies against NF54_SpzPfEMP1 in sera isolated from individuals immunized with infective bites and non-infective bites under chloroquine (CQ) prophylaxis. The sera were used to label HEK293 cells expressing NF54_SpzPfEMP1, schizont egress antigen (PfSEA; positive control), or Etramp14.2 (negative control), which were analyzed by flow cytometry. A second negative control used NF54_SpzPfEMP1 pre-immune sera to probe HEK293 cells expressing NF54_SpzPfEMP1.
(B) An anti-IgM (immunoglobulin M)-FITC (fluorescein isothiocyanate) signal (x axis) indicates specific binding, and a propidium iodide (PI) signal (y axis) indicates dead cells. Gate 3 consists of cells positive for IgM staining for either NF54_SpzPfEMP1, PfSEA, or Etramp.
(C) Quantification of the serum IgG (black) or IgM (gray) response from all individuals in the chloroquine sera set as measured by fluorescence-activated cell sorting (FACS). Individual volunteers are shown on the x axis, and the mean of three technical replicates of serum response is shown on the y axis. Serum response was considered significant when above the red dotted cutoff line, using the Mann-Whitney U test. Serum response was determined as the ratio of the population of cells with bound sera antibodies to the population of transfected cells (transfection efficiency). It is expressed as a percentage illustrated as follows: serum response = (cell population with bound sera antibodies/cell population that is transfected) × 100%.
Lastly, we assessed the immunogenicity of NF54_SpzPfEMP1 during natural infection. Serum of volunteers infected with NF54 sporozoites under chloroquine cover was obtained from a previous clinical trial that demonstrated that protective responses were mainly directed against P. falciparum pre-erythrocytic stages (Roestenberg et al., 2009) and was able to inhibit sporozoite invasion into hepatocytes (Peng et al., 2016). HEK293 cells expressing NF54_SpzPfEMP1, SEA-1 (PF3D7_1021800, positive control), or Etramp 14.2 (PF3D7_1476100, negative control) were incubated with serum and then subjected to fluorescence-assisted cell sorting. As shown in Figures 6B and 6C, the serum of one third of the infected volunteers contained significant antibody levels against NF54_SpzPfEMP1 (N-terminal CIDR region from amino acids 1–737), indicating that this PfEMP1 molecule is exposed on the surface of P. falciparum parasites before parasite blood stage infection develops.
Discussion
Epigenetic regulation of var gene transcription and studies on the role of PfEMP1 in malaria pathogenesis have been focused on blood stage parasites (Guizetti and Scherf, 2013, Smith et al., 2013). Here, we present highly reproducible data from multiple NF54 sporozoite preparations that a specific member of the var multigene family (∼60 members) is expressed in mosquito salivary gland parasites. Although we found that the overall nuclear organization is surprisingly conserved between asexual blood stage parasites and migratory sporozoite stages of P. falciparum, our developed ChIP-seq protocol for salivary gland stage sporozoites uncovered several features that provide insight into the biology of malaria transmission to humans. A striking feature is that subtelomeric heterochromatic region extends to genes that are exported into the host cell in asexual stages, adding a new parasite strategy to silence genes that are not used in free-living stages.
Our genome-wide analysis of PfHP1 and H3K9me3 enrichment predicted the expression of a member of the var gene family: Pf3D7_0809100 or NF54varsporo. Given that a bulk culture of the NF54 blood stage parasites express var genes that are distinct from NF54varsporo, we propose that an epigenetic resetting of var genes occurs during mosquito stage development to select for expression of a specific PfEMP1 on the surface of sporozoites. We detect the NF54varsporo transcript in late oocyst stage parasites, but this does not exclude the possibility that the resetting occurs at an earlier transmission stage. Regardless of when the epigenetic resetting occurs, our data suggest that, similar to asexual blood stage parasites, sporozoites may maintain monoallelic expression of var genes.
Our observation of epigenetic resetting of var genes is supported by transcriptional data obtained from blood stage parasites after passage through the mosquito. Studies with human volunteers have shown that var gene choice is altered upon mosquito transmission (Bachmann et al., 2016). A recent study reported the presence of a specific var gene transcript in sporozoites obtained from P. falciparum-infected patients from a malaria-endemic region in Africa; however, the authors did not report whether the corresponding PfEMP1 was produced (Gómez-Díaz et al., 2017). In light of these published data and our results, genetically different parasite strains may express antigenically variant PfEMP1 proteins in sporozoites.
Thus far, the biological role of the PfEMP1 surface adhesion molecule was believed to be restricted to immune evasion in the asexual blood stage. Our IFA data show that all salivary gland sporozoites express NF54_SpzPfEMP1 on their surface in a manner similar to that of iRBCs. It is currently unclear why a particular var gene member would be specifically targeted for mosquito stage expression. Structural analysis of NF54_SpzPfEMP1 does not reveal any specific features that would make it a particularly good candidate for expression at the sporozoite plasma membrane. One possibility is that the expression of a particular adhesive property may enhance sporozoite migration and hepatocyte infection. This hypothesis may limit the choice of expressed var gene for efficient transmission of parasites. The NF54varsporo gene has not been studied before, and its adhesive features remain unknown.
Given that all asexual blood stage PfEMP1 proteins studied so far display adhesive properties (Smith, 2014), we hypothesized that NF54_SpzPfEMP1 plays a role in sporozoite interaction with various host tissues and infection of hepatocytes. Indeed, antibodies directed against the DBL and CIDR domains of NF54_SpzPfEMP1 resulted in very potent inhibition of hepatocyte infection. In the future, genetically modified parasites that either lack the NF54varsporo var gene or express genetically modified extracellular PfEMP1 domains may help to elucidate the function of NF54_SpzPfEMP1 during the various mosquito stages and, perhaps, even hepatocyte infection.
Our work is highly relevant to vaccine development based on live-attenuated sporozoites. Sterile protection in human volunteers has been reported after challenge with homologous, laboratory-adapted P. falciparum sporozoites (Kublin et al., 2017). Given our discovery that a clonally variant antigen on the surface of sporozoites is expressed in a strain-specific manner, one may speculate that natural protective immune responses target variant sporozoite surface antigens such as PfEMP1. This could impact the efficacy of a sporozoite vaccine in patients from distinct geographic regions, as has been recently reported (Schats et al., 2015). It is noteworthy that PfEMP1 does not exist in rodent models that are often used for sporozoite vaccine evaluation. Thus, prediction of vaccine efficacy in these models does not take into account the expression of P. falciparum species-specific variant molecules.
In conclusion, this study provides evidence for the sporozoite-specific repurposing of the PfEMP1 surface antigen family, the biological function of which was thought to be restricted to immune evasion (antigenic variation) during chronic blood stage infection in the human host. Our discovery of a strain-specific surface antigen may provide a molecular explanation for the reduced protection of human volunteers after challenge with heterologous P. falciparum sporozoites. As our data demonstrate an unprecedented role of PfEMP1 proteins in transmission stages and provide insight into the biology of sporozoites during infection of hepatocytes, this work is highly relevant for vaccine design based on sporozoites.
Experimental Procedures
Human Plasma Samples
Samples were obtained from a previous clinical trial carried out at the Radboud University Nijmegen Medical Center (Nijmegen, the Netherlands), in accordance with principles of good clinical practice and with prior approval from the Central Committee for Research Involving Human Subjects of the Netherlands (NCT00442377). Study subjects (n = 14) were exposed to either infective (n = 9) or non-infective (n = 5) mosquito bites with concurrent chloroquine prophylaxis (Roestenberg et al., 2009).
Isolation and Purification of P. falciparum Sporozoites and Oocysts
Adult Anopheles stephensi females were infected with either P. falciparum strain NF54 (originating from West Africa) or strain NF135, clone C10 (originating from Cambodia) as previously described (Ponnudurai et al., 1989, Teirlinck et al., 2013). For each biological replicate, sporozoites were obtained by aseptically dissecting salivary glands of 200–400 infected mosquitoes 14–21 days after an infective blood meal. Pooled salivary glands were homogenized, filtered with a 40-μm cell strainer, and purified on a 17% Accudenz gradient as previously described (Kennedy et al., 2012). For each biological replicate, oocysts were isolated by dissection of the midguts of 50 infected mosquitoes, with an average of 45 oocysts per mosquito, 8 days post-infection. Biological replicates came from independent rounds of mosquito infection.
ChIP and Next-Generation Sequencing
ChIP was performed as previously described (Lopez-Rubio et al., 2009), using 5 × 106 P. falciparum sporozoites (strain NF54) or 109 asexual ring stage parasites (strain 3D7, clone G7). For each ChIP experiment, 0.5 μg anti-H3K9me3 (Millipore #07-442), anti-H3K9ac (Millipore #07-352), or anti-PfHP1 (Chen et al., 2016) rabbit polyclonal antibodies was used. To generate Illumina-compatible sequencing libraries, the immunoprecipitated DNA was processed using the MicroPlex Library Preparation Kit (Diagenode) according to the manufacturer’s instructions. As a control, DNA corresponding to the ChIP input or DNA immunoprecipitated using rabbit IgG (Diagenode, #C15410206) was processed. Pooled, multiplexed libraries were subjected to 150-bp single-end sequencing on an Illumina NextSeq 500. The resulting data were demultiplexed using bcl2fastq2 (Illumina) to obtain fastq files for downstream analysis. A minimum of three biological replicates was analyzed for each antibody and each parasite stage (Table S6).
ChIP-Seq Data Analysis
Quality control of fastq files was performed using the FastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Sequencing reads were mapped to the P. falciparum genome (Gardner et al., 2002; PlasmoDB, v.3) with the Burrows-Wheeler Alignment tool (BWA-MEM algorithm) using default parameters (Li and Durbin, 2009). Uniquely mapped reads were filtered for duplicates and an alignment quality Phred score (Q)≥ 20 using samtools (Li et al., 2009). The ChIP-seq data were normalized over input. Peak-calling analysis was performed with the MACS2 software (Zhang et al., 2008) with default settings and a false discovery rate cutoff of 5%, using Benjamini and Hochberg’s correction method. For genome-wide representation of PfHP1, H3K9ac, and H3K9me3 distribution, the coverage of each chromatin mark was calculated as average reads per million over bins of 1,000 nt using bamCoverage from the package deepTools2 (Ramírez et al., 2016), unless otherwise specified. Correlation of the different biological replicates was determined using MACS2, bedtools (Quinlan and Hall, 2010), and deepTools2. Circular and linear coverage plots were generated using Circos (Krzywinski et al., 2009) and Integrated Genomics Viewer (Thorvaldsdóttir et al., 2013), respectively.
RNA-Seq Library Preparation and Data Analysis
For RNA-seq library preparation, total RNA from sporozoites, oocysts, or asexual ring stage parasites was extracted using the miRNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions, including on-column DNase digestion. Total RNA was poly(A)-enriched using the Dynabeads mRNA Purification Kit (Life Technologies) and subjected to strand-specific RNA-seq library preparation using the TruSeq Stranded mRNA Library Prep Kit (Illumina). The libraries were multiplexed and subjected to 150-bp single-end sequencing on an Illumina NextSeq 500. The resulting data were demultiplexed using bcl2fastq2 (Illumina) to obtain fastq files for downstream analysis. A minimum of two biological replicates was analyzed for each parasite stage (Table S6).
As described earlier, quality control of fastq files was performed using FastQC software and sequencing reads mapped to the P. falciparum genome (Gardner et al., 2002, PlasmoDB, v.3) with the Burrows-Wheeler Alignment tool (BWA-MEM algorithm) using default parameters. Gene counts (i.e., the number of aligned reads mapping to different genomic loci) were calculated using bedtools (Quinlan and Hall, 2010). Differential gene expression analysis was performed using the R package edgeR (Robinson et al., 2010).
Multidimensional scaling (MDS) plots were also made in edgeR using “plotMDS,” where distances between pairs of RNA samples correspond to the leading log2 fold changes (i.e., average [root-mean-square] of the largest absolute log2 fold changes) (Robinson et al., 2010). To generate coverage plots, the gene counts normalized for gene length and sequencing depth by calculating RPKM values in the R:limma package (Ritchie et al., 2015). Coverage plots were visualized using the Integrated Genomics Viewer (Robinson et al., 2011, Thorvaldsdóttir et al., 2013).
DNA Fluorescence In Situ Hybridization and Immunofluorescence Imaging
Sporozoites were fixed in suspension with 4% paraformaldehyde (PFA) in PBS (pH 7.4) overnight at 4°C. After three PBS washes, parasites were resuspended in PBS, and deposited on poly-L-lysine-coated #1.5 coverslips (Marienfeld Superior). Parasites were permeabilized with 0.1% Triton X-100 for 15 min and subjected to DNA FISH labeling with the TARE6 probe as described previously for blood stage parasites (Mancio-Silva et al., 2008). Immunofluorescent labeling using rabbit anti-PfHP1 antibodies was performed as previously described (Chen et al., 2016). For combined DNA FISH and immunofluorescence, sporozoites were first labeled with rabbit anti-PfHP1 antibodies (diluted 1:2,000 in 3% BSA [Sigma] in PBS) and then with anti-rabbit Alexa Fluor 488 secondary antibodies (Life Technologies). After an additional fixation step with 4% PFA, DNA FISH was performed using a biotinylated locked nucleic acid (LNA) probe (Exiqon) targeting all TARE6 telomeric repeats (5′-Biotin-+AC+T+AACA+TA+GG+T+CT+T+A-Biotin-3′), which was then detected with streptavidin-conjugated Alexa Fluor 568 secondary antibodies (Life Technologies).
NF54_SpzPfEMP1 Protein Expression and Antibody Generation
The DNA sequence encoding the CIDR-alpha of PF3D7_0809100 was cloned into the Baculovirus transfer vector pAcGP67-A (BD Biosciences). Recombinant CIDR product was expressed by infection of insect High Five cells with recombinant Baculovirus. Antibodies to Baculo-expressed CIDR were raised in Wistar rats by subcutaneous injection as previously described (Bengtsson et al., 2013). All experimental animal procedures were approved by The Danish Animal Procedures Committee (“Dyreforsøgstilsynet”) as described in permit no. 2008/561-1498 and according to the guidelines described in Danish acts LBK 1306 (23/11/2007) and BEK 1273 (12/12/2005). For the mouse, anti-NF54_SpzPfEMP1 antibody production, a codon-optimized synthetic gene encoding amino acids 1–737 of PF3D7_0809100 was synthesized by GenScript Biotech and cloned into the pET28 vector. Recombinant protein was produced using SHuffle T7 Express bacteria (New England BioLabs), and soluble protein was purified using Ni-NTA His Bind Superflow Resin (Novagen) according to manufacturer’s instructions. Purified protein (10 μg of >90% purity) was used to immunize C57BL/6 mice. Animal care and experiments involving mice were conducted at the Institute Pasteur with the approval of the Direction Départementale des Services Vétérinaires of Paris, France (permit no. 75-066, issued on September 14, 2009) and performed in compliance with institutional guidelines and European regulations (http://ec.europa.eu/environment/chemicals/lab_animals/index_en.htm). A statement of compliance with the French government’s ethical and animal experiment regulations was issued by the Ministère de l’Enseignement Supérieur et de la Recherche under the number 00218.01.
Western Blot Analysis
5 × 106 P. falciparum NF54 sporozoites were resuspended in 100 μL 2× NuPAGE sample buffer supplemented with reducing agent (Thermo Fisher Scientific) and incubated at 95°C for 5 min, after which DNA was sheared. The resulting sporozoite protein extract was resolved on a 3%–8% Tris-Acetate SDS-polyacrylamide gel (NuPAGE) using MOPS SDS Running Buffer (Thermo Fisher Scientific) and transferred to a PVDF membrane. The membrane was blocked for 1 hr with 5% milk in TBST (50 mM Tris, 150 mM NaCl, 0.1% Tween 20). PfEMP1 proteins were detected using guinea pig anti-ATS antibodies (Nacer et al., 2015) or anti-NF54_SpzPfEMP1 antibodies. PfAldolase was detected with anti-PfAldolase-HRP (horseradish peroxidase) antibodies (Abcam, ab38905). Primary antibodies were detected by goat anti-guinea pig-HRP (Abcam, ab6908), anti-rat-HRP, or anti-mouse-HRP (GE Healthcare Life Sciences, NA935V and NA931V, respectively) secondary antibodies. HRP signal was developed with SuperSignal West Pico chemiluminescent substrate (Thermo Fisher Scientific, #34080) and imaged with a ChemiDoc XRS+ System (Bio-Rad). Images were analyzed using the Fiji image processing package (http://fiji.sc).
Immunofluorescence Assay
NF54 or NF135 sporozoites were placed on poly-L-lysine-coated coverslips for 10 min at room temperature (RT) under one of three conditions: live or fixed in 4% PFA and permeabilized with 0.01% Triton X-100. The cells were blocked with 4% BSA in PBS for 30 min at RT and then incubated with primary antibody for 1 hr at RT; dilutions were 1:1,000 for anti-ATS, 1:1,000 for anti-NF54_SpzPfEMP1, 1:200 for anti-var2CSA (Avril et al., 2011), and 1:15,000 for anti-CSP (Okitsu et al., 2007) antibodies in 3% BSA in PBS. After three washes with PBS, primary antibodies were detected with Alexa Fluor 488- or 568-conjugated secondary antibodies (Life Technologies) diluted 1:1,000 in 3% BSA in PBS. After three final washes in PBS, cells were mounted in Vectashield containing DAPI for nuclear staining. Images were captured using a Deltavision Elite imaging system (GE Healthcare). Image processing of at least two independent replicates was performed using the Fiji package (http://fiji.sc). For trypsinization prior to imaging, 106 living sporozoites were treated with trypsin (GIBCO, 50 mg/mL in 200 μL Leibovitz’s L-15 medium) for 40 min at RT. Sporozoites were then washed three times with Leibovitz’s L-15 medium containing serum and processed as described earlier, with all incubations on ice.
Liver Stage Development Inhibition Assay
Primary human hepatocyte cultures were prepared from two different batches of human hepatocytes following the manufacturer’s instructions. Briefly, cryopreserved human hepatocytes (Biopredic International) were thawed and resuspended in seeding medium according to the manufacturer’s instructions. Cells were seeded in 96-well plates previously coated with rat tail collagen I (BD Biosciences), and after cell attachment, a Matrigel (BD Biosciences) overlay was added. 24 hr after plating, medium was replaced with culture medium (William’s medium E, Life Technologies) supplemented with 10% fetal calf serum (FCS; Perbio), 5 × 10−5 M hydrocortisone hemisuccinate (Upjohn Laboratories SERB, Paris, France), 5 μg/mL human insulin (Sigma), 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin (Life Technologies). The heat-inactivated mouse sera containing anti-NF54_SpzPfEMP1 antibodies (diluted 1:100, 1:500, or 1:1000), the correspondent pre-immune sera (diluted 1:100), or the purified anti-NF54_SpzPfEMP1 IgG (diluted 1:100) were added to 50 μL medium containing 3 × 104 P. falciparum NF54 or NF135 sporozoites. The Matrigel overlay was removed from the infected human hepatocyte culture before addition of the mixture of antibodies and sporozoites. Plates were centrifuged at 2,000 rpm for 10 min at RT to facilitate parasite settling onto the cells. Medium was changed 3 hr post-infection, and Matrigel was added to the infected culture; medium was replaced 48 hr post-addition of sporozoites. Cultures were stopped at day 3 post-infection by fixing with cold methanol. Parasite numbers within hepatocytes were assessed via immunofluorescence using anti-PfHSP70 antibodies as previously described (Rénia et al., 1990). Quantification was performed by microscopy or by using the ArrayScan XTI imaging system (Thermo Fisher Scientific).
Determination of Serum Response against NF54_SpzPfEMP1
Sera isolated from individuals from a previous clinical trial (Roestenberg et al., 2009) were used to probe HEK293 cells transfected for cell-surface expression of P. falciparum proteins. For NF54_SpzPfEMP1, a DNA construct encoding amino acids 1–737 was used for transfection. These HEK293 cells were analyzed by flow cytometry, as previously described (Peng et al., 2016). Serum response was defined as the percentage of transfected cells that show bound serum antibodies (Peng et al., 2016). For sera with positive serum response (>5%), the serum response against var-transfected cells was compared with the serum response against Etramp14.2-transfected cells (negative control), using the Mann Whitney U test, and was found to be significantly higher.
Parasite Panning on Antibody-Coated Magnetic Beads
Sterile Dynabeads Protein G (Life Technologies 10003D) were incubated with 20 μg filtered anti-NF54_SpzPfEMP1 IgG for 10 min at RT under agitation. The Dynabeads were blocked with 1% BSA for 20 min. iRBCs containing trophozoites and schizonts were isolated by plasmagel enrichment and resuspended in 400 μL PBS at a concentration of 5 × 107 iRBCs per milliliter. The iRBCs and the beads were mixed by gently pipetting and allowed to bind for 1 hr with gentle agitation at 37°C. After incubation, the beads were washed 8 times with 800 μL sterile PBS to eliminate all unbound cells. Bound cells were resuspended with culture medium and fresh red blood cells. In total, three antibody pannings were performed. The enrichment was assessed after each panning by IFA using anti-NF54_SpzPfEMP1.
Acknowledgments
We thank Benoit Gamain for providing anti-var2CSA antibodies. This work was supported by a European Research Council advanced grant (PlasmoSilencing 670301) to A.S., by an ANR grant HypEpiC (ANR-14-CE16-0013) to D. Mazier and A.S., and by the French Parasitology consortium ParaFrap (ANR-11-LABX0024) to A.S. and D. Mazier. G.Z. and S.D. were supported by a ParaFrap PhD fellowship, S.S.V. was supported by a Carnot-Pasteur-Maladies Infectieuse fellowship, and J.B. was supported by an EMBO fellowship (ALTF 180-2015).
Author Contributions
Conceptualization, G.Z., S.S.V., D. Mazier, and A.S.; Methodology, G.Z. and S.S.V.; Investigation, G.Z., S.S.V., S.D., S.B., J.G., J.-F.F., M.B., V.S., O. Silvie, R.S., L.R., Y.S.G., and J.M.B. Writing – Original Draft, G.Z., S.S.V., and A.S.; Writing – Review & Editing, G.Z. and A.S.; Resources, D. Mattei, P.C., S.M., C.C.H., O. Scatton, and A.T.R.J.; Funding Acquisition, A.S.
Declaration of Interests
The authors declare no competing interests.
Published: March 13, 2018
Footnotes
Supplemental Information includes six figures and six tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.02.075.
Contributor Information
Dominique Mazier, Email: dominique.mazier@upmc.fr.
Artur Scherf, Email: artur.scherf@pasteur.fr.
Data and Software Availability
The accession number for the ChIP-seq and RNA-seq fastq files generated in this study is NCBI: PRJNA344838. To understand the files, please refer to Table S6.
Supplemental Information
References
- Amit-Avraham I., Pozner G., Eshar S., Fastman Y., Kolevzon N., Yavin E., Dzikowski R. Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. USA. 2015;112:E982–E991. doi: 10.1073/pnas.1420855112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avril M., Hathaway M.J., Srivastava A., Dechavanne S., Hommel M., Beeson J.G., Smith J.D., Gamain B. Antibodies to a full-length VAR2CSA immunogen are broadly strain-transcendent but do not cross-inhibit different placental-type parasite isolates. PLoS ONE. 2011;6:e16622. doi: 10.1371/journal.pone.0016622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann A., Petter M., Krumkamp R., Esen M., Held J., Scholz J.A., Li T., Sim B.K., Hoffman S.L., Kremsner P.G. Mosquito passage dramatically changes var gene expression in controlled human Plasmodium falciparum infections. PLoS Pathog. 2016;12:e1005538. doi: 10.1371/journal.ppat.1005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson A., Joergensen L., Rask T.S., Olsen R.W., Andersen M.A., Turner L., Theander T.G., Hviid L., Higgins M.K., Craig A. A novel domain cassette identifies Plasmodium falciparum PfEMP1 proteins binding ICAM-1 and is a target of cross-reactive, adhesion-inhibitory antibodies. J. Immunol. 2013;190:240–249. doi: 10.4049/jimmunol.1202578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brancucci N.M.B., Bertschi N.L., Zhu L., Niederwieser I., Chin W.H., Wampfler R., Freymond C., Rottmann M., Felger I., Bozdech Z., Voss T.S. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe. 2014;16:165–176. doi: 10.1016/j.chom.2014.07.004. [DOI] [PubMed] [Google Scholar]
- Chen P.B., Ding S., Zanghì G., Soulard V., DiMaggio P.A., Fuchter M.J., Mecheri S., Mazier D., Scherf A., Malmquist N.A. Plasmodium falciparum PfSET7: enzymatic characterization and cellular localization of a novel protein methyltransferase in sporozoite, liver and erythrocytic stage parasites. Sci. Rep. 2016;6:21802. doi: 10.1038/srep21802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning-Ward T.F., Dixon M.W., Tilley L., Gilson P.R. Plasmodium species: master renovators of their host cells. Nat. Rev. Microbiol. 2016;14:494–507. doi: 10.1038/nrmicro.2016.79. [DOI] [PubMed] [Google Scholar]
- Epp C., Li F., Howitt C.A., Chookajorn T., Deitsch K.W. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flueck C., Bartfai R., Volz J., Niederwieser I., Salcedo-Amaya A.M., Alako B.T., Ehlgen F., Ralph S.A., Cowman A.F., Bozdech Z. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5:e1000569. doi: 10.1371/journal.ppat.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Díaz E., Yerbanga R.S., Lefèvre T., Cohuet A., Rowley M.J., Ouedraogo J.B., Corces V.G. Epigenetic regulation of Plasmodium falciparum clonally variant gene expression during development in Anopheles gambiae. Sci. Rep. 2017;7:40655. doi: 10.1038/srep40655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J., Scherf A. Silence, activate, poise and switch! Mechanisms of antigenic variation in Plasmodium falciparum. Cell. Microbiol. 2013;15:718–726. doi: 10.1111/cmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Mu J., Zhang Q., Ni T., Srinivasan P., Rayavara K., Yang W., Turner L., Lavstsen T., Theander T.G. PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature. 2013;499:223–227. doi: 10.1038/nature12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M., Fishbaugher M.E., Vaughan A.M., Patrapuvich R., Boonhok R., Yimamnuaychok N., Rezakhani N., Metzger P., Ponpuak M., Sattabongkot J. A rapid and scalable density gradient purification method for Plasmodium sporozoites. Malar. J. 2012;11:421. doi: 10.1186/1475-2875-11-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin J.G., Mikolajczak S.A., Sack B.K., Fishbaugher M.E., Seilie A., Shelton L., VonGoedert T., Firat M., Magee S., Fritzen E. Complete attenuation of genetically engineered Plasmodium falciparum sporozoites in human subjects. Sci. Transl. Med. 2017;9:eaad9099. doi: 10.1126/scitranslmed.aad9099. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio J.J., Mancio-Silva L., Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5:179–190. doi: 10.1016/j.chom.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio J.J., Siegel T.N., Scherf A. Genome-wide chromatin immunoprecipitation-sequencing in Plasmodium. Methods Mol. Biol. 2013;923:321–333. doi: 10.1007/978-1-62703-026-7_23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancio-Silva L., Rojas-Meza A.P., Vargas M., Scherf A., Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J. Cell Sci. 2008;121:2046–2053. doi: 10.1242/jcs.026427. [DOI] [PubMed] [Google Scholar]
- Nacer A., Claes A., Roberts A., Scheidig-Benatar C., Sakamoto H., Ghorbal M., Lopez-Rubio J.J., Mattei D. Discovery of a novel and conserved Plasmodium falciparum exported protein that is important for adhesion of PfEMP1 at the surface of infected erythrocytes. Cell. Microbiol. 2015;17:1205–1216. doi: 10.1111/cmi.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okitsu S.L., Kienzl U., Moehle K., Silvie O., Peduzzi E., Mueller M.S., Sauerwein R.W., Matile H., Zurbriggen R., Mazier D. Structure-activity-based design of a synthetic malaria peptide eliciting sporozoite inhibitory antibodies in a virosomal formulation. Chem. Biol. 2007;14:577–587. doi: 10.1016/j.chembiol.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Peng K., Goh Y.S., Siau A., Franetich J.F., Chia W.N., Ong A.S., Malleret B., Wu Y.Y., Snounou G., Hermsen C.C. Breadth of humoral response and antigenic targets of sporozoite-inhibitory antibodies associated with sterile protection induced by controlled human malaria infection. Cell. Microbiol. 2016;18:1739–1750. doi: 10.1111/cmi.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Toledo K., Rojas-Meza A.P., Mancio-Silva L., Hernández-Cuevas N.A., Delgadillo D.M., Vargas M., Martínez-Calvillo S., Scherf A., Hernandez-Rivas R. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37:2596–2606. doi: 10.1093/nar/gkp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnudurai T., Lensen A.H., Van Gemert G.J., Bensink M.P., Bolmer M., Meuwissen J.H. Infectivity of cultured Plasmodium falciparum gametocytes to mosquitoes. Parasitology. 1989;98:165–173. doi: 10.1017/s0031182000062065. [DOI] [PubMed] [Google Scholar]
- Prudêncio M., Mota M.M. To migrate or to invade: those are the options. Cell Host Microbe. 2007;2:286–288. doi: 10.1016/j.chom.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph S.A., Bischoff E., Mattei D., Sismeiro O., Dillies M.A., Guigon G., Coppee J.Y., David P.H., Scherf A. Transcriptome analysis of antigenic variation in Plasmodium falciparum - var silencing is not dependent on antisense RNA. Genome Biol. 2005;6:R93. doi: 10.1186/gb-2005-6-11-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rénia L., Mattei D., Goma J., Pied S., Dubois P., Miltgen F., Nüssler A., Matile H., Menégaux F., Gentilini M. A malaria heat-shock-like determinant expressed on the infected hepatocyte surface is the target of antibody-dependent cell-mediated cytotoxic mechanisms by nonparenchymal liver cells. Eur. J. Immunol. 1990;20:1445–1449. doi: 10.1002/eji.1830200706. [DOI] [PubMed] [Google Scholar]
- Richie T.L., Billingsley P.F., Sim B.K., James E.R., Chakravarty S., Epstein J.E., Lyke K.E., Mordmüller B., Alonso P., Duffy P.E. Progress with Plasmodium falciparum sporozoite (PfSPZ)-based malaria vaccines. Vaccine. 2015;33:7452–7461. doi: 10.1016/j.vaccine.2015.09.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roestenberg M., McCall M., Hopman J., Wiersma J., Luty A.J., van Gemert G.J., van de Vegte-Bolmer M., van Schaijk B., Teelen K., Arens T. Protection against a malaria challenge by sporozoite inoculation. N. Engl. J. Med. 2009;361:468–477. doi: 10.1056/NEJMoa0805832. [DOI] [PubMed] [Google Scholar]
- Rovira-Graells N., Gupta A.P., Planet E., Crowley V.M., Mok S., Ribas de Pouplana L., Preiser P.R., Bozdech Z., Cortés A. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 2012;22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schats R., Bijker E.M., van Gemert G.J., Graumans W., van de Vegte-Bolmer M., van Lieshout L., Haks M.C., Hermsen C.C., Scholzen A., Visser L.G., Sauerwein R.W. Heterologous protection against malaria after immunization with Plasmodium falciparum sporozoites. PLoS ONE. 2015;10:e0124243. doi: 10.1371/journal.pone.0124243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.D. The role of PfEMP1 adhesion domain classification in Plasmodium falciparum pathogenesis research. Mol. Biochem. Parasitol. 2014;195:82–87. doi: 10.1016/j.molbiopara.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.D., Rowe J.A., Higgins M.K., Lavstsen T. Malaria’s deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell. Microbiol. 2013;15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teirlinck A.C., Roestenberg M., van de Vegte-Bolmer M., Scholzen A., Heinrichs M.J., Siebelink-Stoter R., Graumans W., van Gemert G.J., Teelen K., Vos M.W. NF135.C10: a new Plasmodium falciparum clone for controlled human malaria infections. J. Infect. Dis. 2013;207:656–660. doi: 10.1093/infdis/jis725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlgren M., Goel S., Akhouri R.R. Variant surface antigens of Plasmodium falciparum and their roles in severe malaria. Nat. Rev. Microbiol. 2017;15:479–491. doi: 10.1038/nrmicro.2017.47. [DOI] [PubMed] [Google Scholar]
- World Health Organization (2017). Fact sheet: world malaria report 2017. http://www.who.int/mediacentre/factsheets/fs094/en/.
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.