Abstract
Rectal transmission accounts for the majority of HIV transmissions among men who have sex with men (MSM). We previously demonstrated a distinct rectal mucosal immune environment in MSM engaging in condomless receptive anal intercourse (CRAI) compared with men who do not engage in anal intercourse, including enrichment of the microbiota for the Prevotellaceae family as compared with Bacteroidaceae. Here, we expand upon these previous findings to determine differences by anatomic site of collection (anal vs. rectal mucosa) and examine associations of the predominant taxa with other genera. We analyzed 16SrRNA gene sequences of the V1–V2 region generated on an Illumina MiSeq® from 35 MSM engaging in CRAI and 20 male controls. Observation by principal coordinates analysis and analysis of similarities test showed differing composition of the microbiota by anatomic site of collection. When analyzing the top 10 abundant genera from each anatomic site by generalized linear models, the predominant genera (Prevotella enrichment among MSM engaging in CRAI vs. Bacteroides among controls) were consistent; however, the two sites shared only four common genera. In addition, associations between the relative abundance of Prevotella and Bacteroides with other prevalent genera, by Spearman's rank correlations, were inconsistent when stratifying by study group. Prevotella versus Bacteroides predominant microbiota may not define a consistent underlying microbial community, and our data underline the importance of anatomic sampling site. Understanding the rectal mucosal immune environment, of which the microbiota is a critical component, will enable a better understanding of rectal HIV transmission.
Keywords: : Prevotella, Bacteroides, microbiome, rectal mucosa, MSM, HIV transmission
Men who have sex with men (MSM) are disproportionally at risk of acquiring HIV,1 in part due to the relative ease of transmission across the rectal mucosa where ∼70% of transmissions occur.2 Recently, we characterized the rectal mucosal immune environment of HIV negative MSM engaging in condomless receptive anal intercourse (CRAI) and showed a distinct cellular and gene expression profile compared with men who had never engaged in anal intercourse (controls).3 In addition, the rectal mucosal microbiota of MSM engaging in CRAI was enriched for the Prevotellaceae family as compared with Bacteroidaceae family for controls. In prior studies, two distinct species belonging to the Prevotellaceae family and Prevotella genus were positively associated with prevalent HIV infection4; however, our previous data analyses were limited to the family level, and it is unclear whether multiple taxa contribute to the enrichment of Prevotellaceae among MSM. In addition, many studies report microbiota data based on anal swabs; yet, this collection site may not be representative of the microbiota at the site of HIV transmission, the rectal mucosa.5 Here, we expand upon the previously reported microbiome data to evaluate whether the anatomic site of specimen collection (anus vs. rectal mucosa) provided consistent results, determine whether multiple genera of Prevotella contributed to the enrichment seen previously among MSM, and examine associations of the relative abundance of Prevotella and Bacteroides with other bacterial taxa in the rectal mucosa. A better understanding of the microbiota at mucosal surfaces may lead to enhanced understanding of HIV tranmission and the design of improved biomedical prevention interventions, including candidate vaccines.
Detailed methods on the clinical cohort were previously published.3 In brief, 41 HIV negative MSM engaging in CRAI at least four times monthly and 21 controls underwent rectal mucosal sampling conducted at two study visits separated by a median of 9 weeks. Clinicians collected mucosal swabs for microbiota sequencing at two anatomic locations: the anus and through a rigid sigmoidoscope ∼8–10 cm from the anal verge (rectal mucosa). Swabs were stored in lysis buffer at −80°C; DNA was extracted and amplified using previously described methods.3 Microbiota data generated by sequencing the V1–V2 region of the 16SrRNA gene on an Illumina MiSeq® instrument (rarefied to 2,215 sequences) were available from 54 participants for these analyses due to missing specimens and adequate sequence generation. The Greengenes database (version 13_8) was used to classify operational taxonomic units (OTUs) to the genus level. Sequences were deposited to the GEO database (accession GSE83245).
Alpha diversity was measured using Chao1 and Shannon indices and statistical significance was assessed with generalized linear models (GLMs) using RStudio® adjusting for age using the generalized estimated equation approach to account for repeated measurements. Beta diversity was measured using Bray–Curtis dissimilarity analysis and observed by principal coordinate analysis (PCoA). Differences in microbial composition between anatomic collection sites were evaluated using the analysis of similarities (ANOSIM) test.6 The 10 most abundant genera in both anatomic sites were identified and compared with GLMs as already described. No differences were seen between visit 1 and visit 2, thus overall model-based means were reported. Spearman's rank correlation coefficients, adjusted for multiple correlations by the Holm–Bonferroni method, were calculated to evaluate the relationships between the OTUs of interest among MSM engaging in CRAI and controls for the rectal mucosa.
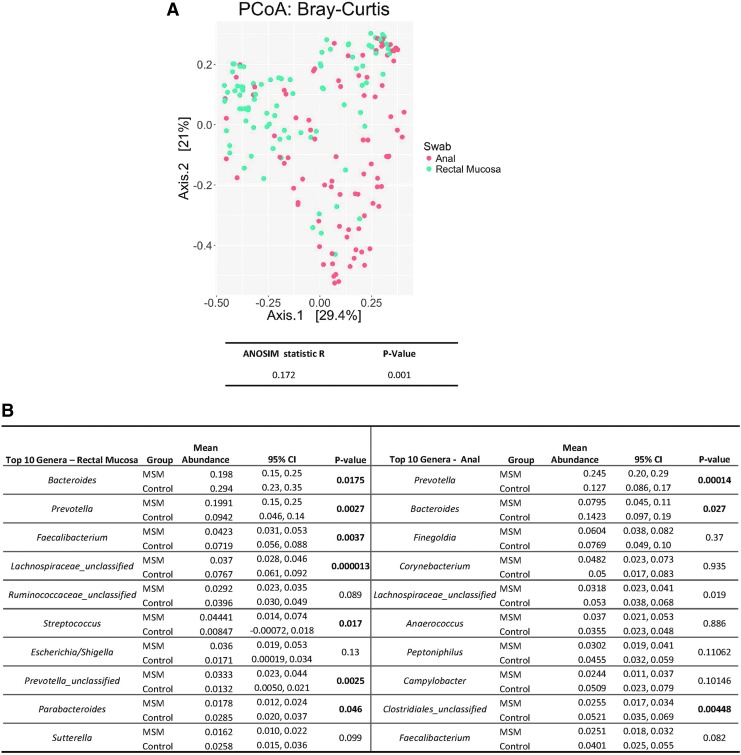
Microbiota sequences from the anus and the rectal mucosa were analyzed from 34 MSM engaging in CRAI and 20 controls. MSM engaging in CRAI were slightly older than controls (30 years vs. 27 years; p = .02); however, there were no differences in self-reported race between MSM engaging in CRAI and controls (78% white, 13% black, 9% other; p = .16). Alpha diversity indices (Shannon and Chao1) did not differ between anatomic sites of collection (p = .078; p = .38, respectively). Beta diversity differed between anatomic sites; the rectal mucosa and anus clustered separately in PCoA (Fig. 1A) and had statistically dissimilar microbial compositions (ANOSIM R = 0.1703, p = .001).
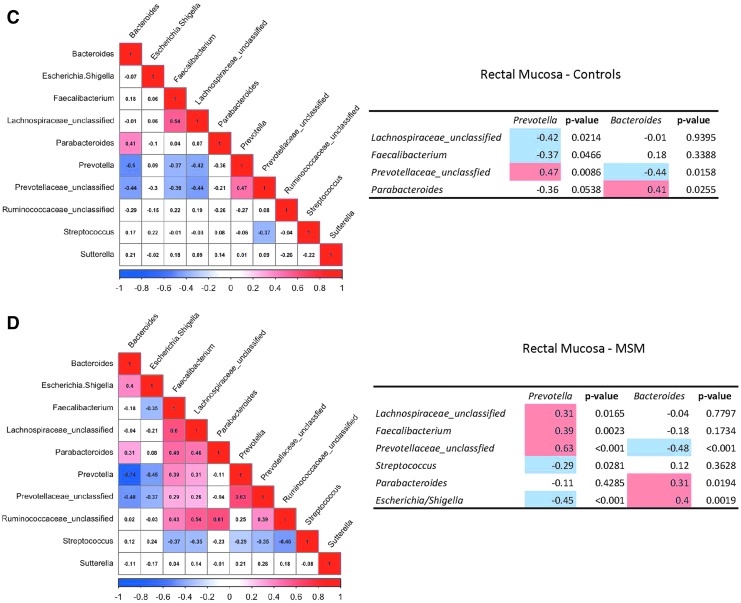
FIG. 1.
Bray–Curtis dissimilarity displayed by principal coordinate analysis shows anal and rectal mucosal swabs clustering independently, and ANOSIM results confirm statistical significance (A). Model-based mean relative abundances of the top 10 genera generated with generalized linear models in rectal mucosal and anal swabs are listed in order of prevalence (B). Statistical comparisons between MSM and controls were calculated with the Wald test. Heat maps of Spearman rank correlation coefficients among men who never engaged in anal intercourse (C) and among MSM engaging in CRAI of top 10 OTUs (D). Red panels report a significant positive correlation and blue panels report a significant negative correlation (adjusted p < .05). Panels in white are nonsignificant (adjusted p > .05). ANOSIM, analysis of similarities; CRAI, condomless receptive anal intercourse; MSM, men who have sex with men; OTU, operational taxonomic unit.
The top 10 genera and relative abundances from each anatomic site are listed in Figure 1B. Although there were prominent similarities between the most abundant genera from both anatomic sites (Prevotella and Bacteroides), only the top four genera from the rectal mucosa were present in the anus. Not surprisingly, Corynebacterium, a component of normal skin flora, was the fourth most prevalent genus from the anus but was absent in the rectal mucosa.7 In addition, taxa with pathogenic potential such as Streptococcus and Escherichia/Shigella were present only in the rectal mucosa.7,8
The microbiota of MSM engaging in CRAI was enriched for taxa from the Prevotella genus in both the rectal mucosa and anus, whereas controls were enriched for the Bacteroides genus at both sites (Fig. 1B). Of interest, MSM engaging in CRAI exhibited a statistically significant increase in an unclassified genus also belonging to the Prevotellaceae family and the pathogenic genera Streptococcus, compared with controls in the rectal mucosa.
We examined associations between the relative abundance of Prevotella and Bacteroides and other genera stratified by study group (Fig. 1C, D) in the rectal mucosa. Both study groups showed a statistically significant negative association between Prevotella and Bacteroides. In rectal mucosa, Prevotella was positively correlated with an unclassified genus from the Prevotellaceae family in both study groups. Interestingly, a significant negative association between Prevotella and both Lachnospiraceae and Faecalibacterium was seen among controls, whereas a positive association was seen among MSM engaging in CRAI. Finally, Prevotella was negatively associated with genera containing pathogenic bacteria such as Escherichia/Shigella and Streptococcus among MSM but not among controls.
In this analysis, we have expanded upon our previously published data that showed rectal mucosal enrichment for Prevotellaceae among MSM engaging in CRAI as contrasted to Bacteroidaceae among controls.3 Here, we compared the microbiota composition at the genus level between specimens collected at the anus and rectal mucosa and demonstrated that although the predominant genera (Prevotella vs. Bacteroides) were consistent, significant differences were apparent in the top OTUs between the anatomic sites. Previous studies have determined that rectal swabs, fecal samples, and rectal biopsies offer different perspectives of the gut microbiota, and our data support these conclusions.9,10 In our study, taxa commonly associated with skin flora7,11 were more prevalent in the anus, and enrichment for a second genus of Prevotella and pathogenic taxa among MSM engaging in CRAI was only detected in the rectal mucosa. The finding of two distinct Prevotella genera also suggests that a single species does not account for the enrichment seen among MSM engaging in CRAI.
Enrichment for Prevotella or Bacteroides in the rectal mucosa was inconsistently associated with other genera when stratifying by study group and was not consistent with previously reported associations from stool specimens.12 In addition, although MSM engaging in CRAI were enriched for the pathogenic genera Streptococcus and Escherichia/Shigella, this was negatively associated with the relative abundance of Prevotella. This suggests that enrichment for either Prevotella or Bacteroides may not represent an underlying microbial community as previously suggested13 and/or that CRAI may operate to influence other taxa independent of its effect on the relative abundance of Prevotella.
This study is limited by its modest sample size and potential confounders such as diet and medications. We were unable to fully characterize the specific species of Prevotella contributing to the enrichment seen among MSM, which will be the goal of future research. Additional research including ex vivo and in vitro experiments will be needed to fully understand the interaction of microbial taxa in the rectal mucosa and implications for mucosal immunity.
Understanding the rectal mucosal immune environment, of which the microbiota is a critical component, will enable a better understanding of rectal HIV transmission and the development of effective biomedical HIV prevention interventions. Based on our data, researchers must carefully ensure that the most relevant anatomic sites are sampled in microbiome investigations and interpret results beyond the predominant genera with caution.
Acknowledgments
We would like to thank the Emory Center for AIDS Research Clinical Virology Core laboratory for its contributions in generating the 16SrRNA sequencing data. This work was supported by the National Institutes of Health: K23AI108335, and a supplement grant to the Emory Center for AIDS Research P30 AI050409.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.HIV Among Gay and Bisexual Men. 2017. Centers for Disease Control and Prevention. Available at: www.cdc.gov/hiv/group/msm/index.html, accessed July1, 2017
- 2.Sullivan PS, Salazar L, Buchbinder S, Sanchez TH: Estimating the proportion of HIV transmissions from main sex partners among men who have sex with men in five US cities. AIDS 2009;23:1153–1162 [DOI] [PubMed] [Google Scholar]
- 3.Kelley CF, Kraft CS, de Man TJ, et al. : The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: Implications for HIV transmission and prevention. Mucosal Immunol 2017;10:996–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone CA, Li M, Campbell TB, et al. : Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013;14:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Louissaint NA, Nimmagadda S, Fuchs EJ, et al. : Distribution of cell-free and cell-associated HIV surrogates in the colon after simulated receptive anal intercourse in men who have sex with men. J Acquir Immune Defic Syndr 2012;59:10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callahan BJ, Sankaran K, Fukuyama JA, McMurdie PJ, Holmes SP: Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. Version 2. F1000Res 2016;5:1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cogen AL, Nizet V, Gallo RL: Skin microbiota: A source of disease or defence? Br J Dermatol 2008;158:442–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D: Role of the normal gut microbiota. World J Gastroenterol 2015;21:8787–8803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Araujo-Perez F, McCoy AN, Okechukwu C, et al. : Differences in microbial signatures between rectal mucosal biopsies and rectal swabs. Gut Microbes 2012;3:530–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Budding AE, Grasman ME, Eck A, et al. : Rectal swabs for analysis of the intestinal microbiota. PLoS One 2014;9:e101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy EC, Morgelin M, Reinhardt DP, Olin AI, Bjorck L, Frick IM: Identification of molecular mechanisms used by Finegoldia magna to penetrate and colonize human skin. Mol Microbiol 2014;94:403–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguera-Julian M, Rocafort M, Guillen Y, et al. : Gut microbiota linked to sexual preference and HIV infection. EBioMedicine 2016;5:135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorvitovskaia A, Holmes SP, Huse SM: Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome 2016;4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]