Abstract
Dyes containing one or more azo linkages are widely applied in cosmetics, tattooing, food and drinks, pharmaceuticals, printing inks, plastics, leather, as well as paper industries. Previously we reported that bacteria living on human skin have the ability to reduce some azo dyes to aromatic amines, which raises potential safety concerns regarding human dermal exposure to azo dyes such as those in tattoo ink and cosmetic colorant formulations. To comprehensively investigate azo dye-induced toxicity by skin bacteria activation, it is very critical to understand the mechanism of metabolism of the azo dyes at the systems biology level. In this study, an LC/MS-based metabolomics approach was employed to globally investigate metabolism of azo dyes by Staphylococcus aureus as well as their effects on the metabolome of the bacterium. Growth of S. aureus in the presence of Sudan III or Orange II was not affected during the incubation period. Metabolomics results showed that Sudan III was metabolized to 4-(phenyldiazenyl) aniline (48%), 1-[(4-aminophenyl) diazenyl]-2-naphthol (4%) and eicosenoic acid Sudan III (0.9%). These findings indicated that the azo bond close to naphthalene group of Sudan III was preferentially cleaved compared with the other azo bond. The metabolite from Orange II was identified as 4-aminobenzene sulfonic acid (35%). A much higher amount of Orange II (~90×) was detected in the cell pellets from the active viable cells compared with those from boiled cells incubated with the same concentration of Orange II. This finding suggests that Orange II was primarily transported into the S. aureus cells for metabolism, instead of the theory that the azo dye metabolism occurs extracellularly. In addition, the metabolomics results showed that Sudan III affected energy pathways of the S. aureus cells, while Orange II had less noticeable effects on the cells. In summary, this study provided novel information regarding azo dye metabolism by the skin bacterium, the effects of azo dyes on the bacterial cells and the important role on the toxicity and/or inactivation of these compounds due to microbial metabolism.
Keywords: Azo dyes, Staphylococcus aureus, Metabolism, Metabolomics
Introduction
Dyes containing one or more azo linkages (R–N=N–R) are widely applied in cosmetics, tattooing, food and drinks, pharmaceuticals, printing inks, plastics, leather, as well as paper industries. Over the last century, epidemiological studies have suggested the link of the azo dye carcinogenesis to the high incidence of bladder cancer among workers in the aniline dye industry [22, 42]. Since these early reports, numerous studies have been conducted to investigate the mutagenicity and carcinogenicity of aromatic amines produced by azo dye metabolism in rats, monkeys or hamsters [17, 26, 31, 40]. The azo dye-induced toxicity is mainly attributed to the reduction of azo dyes to aromatic amines, with many being carcinogenic [3]. Previously, we [27, 32] and others [30] reported that the skin bacterium Staphylococcus aureus has the ability to reduce some azo dyes to aromatic amines, which raises potential safety concerns regarding human dermal exposure to azo dyes such as those in tattoo ink and cosmetic colorant formulations. To comprehensively investigate azo dye-induced toxicity, it is very critical to understand the metabolic activation of azo dyes at the systems biology level.
In the 1950s, Mueller and Miller [25] and others [14] reported that mammalian liver microsomes or other organ homogenate including kidney, lung, heart and brain, reduced azo dyes, which suggested the reduction require one of multiple azoreductases with different reaction mechanisms. Microorganisms including intestinal and skin microbiota in vitro cultures were also reported of having the capability to reduce azo dyes in a fairly general and non-specific mechanism [7, 15, 16, 38], which has been well reviewed [8, 9, 13, 39]. It has been reported that benzidine-based azo dyes are reduced to their corresponding benzidine derivatives after incubation with anaerobic intestinal bacteria isolated from human, monkey and rat feces [1, 2]. Although azo dye metabolism has been widely studied [20, 24], there are still many unanswered questions such as the preferable reduction bond in an azo dye containing multiple azo bonds, and whether the location of microbial enzymatic reduction is intra- or extracellular or on cell membranes. Some studies suggested that bacteria which decolorize azo dyes require the presence of azo dye degrading enzymes and transport systems allowing the uptake of the azo dyes into the bacterial cells [12, 18]. Some other studies also suggested that decolorization of azo dye is mainly an extracellular process [19, 29], although how the extracellular azoreductases access to reduced nicotinamide adenine dinucleotide (NADH) is unclear [12]. Thus, it is still not certain where the reduction reaction occurs in cells although bacterial azoreductases were purified and identified as intracellular flavor proteins [4, 5, 43, 44]. Further, a major concern of the traditional spectrophotometric measurement of azo dye reduction is the non-specific absorption to bacterial cell walls and/or membranes, which varies widely between dyes. Therefore, novel emerging methodological approaches like liquid chromatography coupled to mass spectrometry (LC/MS)-based metabolomics are available to provide new information for assisting to solve these questions.
LC/MS-based metabolomics is one of the major analytical platforms used to semi-quantitatively examine the changes in endogenous and exogenous compounds (i.e., metabolites) in response to treatment or physiological changes. This technique has been successfully applied in drug metabolites profiling to discover novel drug metabolites at a global view [33, 34]. In this study, S. aureus, one of prevalent species of skin bacteria [6, 10], was used to evaluate effects of two azo dyes, lipid-soluble Sudan III and water-soluble Orange II, on skin bacterial metabolome. Most importantly, the azo dye biotransformation by S. aureus was examined as such to provide novel information on metabolite product formation and to understand the enzymatic mechanism using LC/MS-based metabolomics for the analysis of the cell extracts.
Materials and methods
Chemicals
Optima LC/MS grade acetonitrile and water were purchased from Thermo Fisher Scientific (Pittsburgh, PA, USA). Sudan III [1-(4-(phenylazo)phenylazo)-2-naphthol] and Orange II [4-(2-hydroxy-1-naphthylazo) benzenesulfonic acid sodium salt], dimethylsulfoxide (DMSO), formic acid, leucine-enkephalin, L-tryptophan, and all MS standards were obtained from Sigma-Aldrich Corporation (St. Louis, MO, USA).
Bacterial culture and treatment
Staphylococcus aureus ATCC 25923 purchased from the American Type Culture Collection was cultured as previously described [28]. In brief, the bacterium was inoculated into Brain Heart infusion (BHI) broth at a ratio of 1% (v/v) at 37 °C. The cells were treated with DMSO (vehicle, eight replicates). Stock solutions of Sudan III or Orange II were added to the BHI medium at final concentrations of 10 μg/ml (each with quintuplet replicates) at 37 °C for 18 h without agitation [28]. After incubation, four replicates of controls were boiled for 10 min followed by cooling down to the room temperature. Sudan III or Orange II was then added to the cooled culture to 10 μg/ml final concentrations. These samples were used as dead cell controls (DC). The number of bacteria in the culture media was counted by flow cytometry (FCM) on an Accuri C6 flow cytometer [27]. Bacterial cells were harvested by centrifugation at 4 °C for 10 min at 5000×g (Thermo Scientific Sorvall RC 6 + Centrifuge), and the collected bacterial cell pellets were frozen at −80 °C until analysis.
Sample preparation
Cell pellets (~1010 cells) suspended in 200 μL ice-cold water were processed the same as previously described [37]. In brief, an aliquot (120 μL) of the cell suspension was transferred into a tube containing 450 μL methanol, vortexed, and then kept at 4 °C for 15 min. The cell lysate by PRECELLYS® 24 homogenizer (Bertin Co., Rockville, MD, USA) was centrifuged at 16,060×g for 12 min at 4 °C to precipitate proteins. The resulting supernatant was then transferred into a clean tube and evaporated to dryness using a SpeedVac concentrator (Thermo Scientific, Waltham, MA, USA). The samples were reconstituted in 200 μL 95:5 water/acetonitrile, vortexed for 2 min, and kept at 4 °C for 20 min. The resulting solution was then centrifuged at 16,060×g for 12 min at 4 °C. The supernatant was transferred to autosampler vials for LC/MS analysis.
Open metabolic profiling
A 3 μL aliquot of cell pellet extracted supernatant after methanol precipitation was introduced into a Waters Acquity ultra performance liquid chromatography (UPLC)/QT of system (Waters, Milford, MA, USA) equipped with a Waters bridged ethyl hybrid (BEH) C8 column with a dimension of 2.1 mm × 10 cm and 1.7 μm particle size. The same chromatography method was used for metabolite separation as described previously [37]. Mass spectrometric data were collected with a Waters QTof Premier mass spectrometer (Waters, Milford, MA, USA) operated in positive and negative ionization electrospray modes. Mass spectrometer set-up parameters and raw data processing method were the same as described previously [35–37]. Full scan mode from m/z 100 to 900 Da was used for data acquisition. The identity of compounds was based on the combined information of accurate mass measurement, fragmentation mass spectra, and compared data from a free online database (www.hmdb.ca). Some compounds were confirmed by authentic standards.
A quality control (QC) sample composed of 40 common chemicals for LC/MS open profiling, was evaluated after every 10 sample runs. UPLC/QTof-MS spectra of pooled bacterial cells or pooled media were acquired from every 10 sample runs for pooled bacterial cells or pooled media to monitor analytical equipment variability.
Statistics
In the metabolome analyses, the values in the treated groups and the respective control group were compared. The data were analyzed by Student’s t test (MS EXCEL). A value of p < 0.05 was considered statistically significant.
Results
The growth of Staphylococcus aureus in BHI broth in the presence of 10 μg/ml Sudan III or Orange II compared to the controls was evaluated. Bacterial cultures with vehicle were used as viable controls (VC), while boiled bacterial cultures mixed with either of the azo dyes at 10 μg/ml concentration were used as dead cell controls (DC) to take into account for physical absorption of azo dyes onto the cell walls or membrane. Bacterial cultures were incubated at 37 °C for 18 h under static aerobic conditions. No significant changes in cell counts were observed after 18-h incubation with either of the azo dyes compared with their corresponding controls (~109 cells/ml) similar to those reported previously [27]. Therefore, by comparing the azo dye treated to VC and DC samples, the concentration changes in azo dyes and their metabolites would be only due to metabolism by the skin bacteria.
Detection of azo dye-related metabolites
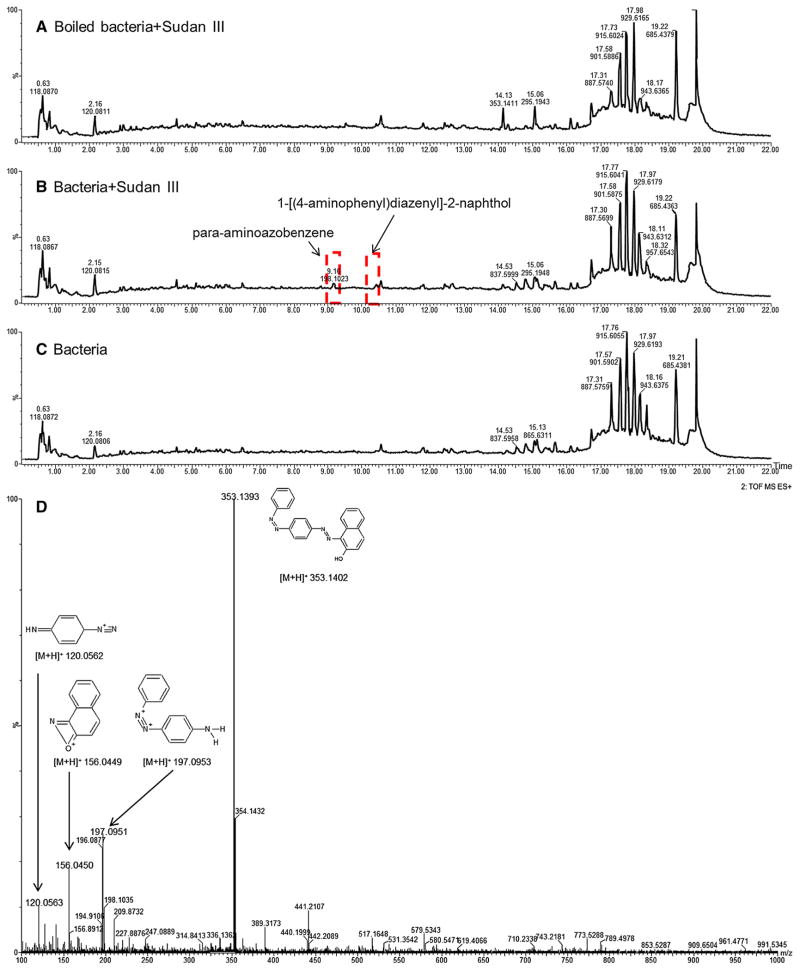
Both positive and negative ionization LC/MS modes were employed to analyze the cell pellet samples for the azo dyes and their metabolites. Interestingly, results from the positive LC/MS data provided more Sudan III metabolite information while LC/MS data from negative ionization mode contained more information for Orange II metabolites. This is due to different physiochemical characteristics of the compounds. Figure 1 displays representative total-ion chromatograms (TIC) of LC/MS from boiled bacteria mixed with Sudan III (A), bacteria treated with Sudan III (B), and bacteria with no treatment (C). From visual observation, there are distinct changes in the chromatogram from treated bacteria compared with DC and VC samples. Peaks at 9.16 and 10.39 min appeared in the treated sample (Fig. 1b) but were absent or very low in the DC or VC samples (Fig. 1a, c). These peaks were detected and identified as para-aminoazobenzene and 1-[(4-ami-nophenyl) diazenyl]-2-naphthol—Sudan III metabolites based on accurate mass, and fragment mass spectra. Figure 1d shows the fragment ions of Sudan III at m/z 197.0953, 156.0449 and 120.0562 along with the fragment structures. This fragmentation information confirms that the ion at m/z 353.1402 is indeed Sudan III. Other Sudan III and Orange II metabolites were identified in similar methods. The identities of the metabolites were based on accurate m/z mass measurement as well as based on the fragment mass spectra.
Fig. 1.
Typical positive ionization mode of total-ion chromatograms of cell pellets from boiled bacteria (a) viable bacteria incubated with 10 μg Sudan III/ml medium (b) and without Sudan III (c) at 37 °C for 18 h. d The fragment mass spectrum of Sudan III. The two chromatogram peaks in the rectangles only show in the viable bacteria incubated with Sudan III
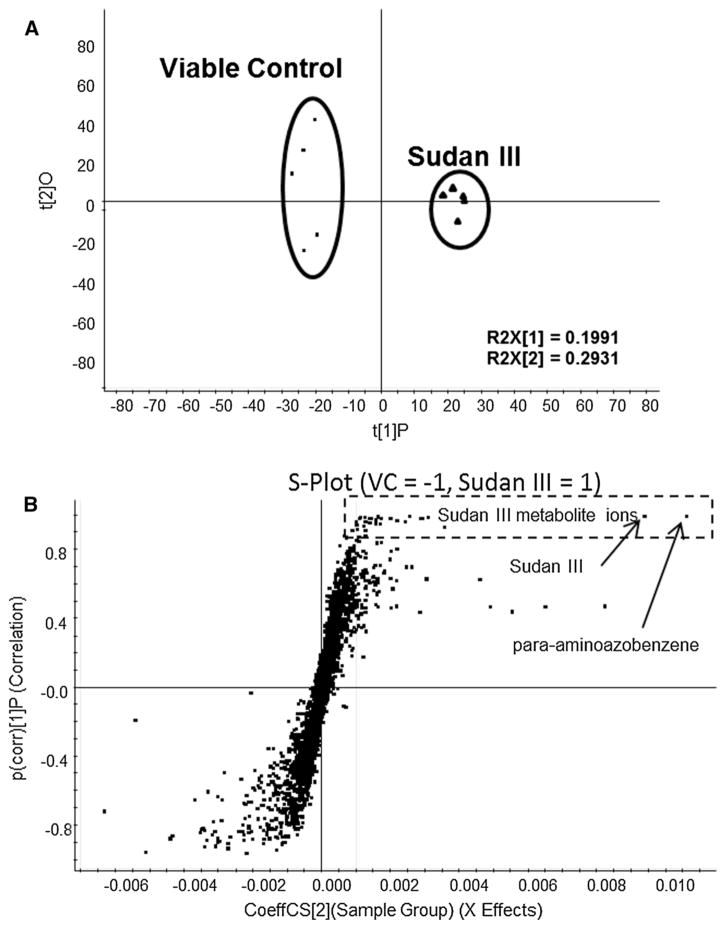
To detect and identify Sudan III metabolites globally, partial least squares discriminant analysis (PLS-DA) of LC/MS data from VC and treated cell pellet samples was used to determine azo dye metabolites. Figure 2 shows the scores plot and S-plot (Fig. 2a, c) from PLS-DA analysis of the positive LC/MS data from Sudan III treatment. The scores plot shows a clear separation of the treated samples from the VC samples. The ion features of the S-plot (Fig. 2b) in the rectangle box are responsible for the group separations, which were identified as Sudan III and its metabolites. Orange II-related metabolites were detected and identified using PLS-DA analysis of negative LC/MS data from Orange II treated samples compared to VC (data not shown).
Fig. 2.
The scores plot (a) and S-plot (b) from PLS-DA analysis of LC/MS data in positive ionization mode from cell extracts after treatment with DMSO (viable control) and 10 μg/ml Sudan III, incubated at 37 °C for 18 h without agitation. Five replicates were included in control and Sudan III treated groups, respectively. The Sudan III-related metabolites were located in the rectangle with dotted lines, and major metabolite ions were labeled
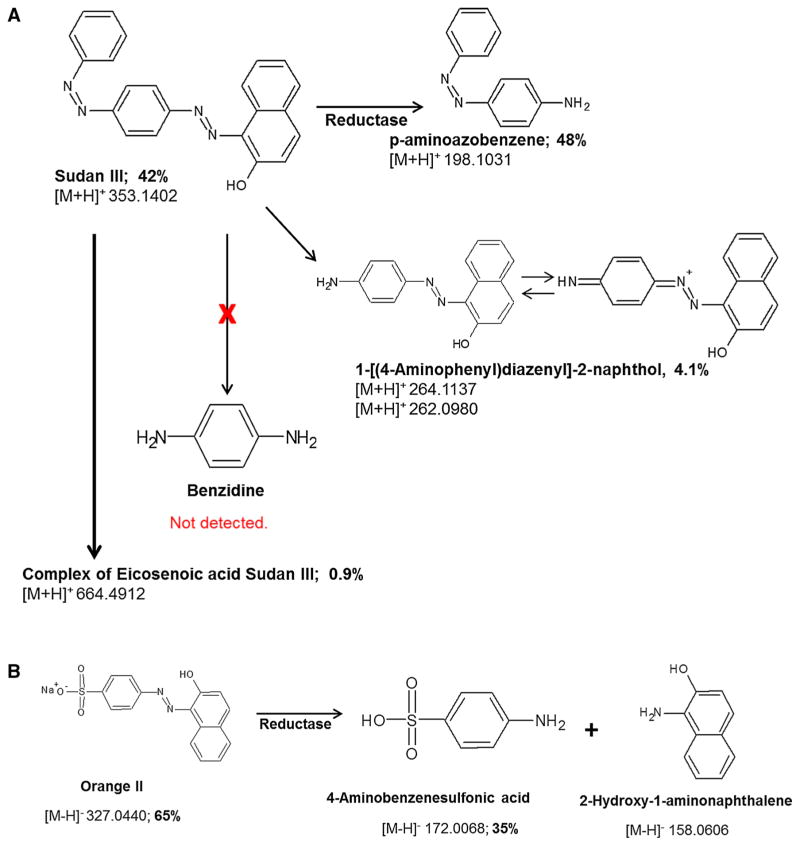
Table 1 summarizes the average intensities of azo dye—related metabolites in DC, treated and VC cell pellets. Three major metabolites of Sudan III were observed as para-aminoazobenzene, 1-[(4-aminophenyl)diazenyl]-2-naphthol and eicosenoic acid coupled with Sudan III in the treated samples. These metabolites were not detected or present at extremely low levels in the VC samples. Comparing with DC, eicosenoic acid coupled with Sudan III was only in the treated samples, and para-aminoazobenzene, 1-[(4-ami-nophenyl)diazenyl]-2-naphthol was found at very high concentration in the treated samples (>9× of DC). The concentration of the total Sudan III (including Sudan III salt) was lower (50%) in the treated samples since the viable bacteria was capable of metabolizing Sudan III. Two major metabolites of Orange II were detected and identified as 4-amin-obenzenesulfonic acid and 2-hydroxy-1-aminonaphthalene in the Orange II treated samples. These metabolites were not detected or present at extremely low levels in both DC and VC samples. The azo dye treated S. aureus cells contained more Orange II (>54×) than the DC samples.
Table 1.
Detected azo dye metabolites and their intensity data from cell pellets
| Azo dye metabolitesa | Average intensity ± STD
|
||
|---|---|---|---|
| Dead control With azo dye |
Sudan III | Viable control No azo dye |
|
| Sudan III metabolites | |||
| para -Aminoazobenzene | 7.15 ± 1.14 | 63.98 ± 4.52 | 0.18 ± 0.40 |
| 1-[(4-aminophenyl)diazenyl]-2-naphthol | 0.06 ± 0.13 | 5.42 ± 0.67 | 0.00 ± 0.00 |
| Sudan III | 93.85 ± 75.67 | 50.00 ± 3.93 | 0.00 ± 0.00 |
| Sudan III K | 2.87 ± 0.59 | 0.89 ± 0.22 | 0.00 ± 0.00 |
| Sudan III sodium | 17.19 ± 2.52 | 4.89 ± 0.65 | 0.00 ± 0.00 |
| Eicosenoic acid Sudan III | 0.00 ± 0.00 | 1.25 ± 0.38 | 0.00 ± 0.00 |
|
| |||
| Orange II metabolites | Dead control With azo dye |
Orange II | Viable control No azo dye |
|
| |||
| 4-aminobenzenesulfonic acid | 0.86 ± 0.85 | 52.15 ± 14.19 | 0.95 ± 0.39 |
| 2-hydroxy-1-aminonaphthalene | 0.00 ± 0.00 | 3.03 ± 0.69 | 0.00 ± 0.00 |
| Orange II | 1.76 ± 0.27 | 95.93 ± 12.80 | 0.00 ± 0.00 |
All of the detected azo dye metabolites were statistically significant compared to either DC or VC
Figure 3 displays the chemical structures of Sudan III and Orange II metabolites (Fig. 3a, b) along with their detected m/z and metabolism percentage produced by S. aureus. Based on the chemical structures, Sudan III and its three major metabolites should have similar positive ionization efficacy since all of them belong to the same class of compounds and contain amine and aromatic groups. It is very common in the metabolomics field [11] to semi-quantitatively assess compound levels using the signal intensity data, which is associated with the concentrations or masses of the compounds in the same chemical class. To determine which metabolism is the major pathway, the metabolite percentage was calculated based on the intensity data (Table 1). Figure 3a shows that 42% of Sudan III was un-metabolized and the percentage of p-aminoazobenzene was 48%, while the percentage of 1-[(4-aminophenyl)diazenyl]-2-naphthol was 4.1%. The big difference between the amounts of the two metabolites produced suggests that, the reduction of the two azo bonds were not equal. The azo bond located close to the naphthol group was preferred to be reduced compared to the other azo bond. The higher percentage of the metabolite indicates that more Sudan III was metabolized in this pathway. Only 0.9% of Sudan III was coupled with eicosenoic acid. No benzidine was detected in this study, which indicates the two azo bonds were not reduced simultaneously. Figures 3b shows that 65% of Orange II was intact and the percentage of 4-aminoben-zenesulfonic acid was 35%. Orange II and 4-aminoben-zenesulfonic acid have similar ionization efficacy since both contain sulfonic acid group which is easily negative charged, while 2-hydroxy-1-aminonapthalene does not contain sulfonic acid group so the ionization efficacy is different from the other two. Therefore, the metabolism extent of Orange II was only based on the intensity data of Orange II and 4-aminobenzenesulfonic acid, while the percentage of 2-hydroxy-1-aminonapthalene was not calculated.
Fig. 3.
Potential metabolic pathways for Sudan III (a) and Orange II (b) by S. aureus, along with structures and monoisotopic [M + H]+ for Sudan III metabolites or [M-H]− for Orange II metabolites
Metabolome changes in S. aureus cells induced by azo dye treatment
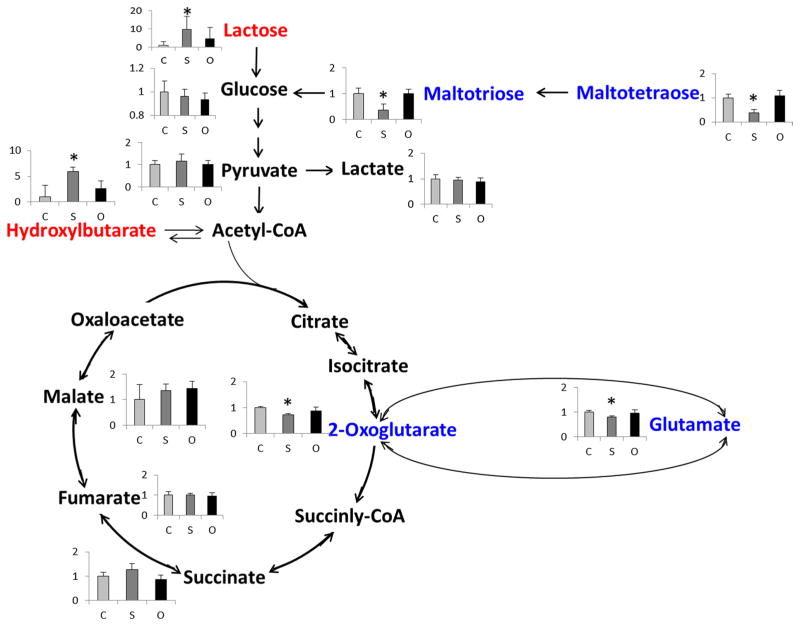
In total, 101 unique endogenous metabolites of S. aureus cells were detected and the intensity ratio data (treated/VC) was reported in the supplemental data (Table S1). The detected metabolites included co-factor, amino acids, phenyl-containing organic acids, nucleotides, short chain fatty acids and other compounds. To examine the azo dye-induced effects on S. aureus cells, the detected metabolites were classified by the pathways that the metabolites are involved. Among the detected 101 metabolites, eight metabolites significantly increased and 17 significantly decreased by Sudan III treatment, while only one metabolite significantly increased and three significantly decreased by Orange II treatment. Relative more metabolites changes in the Sudan III treatment indicate that Sudan III might be more toxic to S. aureus cells compared to Orange II. Furthermore, the most noticeable change in metabolites was that nine energy pathway-related metabolites decreased and three increased after Sudan III treatment. Figure 4 displays the detected metabolites involved in the citric acid cycle (TCA) pathway and its connections with other energy pathways. 2-Oxoglu-tarate (2-OG) and glutamate significantly decreased in the Sudan III treated cells. Polysaccharides including maltotetraose and maltotriose were significantly decreased, while lactose was significantly increased in the Sudan III treatment group. Hydroxybutyric acid, from ketosis, significantly increased to compensate 2-OG depletion in the Sudan III treatment group. No significant changes in the metabolites (Fig. 4) were observed in the Orange II treatment group.
Fig. 4.
Observed intensity changes in metabolites mapped onto the energy metabolism pathway. * p < 0.05, Sudan III or Orange II vs VC. Note: metabolites in red were significantly increased; metabolites in blue were significantly decreased
Discussion
Staphylococcus aureus is one of prevalent species of skin bacteria [6, 10]. Our previous studies [27, 28] have shown that S. aureus is able to metabolize 76% Sudan III and 60% Orange II at 6 μg/ml for 24-h incubation. Spectrophotometric measurements were used to measure the absorbance changes in the cell culture media before and after the incubations. In this study, results showed that ~58% Sudan III and ~35% Orange II was metabolized at 10 μg/ml for 18-h incubation (Fig. 3). LC/MS-based metabolomics was used to analyze the cell pellets extracts to evaluate azo dye metabolites levels changes. Reduction rates of the azo dyes in the current work are comparable to our previous reports [27] considering shorter incubation time (24 vs 18 h), higher concentration levels of azo dye in the culture incubation media and different analytical technologies that were used in this investigation for the measurements, i.e., spectrophotometric measurement vs LC/MS in different biomatrices (cell media vs cell pellet).
Benzidine—a common metabolite of diazo compounds such as Sudan III [2]—was not detected in the cell pellets of S. aureus cultures treated with 10 μg/ml Sudan III. This indicates that the two azo bonds were not reduced simultaneously by S. aureus for 18 h incubation under non-agitation condition. Further, the level of p-aminoazobenzene was approximately >12-fold of 1-[(4-aminophenyl) diazenyl]-2-naphthol, which indicates that the two azo linkages were not equally reduced suggesting the azo bond closest to the naphthol group was preferably reduced by S. aureus. The direction of the reduction depends on the physiochemical structure of Sudan III that the electron-withdrawing capability of the hydroxyl group on the naphthol is stronger which causes different electronegativities of the two nitrogen atoms of the azo bond next to naphthol. Such difference in electronegativities can result in destabilizing the azo linkage next to naphthol. Indeed, much more p-aminoazobenzene (product of the cleavage of the azo bond next to naphthol) was observed. Results from this study provide new information regarding the reduction preference of the two azo linkages, which to our knowledge is unexplored [13, 40].
Approximately 42% Sudan III and 65% Orange II (Fig. 3) remained intact after 18-h incubation with S. aureus. The difference in the metabolism extent could be related to the way the azo dyes are transported onto the cell membranes or into the cells. Since Sudan III is a lipid-soluble dye while Orange II is a water-soluble polar azo dye, it has been reported that understanding the azo dye solubility is very important to explain azo compounds metabolism [38]. Indeed, Gingell et al. [15] reported that the water-soluble neoprontosil, poorly absorbed in the intestine tract was metabolized mainly by intestinal microorganism while the lipid-soluble prontosil, easily absorbed in the intestine, was reduced by both the intestinal bacteria and the liver [15]. This raises a question whether the highly charged sulfonate Orange II can attach to or even pass through the cell membrane into the S. aureus cells. The big difference (~54-fold, Table 1) of the Orange II levels from the viable cells vs the dead cells taking into account the non-specific absorption by the cells suggests that the viable S. aureus cells do absorb Orange II. This result indicates that the azoreduction might occur in the cells or on the cell membrane. However, it is still unclear how Orange II was attached to or getting into the S. aureus cells. In contrast, Sudan III is a lipid-soluble compound, and can be easily attached to or absorbed by the bacteria cells. This is supported by the current investigation that the dead cells had ~twofold levels of Sudan III vs the viable cells (Table 1), which indicates that the cell membranes even from the dead cells can absorb Sudan III. Sudan III contains azo (–N=N–) and hydroxyl groups (–OH), which can form hydrogen bonds with the polar head groups of the cell membrane phospholipids. The detection of the Sudan III coupled to eicosenoic acid (Table 1) provided supportive evidence for the first time that Sudan III can form hydrogen-bonded complex in living bacterial cells which are able to actively break down the dye. As such, it is rational to hypothesize that Sudan III attaches to or penetrates into the cells through hydrogen-bonded complexes. This hypothesis is consistent with the findings by Li et al. [21], who studied the bonding of Sudan II- and Sudan IV- E. coli membrane phospholipid complexes. Li et al. reported that >60% of Sudan II and IV get into the cells while 90% of which is remained in the cell membrane although the apparent reduction of Sudan dyes was not observed in E. coli [41].
The impact of Sudan III and Orange II on the S. aureus cells was also evaluated by measuring the endogenous metabolite changes in the cells. Results showed that more effects caused by Sudan III vs Orange II, which indicates that Sudan III might be comparatively more toxic than Orange II. This result is consistent with our previous study [27] that Sudan III is able to inhibit the bacterial growth at 36 μg/ml in 48-h incubation, while Orange II had no effect on either the cell growth or the cell viability. The apparent changed metabolites (including 2-OG, hydroxybutarate and carbohydrates) by Sudan III were involved in the tricarboxylic acid cycle (TCA) pathway (Fig. 4), critical for cellular energy production. The metabolite 2-OG, an important metabolite for regulation of nitrogen and carbon metabolism as well as a precursor metabolite for reducing factors like reduced nicotinamide dinucleotide and reduced flavin adenine dinucleotide and amino acids, significantly decreased in the cells treated by Sudan III. Decreases in 2-OG indicates that the energy production through aerobic metabolism was inhibited by Sudan III treatment. Increases in hydroxybutarate through anaerobic ketosis metabolism and lactose from carbohydrates indicate the response of the S. aureus cells to compensate the energy loss from the aerobic metabolism. Furthermore, 2-OG has been reported [23] as a key molecule in detoxifying reactive oxygen species (ROS), which depend on reduced nicotinamide dinucleotide phosphate (NADPH). In addition, we [5] and others [43, 44] discovered and purified azoreductases [which requires NAD(P) H as electron source] from S. aureus or other bacteria that can reduce azo dyes. Thus, the decreases in 2-OG could be a unique strategy for the S. aureus to adapt the cellular redox environment changes by Sudan III.
Conclusion
The metabolism of the lipid-soluble Sudan III and water-soluble Orange II by S. aureus was investigated using LC/MS-based metabolomics technique. No benzidine was detected in the cells treated by Sudan III, which indicates that the two azo linkages were not reduced simultaneously. More p-aminoazobenzene (>12-fold) than 1-[(4-aminophe-nyl) diazenyl]-2-naphthol was formed indicating that the azo linkage close to naphthol group of Sudan III was preferably reduced by S. aureus. The finding that viable cells contained more Orange II (~54-fold) vs the dead cells suggests that the viable S. aureus cells do absorb Orange II and this azo dye is reduced on the cell membrane or intracellular. Furthermore, the detection of the complex of Sudan III with eicosenoic acid (Table 1) provided supportive evidence that Sudan III can form hydrogen-bonded complex, which indicates that Sudan III might be transported into the cells or onto the cell walls through formation of a hydrogen-bonded complex. The decrease in 2-OG suggests that Sudan III disturbed the cellular redox environment of the S. aureus cells. Results from this study provide novel information regarding mechanism of the azo dye metabolism by S. aureus as well as the effects of the azo dye on the bacteria at the metabolite levels, which will be useful in the risk assessment process when evaluating azo dye ingredients in cosmetics. Since the human skin microbiome is a diverse and complex ecosystem, future research to evaluate the effects of colorants on the other bacteria living on human skin, is warranted.
Supplementary Material
Acknowledgments
We thank Drs. Li-Rong Yu and Jing Han for their critical review of this manuscript. This study was funded by National Center for Toxicological Research, United States Food and Drug Administration, and supported in part by appointment (JJ) in the Postgraduate Research Fellowship Program by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and the US Food and Drug Administration.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10295-017-1970-8) contains supplementary material, which is available to authorized users.
Disclaimer: The findings and conclusions in this publication are those of the authors and do not represent FDA positions or policies.
References
- 1.Cerniglia CE, Freeman JP, Franklin W, Pack LD. Metabolism of benzidine and benzidine-congener based dyes by human, monkey and rat intestinal bacteria. Biochem Biophys Res Commun. 1982;107:1224–1229. doi: 10.1016/s0006-291x(82)80128-9. [DOI] [PubMed] [Google Scholar]
- 2.Cerniglia CE, Freeman JP, Franklin W, Pack LD. Metabolism of azo dyes derived from benzidine, 3,3′-dimethyl-benzidine and 3,3′-dimethoxybenzidine to potentially carcinogenic aromatic amines by intestinal bacteria. Carcinogenesis. 1982;3:1255–1260. doi: 10.1093/carcin/3.11.1255. [DOI] [PubMed] [Google Scholar]
- 3.Cerniglia CE, Zhuo Z, Manning BW, Federle TW, Heflich RH. Mutagenic activation of the benzidine-based dye direct black 38 by human intestinal microflora. Mutat Res. 1986;175:11–16. doi: 10.1016/0165-7992(86)90138-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Wang RF, Cerniglia CE. Molecular cloning, overexpression, purification, and characterization of an aerobic FMN-dependent azoreductase from Enterococcus faecalis. Protein Expr Purif. 2004;34:302–310. doi: 10.1016/j.pep.2003.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Hopper SL, Cerniglia CE. Biochemical and molecular characterization of an azoreductase from Staphylococcus aureus, a tetrameric NADPH-dependent flavoprotein. Microbiology. 2005;151:1433–1441. doi: 10.1099/mic.0.27805-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen YE, Tsao H. The skin microbiome: current perspectives and future challenges. J Am Acad Dermatol. 2013;69:143–155. doi: 10.1016/j.jaad.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Childs JJ, Nakajima C, Clayson DB. The metabolism of 1-phenylazo-2-naphthol in the rat with reference to the action of the intestinal flora. Biochem Pharmacol. 1967;16:1555–1561. doi: 10.1016/0006-2952(67)90133-5. [DOI] [PubMed] [Google Scholar]
- 8.Chung KT. The significance of azo-reduction in the mutagenesis and carcinogenesis of azo dyes. Mutat Res. 1983;114:269–281. doi: 10.1016/0165-1110(83)90035-0. [DOI] [PubMed] [Google Scholar]
- 9.Chung KT, Stevens SE, Jr, Cerniglia CE. The reduction of azo dyes by the intestinal microflora. Crit Rev Microbiol. 1992;18:175–190. doi: 10.3109/10408419209114557. [DOI] [PubMed] [Google Scholar]
- 10.Cundell AM. Microbial ecology of the human skin. Microb Ecol. 2016 doi: 10.1007/s00248-016-0789-6. [DOI] [PubMed] [Google Scholar]
- 11.Dettmer K, Aronov PA, Hammock BD. Mass spectrometry-based metabolomics. Mass Spectrom Rev. 2007;26:51–78. doi: 10.1002/mas.20108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng J, Heinze TM, Xu H, Cerniglia CE, Chen H. Evidence for significantly enhancing reduction of Azo dyes in Escherichia coli by expressed cytoplasmic Azoreductase (AzoA) of Enterococcus faecalis. Protein Pept Lett. 2010;17:578–584. doi: 10.2174/092986610791112701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng J, Cerniglia CE, Chen H. Toxicological significance of azo dye metabolism by human intestinal microbiota. Front Biosci (Elite Ed) 2012;4:568–586. doi: 10.2741/400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fouts JR, Kamm JJ, Brodie BB. Enzymatic reduction of prontosil and other azo dyes. J Pharmacol Exp Ther. 1957;120:291–300. [PubMed] [Google Scholar]
- 15.Gingell R, Bridges JW, Williams RT. Gut flora and the metabolism of prontosils in the rat. Biochem J. 1969;114:5P–6P. doi: 10.1042/bj1140005p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gingell R, Walker R. Mechanisms of azo reduction by Streptococcus faecalis. II. The role of soluble flavins. Xenobiotica. 1971;1:231–239. doi: 10.3109/00498257109033172. [DOI] [PubMed] [Google Scholar]
- 17.Griswold DP, Jr, Casey AE, Weisburger EK, Weisburger JH. The carcinogenicity of multiple intragastric doses of aromatic and heterocyclic nitro or amino derivatives in young female sprague-dawley rats. Cancer Res. 1968;28:924–933. [PubMed] [Google Scholar]
- 18.Haug W, Schmidt A, Nortemann B, Hempel DC, Stolz A, Knackmuss HJ. Mineralization of the sulfonated azo dye mordant Yellow 3 by a 6-aminonaphthalene-2-sulfonate-degrading bacterial consortium. Appl Environ Microbiol. 1991;57:3144–3149. doi: 10.1128/aem.57.11.3144-3149.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keck A, Klein J, Kudlich M, Stolz A, Knackmuss HJ, Mattes R. Reduction of azo dyes by redox mediators originating in the naphthalenesulfonic acid degradation pathway of Sphingomonas sp. strain BN6. Appl Environ Microbiol. 1997;63:3684–3690. doi: 10.1128/aem.63.9.3684-3690.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levine WG. Metabolism of azo dyes: implication for detoxication and activation. Drug Metab Rev. 1991;23:253–309. doi: 10.3109/03602539109029761. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Gao HW, Ren JR, Chen L, Li YC, Zhao JF, Zhao HP, Yuan Y. Binding of Sudan II and IV to lecithin liposomes and E. coli membranes: insights into the toxicity of hydrophobic azo dyes. BMC Struct Biol. 2007;7:16. doi: 10.1186/1472-6807-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lowry LK, Tolos WP, Boeniger MF, Nony CR, Bowman MC. Chemical monitoring of urine from workers potentially exposed to benzidine-derived azo dyes. Toxicol Lett. 1980;7:29–36. doi: 10.1016/0378-4274(80)90081-8. [DOI] [PubMed] [Google Scholar]
- 23.Mailloux RJ, Beriault R, Lemire J, Singh R, Chenier DR, Hamel RD, Appanna VD. The tricarboxylic acid cycle, an ancient metabolic network with a novel twist. PLoS One. 2007;2:e690. doi: 10.1371/journal.pone.0000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin CN, Kennelly JC. Metabolism, mutagenicity, and DNA binding of biphenyl-based azodyes. Drug Metab Rev. 1985;16:89–117. doi: 10.3109/03602538508991431. [DOI] [PubMed] [Google Scholar]
- 25.Mueller GC, Miller JA. The reductive cleavage of 4-dimeth-ylaminoazobenzene by rat liver; the intracellular distribution of the enzyme system and its requirement for triphosphopyridine nucleotide. J Biol Chem. 1949;180:1125–1136. [PubMed] [Google Scholar]
- 26.Nony CR, Bowman MC, Cairns T, Lowry LK, Tolos WP. Metabolism studies of an azo dye and pigment in the hamster based on analysis of the urine for potentially carcinogenic aromatic amine metabolites. J Anal Toxicol. 1980;4:132–140. doi: 10.1093/jat/4.3.132. [DOI] [PubMed] [Google Scholar]
- 27.Pan H, Feng J, Cerniglia CE, Chen H. Effects of Orange II and Sudan III azo dyes and their metabolites on Staphylococcus aureus. J Ind Microbiol Biotechnol. 2011;38:1729–1738. doi: 10.1007/s10295-011-0962-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan H, Xu J, Kweon OG, Zou W, Feng J, He GX, Cerniglia CE, Chen H. Differential gene expression in Staphylococcus aureus exposed to Orange II and Sudan III azo dyes. J Ind Microbiol Biotechnol. 2015;42:745–757. doi: 10.1007/s10295-015-1599-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearce CI, Christie R, Boothman C, von Canstein H, Guthrie JT, Lloyd JR. Reactive azo dye reduction by Shewanella strain J18 143. Biotechnol Bioeng. 2006;95:692–703. doi: 10.1002/bit.21021. [DOI] [PubMed] [Google Scholar]
- 30.Platzek T, Lang C, Grohmann G, Gi US, Baltes W. Formation of a carcinogenic aromatic amine from an azo dye by human skin bacteria in vitro. Hum Exp Toxicol. 1999;18:552–559. doi: 10.1191/096032799678845061. [DOI] [PubMed] [Google Scholar]
- 31.Rinde E, Troll W. Metabolic reduction of benzidine azo dyes to benzidine in the rhesus monkey. J Natl Cancer Inst. 1975;55:181–182. doi: 10.1093/jnci/55.1.181. [DOI] [PubMed] [Google Scholar]
- 32.Stingley RL, Zou W, Heinze TM, Chen H, Cerniglia CE. Metabolism of azo dyes by human skin microbiota. J Med Microbiol. 2010;59:108–114. doi: 10.1099/jmm.0.012617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun J, Schnackenberg LK, Beger RD. Studies of aceta-minophen and metabolites in urine and their correlations with toxicity using metabolomics. Drug Metab Lett. 2009;3:130–136. doi: 10.2174/187231209789352139. [DOI] [PubMed] [Google Scholar]
- 34.Sun J, Von Tungeln LS, Hines W, Beger RD. Identification of metabolite profiles of the catechol-O-methyl transferase inhibitor tolcapone in rat urine using LC/MS-based metabonomics analysis. J Chromatogr B Anal Technol Biomed Life Sci. 2009;877:2557–2565. doi: 10.1016/j.jchromb.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 35.Sun J, Von Tungeln LS, Hines W, Beger RD. Identification of metabolite profiles of the catechol-O-methyl transferase inhibitor tolcapone in rat urine using LC/MS-based metabonomics analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:2557–2565. doi: 10.1016/j.jchromb.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 36.Sun J, Schnackenberg LK, Hansen DK, Beger RD. Study of valproic acid-induced endogenous and exogenous metabolite alterations using LC–MS-based metabolomics. Bioanalysis. 2010;2:207–216. doi: 10.4155/bio.09.173. [DOI] [PubMed] [Google Scholar]
- 37.Sun J, Jin J, Beger RD, Cerniglia CE, Yang M, Chen H. Metabolomics evaluation of the impact of smokeless tobacco exposure on the oral bacterium Capnocytophaga sputigena. Toxicol Vitro. 2016;36:133–141. doi: 10.1016/j.tiv.2016.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker R. The metabolism of azo compounds: a review of the literature. Food Cosmet Toxicol. 1970;8:659–676. doi: 10.1016/s0015-6264(70)80455-2. [DOI] [PubMed] [Google Scholar]
- 39.Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 2017;179:204–222. doi: 10.1016/j.trsl.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu H, Heinze TM, Chen S, Cerniglia CE, Chen H. Anaerobic metabolism of 1-amino-2-naphthol-based azo dyes (Sudan dyes) by human intestinal microflora. Appl Environ Microbiol. 2007;73:7759–7762. doi: 10.1128/AEM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu H, Heinze TM, Paine DD, Cerniglia CE, Chen H. Sudan azo dyes and Para Red degradation by prevalent bacteria of the human gastrointestinal tract. Anaerobe. 2010;16:114–119. doi: 10.1016/j.anaerobe.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida O. Etiological factors in bladder tumors. Nihon Hinyokika Gakkai Zasshi. 1973;64:707–712. [PubMed] [Google Scholar]
- 43.Zimmermann T, Kulla HG, Leisinger T. Properties of purified Orange II azoreductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur J Biochem. 1982;129:197–203. doi: 10.1111/j.1432-1033.1982.tb07040.x. [DOI] [PubMed] [Google Scholar]
- 44.Zimmermann T, Gasser F, Kulla HG, Leisinger T. Comparison of two bacterial azoreductases acquired during adaptation to growth on azo dyes. Arch Microbiol. 1984;138:37–43. doi: 10.1007/BF00425404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.