Abstract
Background
It has been found that single-nucleotide polymorphisms (SNPs) of microRNA might be involved in the development of inflammatory bowel diseases (IBDs). However, the related retrospective research has not been reported. In this work, we performed a meta-analysis to derive a more precise estimation of the associated relationship.
Methods
We searched the studies on the association of SNPs of microRNA with the hereditary susceptibility of IBD in PubMed and Embase; eligible research was selected by screening the abstract and full text. The meta-analysis was performed based on the statistical software Stata 14.0, and besides, the odds ratio and 95% confidence interval were calculated to evaluate the strength of the association.
Results
159 papers were acquired from the PubMed and Embase databases, and five eligible articles containing nine case-control studies were selected. In the study, we first found that the association between miRNA-196a2 rs11614913 and IBD was insignificant. Then, the susceptibility of miRNA-146a rs2910146 to IBD increased significantly in allelic comparison, homozygote model, heterozygote model, and dominant model. Moreover, a positive relationship between miRNA-499 rs3746444 and IBD was identified in the homozygote model.
Conclusion
Our findings demonstrated that miRNA-146a rs2910146 (G>C) polymorphism was associated with the susceptibility to IBD and miRNA-196a2 rs11614913 (T>C) and miRNA-499 rs3746444 (A>G) did not reveal an obvious relationship with the IBD susceptibility.
1. Introduction
Inflammatory bowel diseases (IBDs) are chronic multifactorial and relapsing inflammatory disorders of the gastrointestinal tract, including Crohn's disease (CD) and ulcerative colitis (UC). In recent years, the population of patients suffering the IBDs shows an increasing trend. The complex interaction among host immune system, intestinal flora, and environmental factors is considered to be the cause of the IBD pathogenesis [1, 2]. Hence, the identification of genetic factors involved in the development of IBD has attracted attention in the current research area [3].
MicroRNAs (miRNAs) are an abundant class of small, noncoding, and single-stranded RNAs, which play an important role in many biological processes, such as cellular proliferation, differentiation, apoptosis, immune response, and signal transduction by regulating gene expression [4]. It has been reported that many diseases, for example, inflammatory diseases and various cancers, will implicate an miRNA dysregulation [5, 6]. Though the majority of studies are focused on the miRNA expression, a considerable number of publications have reported that the single-nucleotide polymorphisms (SNPs) in miRNA genes could be linked to the genetic susceptibility to disease development.
In the literature, the scholars have reported several miRNA-SNPs, including miRNA-196a2 rs11614913, miRNA-146a rs2910146, and miRNA-499 rs3746444, that are associated with the susceptibility to various diseases such as rheumatoid arthritis (RA) [7, 8], systemic lupus erythematosus (SLE), and various cancers [9, 10]. In addition, some research has been proposed to investigate the association between the SNPs and the risk of IBD [11–15], while the results were inconclusive or even conflicting. Therefore, we performed a meta-analysis to examine how miRNA-196a2 rs11614913, miRNA-146a rs2910146, and miRNA-499 rs3746444 polymorphisms will affect the IBD susceptibility.
2. Methods and Materials
In this work, the meta-analysis was performed according to the statement guidelines of Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) [16] and the Cochrane handbook was used to lead the analysis.
2.1. Identification of Eligible Studies
The last search was updated on May 1, 2017. Specifically, we carried out a systematic search in the PubMed and Embase databases with two independent investigators. In the search, the following terms: “microRNA OR mirRNA,” “polymorphism,” and “inflammatory bowel disease OR ulcerative colitis OR Crohn disease,” were adopted and the publication date was not restricted. The reference lists were searched manually to identify potential candidates. Both of the investigators had received professional training in literature review and search, statistics, and evidence-based medicine.
2.2. Inclusion and Exclusion Criteria
In this meta-analysis, we also set up inclusion and exclusion criteria to select useful and/or meaningful literature. The selected research should meet the inclusion criteria as follows: (1) evaluation of the association between miRNA196a2/146a/499 and IBD, (2) independent case-control study for human beings, (3) detailed genotype data could be acquired to calculate the odds ratios (ORs) and 95% confidence intervals (CIs), and (4) if multiple studies from the same population and/or the same group were found, we only include the largest study. On the other hand, we apply the following exclusion criteria to eliminate redundant articles: (1) duplication of previous publications, (2) comments, reviews, and editorials, (3) family-based studies of pedigree, and (4) studies without detailed genotype data. In particular, based on the proposed inclusion and exclusion criteria, two investigators (Ying Liu and Lingxin Xiong) were assigned to select the literature independently by screening the title, abstract, and full text, and any dispute was solved by discussion.
2.3. Data Extraction
The data of the eligible studies were extracted in duplicate by two investigators independently. In each study, we collected the following information: name of the first author, year of publication, country of origin, ethnicity, characteristics of cases and controls, detective sample, genotyping methods, criteria of IBD, Hardy-Weinberg equilibrium (HWE), number of cases and controls, and genotype frequencies of cases and controls for miRNA-196a2, miRNA-146a, and miRNA-499, respectively. Different ethnic descents were classified as Caucasian and Asian. The two investigators had checked the statistical difference of the extracted data, and they should check up on work until consensus was achieved on every item.
2.4. Quality Score Assessment
According to the methodological quality assessment scale modified from previous research (Table 1 [17]), the quality of each selected work was scored by two authors, respectively. Five items were carefully checked based on this scale, including representativeness of cases, source of control, sample size, quality control of genotyping methods, and Hardy-Weinberg equilibrium. We used traditional epidemiological considerations to evaluate the quality scores (range from 0 to 10; a high score indicates the study has a good quality). Again, the two investigators solved the disagreements via discussion.
Table 1.
Characteristics of studies included in the meta-analysis.
| Study | Year | Country | Ethnicity | Control | Diseases | Case | P for HWE | Quality score | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| miR196a2rs11614913 T>C | CC | TC | TT | CC | TC | TT | ||||||
| Ranjha et al. | 2017 | India | Caucasian | 305 | 81 | 55 | UC | 102 | 81 | 14 | <0.0001 | 6 |
| Ciccacci et al. | 2017 | Italy | Caucasian | 101 | 115 | 23 | IBD | 171 | 172 | 38 | 0.234 | 8 |
| UC | 94 | 91 | 21 | |||||||||
| CD | 77 | 81 | 17 | |||||||||
| Zhu et al. | 2016 | China | Asian | 102 | 213 | 135 | IBD | 117 | 214 | 137 | 0.306 | 8 |
| UC | 67 | 108 | 66 | |||||||||
| CD | 50 | 106 | 71 | |||||||||
| Gazouli et al. | 2013 | Greece | Caucasian | 108 | 144 | 48 | IBD | 181 | 199 | 72 | 1 | 7 |
| UC | 98 | 91 | 21 | |||||||||
| CD | 83 | 108 | 51 | |||||||||
| Okubo et al. | 2011 | Japan | Asian | 75 | 206 | 122 | UC | 30 | 82 | 58 | 0.465 | 8 |
| miRNA146a rs2910164 G>C | GG | CG | CC | GG | CG | CC | ||||||
| Ciccacci et al. | 2017 | Italy | Caucasian | 20 | 88 | 144 | IBD | 39 | 188 | 277 | 0.213 | 8 |
| UC | 16 | 71 | 119 | |||||||||
| CD | 23 | 117 | 158 | |||||||||
| Zhu et al. | 2016 | China | Asian | 97 | 202 | 151 | IBD | 62 | 225 | 181 | 0.059 | 7 |
| UC | 32 | 113 | 96 | |||||||||
| CD | 30 | 112 | 85 | |||||||||
| Gazouli et al. | 2013 | Greece | Caucasian | 200 | 90 | 10 | IBD | 231 | 191 | 30 | 0.974 | 7 |
| UC | 126 | 78 | 6 | |||||||||
| CD | 105 | 113 | 24 | |||||||||
| Okubo et al. | 2011 | Japan | Asian | 74 | 178 | 151 | UC | 28 | 67 | 75 | 0.095 | 8 |
| miRNA499 rs3746444 A>G | AA | GA | GG | AA | GA | GG | ||||||
| Ranjha et al. | 2017 | India | Caucasian | 167 | 220 | 54 | UC | 97 | 35 | 65 | 0.154 | 8 |
| Ciccacci et al. | 2017 | Italy | Caucasian | 139 | 98 | 15 | IBD | 209 | 153 | 25 | 0.677 | 8 |
| UC | 108 | 87 | 11 | |||||||||
| CD | 101 | 66 | 14 | |||||||||
| Zhu et al. | 2016 | China | Asian | 339 | 105 | 6 | IBD | 357 | 105 | 6 | 0.504 | 8 |
| UC | 185 | 54 | 2 | |||||||||
| CD | 172 | 51 | 4 | |||||||||
| Okubo et al. | 2011 | Japan | Asian | 272 | 111 | 20 | UC | 102 | 62 | 6 | 0.055 | 7 |
2.5. Statistics Analysis
We followed the guidelines of the PRISMA checklists to conduct the associated meta-analysis. First, the HWE was assessed for each study by the chi-square test in the control group. The odds ratio (OR) and 95% confidence interval (CI) were calculated to evaluate the strength of the association between miRNA196a2/146a/499 SNPs and susceptibility of UC and CD. The pooled ORs were performed for allelic comparison (miR196a2: T versus C, miR146a: C versus G, and miR499: G versus A), homozygote model (miR196a2: TT versus CC, miR146a: CC versus GG, and miR499: GG versus AA), heterozygote model (miR196a2: TC versus CC, miR146a: CG versus GG, and miR499: GA versus AA), dominant model (miR196a2: TT + TC versus CC, miR146a: CC + CG versus GG, and miR499: GG + GA versus AA), and recessive model (miR196a2: TT versus TC + CC, miR146a: CC versus CG + GG, and miR499: GG versus GA + AA). The statistical significance level of the pooled OR was determined by a Z-test with a P value less than 0.05. Heterogeneity was assessed by the Q statistic (significance level of P < 0.1) and I2 statistic (the evidence of significant inconsistency can be revealed when it is greater than 50) [18]. If the heterogeneity was not significant, the fixed effects model was utilized to evaluate the summary OR and 95% CI. Otherwise, the random effects model was implemented.
As a further step, by deleting one study in each turn, the corresponding sensitivity analysis was carried out to evaluate the effect of each study on the combined ORs. Besides, subgroup analyses were stratified based on the ethnicity. All statistical analyses were performed with the software Stata 14.0 (StataCorp, College Station, TX, USA). Note that, except specified conditions, we considered the significance by P < 0.05.
3. Results
3.1. Characteristics of Studies
In this work, we acquired 159 studies in total from the PubMed and Embase databases and the literature review process was summarized as a flow chart shown in Figure 1. In the current study, according to the inclusion and exclusion criteria, 5 eligible case-control studies containing 1761 cases and 1892 controls were chosen in our meta-analysis [11–15]. The characteristics of each study are given in Table 1. Three of them are for Caucasian and the other two are for the Asian population. In each case, the genotype frequencies of miRNA-196a2 rs11614913, miRNA-146a rs2910146 and miRNA-499 rs3746444 were presented and each of them was treated as a separated study. Different genotyping methods were applied to the studies, including polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) [11, 13–15] and allelic discrimination assay by TaqMan® technology [12]. The genotyping distribution was in agreement with the HWE in all studies except the one proposed by Ranjha et al. [11].
Figure 1.
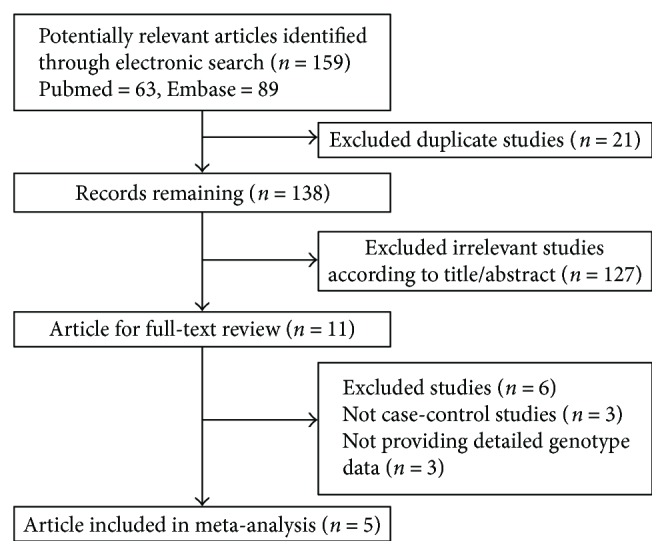
Characteristics of studies included in the meta-analysis.
3.2. Association between miRNA-196a2 and IBD
The pooled ORs among three SNPs and UC are presented in Table 2. We first analyzed the association between the miRNA-196a2 polymorphism and the IBD susceptibility. When the Q-test of heterogeneity was not significant, the fixed effects model was used to conduct analyses. Otherwise, the random effects model was adopted.
Table 2.
Polled ORs of SNPs and IBD.
| n | OR (95% CI) P | P H | OR (95% CI) P | P H | OR (95% CI) P | P H | OR (95% CI) P | P H | OR (95% CI) P | P H | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR196a2 C>T | T/C Allelic model |
TT/CC Homozygote model |
TC/CC Heterozygote model |
TT + TC/CC Dominant model |
TT/TC + CC Recessive model |
||||||
| Ethnicity | |||||||||||
| Overall | 0.98 (0.85–1.13) P = 0.80 |
0.019 | 0.92 (0.76–1.11) P = 0.38 |
0.18 | 1.03 (0.74–1.43) P = 0.87 |
<0.001 | 1.04 (0.79–1.36) P = 0.80 |
<0.001 | 0.97 (0.83–1.14) P = 0.70 |
0.12 | |
| Caucasian | 0.99 (0.78–1.25) P = 0.92 |
0.005 | 0.88 (0.60–1.27) P = 0.46 |
0.099 | 1.10 (0.66–1.83) P = 0.72 |
<0.001 | 1.10 (0.72–1.66) P = 0.67 |
<0.001 | 0.88 (0.59–1.30) P = 0.51 |
0.049 | |
| Asian | 0.97 (0.90–1.07) P = 0.62 |
0.4 | 0.96 (0.73–1.25) P = 0.74 |
0.32 | 0.91 (0.71–1.16) P = 0.43 |
0.58 | 0.92 (0.74–1.16) P = 0.49 |
0.40 | 1.03 (0.83–1.14) P = 0.80 |
0.50 | |
| Diseases | |||||||||||
| UC | 5 | 0.94 (0.76–1.17) P = 0.59 |
0.007 | 0.79 (0.62–1.00) P = 0.06 |
0.23 | 1.07 (0.62–1.84) P = 0.82 |
<0.001 | 1 (0.64–1.53) P = 0.97 |
<0.001 | 0.87 (0.71–1.06) P = 0.16 |
0.11 |
| CD | 3 | 1.06 (0.92–1.22) P = 0.45 |
0.65 | 1.16 (0.86–1.56) P = 0.33 |
0.65 | 0.97 (0.77–1.22) P = 0.8 |
0.95 | 1.14 (0.92–1.41) P = 0.25 |
0.25 | 1.16 (0.9–1.48) P = 0.26 |
0.57 |
| miRNA146a G>C | C/G Allelic model |
CC/GG Homozygote model |
CG/GG Heterozygote model |
CC + CG/GG Dominant model |
CC/CG + GG Recessive model |
||||||
| Ethnicity | |||||||||||
| Overall | 1.34 (1.10–1.62) P = 0.003 |
0.001 | 1.56 (1.10–2.22) P = 0.001 |
0.037 | 1.50 (1.17–1.93) P = 0.04 |
0.081 | 1.54 (1.17–2.02) P = 0.02 |
0.022 | 1.19 (0.95–1.49) P = 0.13 |
0.062 | |
| Caucasian | 1.39 (0.94–2.04) P = 0.097 |
<0.001 | 1.45 (0.68–3.08) P = 0.34 |
0.009 | 1.50 (1.00–2.24) P = 0.001 |
0.051 | 1.46 (0.90–2.34) P = 0.01 |
0.008 | 1.18 (0.72–1.92) P = 0.51 |
0.019 | |
| Asian | 1.29 (1.12–1.48) P < 0.001 |
0.79 | 1.69 (1.27–2.24) P < 0.001 | 0.52 | 1.49 (1.31–1.87) P = 0.005 |
0.198 | 1.61 (1.24–2.08) P = 0.25 |
0.25 | 1.27 (1.04–1.54) P = 0.02 |
0.886 | |
| Diseases | |||||||||||
| UC | 4 | 1.32 (1.15–1.50) P < 0.001 | 0.64 | 1.43 (1.06–1.93) P = 0.02 |
0.39 | 1.32 (1.04–1.67) P = 0.02 |
0.42 | 1.38 (1.10–1.73) P = 0.34 |
0.34 | 1.20 (0.99–1.47) P = 0.07 |
0.65 |
| CD | 3 | 1.37 (0.85–2.22) P = 0.021 |
0 | 1.94 (0.89–4.24) P = 0.10 |
0.009 | 1.94 (1.49–2.52) P < 0.001 | 0.15 | 1.79 (1.10–2.02) P = 0.03 |
0.034 | 1.33 (0.76–2.33) P = 0.32 |
0.006 |
| miRNA499 A>G | G/A | GG/AA | GA/AA | GG + GA/AA | GG/GA + AA | ||||||
| Ethnicity | |||||||||||
| Overall | 1.08 (0.96–1.22) P = 0.20 |
0.75 | 1.43 (1.06–1.93) P = 0.02 |
0.27 | 0.86 (0.56–1.33) P = 0.50 |
<0.001 | 0.97 (0.78–1.20) P = 0.77 |
0.06 | 1.28 (0.64–2.53) P = 0.49 |
0.001 | |
| Caucasian | 1.08 (0.92–1.28) P = 0.37 |
0.38 | 1.72 (1.19–1.49) P = 0.004 |
0.24 | 0.62 (0.29–1.35) P = 0.23 |
<0.001 | 0.81 (0.66–1.00) P = 0.051 |
0.17 | 1.74 (0.68–4.49) P = 0.25 |
0.016 | |
| Asian | 1.09 (0.91–1.30) P = 0.37 |
0.70 | 0.94 (0.54–1.64) P = 0.84 |
0.83 | 1.17 (0.94–1.46) P = 0.16 |
0.28 | 1.14 (0.93–1.42) P = 0.21 |
0.42 | 0.88 (0.51–1.51) P = 0.63 |
0.73 | |
| Diseases | |||||||||||
| UC | 4 | 1.11 (0.96–1.29) P = 0.15 |
0.53 | 1.47 (1.04–2.06) P = 0.03 |
0.1 | 0.82 (0.41–1.63) P = 0.57 |
<0.001 | 0.97 (0.69–1.36) P = 0.15 |
0.02 | 1.19 (0.43–3.30) P = 0.73 |
0 |
| CD | 2 | 1.02 (0.81–1.28) P = 0.89 |
0.9 | 1.29 (0.67–2.5) P = 0.45 |
0.98 | 0.94 (0.71–1.24) P = 0.68 |
0.91 | 0.98 (0.75–1.27) P = 0.86 |
0.99 | 1.33 (0.69–2.54) P = 0.40 |
0.99 |
P: P value with significance. P < 0.05: significant statistical difference among different data. PH: P value with heterogeneity. PH < 0.1: significant statistical heterogeneity among various data. The random effects model should be applied; otherwise, instead, the fixed effects model should be used.
Overall, significant statistical heterogeneity was not identified in the homozygote model and recessive model. Therefore, the fixed effects model was implemented and the random effects model was applied to the other three models. In addition, no significant association was identified in all models (T versus C: OR = 0.98, 95% CI 0.85–1.13, PH = 0.019; TT versus CC: OR = 0.92, 95% CI 0.76–1.11, PH = 0.18; TC versus CC: OR = 1.03, 95% CI 0.74–1.43, PH< 0.001; TT + TC versus CC: OR = 1.04, 95% CI 0.79–1.36, PH< 0.001; and TT versus TC + CC OR = 0.97, 95% CI 0.83–1.14, PH = 0.12).
Next, we conducted subgroup analyses for ethnicity and diseases. In Caucasian subjects, significant statistical heterogeneity was identified in all models and the random effects model was used. However, the reduction of IBD risk was not significant in each model (T versus C: OR = 0.99, 95% CI 0.78–1.25, PH = 0.005; TT versus CC: OR = 0.88, 95% CI 0.60–1.27, PH = 0.099; TC versus CC: OR = 1.10, 95% CI 0.66–1.83, PH< 0.001; TT + TC versus CC: OR = 1.10, 95% CI 0.72–1.66, PH< 0.001; and TT versus TC + CC: OR = 0.88, 95% CI 0.59–1.30, PH = 0.049). On the other hand, in the Asian population, no significant statistical heterogeneity was identified in the models and the fixed effects model was used. In each model, the association was not significant (T versus C: OR = 0.97, 95% CI 0.90–1.07, PH = 0.4; TT versus CC: OR = 0.96, 95% CI 0.73–1.25, PH = 0.32; TC versus CC: OR = 0.91, 95% CI 0.71–1.16, PH = 0.58; TT + TC versus CC: OR = 0.92, 95% CI 0.74–1.16, PH = 0.40; and TT versus TC + CC: OR = 1.03, 95% CI 0.83–1.14, PH = 0.80).
In the UC, significant statistical heterogeneity was observed in all models, while no significant statistical heterogeneity was found in the CD. Association could not be significantly identified in both subgroups (UC: T versus C: OR = 0.94, 95% CI 0.76–1.17, PH = 0.007; TT versus CC: OR 0.79, 95% CI 0.62–1.00, PH = 0.23; TC versus CC: OR = 1.07, 95% CI 0.62–1.84, PH< 0.001; TT + TC versus CC: OR = 1.00, 95% CI 0.64–1.53, PH< 0.001; TT versus TC + CC OR = 0.87, 95% CI 0.71–1.06, PH = 0.11;CD: T versus C: OR = 1.06, 95% CI 0.92–1.22, PH = 0.65; TT versus CC: OR = 1.16, 95% CI 0.86–1.56, PH = 0.65; TC versus CC: OR = 0.97, 95% CI 0.77–1.22, PH = 0.96; TT + TC versus CC: OR1.14, 95% CI 0.92–1.41, PH = 0.25; and TT versus TC + CC: OR = 1.16, 95% CI 0.9–1.48, PH = 0.57).
3.3. Association between miRNA-146a and IBD
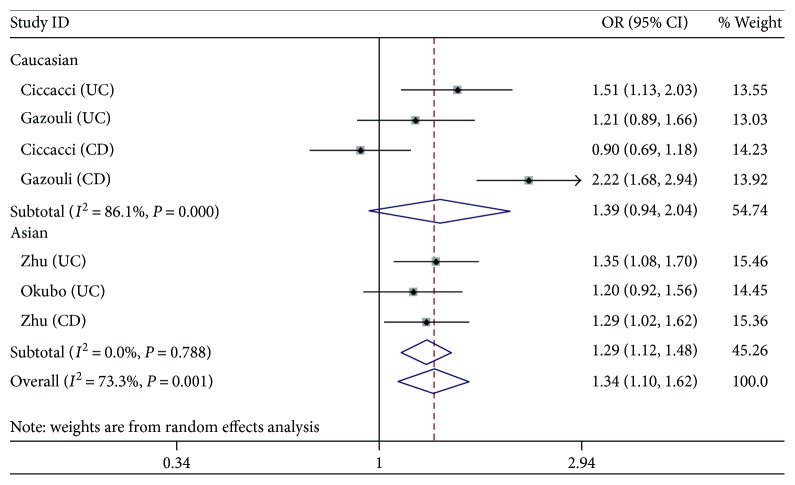
The analyses for the association between miRNA-146a polymorphism and the risk of IBD were realized by 7 independent studies (studies related to both CD and UC can be seen as two studies). Overall, the random effects model was used in all models due to the significant heterogeneity. The risk of IBD was significantly increased and could be observed in all models (C versus G: OR = 1.34, 95% CI 1.10–1.62, PH = 0.001; CC versus GG: OR = 1.56, 95% CI 1.10–2.22; CG versus GG: OR = 1.50, 95% CI 1.17–1.93, PH = 0.081; and CC + CG versus GG: OR = 1.54, 95% CI 1.17–2.02, PH = 0.022), except the recessive model (CC versus CG + GG: OR = 1.19, 95% CI 0.95–1.49, PH = 0.062). Figure 1 depicts a trend of the increased risk.
Then, the subgroup analyses were concluded. In the Caucasian population, significant heterogeneity was found in all models. Except for the heterozygote model (CG versus GG: OR = 1.50, 95% CI 1.00–2.24, PH = 0.051), no significant association was observed in other models (C versus G: OR = 1.39, 95% CI 0.94–2.04, PH< 0.001; CC versus GG: OR = 1.45, 95% CI 0.68–3.08, PH = 0.009; CC + CG versus GG: OR = 1.46, 95% CI 0.90–2.34, PH = 0.008; and CC versus CG + GG: OR = 1.18, 95% CI 0.72–1.92, PH = 0.019), while the trend of increased risk still existed. In the Asian population, heterogeneity was not significant in each model and an increased susceptibility was observed in all models (C versus G: OR = 1.29, 95% CI 1.12–1.48, PH = 0.79; CC versus GG: OR = 1.69, 95% CI 1.27–2.24, PH = 0.52; CG versus GG: OR = 1.49, 95% CI 1.31–1.87, PH = 0.198; CC + CG versus GG: OR = 1.61, 95% CI 1.24–2.08, PH = 0.25; and CC versus CG + GG: OR = 1.27, 95% CI 1.04–1.54, PH = 0.886).
In the UC subjects, statistical heterogeneity was not significant in all models. A significant increased risk of UC could be observed in all models (C versus G: OR = 1.32, 95% CI 1.15–1.50, PH = 0.64; CC versus GG: OR = 1.43, 95% CI 1.06–1.93, PH = 0.39; CG versus GG: OR = 1.32, 95% CI 1.04–1.67, PH = 0.42; and CC + CG versus GG: OR = 1.38, 95% CI 1.10–1.73, PH = 0.34), except the recessive model (CC versus CG + GG: OR = 1.20, 95% CI 0.99–1.47, PH = 0.65). However, the increasing trend of susceptibility was apparent. In the CD subjects, except for the heterozygote model, significant statistical heterogeneity existed. Although the risk reduction still existed in other models (C versus G: OR = 1.37, 95% CI 0.85–2.22, PH < 0.001; CC versus GG: OR = 1.94, 95% CI 0.89–4.24, PH = 0.009; and CC versus CG + GG: OR = 1.33, 95% CI 0.76–2.33, PH = 0.006), an increased risk of CD was found in the heterozygote model (CG versus GG: OR = 1.94, 95% CI 1.49–2.52, PH = 0.15) and dominant model (CC + CG versus GG: OR = 1.79, 95% CI 1.10–2.02, PH = 0.034).
3.4. Association between miRNA-499 and UC
Six independent studies in three articles were included in the assessment of the association between the miRNA-499 polymorphism and the risk of IBD. In a nutshell, significant statistical heterogeneity was identified in the heterozygote model, dominant model, and recessive model, for which the random effects model was used in the three models. A significant increased risk was identified in the homozygote model (GG versus AA: OR = 1.43, 95% CI 1.06–1.93, PH = 0.27), while no obvious association was observed in other models (G versus A: OR = 1.08, 95% CI 0.96–1.22, PH = 0.75; GA versus AA: OR = 0.86, 95% CI 0.56–1.33, PH< 0.001; GG + GA versus AA: OR = 0.97, 95% CI 0.78–1.20, PH = 0.064; and GG versus GA + AA: OR = 1.28, 95% CI 0.64–2.53, PH = 0.001).
In the Caucasian population, the random effects model was adopted in the heterozygote model and recessive model (due to the significant statistical heterogeneity), and the fixed effects model was implemented in other models. A significant increased risk in the homozygote model (GG versus AA: OR = 1.72, 95% CI 1.19–1.49, PH = 0.24) was observed, while no significant association was found in other models (G versus A: OR = 1.08, 95% CI 0.92–1.28, PH = 0.38; GA versus AA: OR = 0.62, 95% CI 0.29–1.35, PH< 0.001; GG + GA versus AA: OR = 0.81, 95% CI 0.66–1.00, PH = 0.17; and GG versus GA + AA: OR = 1.74, 95% CI 0.68–4.49, PH = 0.016). In the Asian population, no significant statistical heterogeneity was identified in all models and therefore the fixed effects model was used. However, no significant association was found (G versus A: OR = 1.09, 95% CI 0.91–1.30, PH = 0.70; GG versus AA: OR = 0.94, 95% CI 0.54–1.64, PH = 0.83; GA versus AA: OR = 1.17, 95% CI 0.94–1.46, PH = 0.28; GG + GA versus AA: OR = 1.14, 95% CI 0.93–1.42, PH = 0.42; and GG versus GA + AA: OR = 0.88, 95% CI 0.51–1.51, PH = 0.73).
In the UC, the random effects model was used in the heterozygote model, dominant model, and recessive model, because of the presence of heterogeneity in the three models. A significant increased risk was identified in the homozygote model (GG versus AA: OR = 1.47, 95% CI 1.04–2.06, PH = 0.1), while no obvious association was observed in other models (G versus A: OR = 1.11, 95% CI 0.96–1.29, PH = 0.53; GA versus AA: OR = 0.82, 95% CI 0.41–1.63, PH< 0.001; GG + GA versus AA: OR = 0.97, 95% CI 0.69–1.36, PH = 0.02; and GG versus GA + AA: OR = 1.19, 95% CI 0.43–3.30, PH< 0.001). In the CD, no significant statistical heterogeneity was identified in all models and hence the fixed effects model was used. No significant association existed (G versus A: OR = 1.02, 95% CI 0.81–1.28, PH = 0.9; GG versus AA: OR = 1.29, 95% CI 0.67–2.5, PH = 0.98; GA versus AA: OR = 0.94, 95% CI 0.71–1.24, PH = 0.91; GG + GA versus AA: OR = 0.98, 95% CI 0.75–1.27, PH = 0.99; and GG versus GA + AA: OR = 1.33, 95% CI 0.69–2.54, PH = 0.99).
3.5. Sensitivity Analysis
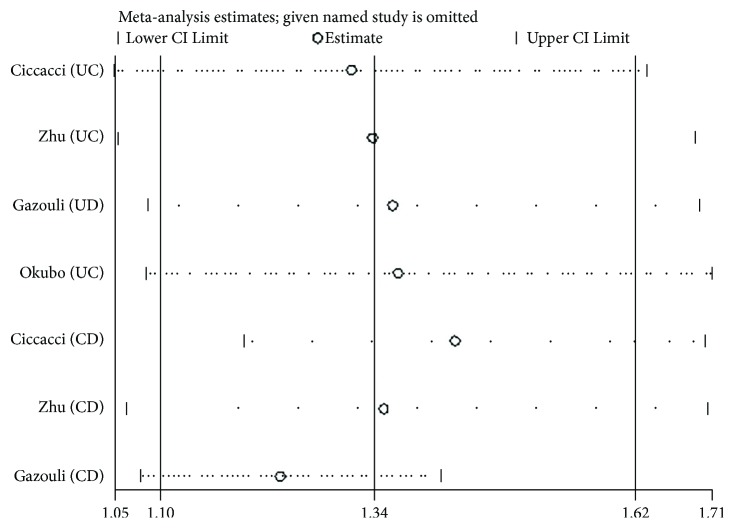
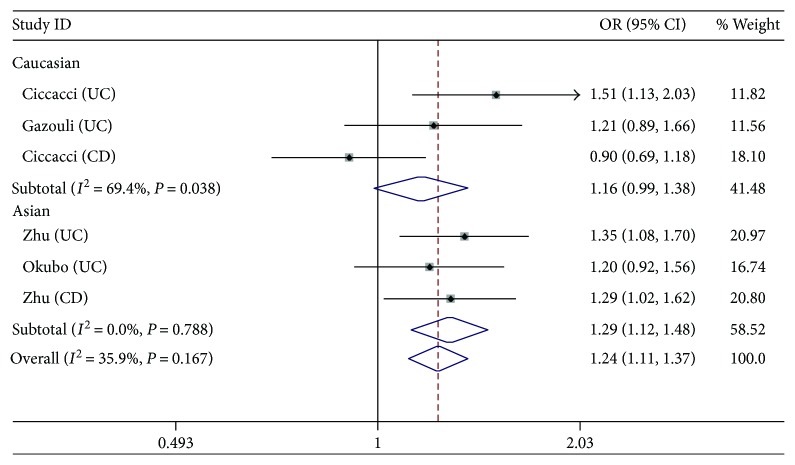
We performed the sensitivity analysis to test the influence of individual study on the pooled ORs by deleting each study every time (Figure 2). For rs11614913 and rs3746444, the pooled estimate indicated that there was no significant difference. For rs2910146, the study from Okubo et al. [15] showed a significant effect on the pooled ORs. After excluding this study, the result of heterogeneity test was negative, and the pooled OR of allelic comparison C versus G turned to be 1.24 (95% CI: 1.11–1.37, P < 0.0001) (Figure 3). Nevertheless, the ORs of the homozygote model, heterozygote model, and dominant model remained significant, which indicated that the study about CD from Gazouli et al. might have a major influence on the pooled estimate (see Figure 4).
Figure 2.
Forest plots of the OR with 95% CI for miRNA-146a rs2910164 (C versus G).
Figure 3.
Sensitivity analyses by deleting one study every time.
Figure 4.
Forest plots of the OR with 95% CI for miRNA-146a rs2910164 after excluding the study from Okubo et al. (C versus G).
3.6. Publication Bias
Publication bias was evaluated by Begg's funnel plot and Egger's test. No publication bias for the association between miRNA-146a rs2910146 (G>C) and IBD susceptibility which was identified by Begg's funnel plot (P = 0.881, C versus G) and Egger's test (P = 0.793, C versus G). Begg's funnel plot (Figure 5) and Egger's test (P = 0.882, C versus T) between miRNA-196a2 rs11614913 (T>C) and IBD susceptibility showed no publication bias. No test was assessed for the association between miRNA-499 rs3746444 (A>G) and IBD susceptibility due to the limited number of selected studies.
Figure 5.
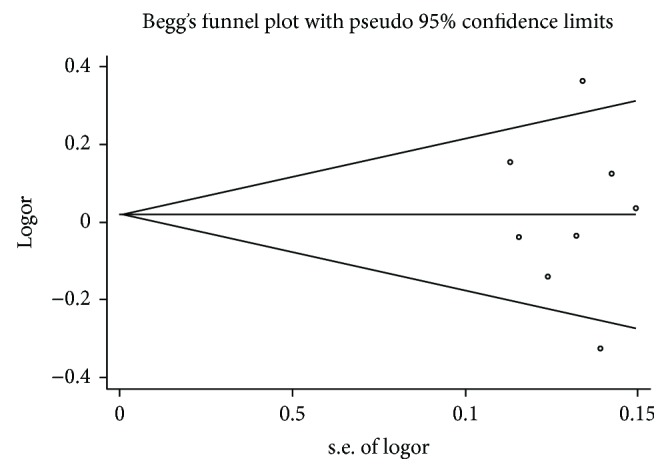
Begg's funnel plot for miRNA-196a2 rs11614913 (T>C) and IBD.
4. Discussion
Accumulating emerged evidences had shown that there was specific relationship between miRNAs and autoimmunity diseases (ADs). A series of meta-analyses [19, 20], published from the year 2013 to the year 2015, summarized the potential association between ADs and miRNAs (miRNA-146a rs2910146 and miRNA-499 rs3746444). However, no investigation was conducted to examine the relationship between IBDs and these three miRNAs. In the present meta-analysis, five eligible articles containing nine case-control studies were selected and 1761 cases and 1892 controls for the association between the three SNPs and IBD were analyzed. In our findings, there was no obvious association between miRNA-196a2 rs11614913 and IBD, even in the subgroup analysis based on ethnicity or diseases. For miRNA-146a rs2910146 and IBD, the associated susceptibility increased significantly. In allelic comparison, homozygote model, heterozygote model, and dominant model, similar results were obtained in Asian subgroups and UC subjects. In the Asian population, significant results were also observed in the recessive model. In addition, significant result existed in the reserved model of subgroup CD. As for miRNA-499 rs3746444 and IBD, a positive relationship between them was identified in the homozygote model, and the same results were observed in subgroups Caucasian and UC.
It was reported that miRNA-146a, mapped to chromosome 5q33, was a nuclear factor-κB- (NF-κB-) dependent gene whose G/C polymorphism is located in the stem region (at position +60 relative to the first nucleotide of pre-miR-146a) opposite to the mature miR-146a precursor and could reduce the expression of tumor necrosis factor-receptor-associated factor-6 and interleukin-1 receptor- (ILR-) associated kinase-1, thus preventing excessive inflammation [21, 22]. Jazdzewski et al. [23] found that the miRNA-146a rs2910146 polymorphism could cause additional generation of mature microRNAs from the passenger strands of the miRNA precursor. Regardless of the approaches the miRNA-146a rs2910146 polymorphism influence the development of diseases, increasing studies had shown that rs2910146 (G>C) played an important role in the pathogenetic process of many diseases, for example, asthma [24] and rheumatoid arthritis [25] (which were both associated with immune disorders). Our results provided powerful evidences for the association between miRNA-146a rs2910146 and susceptibility to IBD, for which research attention should be paid in future investigations. As one can expect, illuminating the inherent association between miRNA-146a rs2910146 and IBD could help search new approaches for diagnosis and treatment [26, 27].
The strengths of our meta-analysis are as follows: (1) this is the first meta-analysis that is focused on the association between SNPs and IBD; (2) more updated studies were included in our research, compared to the previous meta-analyses about the association between SNPs and AD; (3) miRNA-196a2 rs11614913 was investigated in our meta-analysis, which did not receive attention from previous research; and (4) the result remained significant after excluding heterogenic origins.
However, some limitations still exist in our studies. First of all, because of the shortage of original studies, the number of selected studies limited our further analysis. In addition, one of the included studies on miRNA196 did not meet the HWE, though the pooled estimate remained insignificant. Moreover, obvious heterogeneity existed in miRNA146a, even in the subgroup Caucasian population. Although the sensitive analyses were conducted, the results need further supports from more well-designed studies with larger sample sizes. Finally, only three SNPs were included in our study, while there were more SNPs in other genes that could influence the susceptibility to IBD, and interactions among these SNPs and impacts of their network on diseases were not considered in our study.
In conclusion, our results suggested that miRNA-146a polymorphism was significantly associated with the susceptibility to IBD, especially in Asian and in UC. Nevertheless, in the future, more studies with larger samples as well as joint research on SNPs and the roles of SNPs in the pathogenesis of diseases and disorders are needed to further validate our results.
Acknowledgments
Lingxin Xiong was receiving support from the National Science and Technology Major Project for new drug development in China (no. 2010ZX09401-305-26) and Science and Technology Development Program of Jilin Province (Grant no. 20150519015JH).
Conflicts of Interest
The authors declare no conflict of interest.
Authors' Contributions
Ying Liu and Tongjun Liu conducted the literature search, drafted the manuscript, and contributed to the study design, data collection, data analysis, and data interpretation. Lingxin Xiong helped search the literature and participated in writing the manuscript, collecting the data, and analyzing the data. Yan Zhou and Bingzhen Zheng helped collect the data and analyzed the data. Lingxin Xiong and Wei Xie helped revised the manuscript. All authors read and revised the manuscript critically for intellectual content and approved the final manuscript.
References
- 1.Hart A. L., Ng S. C., Mann E., al-Hassi H. O., Bernardo D., Knight S. C. Homing of immune cells: role in homeostasis and intestinal inflammation. Inflammatory Bowel Diseases. 2010;16(11):1969–1977. doi: 10.1002/ibd.21304. [DOI] [PubMed] [Google Scholar]
- 2.Mahida Y.-R. The key role of macrophages in the immunopathogenesis of inflammatory bowel disease. Inflammatory Bowel Diseases. 2000;6(1):21–33. doi: 10.1097/00054725-200002000-00004. [DOI] [PubMed] [Google Scholar]
- 3.Liu J.-Z., van Sommeren S., Huang H., et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nature Genetics. 2015;47(9):979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritchard C.-C., Cheng H.-H., Tewari M. MicroRNA profiling: approaches and considerations. Nature Reviews Genetics. 2012;13(5):358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X., Zhu W., Li H., et al. Diagnostic value of a plasma microRNA signature in gastric cancer: a microRNA expression analysis. Scientific Reports. 2015;5, article 11251(1) doi: 10.1038/srep11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirola C. J., Fernández G. T., Castaño G. O., et al. Circulating microRNA signature in non-alcoholic fatty liver disease: from serum non-coding RNAs to liver histology and disease pathogenesis. Gut. 2015;64(5):800–812. doi: 10.1136/gutjnl-2014-306996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan Z., Xu Z., Dan Y., Wang W., Liu Z. Effects of common polymorphism rs11614913 in Hsa-miR-196a2 on lung cancer risk. PLoS One. 2013;8(4, article e61047) doi: 10.1371/journal.pone.0061047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fu L., Lei L., Lei Y., et al. Comprehensive review of genetic association studies and meta-analysis on miRNA polymorphisms and rheumatoid arthritis and systemic lupus erythematosus susceptibility. Human Immunology. 2016;77(1):1–6. doi: 10.1016/j.humimm.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 9.Xu W., Xu J., Liu S., et al. Effects of common polymorphisms rs11614913 in miR-196a2 and rs2910164 in miR-146a on cancer susceptibility: a meta-analysis. PLoS One. 2011;6(5, article e20471) doi: 10.1371/journal.pone.0020471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Wang Q., Liu H., et al. The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies. Mutagenesis. 2012;27(6):779–788. doi: 10.1093/mutage/ges052. [DOI] [PubMed] [Google Scholar]
- 11.Ranjha R., Meena N. K., Singh A., Ahuja V., Paul J. Association of miR-196a-2 and miR-499 variants with ulcerative colitis and their correlation with expression of respective miRNAs. PLoS One. 2017;12(3, article e0173447) doi: 10.1371/journal.pone.0173447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciccacci C., Politi C., Biancone L., et al. Polymorphisms in MIR122, MIR196A2, and MIR124A genes are associated with clinical phenotypes in inflammatory bowel diseases. Molecular Diagnosis & Therapy. 2017;21(1):107–114. doi: 10.1007/s40291-016-0240-1. [DOI] [PubMed] [Google Scholar]
- 13.Zhu M., Li D., Jin M., Li M. Association between microRNA polymorphisms and the risk of inflammatory bowel disease. Molecular Medicine Reports. 2016;13(6):5297–5308. doi: 10.3892/mmr.2016.5157. [DOI] [PubMed] [Google Scholar]
- 14.Gazouli M., Papaconstantinou I., Stamatis K., et al. Association study of genetic variants in miRNAs in patients with inflammatory bowel disease: preliminary results. Digestive Diseases and Sciences. 2013;58(8):2324–2328. doi: 10.1007/s10620-013-2640-y. [DOI] [PubMed] [Google Scholar]
- 15.Okubo M., Tahara T., Shibata T., et al. Association study of common genetic variants in pre-microRNAs in patients with ulcerative colitis. Journal of Clinical Immunology. 2011;31(1):69–73. doi: 10.1007/s10875-010-9461-y. [DOI] [PubMed] [Google Scholar]
- 16.Moher D., Liberati A., Tetzlaff J., Altman D. G., The PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. International Journal of Surgery. 2010;8(5):336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Wang P., Xie S., Cui A., Zhang Y., Jiang B. miR-196a2 polymorphisms and susceptibility to cancer: a meta-analysis involving 24,697 subjects. Experimental and Therapeutic Medicine. 2012;3(2):324–330. doi: 10.3892/etm.2011.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins J. P. T., Thompson S. G. Quantifying heterogeneity in meta-analysis. Statistics in Medicine. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 19.Chen H.-F., Hu T.-T., Zheng X.-Y., et al. Association between miR-146a rs2910164 polymorphism and autoimmune diseases susceptibility: a meta-analysis. Gene. 2013;521(2):259–264. doi: 10.1016/j.gene.2013.03.073. [DOI] [PubMed] [Google Scholar]
- 20.Li C., Fu W., Zhang Y., et al. Meta-analysis of microRNA-146a rs2910164 G > C polymorphism association with autoimmune diseases susceptibility, an update based on 24 studies. PLoS One. 2015;10(4, article e0121918) doi: 10.1371/journal.pone.0121918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taganov K. D., Boldin M. P., Chang K. J., Baltimore D. NF-κB-dependent induction of microRNA MiR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akkiz H., Bayram S., Bekar A., Akgöllü E., Üsküdar O., Sandıkçı M. No association of pre-microRNA-146a rs2910164 polymorphism and risk of hepatocellular carcinoma development in Turkish population: a case-control study. Gene. 2011;486(1-2):104–109. doi: 10.1016/j.gene.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Jazdzewski K., Liyanarachchi S., Swierniak M., et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(5):1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiménez-Morales S., Gamboa-Becerra R., Baca V., et al. MiR-146a, polymorphism is associated with asthma but not with systemic lupus erythematosus and juvenile rheumatoid arthritis in Mexican patients. Tissue Antigens. 2012;80(4):317–321. doi: 10.1111/j.1399-0039.2012.01929.x. [DOI] [PubMed] [Google Scholar]
- 25.Hashemi M., Eskandari-Nasab E., Zakeri Z., et al. Association of pre-miRNA-146a rs2910164 and pre‑miRNA-499 rs3746444 polymorphisms and susceptibility to rheumatoid arthritis. Molecular Medicine Reports. 2013;7(1):287–291. doi: 10.3892/mmr.2012.1176. [DOI] [PubMed] [Google Scholar]
- 26.Schaefer J. S., Attumi T., Opekun A. R., et al. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunology. 2015;16(1):p. 5. doi: 10.1186/s12865-015-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luchuan L., Bin L., Bo C., et al. Inhibition of miR-146b expression increases radioiodine-sensitivity in poorly differential thyroid carcinoma via positively regulating NIS expression. Biochemical and Biophysical Research Communications. 2015;462(4):314–321. doi: 10.1016/j.bbrc.2015.04.134. [DOI] [PubMed] [Google Scholar]