Abstract
Background:
The aim of the current study was to detect hepatitis C virus (HCV) RNA in blood and saliva of a population of patients with thalassemia who have HCV antibody in their serum.
Materials and Methods:
In this cross-sectional study, blood and saliva samples were collected and were analyzed with quantitative reverse transcription polymerase chain reaction (RT-PCR) for the detection of HCV RNA. In addition, liver-related blood tests were performed, and patients’ medical history was recorded. Data were analyzed by independent samples t-test and Chi-square with a significant level of 0.05.
Results:
Overall, 62 adult patients (29 males and 33 females) were included. Most (87%) of the patients had major thalassemia and genotype 1a was the most common (42%) type. HCV RNA was detected in 71 and 16% of blood and saliva samples, respectively. HCV RNA was detected more in female patients (31%) (P = 0.003) and in intermediate thalassemia (50%) (P < 0.005). The mean age of the patients with positive saliva was almost 10 years older (P < 0.001), and the mean number of blood transfusion was fewer in positive saliva group (P = 0.037). The sensitivity, specificity, and positive and negative predictive values of saliva PCR was calculated to be 18%, 88%, 80%, and 69%, respectively.
Conclusion:
Saliva contained HCV RNA in 16% of the assessed population. The probability of detection of HCV RNA in saliva increased in older patients, less number of blood transfusions, females and intermediate thalassemia. Saliva RT-PCR demonstrated low sensitivity and high specificity with high positive predictive value in the assessed population.
Keywords: Blood, diagnosis, hepatitis C, hepatitis C antigens, saliva, thalassemia
Introduction
Hepatitis C virus (HCV) is one of the common causes of chronic hepatic diseases such as cirrhosis and hepatocellular carcinoma (HCC) and is regarded as a major indication of liver transplant.[1] Principal risk factors for the transmission of HCV include blood transfusion, drug addiction, unprotected sexual contact, and low socioeconomic level.[2] Transfusion-dependent thalassemia is a common genetic hemoglobinopathic disease which necessitates multiple blood transfusions during the life of a patient. These transfusions carry the risk for several diseases including HCV infection.[3] Iran is located on the so-called “Thalassemia belt,” and there are more than 25,000 diagnosed cases of thalassemia in this country.[4]
Previously, it was stated that saliva analysis could be a simple, reliable, and valid screening test for the detection of HCV infection and could be used in epidemiological studies.[5,6] However, some authors reported that HCV was not detectable in saliva.[7,8] Furthermore, there is no general consensus regarding the possibility of transmission of HCV through saliva.[9]
Dental staff and patient's family are exposed to saliva. Therefore, it is imperative for the health of the dentists and the oral hygienists as well as the patient's family to determine the presence of HCV in patient's saliva so that necessary precautions could be taken to control the transmission of the virus. The aim of the current study was to determine the presence of HCV RNA in blood and saliva of a population of Iranian transfusion-dependent thalassemia patients who are HCV antibody (anti-HCV) positive.
Patients
This cross-sectional study was performed in a public Gasterohepatology clinic of Baghiatollah Hospital and Iranian Blood Transfusion Organization, Tehran, Iran, in 2014–2015. The study protocol was approved by ethical committee of Shahid Beheshti University of Medical Sciences. In addition, informed consent was taken from all included patients.
The inclusion criteria were transfusion-dependent thalassemia, positive anti-HCV, older than 15 years, and at least 6 months after the last treatment for HCV infection. Exclusion criteria were pregnancy and lactation, history of HCC, alcoholic liver, autoimmune hepatitis, metabolic hepatic diseases, hemochromatosis, severe psoriasis, scleroderma, autoimmune diseases, gastrointestinal inflammatory diseases, cardiovascular and cerebrovascular diseases, major depression and consumption of drugs that affect salivary flow.
Demographic data, type of the disease, duration and number of blood transfusion, and type of the treatment (if any) of the included patients were recorded. Furthermore, hepatitis grade and stage of those patients who had undergone liver biopsy were recorded. Staging and grading were determined based on the modified Scheuer system[10] including five-point grading (0 = no inflammation, 1 = portal inflammation, 2 = mild piecemeal necrosis, 3 = moderate piecemeal necrosis, and 4 = severe piecemeal necrosis) and scaling (0 = normal connective tissue, 1 = fibrous portal expansion, 2 = periportal or rare portal septa, 3 = fibrous septa with architectural distortion, and 4 = cirrhosis).
Samples collection
Finally, 2 ml of unstimulated whole saliva secreted from parotid gland was collected using buccal collection cups (Quality Biological, Gaithersburg, MD) and stored at −70°C until analysis. Furthermore, 5 ml of vascular blood of each patient was collected in acid-citrate-dextrose anticoagulated tubes.
Assessments
All assessments were performed following manufacturers’ guidelines. The presence of anti-HCV in blood was assessed by HCV ELISA Test System (ELISA3.0 HCV; Ortho Clinical Systems, Raritan, NJ, USA). Then, liver-related blood markers including aspartate aminotransferase (AST), alanine transaminase (ALT), and total and direct bilirubin were assessed using AST Activity Assay Kit (ab105135, Abcam, Cambridge, MA, USA), ALT Activity Assay Kit (Colorimetric/Fluorometric: Ab105134, Abcam), and Bilirubin Assay Kit (Sigma Diagnostics, St Louis, MO, USA), respectively. Finally, the presence of HCV RNA in blood and saliva samples was detected using quantitative reverse transcription polymerase chain reaction (RT-PCR) (TaqMan RT-PCR system, Applied Biosystems, Foster City, CA, USA). The priming and genotyping were performed as described in literature.[11]
Statistical analysis
Due to normal distribution of the data as evaluated by Kolmogorov–Smirnov test (P > 0.05), independent samples t-test, Pearson's correlation coefficient, one-way ANOVA, and Chi-square were used to analyze the results. Data analysis was performed using SPSS version 21 (SPSS, Chicago, IL, USA) with a significant level of 0.05.
Results
A total number of 62 patients including 29 (46.8%) males and 33 (53.2%) females with the mean age of 31.66 ± 7.8 years (range: 21–55 years) were included. Most patients suffered from major thalassemia (54 patients) while others had intermediate thalassemia (12.9%). The mean duration of blood transfusion was 324.59 ± 69.38 weeks, and the mean number of blood transfusion per year was 13.15 ± 7.4 times. Among the 62 patients, 30 had a history of liver biopsy. In these patients, the mean grade and stage of the hepatitis was 1.77 ± 0.76 and 1.31 ± 0.52, respectively. Forty-seven out of 62 patients (75.8%) had a history of pharmacologic hepatitis treatment out of which 11 (17.7%) had peginterferon alfa-2a, 27 (43.5%) had peginterferon alfa-2a plus Ribavirin, and 9 (14.5%) had interferon. There was no significant difference in the duration of blood transfusion and numbers between various types of thalassemia and the types of hepatitis treatment (P > 0.05).
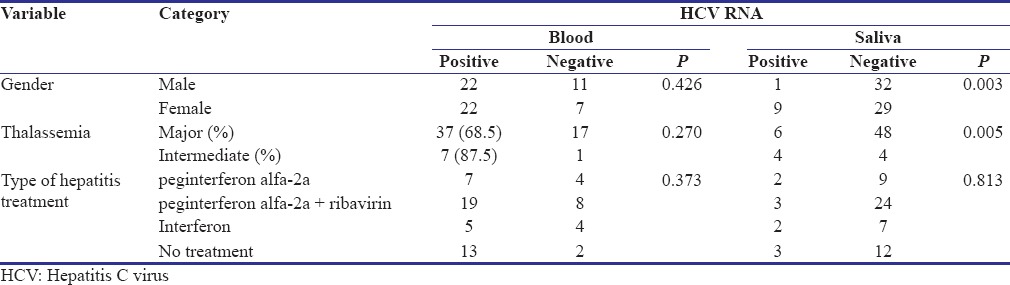
The mean AST, ALT, total and direct bilirubin was 53.03 ± 25.9, 57.24 ± 41.5, 2.95 ± 2.03 and 23.46 ± 12.5, respectively. Although all the patients were anti-HCV positive, HCV RNA was detected in the blood of 44 (71%) patients. The mean viral load was 413121.6 ± 5.03. Table 1 compares the evaluated variables among seropositive and seronegative patients. There was no significant association between detection of HCV RNA in blood with gender, type of thalassemia and the type of previous treatment [Table 1].
Table 1.
Distribution of the different parameters based on the presence of hepatitis C virus RNA in blood and saliva in a population of Iranian anti-hepatitis C virus positive thalassemia-dependent to transfusion patients (sample size: 62)
The most common genotype of HCV was 1a (41.9%), followed by 1b (19.4%) and 3a (9.7%). Table 2 demonstrates the distribution of HCV genotypes based on gender, type of thalassemia, and presence of HCV RNA in saliva.
Table 2.
Distribution of hepatitis C virus genotype based on different parameters in a population of Iranian anti-hepatitis C virus positive, hepatitis C virus RNA positive, thalassemia-dependent to transfusion patients (sample size: 44)

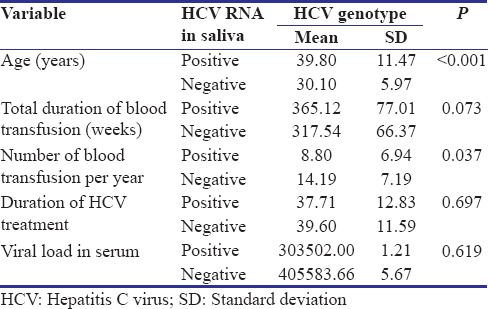
Assessment of salivary samples showed that HCV RNA was present in the saliva of 10 (16%) patients. Table 1 compares the evaluated variables among patients with positive and negative HCV RNA in the saliva. There was no significant difference between the type of previous treatment and detection of HCV RNA in saliva [Table 1]. HCV RNA was detected more in the saliva of female patients (31%) compared to males (3%) (P = 0.003) and in intermediate thalassemia (50%) compared to major thalassemia (11.1%) (P = 0.005). To further investigate the effect of possible confounding variables on the possibility of detection of HCV RNA in saliva, quantitative variables were compared between patients with positive and negative saliva [Table 3]. The mean age of the patients with positive saliva was almost 10 years older than patients with negative saliva (P < 0.001). Furthermore, the mean number of blood transfusion was 8.8 times in a year in positive saliva group compared to 14.2 times in negative saliva group (P = 0.037).
Table 3.
Comparison of different parameters in a population of Iranian anti-hepatitis C virus positive thalassemia-dependent to transfusion patients based on detection of hepatitis C virus RNA in saliva (sample size: 62)
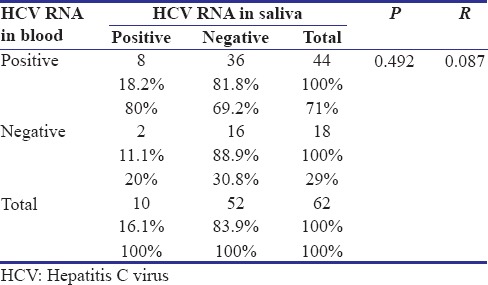
There was an insignificant (P = 0.492), very weak (Pearson's r = 0.087) correlation between the presence of HCV RNA in blood and saliva [Table 4]. Considering blood PCR as the gold standard method for the diagnosis of current HCV infection,[2] the sensitivity, specificity, positive and negative predictive values of saliva PCR were calculated to be 18, 88, 80 and 69%, respectively.
Table 4.
Relationship between polymerase chain reaction results of saliva and blood in a population of Iranian anti-hepatitis C virus positive thalassemia-dependent to transfusion patients based on detection of hepatitis C virus RNA in saliva (sample size: 62)
Discussion
The aim of the current study was to evaluate the presence of HCV in the saliva of transfusion-dependent thalassemia patients with a history of HCV infection to inform patients’ families and dental staff to use appropriate protections for infection control. Thalassemia patients are prone to HCV infection and need special dental care.[12] Epidemiologic studies revealed that 4.5% (Turkey[13]) to 91.8% (Romani[14]) of thalassemia patients have HCV infection.[15,16,17,18,19] In Iran, the prevalence of thalassemia is relatively high and previous studies reported HCV infection in 13.6%,[20] 19.3%,[21] 26.9%,[22] to 39.7%[23] in Iranian thalassemia patients.
The most common treatment protocol in patients of the current study was combination of peginterferon alfa-2a and Ribavirin which has been reported to be successful in almost half of the patients.[12] Similar to previous studies in Iranian population,[23] the most common genotype of HCV in the present study was type 1a. However, the prevalence of thalassemia genotypes has been different in other populations. In Pakistan, 3a and 3b were the most common genotypes.[24] In Greece[25] and in Italy,[26] genotype 3a and 1b, respectively, were more common.
Using RT-PCR, the HCV RNA was detectable in blood samples of 71% of the population of anti-HCV seropositive transfusion-dependent thalassemia patients. Previously, Rosenthal et al.[16] reported that 47% of positive anti-HCV had serum HCV RNA.
The present study revealed that HCV RNA was detectable in the saliva of 16% of the population of anti-HCV+ transfusion-dependent thalassemia patients. On the one hand, saliva has been used in epidemiologic studies for screening HCV infection.[5,6] Hermida et al.[27] detected HCV RNA in the saliva of 52.4% seropositive patients. Wang et al.[28] reported that HCV RNA could be detected in the saliva of nearly half of the patients 1–38 weeks after seroconversion. On the contrary, some authors revealed low or no presence of HCV in the saliva of seropositive patients.[7,8] Fried et al.[8] reported that HCV RNA could not be detected in saliva and semen samples of seropositive patients.
Similar to some previous studies,[29,30] the results of this study showed no significant relation between serum viral load and detection of HCV in the saliva. A notable finding was that the serum viral load in patients with positive-HCV RNA in their saliva was less than patients with negative HCV RNA in their saliva; however, it was not statistically significant. On the contrary, Suzuki et al.[31] showed that the probability of detection of HCV in the saliva of patients with lower serum viral load was less than those with high serum viral load. Although their results indicated that this difference was not significant, some other investigators showed significant positive correlation between serum viral load and probability of detection of HCV in saliva.[27,28,32]
No significant correlation was found between detection of HCV RNA in serum and saliva; however, most (80%) of the patients who had had HCV RNA in their saliva were seropositive. Detection of HCV in the other 20% who were seronegative could be explained in two ways. The primary reason could be the probability of virus colonization in the salivary gland or peripheral blood mononuclear cells.[29,33] The other reason might be due to the detection method which was quantitative RT-PCT in the present study. Suzuki et al.[31] showed that in 4 patients from their study, HCV RNA was detectable in their saliva while no HCV was detected in their serum using RT-PCR. However, using nested RT-PCR, they demonstrated that all these 4 patients were seropositive.
Considering RT-PCR with its limitations as the gold standard method for the detection of HCV in the blood,[2] saliva RT-PCR showed low sensitivity (18%) and relatively high specificity (88%). Furthermore, the positive predictive value of saliva RT-PCR was 80%. Therefore, similar to the study of Cameron et al.[34] the sensitivity of saliva for the detection of HCV infection was low, and this method could not be applied as a screening tool. However, González et al.[6] reported a sensitivity of 90% and specificity of 100% for anti-HCV in saliva.
Finally, detection of HCV RNA in the saliva of 16% of anti-HCV+ transfusion-dependent thalassemia patients reveals the necessity of infection control precautions during dental treatment. Furthermore, the family members should be aware of the probability of the presence of HCV in saliva. However, a study in Israel[16] and China[28] revealed low occurrence of HCV infection in family members of anti-HCV+ thalassemia patients. On the other hand, the study of Said et al.[35] in Egypt revealed that 19.2% of family members of anti-HCV+ thalassemia patients are also seropositive. Therefore, the transmission of HCV might be through saliva and as a result is dependent on the lifestyle of the patient and his/her family.
Conclusion
Within the limitations of the current study, it could be concluded that saliva could contain HCV in 16% of the assessed population of adult anti-HCV+ transfusion-dependent thalassemia patients. The probability of detection of HCV in saliva increases as the patient ages and with less number of blood transfusions. Furthermore, positive HCV in saliva was more common in females and intermediate thalassemia compared to males and major thalassemia, respectively.
In addition, saliva RT-PCR showed low sensitivity and high specificity with high positive predictive value in the assessed population when serum RT-PCR was considered as the gold standard.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Mukherjee R, Burns A, Rodden D, Chang F, Chaum M, Garcia N, et al. Diagnosis and management of hepatitis C virus infection. J Lab Autom. 2015;20:519–38. doi: 10.1177/2211068214563794. [DOI] [PubMed] [Google Scholar]
- 2.Ansaldi F, Orsi A, Sticchi L, Bruzzone B, Icardi G. Hepatitis C virus in the new era: Perspectives in epidemiology, prevention, diagnostics and predictors of response to therapy. World J Gastroenterol. 2014;20:9633–52. doi: 10.3748/wjg.v20.i29.9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kountouras D, Tsagarakis NJ, Fatourou E, Dalagiorgos E, Chrysanthos N, Berdoussi H, et al. Liver disease in adult transfusion-dependent beta-thalassaemic patients: Investigating the role of iron overload and chronic HCV infection. Liver Int. 2013;33:420–7. doi: 10.1111/liv.12095. [DOI] [PubMed] [Google Scholar]
- 4.Hashemieh M, Timori Naghadeh H, Tabrizi Namini M, Neamatzadeh H, Hadipour Dehshal M. The Iran thalassemia prevention program: Success or failure? Iran J Ped Hematol Oncol. 2015;5:161–6. [PMC free article] [PubMed] [Google Scholar]
- 5.Bello PY, Pasquier C, Gourney P, Puel J, Izopet J. Assessment of a hepatitis C virus antibody assay in saliva for epidemiological studies. Eur J Clin Microbiol Infect Dis. 1998;17:570–2. doi: 10.1007/BF01708621. [DOI] [PubMed] [Google Scholar]
- 6.González V, Martró E, Folch C, Esteve A, Matas L, Montoliu A, et al. Detection of hepatitis C virus antibodies in oral fluid specimens for prevalence studies. Eur J Clin Microbiol Infect Dis. 2008;27:121–6. doi: 10.1007/s10096-007-0408-z. [DOI] [PubMed] [Google Scholar]
- 7.Hsu HH, Wright TL, Luba D, Martin M, Feinstone SM, Garcia G, et al. Failure to detect hepatitis C virus genome in human secretions with the polymerase chain reaction. Hepatology. 1991;14:763–7. doi: 10.1002/hep.1840140504. [DOI] [PubMed] [Google Scholar]
- 8.Fried MW, Shindo M, Fong TL, Fox PC, Hoofnagle JH, Di Bisceglie AM, et al. Absence of hepatitis C viral RNA from saliva and semen of patients with chronic hepatitis C. Gastroenterology. 1992;102:1306–8. [PubMed] [Google Scholar]
- 9.Ferreiro MC, Dios PD, Scully C. Transmission of hepatitis C virus by saliva? Oral Dis. 2005;11:230–5. doi: 10.1111/j.1601-0825.2005.01076.x. [DOI] [PubMed] [Google Scholar]
- 10.Batts KP, Ludwig J. Chronic hepatitis. An update on terminology and reporting. Am J Surg Pathol. 1995;19:1409–17. doi: 10.1097/00000478-199512000-00007. [DOI] [PubMed] [Google Scholar]
- 11.Ohno O, Mizokami M, Wu RR, Saleh MG, Ohba K, Orito E, et al. New hepatitis C virus (HCV) genotyping system that allows for identification of HCV genotypes 1a, 1b, 2a, 2b, 3a, 3b, 4, 5a, and 6a. J Clin Microbiol. 1997;35:201–7. doi: 10.1128/jcm.35.1.201-207.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Butensky E, Pakbaz Z, Foote D, Walters MC, Vichinsky EP, Harmatz P, et al. Treatment of hepatitis C virus infection in thalassemia. Ann N Y Acad Sci. 2005;1054:290–9. doi: 10.1196/annals.1345.038. [DOI] [PubMed] [Google Scholar]
- 13.Ocak S, Kaya H, Cetin M, Gali E, Ozturk M. Seroprevalence of hepatitis B and hepatitis C in patients with thalassemia and sickle cell anemia in a long-term follow-up. Arch Med Res. 2006;37:895–8. doi: 10.1016/j.arcmed.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Antipa C, Popescu A, Teleguţă M, Ruţă S, Cernescu C, Târdei G, et al. Seroprevalence of hepatitis C virus among the multiply transfused. Rom J Virol. 1993;44:9–15. [PubMed] [Google Scholar]
- 15.Isahak I, Baharin R, Hakim AS, Abu Bakar M, George E. Antibody to hepatitis C virus in thalassemia patients. Malays J Pathol. 1993;15:85–7. [PubMed] [Google Scholar]
- 16.Rosenthal E, Hazani A, Segal D, Koren A, Kamal S, Rimon N, et al. Lack of transmission of hepatitis C virus in very close family contacts of patients undergoing multitransfusions for thalassemia. J Pediatr Gastroenterol Nutr. 1999;29:101–3. doi: 10.1097/00005176-199907000-00025. [DOI] [PubMed] [Google Scholar]
- 17.Katsanos K, Chaidos A, Christodoulou D, Tzambouras N, Zervou E, Bourandas K, et al. Epidemiological and clinical characteristics of HCV infection in transfusion-dependent thalassemia. Ann Gastroenterol. 2005;18:56–64. [Google Scholar]
- 18.Saeed U, Waheed Y, Ashraf M, Waheed U, Anjum S, Afzal MS, et al. Estimation of hepatitis B virus, hepatitis C virus, and different clinical parameters in the thalassemic population of capital twin cities of Pakistan. Virology (Auckl) 2015;6:11–6. doi: 10.4137/VRT.S31744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahmoud RA, El-Mazary AA, Khodeary A. Seroprevalence of hepatitis C, hepatitis B, cytomegalovirus, and human immunodeficiency viruses in multitransfused thalassemic children in upper Egypt. Adv Hematol. 2016;2016:9032627. doi: 10.1155/2016/9032627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jafroodi M, Davoudi-Kiakalayeh A, Mohtasham-Amiri Z, Pourfathollah AA, Haghbin A. Trend in prevalence of hepatitis C virus infection among beta-thalassemia major patients: 10 years of experience in Iran. Int J Prev Med. 2015;6:89. doi: 10.4103/2008-7802.164832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karimi M, Ghavanini AA. Seroprevalence of hepatitis B, hepatitis C and human immunodeficiency virus antibodies among multitransfused thalassaemic children in Shiraz, Iran. J Paediatr Child Health. 2001;37:564–6. doi: 10.1046/j.1440-1754.2001.00709.x. [DOI] [PubMed] [Google Scholar]
- 22.Bastani MN, Bokharaei-Salim F, Keyvani H, Esghaei M, Monavari SH, Ebrahimi M, et al. Prevalence of occult hepatitis C virus infection in Iranian patients with beta thalassemia major. Arch Virol. 2016;161:1899–906. doi: 10.1007/s00705-016-2862-3. [DOI] [PubMed] [Google Scholar]
- 23.Alavian SM, Miri SM, Keshvari M, Elizee PK, Behnava B, Tabatabaei SV, et al. Distribution of hepatitis C virus genotype in Iranian multiply transfused patients with thalassemia. Transfusion. 2009;49:2195–9. doi: 10.1111/j.1537-2995.2009.02252.x. [DOI] [PubMed] [Google Scholar]
- 24.Akhtar S, Moatter T, Azam SI, Rahbar MH, Adil S. Prevalence and risk factors for intrafamilial transmission of hepatitis C virus in Karachi, Pakistan. J Viral Hepat. 2002;9:309–14. doi: 10.1046/j.1365-2893.2002.00350.x. [DOI] [PubMed] [Google Scholar]
- 25.Diamantis I, Vafiadis I, Voskaridou E, Dellatetsima J, Jäggi N, Gyr K, et al. Genotype distribution of hepatitis C virus infection in Greece: Correlation with different risk factors and response to interferon therapy. Eur J Gastroenterol Hepatol. 1998;10:75–9. doi: 10.1097/00042737-199801000-00014. [DOI] [PubMed] [Google Scholar]
- 26.Di Marco V, Lo Iacono O, Almasio P, Ciaccio C, Capra M, Rizzo M, et al. Long-term efficacy of alpha-interferon in beta-thalassemics with chronic hepatitis C. Blood. 1997;90:2207–12. [PubMed] [Google Scholar]
- 27.Hermida M, Ferreiro MC, Barral S, Laredo R, Castro A, Diz Dios P. Detection of HCV RNA in saliva of patients with hepatitis C virus infection by using a highly sensitive test. J Virol Methods. 2002;101:29–35. doi: 10.1016/s0166-0934(01)00417-7. [DOI] [PubMed] [Google Scholar]
- 28.Wang JT, Wang TH, Sheu JC, Lin JT, Chen DS. Hepatitis C virus RNA in saliva of patients with posttransfusion hepatitis and low efficiency of transmission among spouses. J Med Virol. 1992;36:28–31. doi: 10.1002/jmv.1890360106. [DOI] [PubMed] [Google Scholar]
- 29.Young KC, Chang TT, Liou TC, Wu HL. Detection of hepatitis C virus RNA in peripheral blood mononuclear cells and in saliva. J Med Virol. 1993;41:55–60. doi: 10.1002/jmv.1890410112. [DOI] [PubMed] [Google Scholar]
- 30.Lins L, Almeida H, Vitvisk L, Carmo T, Parana R, Reis MG. Detection of hepatitis C virus RNA in saliva is not related to oral health status or viral load. J Med Virol. 2005;77:216–20. doi: 10.1002/jmv.20438. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Omata K, Satoh T, Miyasaka T, Arai C, Maeda M, et al. Quantitative detection of hepatitis C virus (HCV) RNA in saliva and gingival crevicular fluid of HCV-infected patients. J Clin Microbiol. 2005;43:4413–7. doi: 10.1128/JCM.43.9.4413-4417.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eirea M, Dios PD, Hermida M, Rodríguez I, Castro A, Ocampo A, et al. Detection of HCV-RNA in saliva of HIV-HCV coinfected patients. AIDS Res Hum Retroviruses. 2005;21:1011–5. doi: 10.1089/aid.2005.21.1011. [DOI] [PubMed] [Google Scholar]
- 33.Chen M, Yun ZB, Sällberg M, Schvarcz R, Bergquist I, Berglund HB, et al. Detection of hepatitis C virus RNA in the cell fraction of saliva before and after oral surgery. J Med Virol. 1995;45:223–6. doi: 10.1002/jmv.1890450219. [DOI] [PubMed] [Google Scholar]
- 34.Cameron SO, Wilson KS, Good T, McMenamin J, McCarron B, Pithie A, et al. Detection of antibodies against hepatitis C virus in saliva: A marker of viral replication. J Viral Hepat. 1999;6:141–4. doi: 10.1046/j.1365-2893.1999.00142.x. [DOI] [PubMed] [Google Scholar]
- 35.Said F, El Beshlawy A, Hamdy M, El Raziky M, Sherif M, Abdel kader A, et al. Intrafamilial transmission of hepatitis C infection in Egyptian multitransfused thalassemia patients. J Trop Pediatr. 2013;59:309–13. doi: 10.1093/tropej/fmt017. [DOI] [PubMed] [Google Scholar]


