Abstract
The vast number of detectable odors makes matching olfactory receptors (ORs) to their ligands a daunting task. Krautwurst and colleagues have hypothesized that this process can be simplified by focusing on those odorants that are perceptually relevant food odors. In this issue of Chemical Senses, they use this framework to identify highly sensitive receptors for 2 key food odorants found in red wine and onions, that activate broadly tuned OR1A1 and narrowly tuned OR2M3, respectively. This work provides further evidence for the advantage of screening receptors against ecologically relevant odors, and we discuss it in the context of current limitations in OR screening methods.
Keywords: genetic variability, key food odors, odor, olfaction, onion, wine
Olfactory receptors (ORs) provide the main mechanism for sensing volatile chemicals, but relating OR activation patterns to odor perception has been a challenging problem. First, the number of molecules that have an odor is vast—likely greater than 27 billion (Yu et al. 2015). These can then be mixed in varying concentrations to make a huge variety of odors. Second, the number of ORs is also large. While humans have only 3 types of receptors that mediate color vision, we have more than 400 types of receptors that mediate olfaction, compounding the problem of cataloging odor/receptor interactions. Furthermore, while we can organize light by wavelength, allowing us to methodically test representative stimuli, we have little idea how to organize odors. In combination, these complications have led many researchers to despair—how can we gain a foothold in this immense landscape of odors and receptors?
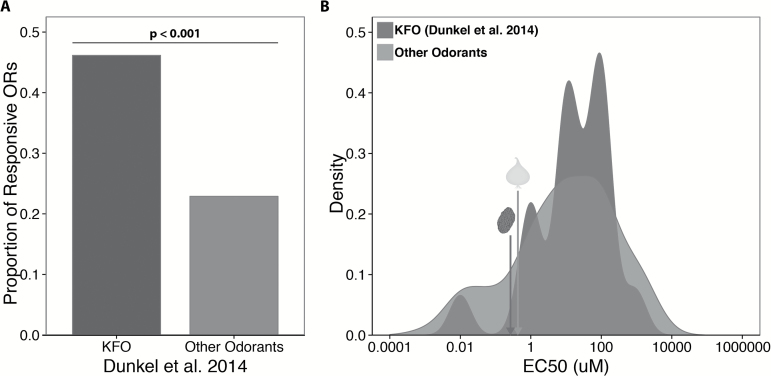
Krautwurst and colleagues have attempted to carve out a foothold by focusing on food odors, hypothesizing that although the olfactory system is able to respond to a wide variety of chemicals, the best ligands will be those with behavioral and evolutionary significance. At first glance, this focus seems to add another layer of complexity to the problem in that food odors are complex mixtures that typically contain hundreds of volatile molecules. Recent work by Krautwurst and colleagues suggests, however, that this mixture landscape can be vastly simplified by identifying key food odors (KFOs) that are the main components of food flavor. This simplification is striking—of the roughly 10000 volatile molecules present in food, reconstruction of food aromas suggests that only 230 are necessary and sufficient to reconstitute the perception of most foods and beverages (Dunkel et al. 2014). Krautwurst and colleagues hypothesize that the size of the food-essential chemical space suggests that the OR gene family may have evolved to recognize a specific, biologically relevant group of odorants. Focusing on KFOs that are relevant to food perception effectively reduces the number of odorants they need to screen by 2 orders of magnitude. This is in contrast to the approach of choosing a representative set of odors by their physiochemical properties and echoes Larry Katz’s observation that “The olfactory system did not evolve to decode the catalog of Sigma-Aldrich; it evolved to decode the world around us.” Indeed, Dunkel et al. (2014) found that in the published literature, odorant receptors are more likely to respond to KFOs than other odors (Figure 1A). Furthermore, although ORs seem to respond to KFOs and non-KFOs with similar sensitivity (similar EC50) (Figure 1B), additional work may demonstrate greater sensitivity in the response of ORs to KFOs.
Figure 1.
KFOs as the best ligands for ORs. (A) Proportion of receptors responding in the published literature when tested against KFOs or other odors. A chi-square test demonstrates a significant difference between these 2 groups (P < 0.001). (B) Distribution of EC50s for published OR/odor pairs, colored by whether Dunkel et al. (2014) identify the odor as a KFO. Arrows indicate EC50s for the “prune” and “onion” odors identified in this issue.
In this issue of Chemical Senses, Krautwurst’s group takes this philosophy forward using what the authors call a bidirectional assay system to probe OR/odorant response relationships in the KFO stimulus space. This method examines 1) the response of an OR to the suite of 190 KFOs available for screening and 2) the activation pattern of 391 ORs in response to specific KFOs of interest. Geithe et al. (2016) started with 2 broadly tuned ORs—OR1A1 and OR2W1—and examined their response to 190 KFOs and other compounds. They found that although OR1A1 responded to a large number of molecules, it was particularly sensitive to a single KFO, 3-methyl-2,4-nonanedione, that smells of prunes. Although OR1A1 was thought to have a very broad role in odor coding, this high sensitivity to a behaviorally relevant odor suggests that the receptor may play multiple roles in representing odors. In addition, 17 new agonists were identified for OR1A1 (14 of which were KFOs), adding to the 30 known odorant ligands for this OR. Although the authors did not identify a high affinity agonist for OR2W1, they found it responded to 21 new ligands (18 KFOs), increasing its response profile to 60 odorants.
In a complimentary approach, Noe et al. (2016) started with a KFO that has a characteristic onion odor, 3-mercapto-2-methylpentan-1-ol, and explored the molecular mechanism of its sensitive detection. Humans can detect this odor at very low levels, as they can with many sulfur-containing odors. By screening with this odor, the authors deorphaned OR2M3. Sulfur-containing molecules have been a puzzle for the field—although behaviorally we can detect many thiols at very low concentrations and they represent important signals of spoiled food and predators, the receptor mechanisms have largely remained a mystery. OR2M3 is the most sensitive receptor for thiols to date, with the recently deorphaned OR2T11 (Li et al. 2016) not far behind. The success of this bidirectional screening approach in identifying high-affinity ligands for both narrowly and broadly tuned ORs makes a strong case for the advantage of using a KFO library to probe OR response profiles and suggests that even for receptors that have been previously deorphaned, focusing on KFOs may allow us to identify particularly potent ligands.
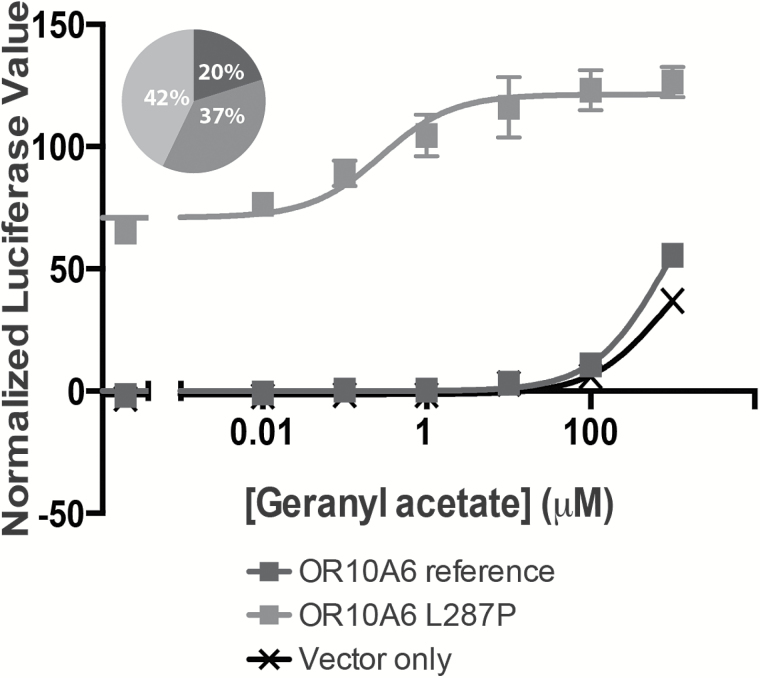
Krautwurst and colleagues address a crucial problem in the field: matching ORs to odor ligands. With these publications, the number of published human ORs with known ligands has now reached 53 (de March et al. 2015), a mere 13% of the full receptor repertoire. As the field begins to expand its efforts to screen odors against receptors, it is important to keep a few caveats in mind. First, many of these screens use one version of a given receptor despite the fact that there are on average 5 common variants for a given receptor in the population (Mainland et al. 2014). As the authors note, if the screen happens to include a nonfunctional variant, then the receptor will, of course, not respond to a given odor, despite the fact that the majority of the population may express the functional receptor. Along these lines, many investigators use the reference version of the OR, assuming that this is the most common and functional version of the receptor. The reference designation, however, has nothing to do with function, and is not necessarily the most common version in the global population. An example of this is the hg19 reference variant for OR10A6, which is annotated as a pseudogene in NCBI; another version of OR10A6 with a single amino acid difference (L287P) is not only more common than the reference version, but also functional whereas the reference is not (Figure 2) (Abecasis et al. 2010; Mainland et al. 2014; NCBI 2016). Functional variability in this gene family reduces the ability of the assay to find the receptors for a given ligand and makes interpretation of each receptor’s role in representing perceptual qualities difficult.
Figure 2.
ORs in the reference genome may not be the most common or most functional variant. The version of OR10A6 in the hg19 reference genome is nonfunctional in cell culture and has an allele frequency of 20% in the 1000 genomes population (Abecasis et al. 2010). In contrast, a variant with a single amino acid change (L287P) responds to geranyl acetate in cell culture and occurs more frequently in the population (Mainland J, unpublished data).
Second, our understanding of how a given receptor alters perception is in its infancy. Although we know ligands for some odorant receptors from cell culture work, we do not know if these are the best ligands for the receptors. In addition, due to differences between in vitro and in vivo conditions, we cannot be certain that the concentrations that activate the receptor in cell culture are relevant to behavior. For some ORs, researchers have shown that genetic variation in the receptor alters the intensity, pleasantness, or character of the receptor’s ligand, but in other cases, genetic variation in a receptor appears to have little to no effect on perception. Noe et al. (2016) relate the sensitivity of OR2M3 to 3-mercapto-2-methylpentan-1-ol to the odorant’s extremely low detection threshold, but the rarity of genetic variation in this OR in the general population means the authors would need a very large study population to directly examine the consequences of OR2M3 loss of function on odor perception. Without direct evidence that loss of OR2M3 alters odor perception, concluding that a nonfunctional OR2M3 will have major perceptual consequences is difficult, particularly in extinct human populations and other species. In addition, we have no way of being certain that alternate ORs in these species are incapable of responding to 3-mercapto-2-methylpentan-1-ol, a function which may have been lost in the human population and therefore undetectable using this screening method.
Despite these limitations in current screening technology, in this issue of Chemical Senses, Krautwurst and colleagues describe the successful application of a cell-based assay system using a group of the ~230 odorants they previously demonstrated to be important building blocks for food and beverages. The authors tested their hypothesis that ORs evolved to detect this group of key food odorants; in doing so, this provides a focused, biologically relevant screening library for exploring receptor/odor interactions that the field has been missing. While food odors are not the only important odors, progress in understanding the composition of odors outside of food is largely nonexistent. While the field has created large databases summarizing chemical analysis of foods, there is nothing comparable for non-food odors and no odor snapshots in a natural environment to develop a picture of what humans and other animals smell throughout a typical day. Shrinking the odor landscape by focusing on KFOs provides a starting point for examining ecologically relevant odors; indeed, the advantage of this approach is shown by the success of Krautwurst et al. in identifying high-affinity ligands for 2 different ORs. Figuring out how we detect the odorants that make up the foods we eat will provide crucial insight into both how the olfactory system works and potentially how to improve the flavor of our food. Given the importance of food in human culture and society, this would be no small achievement. As Jean Brillat-Savarin noted in 1862, “The discovery of a new dish does more for human happiness than the discovery of a new star.”
Funding
NIH R01DC013339.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. 2010. A map of human genome variation from population-scale sequencing. Nature. 467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkel A, Steinhaus M, Kotthoff M, Nowak B, Krautwurst D, Schieberle P, Hofmann T. 2014. Nature’s chemical signatures in human olfaction: a foodborne perspective for future biotechnology. Angew Chem Int Ed Engl. 53:7124–7143. [DOI] [PubMed] [Google Scholar]
- Geithe C, Noe F, Kreissl J, Krautwurst D. 2016. The broadly tuned odorant receptor OR1A1 is highly selective for 3-methyl-2,4-nonanedione, a key food odorant in aged wines, tea, and other foods. Chem Senses. 42: 181–194. [DOI] [PubMed] [Google Scholar]
- Li S, Ahmed L, Zhang R, Pan Y, Matsunami H, Burger JL, Block E, Batista VS, Zhuang H. 2016. Smelling sulfur: copper and silver regulate the response of human odorant receptor OR2T11 to low-molecular-weight thiols. J Am Chem Soc. 138:13281–13288. [DOI] [PubMed] [Google Scholar]
- Mainland JD, Keller A, Li YR, Zhou T, Trimmer C, Snyder LL, Moberly AH, Adipietro KA, Liu WL, Zhuang H, et al. 2014. The missense of smell: functional variability in the human odorant receptor repertoire. Nat Neurosci. 17(1):114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de March CA, Ryu S, Sicard G, Moon C, Golebiowski J. 2015. Structure-odour relationships reviewed in the postgenomic era. Flavour Fragr J. 30:342–361. [Google Scholar]
- NCBI. 2016. National Center for Biotechnology Information, Gene Search Tool. Bethesda (MD): NCBI (US). [Google Scholar]
- Noe F, Polster J, Geithe C, Kotthoff M, Schieberle P, Krautwurst D. 2016. OR2M3: a highly specific and narrowly tuned human odorant receptor for the sensitive detection of onion key food odorant 3-mercapto-2-methylpentan-1-ol. Chem Senses. 42:195–210. [DOI] [PubMed] [Google Scholar]
- Yu CW, Prokop-Prigge KA, Warrenburg LA, Mainland JD. 2015. Drawing the borders of olfactory space. Chem Senses. 40:565.26304508 [Google Scholar]