Abstract
Activity-dependent processes are important to olfactory sensory neurons (OSNs) in several ways, such as cell survival and the specificity of axonal convergence. The identification of activity-dependent mRNAs has contributed to our understanding of OSN axon convergence, but has revealed surprisingly little about other processes. Published studies of activity-dependent mRNAs in olfactory mucosae overlap poorly, but by combining these agreements with meta-analysis of existing data we identify 443 mRNAs that respond to methods that alter OSN activity. Three hundred and fifty of them are expressed in mature OSNs, consistent with the expectation that activity-dependent responses are cell autonomous and driven by odor stimulation. Many of these mRNAs encode proteins that function at presynaptic terminals or support electrical activity, consistent with hypotheses linking activity dependence to synaptic plasticity and energy conservation. The lack of agreement between studies is due largely to underpowered experiments. In addition, methods used to alter OSN activity are susceptible to indirect or off-target effects. These effects deserve greater attention, not only to rigorously identify OSN mRNAs that respond to altered OSN activity, but also because these effects are of significant interest in their own right. For example, the mRNAs of some sustentacular cell enzymes believed to function in odorant clearance (Cyp2a4 and Cyp2g1) are sensitive to unilateral naris occlusion used to reduce odorant stimulation of the ipsilateral olfactory epithelium. Also problematic are odorant receptor mRNAs, which show little agreement across studies and are susceptible to differences in frequency of expression that masquerade as activity-dependent changes in mRNA abundance.
Keywords: odorant receptor, odorant clearance, smell, synaptic plasticity
Introduction
Activity-dependent plasticity is a fundamental property of the nervous system (Flavell and Greenberg 2008; West and Greenberg 2011; Ganguly and Poo 2013). During development, it helps determine which synapses form and persist (Bleckert and Wong 2011). For example, the ocular dominance columns and the orientation columns in the visual cortex are both refined by visual stimulation during a sensitive period in early postnatal life (Wiesel and Hubel 1974; Stryker and Harris 1986). Spontaneous neural activity can also be responsible for refining connectivity during development. For example, dorsal lateral geniculate neurons initially receive inputs from about 20 retinal ganglion cells but refine this down to 1–3 ganglion cells in a process that requires only spontaneous neurotransmission (Hooks and Chen 2006). Similarly, the complexity of dendritic arbors of cultured hippocampal neurons and axon bouton maturation at Drosophila neuromuscular junctions depend on spontaneous activity (Choi et al. 2014; Andreae and Burrone 2015).
In addition to its role in development, activity-dependent mechanisms alter synaptic strengths and establish patterns of enhanced connectivity in the mature nervous system, forming the bases for several types of learning and memory. Early hypotheses about the possible importance of neural connection strength in learning and memory (Cajal 1913) were transformed into a major focus of modern neuroscience with the discovery of long-term potentiation (Bliss and Lomo 1973). We now understand that associative events triggering long term potentiation or long term depression at specific synapses strengthen patterns of neural activity that then serve to represent these events over long periods of time (Bailey et al. 2004; Flavell and Greenberg 2008; Ganguly and Poo 2013).
The alteration of synaptic connectivity between existing neurons is not the only way in which activity-dependent plasticity mediates learning, however. For example, the turnover of interneurons makes possible another form of plasticity in the olfactory bulb and the dentate gyrus of the hippocampus where adult neurogenesis continuously provides new interneurons (Whitman and Greer 2009). New interneurons survive better when their synapses integrate into circuits that become strengthened by associative events, thereby allowing new interneurons to contribute to modified circuits that aid the learning of olfactory and spatial tasks, respectively (Alonso et al. 2006; Jessberger et al. 2009; Sultan et al. 2010; Sultan et al. 2011).
All of these changes in neural connectivity are able to long outlast the initiating stimuli because they become solidified by altered expression of specific genes. Given this fact, the study of changes in gene expression provides clues about mechanisms of activity dependence and can also identify previously unrecognized activity-dependent phenomena. In this perspective review, we focus on what studies of activity-dependent gene expression in the olfactory periphery have revealed about the biology of olfactory sensory neurons (OSNs).
Why should OSNs show activity dependence?
Activity-dependence of OSN axon coalescence and OSN synapses
The axons of OSNs selectively coalesce into glomeruli in the olfactory bulb according to which odorant receptor (OR) each OSN expresses; an organization made possible because each OSN strongly expresses only one OR gene (Chess et al. 1994; Mombaerts et al. 1996; Malnic et al.1999; Rawson et al. 2000; Saraiva et al. 2015; Scholz et al. 2016). Activity-dependent, complementary expression of the axon guidance factors Kirrel2 and Kirrel3 contributes to the specificity of OSN axon coalescence by mediating homophilic adhesion (Imai et al. 2006; Serizawa et al. 2006). Forcing mosaic expression of Kirrel2 in OSNs increases the number of glomeruli innervated by OSNs expressing an OR because the axons of OSNs overexpressing Kirrel2 segregate from the axons of unaffected OSNs that express the same OR. Activity-dependent, complementary expression of EphA5 and ephrin-A5, which mediate contact repulsion of axons, suggests that these factors may similarly contribute to OSN axon segregation (Imai et al. 2006; Serizawa et al. 2006). Interpreting the altered expression of these axon guidance proteins is complicated by evidence that the coalescence of OSN axons into glomeruli is relatively insensitive to odor-stimulated electrical activity (Lin et al. 2000; Zheng et al. 2000). This seems to argue that these factors play only a minor role in OSN axon coalescence. However, a possible explanation is that the constitutive activity of ORs (Imai et al. 2006; Reisert 2010) elevates 3',5'-cyclic adenosine monophosphate (cAMP) and downstream ion channel activity enough to drive activity-dependent mechanisms necessary for OSN axon coalescence.
The locations of glomeruli in the olfactory bulb have also been reported to depend on OR signaling and activity-dependent gene expression. Glomerular position along the anterior-posterior axis has been claimed to depend on gradients of expression of the activity-dependent gene, Nrp1 (Nishizumi and Sakano 2015). This model postulates that differing levels of cAMP signaling characteristic of each OR produce graded amounts of Nrp1, which helps determine the anterior-posterior positions of glomeruli (Imai et al. 2006; Imai et al. 2009). However, recent data reveal that Nrp1 levels in OSN axons in olfactory bulb glomeruli have a mosaic pattern rather than an anterior-posterior gradient as first reported, and that glomerular position in mice lacking Nrp1 is not shifted anteriorly as first reported (Zapiec et al. 2016). These findings call into question whether activity-dependent expression of Nrp1 and its ligand Sema3A in OSNs contributes to determining the anterior–posterior positions of glomeruli. If the anterior–posterior position of glomeruli is indeed independent of OSN activity, this would parallel what is known about position along the dorso-ventral axis. Dorso-ventral position is regulated independently of OSN activity by 2 ligands repulsive to axons, Sema3F interacting with Nrp2 and Slit ligands interacting with Robo2 (Nishizumi and Sakano 2015).
Looking more closely at the connections individual OSNs make with bulbar neurons inside olfactory bulb glomeruli, substantial evidence supports the conclusion that these synapses are modulated in an activity-dependent fashion. Unilateral naris occlusion (UNO), which greatly reduces odor stimulation of the ipsilateral olfactory epithelium, does not affect the number of ipsilateral OSN presynaptic terminals but does decrease their rate of turnover (Cheetham et al. 2016). UNO also causes ipsilateral increases in the probability of glutamate release at these synapses and increased amplitude of quantal synaptic currents mediated by glutamate receptors; effects that are detectable 3 days after UNO and continue for at least a few weeks (Tyler et al. 2007).
The increased strength of ipsilateral OSN synapses after UNO can be viewed as part of a homeostatic compensatory plasticity response in OSNs (Barber and Coppola 2015). In compensatory plasticity, OSN sensitivity to odors increases when odor stimulation is blocked. For example, within 2 weeks of UNO, the ipsilateral olfactory epithelium becomes more sensitive to odor stimulation (Barber and Coppola 2015). Importantly, this effect is reversible. It is also correlated with changes in the abundance of mRNAs encoding proteins involved in olfactory transduction (Coppola and Waggener 2012). These data are consistent with activity-dependent expression of these genes and the fundamental principle that neurons have mechanisms allowing adjustments in sensitivity according to their history of activity.
Activity-dependent survival of OSNs
Activity-dependent survival of OSNs has been well-documented (Zhao and Reed 2001; Watt et al. 2004; Santoro and Dulac 2012; Zhao et al. 2013). Silenced OSNs are unable to compete with active OSNs for connections to targets in the olfactory bulb, leading to loss of silenced OSNs and demonstrating that OSN activity is critical for organizing and maintaining OSN input patterns to the olfactory bulb (Zhao and Reed 2001). These effects could contribute to the development of increased sensitivity to an odorant as stimulated OSNs survive longer and accumulate preferentially. Both animals and humans can become more sensitive to an odorant and discriminate it better after training (Wysocki et al. 1989; Wang et al. 1993; Dalton and Wysocki 1996; Mandairon et al. 2006b; Mandairon et al. 2006a; Parma et al. 2015). This idea depends on OSN lifespan being relatively brief, and though OSN lifespan has proven difficult to measure accurately, this may be true. Lineage tracing data from rats suggest that a typical OSN lifespan is about a month (Caggiano et al. 1994). Given short OSN lifespans, it does seem possible that repeated exposure to an odorant could result in an increase in the number of OSNs expressing the ORs responsive to this odorant. Stimulation with the Olfr151 (M71) agonist acetophone in a fear-conditioning paradigm causes increased numbers of OSNs expressing Olfr151, but this effect must require feedback to OSNs because non-associative stimulation with acetophone fails to evoke the effect (Jones et al. 2008). Interestingly, extinction of the conditioned fear response causes reversal of the increased frequency of OSNs expressing Olfr151 (Morrison et al. 2015).
In summary, OSN survival, the development and maintenance of synaptic connections, compensatory plasticity, and increasing sensitivity to commonly encountered odors are important reasons behind the activity-dependent properties of OSNs. These may not be the only reasons for activity-dependent processes in OSNs, however. Other forms of cellular and tissue homeostasis in the olfactory epithelium, as well as energy conservation, might also be sensitive to OSN activity.
Methods for generating differences in OSN activity
Activity-dependent processes often involve responses that are relatively long-lived and depend on changes in gene expression. In neurons, the mechanisms that lead to changes in gene expression appear to depend largely upon calcium influx (Flavell and Greenberg 2008; West and Greenberg 2011). In OSNs, calcium influx is normally triggered by odorants, so differential exposure to odorants is one way to experimentally manipulate OSN activity. Alternatively, one can bypass the odorant-OR interaction and manipulate calcium influx by preventing odor-stimulated electrical activity or causing constitutive electrical activity. In actual practice, 4 types of methods have been used to experimentally manipulate OSN activity for the purpose of identifying activity-dependent mRNAs (Table 1).
Table 1.
Types of methods used to alter OSN activity
| Odor stimulation |
| Unilateral naris occlusion (UNO) |
| Genetic silencing of OSNs |
| Genetic activation of OSNs |
Direct stimulation with odorants has only rarely been used to drive activity in OSNs in order to search for activity-dependent mRNAs, largely because these experiments are difficult. For example, simply stimulating with a complex odor for 4 hrs failed to detect any changes in the abundance of well-known activity-dependent mRNAs in olfactory epithelium samples from wild-type mice, though some changes were detected in Mecp2 mutant mice (Degano et al. 2014). Effects of odor exposure in the normal cage environment have been tested successfully only when doing RT-PCR analysis of single OSNs (Cadiou et al. 2014). To successfully detect odor-stimulated changes in mRNA abundance in samples of whole olfactory mucosae one must first establish a minimal odor background—housing animals under positive pressure using filtered air, for example—against which odor stimulation effects can be measured (Bennett et al. 2010; Fischl et al. 2014). This is most useful for rapid events, and therefore very likely to identify responses driven directly by odor stimulation. Only 3 mRNAs have been shown to change rapidly, meaning within 30–40 min of odor stimulation: S100a5, Lrrc3b, and Kirrel2. Slower events, which may be equally important, are probably beyond the reach of direct odorant stimulation experiments, so other methods are needed. In fact, all other experiments designed to identify activity-dependent differences in mRNAs have measured mRNA abundance at least 6 days after the initiation of altered OSN activity, leaving a 5-day gap in our knowledge of activity-dependent responses.
Much easier than direct odorant stimulation, more effective at detecting slower effects, and much more commonly used, is UNO. UNO is designed to prevent odor stimulation of the ipsilateral olfactory epithelium behind a blocked naris while the contralateral olfactory epithelium behind the open naris continues to experience stimulation. This strategy has several advantages. It is a powerful experimental design due to the internal comparison between ipsilateral and contralateral olfactory epithelia. When performed on neonates aged 7 days (P7) or younger, it is a very simple surgical procedure. With more effort it can be performed on adults, and it can also be done in ways that are reversible (Cummings and Brunjes 1997). Retronasal olfaction requires expiration through the nostril for normal concentrations of odor molecules to reach the olfactory epithelium (Masaoka et al. 2010), so the retronasal pathway and the septal window, an opening in the septum found in rodents but not in most other mammals (Kelemen 1947), appear to allow only small amounts of odor molecules to reach the ipsilateral olfactory epithelium after UNO. The evidence that detectable levels of odor molecules do reach the ipsilateral olfactory epithelium come from experiments where rodents subjected to both UNO and a contralateral olfactory bulbectomy prove able to perform olfactory tasks (Slotnick and Pazos 1990; Coppola et al. 1994). Nevertheless, the evidence is overwhelming that UNO results in greatly reduced odor stimulation of the ipsilateral olfactory epithelium, even causing effects of the same magnitude as genetic silencing of OSNs (Fischl et al. 2014).
UNO does have some disadvantages. Effects on mRNA abundance within the first few days of occlusion have never been measured, due in part to the risk of confounding effects of the surgery and consequent recovery processes. Information on the temporal profiles of change in affected mRNAs, whether they are gradual or abrupt, is therefore lacking. This absence of temporal information contributes to a complete lack of understanding of the mechanisms that lead to changes in mRNA abundance. Whether most OSN responses to reduced activity are cell autonomous and slow, or whether these responses are slow because they are indirect effects of secondary processes, is unknown. The risk of indirect effects probably increases with the length of the survival period after UNO. Foremost among these potential indirect effects are changes in the cellular composition of the olfactory epithelium. The cellular composition of the olfactory epithelium is stable in rodents through at least 6 days after UNO (Suh et al. 2006; Sammeta and McClintock 2010; Fischl et al. 2014). After 10 days, the number of mature OSNs decreases according to several studies (Maruniak et al. 1990; Cavallin et al. 2010; Cummings and Belluscio 2010), but other studies find no change, or changes only in the numbers of immature neurons (Benson et al. 1984; Farbman et al. 1988; Brunjes and Shurling 2003; Cheetham et al. 2016). Another potential indirect effect is altered feedback from the olfactory bulb. OSN survival depends on feedback from the olfactory bulb (Schwob et al. 1992), and because some bulbar interneurons show activity-dependent responses themselves (Baker et al. 1983), including increased cell death when OSN activity is reduced (Frazier-Cierpial and Brunjes 1989; Najbauer and Leon 1995), it is possible that these are linked and lead to indirect effects on mRNA abundance in OSNs. Yet other potential sources of indirect effects are signaling events between OSNs and neighboring cells in the olfactory epithelium. For example, signaling between OSNs and basal cells helps control OSN replacement (Wu et al. 2003; Lander et al. 2009). Whether this signaling is altered by UNO is unknown. Finally, odor molecules are not the only agents whose access is blocked by UNO. Damaging agents, especially pathogens and particulates, are also blocked. UNO presumably increases the amount of air passing over the contralateral olfactory epithelium and this would contribute to large differences in the amount of damage experienced by the ipsilateral and contralateral olfactory epithelia. Direct evidence of this effect is a demonstration that OSNs of the ipsilateral and contralateral epithelia experience differential chemical stress after UNO (Sammeta and McClintock 2010). For more detail on UNO, see review articles by Brunjes and Coppola (Brunjes 1994; Coppola 2012).
Genetic silencing is also an effective way to investigate the activity dependence of OSNs. Most often this has been done with mice lacking a subunit of the cyclic nucleotide-gated channel; typically the Cnga2 subunit (Serizawa et al. 2006; Kaneko-Goto et al. 2008; Bennett et al. 2010; Williams et al. 2011; Oztokatli et al., 2012; Santoro and Dulac 2012; Fischl et al. 2014). Mice with a germ line targeted deletion of Cnga2 have less disorganization in OSN axon convergence to glomeruli in the olfactory bulb than other germ line mutant mice in which OSNs lack electrical responses to odors, such as mice with targeted deletion of Adcy3 (Col et al. 2007; Zou et al. 2007). Mice lacking Cnga2 also have the advantage that while electrical activity of OSNs is prevented, the biochemical steps in olfactory transduction are intact, so effects must be due to differences in electrical activity and the consequent reduction in the flux of calcium ions. Because genetic silencing by germ line targeted gene deletion is lifelong and male Cnga2(–/0) and female Cnga2(–/–) neonates typically show delayed growth, these experiments have used juvenile or adult mice, so that the period of differential OSN activity is usually measured in weeks. The main disadvantages of genetic silencing are therefore those related to indirect secondary effects that might arise during long periods without OSN activity, disadvantages shared with studies that have used UNO.
Other genetic manipulations that constitutively increase or decrease OSN activity are also possible and are effective ways to investigate activity-dependent processes in OSNs. A clever strategy to alter OSN activity of specific populations of OSNs involved introducing transgenes causing expression of either an OR deficient in G-protein coupling to reduce downstream signaling, or a constitutively active Gαs to force continuous activity in the affected OSNs (Imai et al. 2006; Imai et al. 2009). These strategies alter OSN activity while avoiding differences in external stressors and odor stimulation that might act through confounding mechanisms. As a genetic manipulation, these strategies have the disadvantage that effects are measured after waiting several weeks for the mutant mice to become at least juveniles, if not adults. This increases the possibility that secondary factors contribute to differences found, or that compensatory changes might mask some effects.
Methods used to trigger changes in OSN activity in order to identify activity-dependent mRNAs need to reliably generate differences in activity without introducing confounding variables or allowing indirect processes that cause changes masquerading as activity-dependent effects. None of the methods that have been used fully satisfy these criteria. We therefore question the reliability of existing data and ponder the extent to which these data capture the full set of activity-dependent mRNAs in OSNs. We can now begin to more carefully evaluate these questions because the breadth and depth of data has become sufficient to allow meaningful comparisons across multiple studies, and because additional evidence, such as cell type expression patterns, can be brought to bear.
Reliable identification of activity-dependent mRNAs
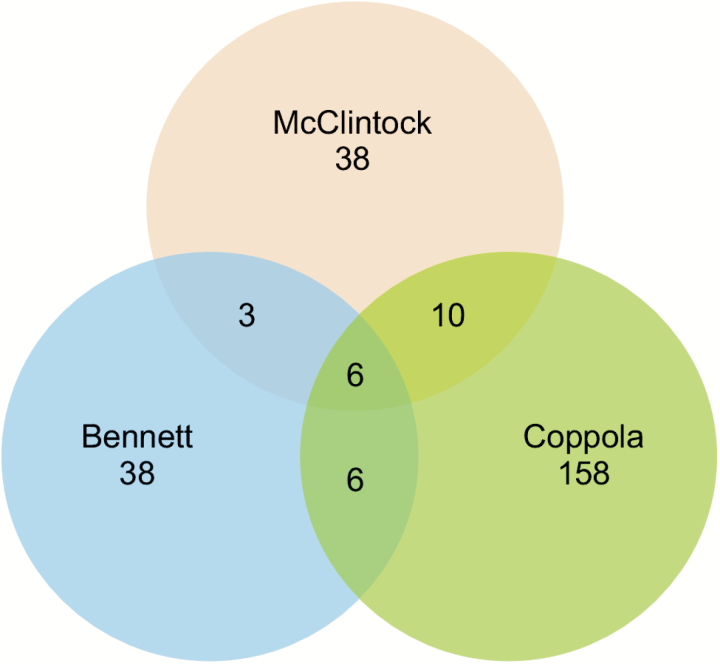
In assessing claims of activity-dependent gene expression, the best supporting evidence is consistency and repeatability, especially when the experiments are done with different methods by different laboratories. A total of 302 mouse mRNAs (not including OR mRNAs) have been identified as being activity-dependent in samples from olfactory mucosae. Unfortunately, when we compare the mRNAs identified by different transcriptome-wide expression profiling studies, the degree of overlap is poor (Figure 1). When we include data from studies targeting subsets of mRNAs to the expression profiling data sets depicted in Figure 1, we increase the number of mRNAs identified in at least 2 studies from 25 to 35 (Table 2); still a disappointingly small number. The poor agreement between studies indicates that our understanding of activity-dependent gene expression in OSNs is insufficient in both rigor and extent. Herein, we explore possible explanations and do additional analyses to further evaluate consistency and repeatability.
Figure 1.
Expression profiling studies that assessed the entire mouse transcriptome after altering OSN activity show poor overlap. Only 25 mRNAs are differentially abundant mRNAs in more than one data set. Data sets from: Bennett et al. 2010; Coppola and Waggener 2012; McClintock data are the merger of published data (Fischl et al. 2014) with an RNA-seq data set described herein (Gene Expression Omnibus accession number GSE89460). OR mRNAs are excluded.
Table 2.
Transcripts found to be activity-dependent in multiple studies.
| Gene Symbol | Cell type | Earliest response | Activity response | Reference # |
|---|---|---|---|---|
| Bace1 | mOSN | 10 days | Down | 12,14 |
| Calb2 | mOSN | 6 days | Down | 1,2,11,13 |
| Cdh15 | mOSN | >14 days | Down | 4,13 |
| Chil3 | ? | 25 days | Down | 1,13 |
| Cnga2 | mOSN | 10 days | Up | 1,12 |
| Cyp26b1 | mOSN | 2 days | Up | 6,10,11,12 |
| Dlg2 | mOSN | 25 days | Up | 1,13 |
| Efna5 | mOSN | 2 days | Down | 1,5,6,8,11,13 |
| Entpd2 | mOSN | 6 days | Down | 11,13 |
| Epha5 | mOSN | 2 days | Up | 8,11 |
| Etv3 | mOSN | 6 days | Down | 1,11 |
| Etv5 | mOSN | 25 days | Up | 4,13 |
| Fos | mOSN | 6 days | Up | 11,13 |
| Gpr158 | mOSN | 6 days | Up | 2,13 |
| Kirrel2 | both | 30–40 min | Up | 2,4,5,6,8,9,11,13 |
| Kirrel3 | mOSN | 2 days | Down | 1,2,8, 9,11,13 |
| Lrrc3b | mOSN | 30–40 min | Up | 1,2,11,12,13 |
| Nphs1 | mOSN | 6 days | Up | 2,11 |
| Nrp1 | both | 2 days | Up | 3,4,6,11 |
| Nxph3 | mOSN | 6 days | Up | 3,11 |
| Pcdh10 | mOSN | 6 days | Up | 2,9,11,13 |
| Pcp4l1 | mOSN | 6 days | Up | 1,2,3, 4,11,13 |
| Plxna3 | mOSN | >14 days | Up | 3,4 |
| Ppp3ca | mOSN | 25 days | Down | 1,13 |
| Ptchd1 | mOSN | 6 days | Up | 1,11,13 |
| Ptprn | mOSN | 6 days | Up | 2,3,4,11 |
| Rasgrp4 | mOSN | 6 days | Up | 2,11 |
| Rims3 | mOSN | 6 days | Down | 11,13 |
| S100a5 | mOSN | 30–40 min | Up | 1,2,4,8,11,12,13 |
| Sema3e | mOSN | 6 days | Up | 11,13 |
| Sema7a | mOSN | 6 days | Up | 1, 11 |
| Slc8a1 | mOSN | 6 days | Up | 1,2,11,13 |
| Slc17a6 | mOSN | 6 days | Up | 1, 2 |
| Snca | mOSN | 25 days | Up | 1,13 |
| Syt4 | mOSN | 6 days | Up | 4,11,13 |
The mRNAs from these genes are significant in at least 2 independent studies. Cell type: both, immature and mature OSN; mOSN, mature OSN; Other, cell types other than OSNs;?, unknown. References: 1. (Bennett et al. 2010), 2. (Fischl et al. 2014), 3. (Imai et al. 2006), 4. (Imai et al. 2009), 5. (Kaneko-Goto et al. 2008), 6. (Oztokatli et al. 2012), 7. (Santoro and Dulac 2012), 8. (Serizawa et al. 2006), 9. (Williams et al. 2011), 10. (Login et al. 2015b), 11. Unpublished UNO RNA-seq data (W.B. Titlow and T.S. McClintock; Gene Expression Omnibus accession number GSE89460), 12. (Login et al. 2015a), 13. (Coppola and Waggener 2012), 14. (Cao et al. 2012).
The requirement that mRNA abundance differences are detected in multiple experiments would be a stronger measure of reliability if it were not for the fact that the data come either from underpowered expression profiling experiments or from studies that investigated only a small set of mRNAs. Due to the high cost of expression profiling experiments, investigators often use fewer replications than needed to fully power their experimental designs, thereby limiting themselves to consistently identifying only the most differentially abundant mRNAs, often along with a small subset of the mRNAs that differ less strongly. This makes for poor congruence between studies. However, because multiple transcriptome-wide expression profiling data sets now exist we can improve upon this by taking advantage of the fact that false positives have randomly distributed P-values across studies while true positives are nonrandomly distributed at the low end of the P-value distribution. We analyzed the P-value distributions of all 302 mRNAs using the 3 UNO expression profiling data sets that are available to us: a microarray data set (Coppola and Waggener 2012), a second microarray data set (Fischl et al. 2014), and an unpublished RNA-seq data set that we had previously generated (n = 3 mice; see Supplemental Methods file). As a group, the 302 mRNAs show nonrandom P-value distributions across the 3 data sets, and these nonrandom distributions are weighted toward low P-values. To extend this approach to the level of individual mRNAs, we required that mRNAs have similar P-values (standard deviation < 0.2) that are consistently low (average P-value < 0.2) across studies. This identified 49 additional mRNAs that are significant in only one study but have consistently low P-values, indicative of nonrandom behavior (Table 3).
Table 3.
Transcripts that have consistently low P-values across expression profiling studies.
| Gene symbol | Cell type | Earliest response | Activity response | Reference # |
|---|---|---|---|---|
| 1500017E21Rik | mOSN | 6 days | Up | 11 |
| 1700012B09Rik | mOSN | 6 days | Up | 4 |
| 2410004P03Rik | mOSN | 25 days | Down | 13 |
| Adcy3 | mOSN | 25 days | Down | 13 |
| Ano2 | mOSN | 6 days | Up | 13 |
| Atp8b3 | mOSN | 6 days | Down | 13 |
| B830017H08Rik | mOSN | 6 days | Down | 11 |
| Bglap3 | ? | 6 days | Down | 11 |
| Bmp6 | mOSN | 6 days | Down | 13 |
| Ccdc126 | mOSN | 6 days | Down | 13 |
| Cdh22 | mOSN | 6 days | Down | 13 |
| Cfap69 | mOSN | >30 days | Down | 1 |
| Chil4 | ? | 6 days | Up | 13 |
| Cntn2 | ? | 25 days | Up | 13 |
| Cntnap4 | mOSN | 6 days | Up | 13 |
| Dcdc2a | mOSN | 6 days | Down | 11 |
| Ear2 | Other | 6 days | Up | 13 |
| Edn2 | Both | 25 days | Up | 13 |
| Ephx4 | ? | 6 days | Up | 11 |
| Fam221a | mOSN | 6 days | Down | 13 |
| Fam78a | Both | 6 days | Down | 13 |
| Galnt15 | mOSN | 6 days | Up | 4 |
| Hcn1 | mOSN | 6 days | Up | 13 |
| Hcn2 | mOSN | 6 days | Down | 13 |
| Ildr2 | ? | 25 days | Up | 13 |
| Impdh1 | mOSN | 25 days | Down | 13 |
| Inpp5f | mOSN | 6 days | Up | 13 |
| Jph3 | mOSN | 6 days | Down | 13 |
| Kcnn2 | mOSN | 6 days | Up | 2 |
| Mgst2 | Other | 25 days | Up | 13 |
| Necab3 | mOSN | 6 days | Down | 11 |
| Ntf3 | mOSN | 6 days | Up | 13 |
| Nwd1 | mOSN | 25 days | Down | 13 |
| Olfm1 | mOSN | 25 days | Up | 1 |
| Pappa | mOSN | 6 days | Up | 13 |
| Pcgf5 | mOSN | 25 days | Down | 13 |
| Pde4a | mOSN | 6 days | Down | 13 |
| Pde7b | mOSN | 6 days | Down | 13 |
| Pigr | ? | 25 days | Up | 13 |
| Retnlg | ? | 6 days | Up | 13 |
| Rgs7 | mOSN | 6 days | Up | 11 |
| S100A3 | mOSN | 6 days | Up | 11 |
| Scgb1b27 | Both | 6 days | Up | 13 |
| Scn3b | mOSN | 6 days | Up | 1 |
| Scn4b | mOSN | 6 days | Up | 1 |
| Sh3glb2 | mOSN | 6 days | Down | 13 |
| Trim45 | mOSN | 6 days | Up | 13 |
| Tusc5 | mOSN | 6 days | Down | 13 |
| Usp21 | mOSN | 6 days | Down | 13 |
The mRNAs from these genes are significant in only one study (Reference column) but have consistently low P-values across UNO expression profiling data sets. Cell type: both, immature and mature OSN; mOSN, mature OSN;?, unknown. References: 1. (Bennett et al. 2010), 2. (Fischl et al. 2014), 4. (Imai et al. 2009); 11. Unpublished UNO RNA-seq data; 13. (Coppola and Waggener 2012)
Taken together, these analyses bring the total of reliably identified activity-dependent mRNAs in the olfactory epithelium to 84. However, given the importance of activity-dependent plasticity to OSNs, this still seems insufficient. We therefore embarked on a meta-analysis using the 3 UNO data sets described above. Requiring that data for each mRNA be represented in at least 2 of the studies, we calculated the differences between ipsilateral and contralateral olfactory epithelia in each mouse. We ranked the differences, standardized the ranks, and compared to a null hypothesis of no difference by one sample t-tests using P < 0.01 as our criterion for significance, which corresponds to an FDR of 4.8% (see Supplementary Methods file). This analysis identified 433 differentially abundant mRNAs: 220 that are higher on the open side (Table 4) and 213 that are higher on the occluded side (Table 5). Merging these mRNAs with those listed in Tables 2 and 3 produces a list of 443 mRNAs (ORs excluded) that show evidence of consistent responses to altered OSN activity (Supplementary Table 1).
Table 4.
Meta-analysis: Increased with OSN activity
| Gene Symbol | P-value | Cell type | Gene symbol | P-value | Cell type | Gene symbol | P-value | Cell type |
|---|---|---|---|---|---|---|---|---|
| 4732456N10Rik | 0.0003 | mOSN | Abcg1 | 0.0001 | Both | Snca | 0.0000 | both |
| 6430548M08Rik | 0.0023 | mOSN | Adam9 | 0.0021 | Both | Spata6 | 0.0008 | both |
| Acoxl | 0.0005 | mOSN | Ahcyl2 | 0.0000 | Both | Spty2d1 | 0.0000 | both |
| Akap3 | 0.0072 | mOSN | Amica1 | 0.0099 | Both | Ssx2ip | 0.0000 | both |
| Ankrd33b | 0.0005 | mOSN | Ank1 | 0.0007 | Both | Stk38l | 0.0000 | both |
| Bcl6 | 0.0000 | mOSN | Btc | 0.0067 | Both | Syngr3 | 0.0017 | both |
| Chgb | 0.0003 | mOSN | Cadps2 | 0.0080 | Both | Tle3 | 0.0014 | both |
| Cntnap4 | 0.0002 | mOSN | Cask | 0.0013 | Both | Tm7sf3 | 0.0067 | both |
| Cyp26b1 | 0.0000 | mOSN | Cldn12 | 0.0032 | Both | Trafd1 | 0.0000 | both |
| Dgkg | 0.0001 | mOSN | Cplx2 | 0.0090 | Both | Uap1l1 | 0.0031 | both |
| Dgkk | 0.0004 | mOSN | Cxadr | 0.0012 | Both | Vat1l | 0.0028 | both |
| Dlg2 | 0.0000 | mOSN | Daam1 | 0.0092 | Both | Xrcc5 | 0.0003 | both |
| Dyrk4 | 0.0000 | mOSN | Ddx31 | 0.0019 | Both | Cxcr4 | 0.0021 | iOSN |
| Eno2 | 0.0022 | mOSN | Dio2 | 0.0002 | Both | Dclk1 | 0.0081 | iOSN |
| Epha5 | 0.0082 | mOSN | Dtna | 0.0013 | Both | Dlx6 | 0.0011 | iOSN |
| Epha7 | 0.0014 | mOSN | Dtx3l | 0.0089 | Both | Tlcd1 | 0.0002 | iOSN |
| Fetub | 0.0000 | mOSN | Dus4l | 0.0011 | Both | Zfp39 | 0.0100 | iOSN |
| Fgf12 | 0.0012 | mOSN | Dusp1 | 0.0002 | Both | Atp6v1c2 | 0.0052 | other |
| Gpr158 | 0.0000 | mOSN | Dusp16 | 0.0001 | Both | Cyp2a4 | 0.0008 | other |
| Gpr162 | 0.0078 | mOSN | Emb | 0.0033 | Both | Cyp2g1 | 0.0001 | other |
| Jph4 | 0.0030 | mOSN | Etv4 | 0.0006 | Both | Ghr | 0.0081 | other |
| Kcnn2 | 0.0000 | mOSN | Etv5 | 0.0003 | Both | Lmo7 | 0.0007 | other |
| Kirrel2 | 0.0000 | mOSN | Exoc6b | 0.0027 | Both | Maf | 0.0083 | other |
| Lrrc3b | 0.0000 | mOSN | Exoc8 | 0.0004 | Both | Rnase4 | 0.0001 | other |
| Mustn1 | 0.0007 | mOSN | Hcn1 | 0.0001 | Both | Slc14a1 | 0.0010 | other |
| Ndrg3 | 0.0003 | mOSN | Hexim2 | 0.0045 | Both | Tbc1d8b | 0.0021 | other |
| Nphs1 | 0.0000 | mOSN | Inpp5f | 0.0000 | Both | Tmprss2 | 0.0074 | other |
| Nppa | 0.0013 | mOSN | Ints8 | 0.0047 | Both | Adamdec1 | 0.0006 | ? |
| Nppc | 0.0067 | mOSN | Itpkb | 0.0019 | Both | Adamts20 | 0.0036 | ? |
| Nptx2 | 0.0000 | mOSN | Klhl9 | 0.0097 | Both | Alox12e | 0.0000 | ? |
| Nrn1l | 0.0004 | mOSN | Lmbr1 | 0.0017 | Both | Aloxe3 | 0.0100 | ? |
| Olfm1 | 0.0000 | mOSN | Lmtk2 | 0.0024 | Both | Aox1 | 0.0002 | ? |
| Palm2 | 0.0002 | mOSN | Lsg1 | 0.0026 | Both | Arsj | 0.0089 | ? |
| Pappa | 0.0049 | mOSN | Mbl2 | 0.0043 | Both | BC016548 | 0.0004 | ? |
| Pcdhb4 | 0.0009 | mOSN | Mcoln3 | 0.0019 | Both | Cbln4 | 0.0089 | ? |
| Pcp4l1 | 0.0000 | mOSN | Mllt11 | 0.0000 | Both | Ccdc129 | 0.0074 | ? |
| Pcsk1 | 0.0029 | mOSN | Nap1l3 | 0.0084 | Both | Cd48 | 0.0071 | ? |
| Ppa1 | 0.0000 | mOSN | Nckap1 | 0.0014 | Both | Chrnb1 | 0.0018 | ? |
| Ppargc1a | 0.0000 | mOSN | Ndfip2 | 0.0001 | Both | Crhbp | 0.0085 | ? |
| Psen2 | 0.0011 | mOSN | Nfatc2 | 0.0001 | Both | Crybb3 | 0.0010 | ? |
| Ptchd1 | 0.0061 | mOSN | Nipa2 | 0.0064 | Both | Dnajc25 | 0.0032 | ? |
| Ptprn | 0.0000 | mOSN | Nkiras1 | 0.0001 | Both | Dnase2b | 0.0077 | ? |
| Rasgrp4 | 0.0000 | mOSN | Nmnat3 | 0.0039 | Both | Dsc3 | 0.0029 | ? |
| Rasl11a | 0.0000 | mOSN | Nsf | 0.0000 | Both | Dusp9 | 0.0024 | ? |
| Rgs7 | 0.0000 | mOSN | Nsg2 | 0.0001 | Both | Eda | 0.0026 | ? |
| S100a3 | 0.0008 | mOSN | Ntf3 | 0.0000 | Both | Edaradd | 0.0042 | ? |
| S100a5 | 0.0000 | mOSN | Nyx | 0.0054 | Both | Fxyd4 | 0.0020 | ? |
| Scn3b | 0.0000 | mOSN | Parp9 | 0.0010 | Both | Gngt1 | 0.0070 | ? |
| Scn4b | 0.0000 | mOSN | Pcsk2 | 0.0000 | Both | Grhl3 | 0.0071 | ? |
| Sdr42e1 | 0.0003 | mOSN | Pgrmc1 | 0.0039 | Both | Hsf4 | 0.0058 | ? |
| Sema3e | 0.0016 | mOSN | Phactr1 | 0.0006 | Both | Iapp | 0.0056 | ? |
| Sema7a | 0.0008 | mOSN | Polg2 | 0.0028 | Both | Krt80 | 0.0008 | ? |
| Serinc2 | 0.0002 | mOSN | Ppapdc2 | 0.0083 | Both | Maml2 | 0.0009 | ? |
| Slc17a6 | 0.0000 | mOSN | Ppfia2 | 0.0010 | Both | Mbtps2 | 0.0060 | ? |
| Slc24a2 | 0.0001 | mOSN | Prepl | 0.0003 | Both | Muc15 | 0.0001 | ? |
| Slc8a1 | 0.0000 | mOSN | Prkacb | 0.0000 | Both | Nr1i2 | 0.0030 | ? |
| Spin1 | 0.0008 | mOSN | Psme4 | 0.0038 | Both | Pde6h | 0.0071 | ? |
| Srxn1 | 0.0017 | mOSN | Pten | 0.0021 | Both | Pkp1 | 0.0050 | ? |
| Syn1 | 0.0001 | mOSN | Pvrl3 | 0.0024 | Both | Pla1a | 0.0018 | ? |
| Syn2 | 0.0003 | mOSN | Qrich1 | 0.0056 | Both | Pnliprp2 | 0.0016 | ? |
| Syngr4 | 0.0091 | mOSN | Rab10 | 0.0009 | Both | Saa2 | 0.0048 | ? |
| Synj2 | 0.0050 | mOSN | Rab30 | 0.0065 | Both | Scrg1 | 0.0080 | ? |
| Syt3 | 0.0015 | mOSN | Rell1 | 0.0009 | Both | Sdcbp2 | 0.0000 | ? |
| Syt4 | 0.0000 | mOSN | Rgs7bp | 0.0099 | Both | Serpinb10 | 0.0063 | ? |
| Tmem163 | 0.0040 | mOSN | Rragd | 0.0001 | Both | Sim2 | 0.0066 | ? |
| Tmtc1 | 0.0072 | mOSN | Sec23ip | 0.0018 | Both | Slc27a3 | 0.0040 | ? |
| Trim45 | 0.0000 | mOSN | Sec31a | 0.0062 | Both | Slc39a5 | 0.0045 | ? |
| Ubl3 | 0.0000 | mOSN | Senp6 | 0.0003 | Both | Slc6a19 | 0.0037 | ? |
| Ubl4b | 0.0010 | mOSN | Sgpp2 | 0.0003 | Both | Sorcs3 | 0.0061 | ? |
| Vsnl1 | 0.0001 | mOSN | Slc20a2 | 0.0061 | Both | Spp2 | 0.0055 | ? |
| Wdr17 | 0.0009 | mOSN | Slc22a17 | 0.0001 | Both | Tktl1 | 0.0064 | ? |
| Slc6a17 | 0.0000 | Both | Tnfaip3 | 0.0094 | ? | |||
| Slc7a3 | 0.0002 | Both | Trp73 | 0.0006 | ? |
Table 5.
Meta-analysis: decreased with OSN activity
| Gene Symbol | p-value | Cell type | Gene symbol | P-value | Cell type | Gene symbol | P-value | Cell type |
|---|---|---|---|---|---|---|---|---|
| 1700001L19Rik | 0.0002 | mOSN | E2f1 | 0.0076 | Both | Spsb1 | 0.0004 | Both |
| 1700012B09Rik | 0.0004 | mOSN | Pigb | 0.0000 | Both | Stk11 | 0.0042 | Both |
| 1700023F06Rik | 0.0015 | mOSN | 1500011B03Rik | 0.0000 | Both | Tars2 | 0.0034 | Both |
| 4430402I18Rik | 0.0001 | mOSN | 9130401M01Rik | 0.0033 | Both | Tas1r1 | 0.0017 | Both |
| 4933413G19Rik | 0.0000 | mOSN | Adam15 | 0.0012 | Both | Tbrg4 | 0.0003 | Both |
| 9530077C05Rik | 0.0078 | mOSN | Adprhl2 | 0.0027 | Both | Thap7 | 0.0000 | Both |
| Acsl6 | 0.0030 | mOSN | Alkbh7 | 0.0000 | Both | Timm22 | 0.0021 | Both |
| Adam23 | 0.0033 | mOSN | Arfip2 | 0.0054 | Both | Tmem14a | 0.0088 | Both |
| Art5 | 0.0000 | mOSN | Arhgef3 | 0.0038 | Both | Tmem55b | 0.0079 | Both |
| Atp2c2 | 0.0045 | mOSN | Arl3 | 0.0072 | Both | Tpd52 | 0.0055 | Both |
| Atp8b3 | 0.0000 | mOSN | Bace1 | 0.0001 | Both | Tpd52l2 | 0.0036 | Both |
| B830017H08Rik | 0.0000 | mOSN | Bbs10 | 0.0001 | Both | Vstm2a | 0.0022 | Both |
| C130036L24Rik | 0.0020 | mOSN | BC025920 | 0.0015 | Both | Xrcc6 | 0.0005 | Both |
| Cacnb3 | 0.0003 | mOSN | Bhlhe41 | 0.0043 | Both | Alox5 | 0.0000 | Other |
| Capn8 | 0.0001 | mOSN | Bmp6 | 0.0004 | Both | Cox6b2 | 0.0013 | Other |
| Ccdc126 | 0.0000 | mOSN | Bok | 0.0002 | Both | E2f2 | 0.0009 | Other |
| Ccdc151 | 0.0071 | mOSN | Calb2 | 0.0000 | Both | Ifrd2 | 0.0001 | Other |
| Cngb1 | 0.0078 | mOSN | Ccdc32 | 0.0067 | Both | Ngp | 0.0001 | Other |
| Dcdc2a | 0.0021 | mOSN | Ccdc74a | 0.0012 | Both | Smyd2 | 0.0042 | Other |
| Dhrs7b | 0.0001 | mOSN | Cd274 | 0.0001 | Both | 1700106J16Rik | 0.0035 | ? |
| Efna3 | 0.0000 | mOSN | Cdh22 | 0.0070 | Both | Adamtsl2 | 0.0072 | ? |
| Elmod1 | 0.0079 | mOSN | Cmpk2 | 0.0018 | Both | Amtn | 0.0059 | ? |
| Eml1 | 0.0002 | mOSN | Crcp | 0.0001 | Both | Apbb1ip | 0.0040 | ? |
| Entpd2 | 0.0000 | mOSN | Dcbld2 | 0.0057 | Both | Bglap2 | 0.0072 | ? |
| Epb4.1l4b | 0.0000 | mOSN | Ddx41 | 0.0067 | Both | Chid1 | 0.0018 | ? |
| Fam178b | 0.0005 | mOSN | Dtnb | 0.0021 | Both | Chodl | 0.0058 | ? |
| Fam179a | 0.0000 | mOSN | Edn2 | 0.0096 | Both | Cstad | 0.0005 | ? |
| Fbxo44 | 0.0003 | mOSN | Efna5 | 0.0001 | Both | Glp1r | 0.0090 | ? |
| Fn3k | 0.0007 | mOSN | Etv3 | 0.0004 | Both | Gngt2 | 0.0048 | ? |
| Gbgt1 | 0.0001 | mOSN | Fam78a | 0.0080 | Both | Hcls1 | 0.0010 | ? |
| Glb1l2 | 0.0008 | mOSN | Farsa | 0.0054 | Both | Mylk2 | 0.0086 | ? |
| Glo1 | 0.0001 | mOSN | Fbxo16 | 0.0054 | Both | Ninj1 | 0.0000 | ? |
| Gramd1c | 0.0000 | mOSN | Fis1 | 0.0003 | Both | Nme6 | 0.0001 | ? |
| Hfe | 0.0000 | mOSN | Fpgt | 0.0033 | Both | Nostrin | 0.0085 | ? |
| Hmox1 | 0.0021 | mOSN | Galt | 0.0003 | Both | Npas3 | 0.0003 | ? |
| Hrasls | 0.0084 | mOSN | Gins4 | 0.0031 | Both | Ntf5 | 0.0022 | ? |
| Hs6st3 | 0.0000 | mOSN | Inpp5k | 0.0002 | Both | Papolb | 0.0018 | ? |
| Htatip2 | 0.0006 | mOSN | Iqcd | 0.0062 | Both | Pgf | 0.0000 | ? |
| Impdh1 | 0.0002 | mOSN | Jam3 | 0.0052 | Both | Smox | 0.0008 | ? |
| Ipo13 | 0.0078 | mOSN | Kctd17 | 0.0010 | Both | Timm10 | 0.0075 | ? |
| Itpka | 0.0000 | mOSN | Kptn | 0.0000 | Both | |||
| Jakmip1 | 0.0004 | mOSN | Lrwd1 | 0.0039 | Both | |||
| Jph3 | 0.0002 | mOSN | Lypla2 | 0.0027 | Both | |||
| Kcnq1 | 0.0093 | mOSN | Metap1 | 0.0060 | Both | |||
| Kif9 | 0.0090 | mOSN | Mgat4b | 0.0002 | Both | |||
| Klhl5 | 0.0082 | mOSN | Mrpl43 | 0.0071 | Both | |||
| Lrrn2 | 0.0076 | mOSN | Muc13 | 0.0009 | Both | |||
| Macrod1 | 0.0000 | mOSN | Mvk | 0.0000 | Both | |||
| Manea | 0.0049 | mOSN | Ndp | 0.0035 | Both | |||
| Mgmt | 0.0014 | mOSN | Ndufa13 | 0.0002 | Both | |||
| Mmp16 | 0.0078 | mOSN | Ndufv3 | 0.0017 | Both | |||
| Necab3 | 0.0000 | mOSN | Ngrn | 0.0073 | Both | |||
| Neu2 | 0.0000 | mOSN | Npdc1 | 0.0047 | Both | |||
| Olfr9 | 0.0092 | mOSN | Pafah1b3 | 0.0091 | Both | |||
| Pde4a | 0.0022 | mOSN | Pard6b | 0.0038 | Both | |||
| Pde6d | 0.0000 | mOSN | Pgls | 0.0019 | Both | |||
| Ppp1r1a | 0.0000 | mOSN | Pknox2 | 0.0006 | Both | |||
| Ppp2r2b | 0.0000 | mOSN | Pllp | 0.0025 | Both | |||
| Ptp4a3 | 0.0001 | mOSN | Pold4 | 0.0071 | Both | |||
| Rab11fip5 | 0.0006 | mOSN | Ppan | 0.0006 | Both | |||
| Rassf7 | 0.0023 | mOSN | Ppp3ca | 0.0070 | Both | |||
| Rfx4 | 0.0004 | mOSN | Ppp3r1 | 0.0001 | Both | |||
| Ribc2 | 0.0039 | mOSN | Psmf1 | 0.0071 | Both | |||
| Rnf152 | 0.0028 | mOSN | Pygo2 | 0.0086 | Both | |||
| Scgn | 0.0000 | mOSN | Rab40b | 0.0025 | Both | |||
| Sgpl1 | 0.0032 | mOSN | Rassf4 | 0.0005 | Both | |||
| Slc1a2 | 0.0003 | mOSN | Rims3 | 0.0000 | Both | |||
| Slc23a3 | 0.0001 | mOSN | Rit1 | 0.0016 | Both | |||
| Spef2 | 0.0046 | mOSN | Rnf135 | 0.0059 | Both | |||
| Stoml3 | 0.0002 | mOSN | Rnf19b | 0.0073 | Both | |||
| Sytl1 | 0.0000 | mOSN | Rogdi | 0.0028 | Both | |||
| Tesc | 0.0008 | mOSN | Saal1 | 0.0026 | Both | |||
| Tusc5 | 0.0000 | mOSN | Sdf2 | 0.0001 | Both | |||
| Umodl1 | 0.0001 | mOSN | Sec13 | 0.0096 | Both | |||
| Usp21 | 0.0000 | mOSN | Sh3glb2 | 0.0096 | Both | |||
| Xylt2 | 0.0045 | mOSN | Siva1 | 0.0001 | Both | |||
| Zfp81 | 0.0027 | mOSN | Slco3a1 | 0.0080 | Both |
Using the NIH DAVID bioinformatics tool (https://david.ncifcrf.gov/gene2gene.jsp) we assessed the functional relationships between the proteins encoded by these 443 mRNAs. Specifically, we used the gene functional classification tool at the high stringency setting and the whole mouse genome as the background for comparison to analyze 3 groups of mRNAs: (1) The 350 mRNAs expressed in mature OSNs, (2) the 236 mRNAs that increase with OSN activity, and (3) the 207 mRNAs that decrease with OSN activity. Table 6 summarizes the significant clusters of over-represented categories detected in these analyses.
Table 6.
Over-represented annotation ontologies.
| Ontology term | Enrichment score | # of genes |
|---|---|---|
| Expressed in mature OSNs | ||
| Synapse (5) | 4.3 | 36 |
| Membrane proteins (10) | 2.8 | 200 |
| Phosphatidylinositol signaling (4) | 2.5 | 9 |
| Cation channels, transporters (21) | 2.0 | 47 |
| Calmodulin binding (3) | 1.9 | 11 |
| Calcium binding proteins (12) | 1.7 | 15 |
| Endoplasmic reticulum (3) | 1.7 | 33 |
| Nucleotydiltransferase (3) | 1.6 | 6 |
| Cyclic nucleotide-related proteins (31) | 1.5 | 10 |
| Increased by OSN activity | ||
| Synapse (5) | 6.5 | 32 |
| Membrane glycoproteins (19) | 3.7 | 160 |
| Cation channels, transporters (21) | 1.9 | 39 |
| Axon guidance (3) | 1.9 | 11 |
| Differentiation/development (4) | 1.6 | 27 |
| MAPK signalling (11) | 1.6 | 10 |
| Sterile alpha motif domain (4) | 1.6 | 5 |
| Decreased by OSN activity | ||
| Calcium binding proteins (13) | 1.4 | 22 |
| Endoplasmic reticulum (5) | 1.3 | 18 |
DAVID functional bioinformatics analysis. Enrichment scores >1.3 identify significantly overrepresented clusters of annotation terms. Numbers in parentheses are the number of related terms grouped together under one heading.
The annotation terms most strongly affected by experiments designed to alter OSN activity revolve around synapses and functions critical to synapses and axons, such as exocytosis, ion channels, transporters, calcium handling, and signaling pathways. Examples of the mRNAs in these categories include synaptotagmins, Rims3, synapsins, a synaptojanin, synuclein, the vesicular glutamate transporter Slc17A6 (Vlgut2), the synaptic glutamate transporter Slc1a2, the sodium/calcium exchanger Slc8a1 (Ncx1), sodium channel regulatory subunits, and the potassium channels Kcnn2 and Kcnq1. These molecular responses are consistent with functional and anatomical data showing that OSN presynaptic terminals respond to the level of activity in OSNs (Tyler et al. 2007; Kikuta et al. 2015; Cheetham et al. 2016). The membrane protein, glycoprotein, and differentiation/development categories consist of many of these same synaptic and axonal proteins genes, so their over-representation also appears to be driven by the activity-dependence of synaptic functions, ion channels, and transporters.
At the other pole of the OSN, olfactory transduction occurs and it is represented by the cyclic nucleotide-related protein category. These include ion channels gated or regulated by cyclic nucleotides, phosphodiesterases, and the adenylyl cyclase Adcy3. These mRNAs do not respond similarly to conditions that alter odor stimulation; some are increased by UNO (e.g., Adcy3, Cng1), others decreased by it (Cnga2), so this category is not over-represented when we separately analyze the mRNAs increased or decreased by OSN activity. Whether this should raise doubts about the concept of compensatory plasticity in OSNs (Barber and Coppola 2015) is uncertain. Perhaps only certain rate-limiting components of the olfactory transduction pathway, such as Adcy3 and Cngb1, need be upregulated in the absence of OSN activity in order to support compensatory plasticity.
The over-represented calcium binding protein category in the mature OSN list results from a group of calcium binding proteins either involved in diverse functions such as modulating neuronal excitability (Calb2, Tesc, Vsnl1), cilia (Spef2), lipid signaling (Dgkg), amyloid protein precursor metabolism (Necab3), linking the cytoskeleton to the extracellular matrix (Dtna, Dtnb), regulating hormone release (Scgn); or having poorly known functions (1700023F06RIK, Capn8, S100a5, S100a3). The majority of these calcium binding protein mRNAs increase with OSN activity, so this group of mRNAs has little overlap with the calcium binding proteins whose mRNAs comprise the over-represented category under mRNAs decreased by OSN activity. Perhaps the appropriate interpretation of these data is that these responses are evidence of homeostatic effects related to the central role of calcium influx in regulating neuronal functions.
Phosphatidylinositol signaling is a category comprised of mRNAs whose response to altered OSN activity is not consistently correlated with the functions of the encoded proteins. The mRNAs encoding 2 kinases that convert inositol-1,4,5-trisphosphate (IP3) into inositol-1,3,4,5-trisphosphate are significant, but they respond oppositely to OSN activity. Itpkb increases while Itpka decreases. Transcripts that encode phosphatases responsible for dephosphorylating IP3 show similarly opposite responses. Inpp5f mRNA increases with OSN activity but Inpp5k mRNA decreases. Why each of these pairs of enzymes should show opposing changes when OSN activity is manipulated is unclear. However, Itpka and Itpkb are F-actin binding proteins whose effects are often restricted to specific subcellular locations (Erneux et al. 2016). Different subcellular locations could explain their opposing responses to UNO, but this has not yet been investigated in OSNs. Interestingly, these 2 enzymes act to antagonize phosphoinositide 3-kinase signaling, which has been implicated in downregulating odor responses in OSNs (Ukhanov et al. 2016; Westernberg et al. 2016). IP3 phosphatases such as Inpp5f and Inpp5k have much higher Vmax values than the IP3 kinases, so their activity may help to restrict the effects of Iptka and Iptkb to specific cellular compartments. Inppf5, which was recently discovered to suppresses axon growth and have better activity on phosphate at the 4 position rather than the 5 position of the inositol ring, is a cytoplasmic protein found throughout neural cell bodies, axons and dendrites (Hsu et al. 2015; Nakatsu et al. 2015; Zou et al. 2015). We hypothesize that OSNs use phosphoinositide signaling in complex ways, perhaps in ways that are specific to different subcellular compartments.
A more consistent regulation of signaling pathways is apparent in the over-represented Mapk signaling category that is specific to mRNAs that increase with OSN activity. The mRNAs in this category include Dusp1, Dusp9, and Dusp16; which negatively regulate Mapk, Erk, and Junk, respectively. These data suggest that OSN activity broadly downregulates tyrosine kinase signaling pathways.
Overall, the bioinformatics analyses indicate that the relationship between neural activity and transcription in OSNs is largely one of activity increasing the abundance of mRNAs encoding proteins critical for synaptic and axonal function. The mRNAs that decrease with increased OSN activity are numerous but more functionally diverse, such that they form few over-represented functional annotation categories in our analyses.
Activity-dependent mRNAs should be expressed by OSNs
By definition, activity-dependent gene expression occurs in cells capable of generating action potentials, meaning neurons and muscle cells. In addition, activity-dependent genes must respond specifically to changes in the electrical activity of these cells. In the olfactory epithelium, we should therefore find that activity-dependent mRNAs are located in mature OSNs. To test this hypothesis, we analyzed the expression patterns of these mRNAs using data sets from experiments that assess OSN gene expression patterns in 3 different ways. (1) Combining expression profiling data on purified mature and immature OSNs allowed Nickell and colleagues (2012) to generate probabilities of expression in 3 categories of cells in olfactory mucosal samples: mature OSNs, immature OSNs, and the grouped population of all other types of cells. (2) Bulbectomy (OBX) can be used to identify mRNAs expressed primarily in mature OSNs. The temporary loss of mature OSNs after OBX causes mature OSN mRNAs to decrease, while at the same time the proliferation of new immature OSNs causes immature OSN mRNAs to increase. We used the data of Heron and colleagues (2013) to assess whether putative activity-dependent mRNAs are decreased at 5 days after OBX, indicative of expression primarily in mature OSNs. (3) We also generated de novo a third measure of mature OSN expression, a mature OSN index, from an RNA-seq data set (Saraiva et al. 2015). RNA-seq counts of mRNAs from dissociated cell samples enriched in mature OSNs were divided by RNA-seq counts of mRNAs in samples of whole olfactory mucosae to generate a simple index where values > 1 indicate expression primarily in mature OSNs. Taken together, these 3 measures provide a robust means of assessing whether a mRNA is expressed mostly in mature OSNs, immature OSNs, both mature and immature OSNs, cell types other than OSNs, or unknown due to absent or equivocal data.
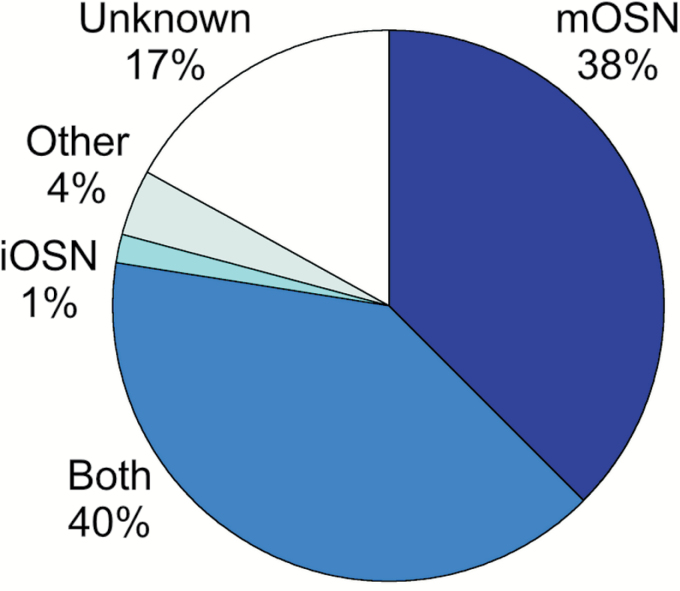
We find that the majority of the 443 significant mRNAs either are expressed primarily in mature OSNs or approximately equally in mature and immature OSNs (Figure 2). This general finding is consistent across all 4 methods used to evoke activity-dependent effects. It increases confidence, but does not prove, that the affected mRNAs respond directly to changes in OSN activity or OSN damage. Only 18 of these 443 mRNAs are known to be expressed primarily in non-neuronal cell types in or near the olfactory epithelium. The expression patterns of another 75 of these mRNAs are as yet unknown, but given the extent of attention given to measuring mRNAs in samples enriched in OSNs (Sammeta et al. 2007; Nickell et al. 2012; Saraiva et al. 2015), most are probably expressed in cell types other than OSNs. This evidence of effects on mRNAs in cell types other than OSNs raises the question of why other cell types might respond to manipulations designed to alter OSN activity.
Figure 2.
Expression patterns of mRNAs identified as differentially abundant after altering OSN activity. The majority are expressed primarily in mature OSNs (mOSN) or in both mature and immature OSNs (Both). Only 5% are expressed primarily in immature OSNs (iOSNs) or in non-neuronal cell types (Other). The expression patterns of the remaining mRNAs are not yet known (Unknown). OR mRNAs are excluded.
An example of an effect on other cell types that hints of a broader response is the ability of UNO to have effects on sustentacular cells. Sustentacular cells interact with odorants in order to clear them from the olfactory epithelium (Strotmann and Breer 2011), so after UNO they unilaterally experience reduced processing of odorant chemicals through their xenobiotic chemical metabolism networks. Quantitative RT-PCR after UNO reveals a significant reduction in the abundance of Cyp2a5/Cyp2a4, 2 nearly identical sustentacular cell mRNAs encoding xenobiotic metabolism enzymes (Sammeta and McClintock 2010). This effect appears to be cell autonomous because these mRNAs are not affected when OSNs are genetically silenced. Our meta-analysis predicts that Cyp2g1, also expressed in sustentacular cells, behaves similarly. Taken together, these data argue that at least some elements of the biochemical pathways sustentacular cells use to clear odorants from the olfactory epithelium are sensitive to odor stimulation. This conclusion highlights the need for renewed attention given to how experimental manipulations designed to alter OSN activity affect cells other than mature OSNs. Investigating responses in cells other than mature OSNs may reveal previously unknown properties of non-neuronal cells in the olfactory epithelium.
As mentioned above, signaling between OSNs and neighboring cells could be sensitive to odor stimulation and thereby cause changes in mRNA abundance in cells other than OSNs. A potentially related indirect effect might arise from reduced basal cell proliferation after UNO (Farbman et al. 1988; Cummings and Brunjes 1994), possibly leading to effects on mRNAs sensitive to the state of proliferation in the olfactory epithelium. These indirect effects could cause some mRNAs to appear to be activity-dependent even though they are not expressed in OSNs. However, at present none of the affected mRNAs expressed primarily in non-neuronal cell types are known to encode proteins whose functions indicate a role in OSN replacement by basal progenitor cells.
The special case of OR mRNAs
We believe that OR mRNA abundance should be assessed more cautiously than the responses of most other mRNAs. This is not because OR mRNAs cannot be demonstrated to change in abundance in samples from olfactory mucosae after experimental manipulations that alter OSN activity; indeed, there is substantial evidence that they can be sensitive to these manipulations (Zhao et al. 2013; Fischl et al. 2014; von der Weid et al. 2015). Instead, we should be cautious because the unusual expression patterns of ORs are potential confounds in expression profiling experiments, including those testing activity-dependent gene expression in OSNs.
One reason for caution arises from the fact that the randomness inherent in the choice of a single OR gene for high expression in each OSN is a source of variation in mRNA abundance that is not possible for genes expressed in all OSNs. Indeed, cell counts of OSNs expressing individual ORs show that the frequencies of expression of some ORs vary measurably between identically treated mice (Bressel et al. 2016). This predicts that OR mRNA abundance should be more variable than mRNA species expressed in all OSNs, and GeneChip microarray measurements do show that on average OR mRNA abundance is more variable than the abundance of most other mRNAs in samples of olfactory mucosae (Fischl et al. 2014). Greater variation increases the likelihood that OR mRNAs may act as false positives relative to other mRNAs in expression profiling experiments.
A second reason for caution is that experimentally-induced changes in OR expression frequency can be mistaken for activity-dependent responses of individual OSNs. Due to monogenic expression of OR genes, an OR mRNA can differ in samples of whole olfactory mucosae if the frequency of OSNs expressing an OR changes even if the amount of OR mRNA per OSN does not change (McIntyre et al. 2008). Given the evidence that differential survival of OSNs may be linked to odor stimulation, there is reason to expect changes in OR mRNA abundance that arise from differential OSN survival in experiments that alter OSN activity. Indeed, this type of effect has been observed after UNO. Four weeks after neonatal UNO, the frequencies of OSNs expressing several ORs are altered, some increasing, others decreasing (Zhao et al. 2013). Altered OSN activity might even affect OR expression frequencies by altering OR gene choice. The activity-dependent mRNA Cyp26b1 encodes an enzyme that inactivates retinoic acid, which has been shown to control expression of at least one gene, Bace1, in OSNs (Login et al. 2015a). When mutant mice that overexpress Cyp26b1 in OSNs were made, they proved to have altered frequencies of OR expression, including shifts in the patterns of zonal expression of some ORs (Login et al. 2015b).
Our meta-analysis identifies 20 OR mRNAs sensitive to manipulations that alter OSN activity, 2 that increase with OSN activity and 18 that decrease. Only one of these OR mRNAs was previously shown to be sensitive to UNO in multiple experiments: Olfr855, which is increased by OSN activity (Fischl et al. 2014). Whether Olfr855 mRNA increases because OSN activity alters its frequency of expression or the amount of Olfr855 mRNA per OSN is as yet unknown. Altered frequencies of OR expression after altering OSN activity by UNO clearly occur, as cited above, but examples of changes in OR mRNA abundance within individual OSNs also exist. Intermittent exposure to lyral, an odorant agonist for Olfr16, for 3 weeks caused individual OSNs to increase the amount of Olfr16 mRNA they contain (Cadiou et al. 2014). Conversely, mouse and rat OSNs that respond to an odor show decreases in the OR mRNAs they express within hours of odor exposure, rapidly enough that altered frequencies of expression are improbable (von der Weid et al. 2015). Overall, the diversity of OR mRNA responses to situations where OSN activity is altered and the poor agreement between our meta-analysis and studies that show altered frequencies of OR expression after altering OSN activity are worrisome (Zhao et al. 2013; Login et al. 2015b). We conclude that measuring OR mRNA abundance in whole olfactory mucosal samples is insufficient to demonstrate true activity dependence of OR mRNAs. Additional experiments that test for changes in frequency of expression and amount of OR mRNA per individual OSN are required.
Summary
Experiments seeking to identify activity-dependent mRNAs in OSNs have made intriguing discoveries but not yet realized their full potential for furthering our understanding of olfactory biology. The methods used thus far to generate differences in OSN activity often fail to fully eliminate indirect effects that may be misinterpreted as true activity dependence. Furthermore, these studies all too often have shortcomings in experimental design. For example, the relatively small changes in most activity-dependent mRNAs detected thus far and the paucity of reliably identified changes in mRNA abundance suggest that previous expression profiling experiments have not been sufficiently powered to measure the full extent of activity-dependent mRNAs. Our meta-analysis improves this situation, but does not solve other reliability issues.
We now identify 443 mRNAs sensitive to manipulations that alter OSN activity. They respond consistently across multiple experiments and at least 350 of them are expressed in mature OSNs. How many of them are activity-dependent in the strict sense of being directly sensitive to odor stimulation is still not known, however. Largely because the period between altered OSN activity and measurement of mRNA abundance is several days or weeks in most cases, we cannot be certain that some are not responding to indirect processes triggered by the experimental manipulations. Potential indirect processes include changes in OSN survival, altered local signaling within the olfactory epithelium, and altered feedback from the olfactory bulb. These homeostatic processes are interesting in their own right and worthy of further study. Identifying OSN mRNAs specifically responsive to indirect effects may prove to be the key to understanding these processes.
Except for the activity-dependence of axonal coalescence that involves Kirrel2, Kirrel3, and ephrinA5-EphA5 signaling (Nishizumi and Sakano 2015), and the related evidence developed herein about the activity-dependent expression of synaptic and axonal genes, the identification of activity-dependent mRNAs has not had a significant impact on our understanding of OSN biology. For example, we have little understanding of how OSN activity causes changes in mRNA abundance. Furthermore, whether activity-dependent changes in mRNA abundance are critical for OSN survival is not yet known. Even the 2 mRNAs most strongly affected by altered OSN activity and directly responsive to odor stimulation do not yet have known functions. Targeted deletion of neither S100a5 nor Lrrc3b causes readily detectable deficiencies in OSNs (Bennett et al. 2010; McClintock et al. 2014). This lack of impact reveals a need for further study, especially studies that investigate the full extent of activity-dependent gene expression in OSNs and the identification of factors necessary for activity-dependent survival of OSNs.
Supplementary Material
Supplementary material are available at Chemical Senses online.
Funding
This work was supported by grant awards from the National Institute on Deafness and Other Communication Disorders to T.M. (K18 DC014050, R21 DC013343, R01 DC014468).
Supplementary Material
Acknowledgement
We thank Dr. David Coppola for helpful discussions.
References
- Alonso M, Viollet C, Gabellec MM, Meas-Yedid V, Olivo-Marin JC, Lledo PM. 2006. Olfactory discrimination learning increases the survival of adult-born neurons in the olfactory bulb. J Neurosci. 26:10508–10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreae LC, Burrone J. 2015. Spontaneous neurotransmitter release shapes dendritic arbors via long-range activation of NMDA receptors. Cell Rep. 10:873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey CH, Kandel ER, Si K. 2004. The persistence of long-term memory: a molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron. 44:49–57. [DOI] [PubMed] [Google Scholar]
- Baker H, Kawano T, Margolis FL, Joh TH. 1983. Transneuronal regulation of tyrosine hydroxylase expression in olfactory bulb of mouse and rat. J Neurosci. 3:69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber CN, Coppola DM. 2015. Compensatory plasticity in the olfactory epithelium: age, timing, and reversibility. J Neurophysiol. 114:2023–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MK, Kulaga HM, Reed RR. 2010. Odor-evoked gene regulation and visualization in olfactory receptor neurons. Mol Cell Neurosci. 43:353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson TE, Ryugo DK, Hinds JW. 1984. Effects of sensory deprivation on the developing mouse olfactory system: a light and electron microscopic, morphometric analysis. J Neurosci. 4:638–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleckert A, Wong RO. 2011. Identifying roles for neurotransmission in circuit assembly: insights gained from multiple model systems and experimental approaches. Bioessays. 33:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Lomo T. 1973. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J Physiol. 232:331–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressel OC, Khan M, Mombaerts P. 2016. Linear correlation between the number of olfactory sensory neurons expressing a given mouse odorant receptor gene and the total volume of the corresponding glomeruli in the olfactory bulb. J Comp Neurol. 524:199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunjes P, Shurling DC. 2003. Cell death in the nasal septum of normal and naris-occluded rats. Brain Res Dev Brain Res. 146(1-2):25–28. [DOI] [PubMed] [Google Scholar]
- Brunjes PC. 1994. Unilateral naris closure and olfactory system development. Brain Res Brain Res Rev. 19:146–160. [DOI] [PubMed] [Google Scholar]
- Cadiou H, Aoudé I, Tazir B, Molinas A, Fenech C, Meunier N, Grosmaitre X. 2014. Postnatal odorant exposure induces peripheral olfactory plasticity at the cellular level. J Neurosci. 34(14):4857–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caggiano M, Kauer JS, Hunter DD. 1994. Globose basal cells are neuronal progenitors in the olfactory epithelium: a lineage analysis using a replication-incompetent retrovirus. Neuron. 13:339–352. [DOI] [PubMed] [Google Scholar]
- Cajal SR. (1913). Estudios sobre la degeneracio n y regeneracio n del sistema nervioso. In: Degeneracio n y regeneracio n de los nervios. Madrid: Moya. [Google Scholar]
- Cao L, Rickenbacher GT, Rodriguez S, Moulia TW, Albers MW. 2012. The precision of axon targeting of mouse olfactory sensory neurons requires the BACE1 protease. Sci Rep. 2:231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallin MA, Powell K, Biju KC, Fadool DA. 2010. State-dependent sculpting of olfactory sensory neurons is attributed to sensory enrichment, odor deprivation, and aging. Neurosci Lett. 483:90–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham CE, Park U, Belluscio L. 2016. Rapid and continuous activity-dependent plasticity of olfactory sensory input. Nat Commun. 7:10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. 1994. Allelic inactivation regulates olfactory receptor gene expression. Cell. 78:823–834. [DOI] [PubMed] [Google Scholar]
- Choi BJ, Imlach WL, Jiao W, Wolfram V, Wu Y, Grbic M, Cela C, Baines RA, Nitabach MN, McCabe BD. 2014. Miniature neurotransmission regulates Drosophila synaptic structural maturation. Neuron. 82:618–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola DM. (2012). Studies of olfactory system neural plasticity: The contribution of the unilateral naris occlusion technique. Neural Plasticity 2012:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola DM, Waggener CT. 2012. The effects of unilateral naris occlusion on gene expression profiles in mouse olfactory mucosa. J Mol Neurosci. 47:604–618. [DOI] [PubMed] [Google Scholar]
- Coppola DM, Coltrane JA, Arsov I. 1994. Retronasal or internasal olfaction can mediate odor-guided behaviors in newborn mice. Physiol Behav. 56:729–736. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Brunjes PC. 1994. Changes in cell proliferation in the developing olfactory epithelium following neonatal unilateral naris occlusion. Exp Neurol. 128:124–128. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Brunjes PC. 1997. The effects of variable periods of functional deprivation on olfactory bulb development in rats. Exp Neurol. 148:360–366. [DOI] [PubMed] [Google Scholar]
- Cummings DM, Belluscio L. 2010. Continuous neural plasticity in the olfactory intrabulbar circuitry. J Neurosci. 30(27):9172–9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Col JA, Matsuo T, Storm DR, Rodriguez I. 2007. Adenylyl cyclase-dependent axonal targeting in the olfactory system. Development. 134(13):2481–2489. [DOI] [PubMed] [Google Scholar]
- Dalton P, Wysocki CJ. 1996. The nature and duration of adaptation following long-term odor exposure. Percept Psychophys. 58:781–792. [DOI] [PubMed] [Google Scholar]
- Degano AL, Park MJ, Penati J, Li Q, Ronnett GV. 2014. MeCP2 is required for activity-dependent refinement of olfactory circuits. Mol Cell Neurosci. 59:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erneux C, Ghosh S, Koenig S. 2016. Inositol(1,4,5)P3 3-kinase isoenzymes: catalytic properties and importance of targeting to F-actin to understand function. Adv Biol Regul. 60:135–143. [DOI] [PubMed] [Google Scholar]
- Farbman AI, Brunjes PC, Rentfro L, Michas J, Ritz S. 1988. The effect of unilateral naris occlusion on cell dynamics in the developing rat olfactory epithelium. J Neurosci. 8:3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl AM, Heron PM, Stromberg AJ, McClintock TS. 2014. Activity-dependent genes in mouse olfactory sensory neurons. Chem Senses. 39:439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. 2008. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 31:563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier-Cierpial L, Brunjes PC. 1989. Early postnatal cellular proliferation and survival in the olfactory bulb and rostral migratory stream of normal and unilaterally odor-deprived rats. J Comp Neurol. 289:481–492. [DOI] [PubMed] [Google Scholar]
- Ganguly K, Poo MM. 2013. Activity-dependent neural plasticity from bench to bedside. Neuron. 80:729–741. [DOI] [PubMed] [Google Scholar]
- Heron PM, Stromberg AJ, Breheny P, McClintock TS. 2013. Molecular events in the cell types of the olfactory epithelium during adult neurogenesis. Mol Brain. 6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooks BM, Chen C. 2006. Distinct roles for spontaneous and visual activity in remodeling of the retinogeniculate synapse. Neuron. 52:281–291. [DOI] [PubMed] [Google Scholar]
- Hsu F, Hu F, Mao Y. 2015. Spatiotemporal control of phosphatidylinositol 4-phosphate by Sac2 regulates endocytic recycling. J Cell Biol. 209:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. 2006. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 314(5799):657–661. [DOI] [PubMed] [Google Scholar]
- Imai T, Yamazaki T, Kobayakawa R, Kobayakawa K, Abe T, Suzuki M, Sakano H. 2009. Pre-target axon sorting establishes the neural map topography. Science. 325(5940):585–590. [DOI] [PubMed] [Google Scholar]
- Jessberger S Clark RE Broadbent NJ Clemenson GD Jr, Consiglio A Lie DC Squire LR Gage FH. 2009. Dentate gyrus-specific knockdown of adult neurogenesis impairs spatial and object recognition memory in adult rats. Learn Mem. 16:147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SV, Choi DC, Davis M, Ressler KJ. 2008. Learning-dependent structural plasticity in the adult olfactory pathway. J Neurosci. 28(49):13106–13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko-Goto T, Yoshihara S, Miyazaki H, Yoshihara Y. 2008. BIG-2 mediates olfactory axon convergence to target glomeruli. Neuron. 57:834–846. [DOI] [PubMed] [Google Scholar]
- Kelemen G. 1947. The junction of the nasal cavity and the pharyngeal tube in the rat. Arch Otolaryngol. 45:159–168. [DOI] [PubMed] [Google Scholar]
- Kikuta S, Sakamoto T, Nagayama S, Kanaya K, Kinoshita M, Kondo K, Tsunoda K, Mori K, Yamasoba T. 2015. Sensory deprivation disrupts homeostatic regeneration of newly generated olfactory sensory neurons after injury in adult mice. J Neurosci. 35:2657–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander AD, Gokoffski KK, Wan FY, Nie Q, Calof AL. 2009. Cell lineages and the logic of proliferative control. PLoS Biol. 7:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DM, Wang F, Lowe G, Gold GH, Axel R, Ngai J, Brunet L. 2000. Formation of precise connections in the olfactory bulb occurs in the absence of odorant-evoked neuronal activity. Neuron. 26:69–80. [DOI] [PubMed] [Google Scholar]
- Login H, Butowt R, Bohm S. 2015a. Activity-dependent and graded BACE1 expression in the olfactory epithelium is mediated by the retinoic acid metabolizing enzyme CYP26B1. Brain Struct Funct. 220:2143–2157. [DOI] [PubMed] [Google Scholar]
- Login H, Håglin S, Berghard A, Bohm S. 2015b. The stimulus-dependent gradient of Cyp26B1+ olfactory sensory neurons is necessary for the functional integrity of the olfactory sensory map. J Neurosci. 35(40):13807–13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. 1999. Combinatorial receptor codes for odors. Cell. 96:713–723. [DOI] [PubMed] [Google Scholar]
- Mandairon N, Stack C, Linster C. 2006a. Olfactory enrichment improves the recognition of individual components in mixtures. Physiol Behav. 89:379–384. [DOI] [PubMed] [Google Scholar]
- Mandairon N, Stack C, Kiselycznyk C, Linster C. 2006b. Enrichment to odors improves olfactory discrimination in adult rats. Behav Neurosci. 120:173–179. [DOI] [PubMed] [Google Scholar]
- Maruniak JA, Henegar JR, Sweeney TP. 1990. Effects of long-term unilateral naris closure on the olfactory epithelia of adult mice. Brain Res. 526:65–72. [DOI] [PubMed] [Google Scholar]
- Masaoka Y, Satoh H, Akai L, Homma I. 2010. Expiration: the moment we experience retronasal olfaction in flavor. Neurosci Lett. 473:92–96. [DOI] [PubMed] [Google Scholar]
- McClintock TS, Adipietro K, Titlow WB, Breheny P, Walz A, Mombaerts P, Matsunami H. 2014. In vivo identification of eugenol-responsive and muscone-responsive mouse odorant receptors. J Neurosci. 34(47):15669–15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre JC, Bose SC, Stromberg AJ, McClintock TS. 2008. Emx2 stimulates odorant receptor gene expression. Chem Senses. 33:825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. 1996. Visualizing an olfactory sensory map. Cell. 87:675–686. [DOI] [PubMed] [Google Scholar]
- Morrison FG, Dias BG, Ressler KJ. 2015. Extinction reverses olfactory fear-conditioned increases in neuron number and glomerular size. Proc Natl Acad Sci USA. 112(41):12846–12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najbauer J, Leon M. 1995. Olfactory experience modulated apoptosis in the developing olfactory bulb. Brain Res. 674:245–251. [DOI] [PubMed] [Google Scholar]
- Nakatsu F, Messa M, Nández R, Czapla H, Zou Y, Strittmatter SM, De Camilli P. 2015. Sac2/INPP5F is an inositol 4-phosphatase that functions in the endocytic pathway. J Cell Biol. 209:85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell MD, Breheny P, Stromberg AJ, McClintock TS. 2012. Genomics of mature and immature olfactory sensory neurons. J Comp Neurol. 520(12):2608–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizumi H, Sakano H. 2015. Developmental regulation of neural map formation in the mouse olfactory system. Dev Neurobiol. 75:594–607. [DOI] [PubMed] [Google Scholar]
- Öztokatli H, Hörnberg M, Berghard A, Bohm S. 2012. Retinoic acid receptor and CNGA2 channel signaling are part of a regulatory feedback loop controlling axonal convergence and survival of olfactory sensory neurons. FASEB J. 26:617–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parma V, Ferraro S, Miller SS, Åhs F, Lundström JN. 2015. Enhancement of odor sensitivity following repeated odor and visual fear conditioning. Chem Senses. 40:497–506. [DOI] [PubMed] [Google Scholar]
- Rawson NE, Eberwine J, Dotson R, Jackson J, Ulrich P, Restrepo D. 2000. Expression of mRNAs encoding for two different olfactory receptors in a subset of olfactory receptor neurons. J Neurochem. 75:185–195. [DOI] [PubMed] [Google Scholar]
- Reisert J. 2010. Origin of basal activity in mammalian olfactory receptor neurons. J Gen Physiol. 136:529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammeta N, McClintock TS. 2010. Chemical stress induces the unfolded protein response in olfactory sensory neurons. J Comp Neurol. 518(10):1825–1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammeta N, Yu TT, Bose SC, McClintock TS. 2007. Mouse olfactory sensory neurons express 10,000 genes. J Comp Neurol. 502:1138–1156. [DOI] [PubMed] [Google Scholar]
- Santoro SW, Dulac C. 2012. The activity-dependent histone variant H2BE modulates the life span of olfactory neurons. Elife. 1:e00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva LR, Ibarra-Soria X, Khan M, Omura M, Scialdone A, Mombaerts P, Marioni JC, Logan DW. 2015. Hierarchical deconstruction of mouse olfactory sensory neurons: from whole mucosa to single-cell RNA-seq. Sci Rep. 5:18178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz P, Kalbe B, Jansen F, Altmueller J, Becker C, Mohrhardt J, Schreiner B, Gisselmann G, Hatt H, Osterloh S. 2016. Transcriptome analysis of murine olfactory sensory neurons during development using single Cell RNA-Seq. Chem Senses. 41:313–323. [DOI] [PubMed] [Google Scholar]
- Schwob JE, Szumowski KE, Stasky AA. 1992. Olfactory sensory neurons are trophically dependent on the olfactory bulb for their prolonged survival. J Neurosci. 12(10):3896–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. 2006. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 127:1057–1069. [DOI] [PubMed] [Google Scholar]
- Slotnick BM, Pazos AJ. 1990. Rats with one olfactory bulb removed and the contralateral naris closed can detect odors. Physiol Behav. 48:37–40. [DOI] [PubMed] [Google Scholar]
- Strotmann J, Breer H. 2011. Internalization of odorant-binding proteins into the mouse olfactory epithelium. Histochem Cell Biol. 136:357–369. [DOI] [PubMed] [Google Scholar]
- Stryker MP, Harris WA. 1986. Binocular impulse blockade prevents the formation of ocular dominance columns in cat visual cortex. J Neurosci. 6:2117–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh KS, Kim SY, Bae YC, Ronnett GV, Moon C. 2006. Effects of unilateral naris occlusion on the olfactory epithelium of adult mice. Neuroreport. 17(11):1139–1142. [DOI] [PubMed] [Google Scholar]
- Sultan S, Lefort JM, Sacquet J, Mandairon N, Didier A. 2011. Acquisition of an olfactory associative task triggers a regionalized down-regulation of adult born neuron cell death. Front Neurosci. 5:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan S, Mandairon N, Kermen F, Garcia S, Sacquet J, Didier A. 2010. Learning-dependent neurogenesis in the olfactory bulb determines long-term olfactory memory. FASEB J. 24:2355–2363. [DOI] [PubMed] [Google Scholar]
- Tyler WJ, Petzold GC, Pal SK, Murthy VN. 2007. Experience-dependent modification of primary sensory synapses in the mammalian olfactory bulb. J Neurosci. 27(35):9427–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukhanov K, Corey E, Ache BW. 2016. Phosphoinositide-3-kinase is the primary mediator of phosphoinositide-dependent inhibition in mammalian olfactory receptor neurons. Front Cell Neurosci. 10:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Weid B, Rossier D, Lindup M, Tuberosa J, Widmer A, Col JD, Kan C, Carleton A, Rodriguez I. 2015. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat Neurosci. 18(10):1455–1463. [DOI] [PubMed] [Google Scholar]
- Wang HW, Wysocki CJ, Gold GH. 1993. Induction of olfactory receptor sensitivity in mice. Science. 260(5110):998–1000. [DOI] [PubMed] [Google Scholar]
- Watt WC, Sakano H, Lee ZY, Reusch JE, Trinh K, Storm DR. 2004. Odorant stimulation enhances survival of olfactory sensory neurons via MAPK and CREB. Neuron. 41:955–967. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME. (2011). Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol 3:a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westernberg L, Conche C, Huang YH, Rigaud S, Deng Y, Siegemund S, Mukherjee S, Nosaka L, Das J, Sauer K. (2016). Non-canonical antagonism of PI3K by the kinase Itpkb delays thymocyte beta-selection and renders it Notch-dependent. Elife 5:e10786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman MC, Greer CA. 2009. Adult neurogenesis and the olfactory system. Prog Neurobiol. 89:162–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel TN, Hubel DH. 1974. Ordered arrangement of orientation columns in monkeys lacking visual experience. J Comp Neurol. 158:307–318. [DOI] [PubMed] [Google Scholar]
- Williams EO, Sickles HM, Dooley AL, Palumbos S, Bisogni AJ, Lin DM. 2011. Delta protocadherin 10 is regulated by activity in the mouse main olfactory system. Front Neural Circuits. 5:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HH, Ivkovic S, Murray RC, Jaramillo S, Lyons KM, Johnson JE, Calof AL. 2003. Autoregulation of neurogenesis by GDF11. Neuron. 37:197–207. [DOI] [PubMed] [Google Scholar]
- Wysocki CJ, Dorries KM, Beauchamp GK. 1989. Ability to perceive androstenone can be acquired by ostensibly anosmic people. Proc Natl Acad Sci USA. 86(20):7976–7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapiec B, Bressel OC, Khan M, Walz A, Mombaerts P. (2016). Neuropilin-1 and the positions of glomeruli in the mouse olfactory bulb. eNeuro. 3(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Reed RR. 2001. X inactivation of the OCNC1 channel gene reveals a role for activity-dependent competition in the olfactory system. Cell. 104:651–660. [DOI] [PubMed] [Google Scholar]
- Zhao S, Tian H, Ma L, Yuan Y, Yu CR, Ma M. 2013. Activity-dependent modulation of odorant receptor gene expression in the mouse olfactory epithelium. PLoS One. 8:e69862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Feinstein P, Bozza T, Rodriguez I, Mombaerts P. 2000. Peripheral olfactory projections are differentially affected in mice deficient in a cyclic nucleotide-gated channel subunit. Neuron. 26:81–91. [DOI] [PubMed] [Google Scholar]
- Zou DJ, Chesler AT, Le Pichon CE, Kuznetsov A, Pei X, Hwang EL, Firestein S. 2007. Absence of adenylyl cyclase 3 perturbs peripheral olfactory projections in mice. J Neurosci. 27(25):6675–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Stagi M, Wang X, Yigitkanli K, Siegel CS, Nakatsu F, Cafferty WB, Strittmatter SM. 2015. Gene-silencing screen for mammalian axon regeneration identifies Inpp5f (Sac2) as an endogenous suppressor of repair after spinal cord injury. J Neurosci. 35(29):10429–10439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.