Abstract
Dietary fats serve multiple essential roles in human health but may also contribute to acute and chronic health complications. Thus, understanding mechanisms that influence fat ingestion are critical. All sensory systems may contribute relevant cues to fat detection, with the most recent evidence supporting a role for the sense of taste. Taste detection thresholds for fat vary markedly between individuals and responses are not normally distributed. Genetics may contribute to these observations. Using crowdsourced data obtained from families visiting the Denver Museum of Nature & Science, our objective was to estimate the heritability of fat taste (oleogustus). A pedigree analysis was conducted with 106 families (643 individuals) who rated the fat taste intensity of graded concentrations of linoleic acid (LA) embedded in taste strips. The findings estimate that 19% (P = 0.043) of the variability of taste response to LA relative to baseline is heritable at the highest concentration tested.
Keywords: citizen science, crowdsourcing, fat taste, heredity estimate, oleogustus
Introduction
Dietary fat provides energy and transports fat-soluble vitamins, while its fatty acids are precursors for various hormones, serve as signaling molecules, and are critical components of cell membranes. Linoleic acid and alpha-linolenic acid are essential fatty acids and must be consumed to ensure optimal health. At the same time, ingestion of rancid fats may be toxic, resulting in acute gastrointestinal disturbances, and high intake of various fats/fatty acids increases risk of chronic disorders such as cardiovascular disease, obesity, diabetes, and cancers of the reproductive organs (Lawrence 2013; Schwab et al. 2014). Thus, systems to appropriately guide food choice, and fat intake in particular, would be highly adaptive. Unsurprisingly, multiple pre and postingestive cues such as mouth-coating, viscosity, emulsion stability, heat transfer, papillae density, postoral receptors have been described as contributors to oral fat detection (Ramirez 1993; Greenberg and Smith 1996; Tepper and Nurse 1997).
Most of the appealing sensory properties of dietary fats are attributable to their form as triglycerides as they contribute desirable mouthfeel characteristics. The detection and sensory impression of dietary fats are largely mediated by the somatosensory system. In contrast, nonesterified fatty acids generally have objectionable sensory properties, and a growing body of evidence indicates that this form is detected, in part, through the gustatory system (DiPatrizio 2014; Gilbertson and Khan 2014; Passilly-Degrace et al. 2014; Tucker et al. 2014; Keast and Costanzo 2015). This modality is referred to as oleogustus (Running et al. 2015). However, threshold sensitivity for oleogustus is highly variable, often ranging over 4 orders of magnitude within a study (Mattes 2009; Stewart et al. 2010; Chevrot et al. 2014), and the distributions are not normally distributed (Mattes 2009). Multiple explanations for this variability, including contributions of sex, number of taste testing occasions, adiposity, and genetics have been proposed (Running et al. 2013).
The limited evidence supporting a genetic basis for the traditionally recognized primary taste qualities of sweet, sour, salty, and bitter is highly variable. Estimates range from very low for salt (Beauchamp et al. 1985; Wise et al. 2007), to moderate for sweet (Hwang et al. 2015) and possibly sour (Wise et al. 2007), up to high for selected bitter compounds (Kaplan et al. 1967; Hansen et al. 2006). The largest estimates are observed where there are marked interindividual differences; the classic example being for phenylthiocarbamide or propylthiouracil (Kaplan et al. 1967; Hansen et al. 2006). In such cases, the distinct groups are classified as tasters and nontasters. Similar claims have been made for oleogustus (Running et al. 2013) supporting further study of the heritable component of this sensation. A first step in assessing the role of genetics in oleogustus is to estimate heritability through the use of family data; however, no study to-date has been large enough to provide the statistical power to do so. Therefore, our main objective was to utilize crowdsourced taste data to establish an estimate of the heritability for oleogustus.
Crowdsourcing, the practice of enlisting the help of a large population towards a task, is well suited to this purpose. It facilitates the collection of data from large samples while building ownership of science in communities. When 2 iterations of crowdsourcing are combined as we have done here, that is self-contributed data from participants (human subjects) and citizen science (which engages members of the community in the research enterprise itself), it helps to build a trust for science and scientists and a foundation of strong public support for scientific research. This evolving approach is amenable to the missions of many nontraditional research sites within communities, such as museums like the Denver Museum of Nature & Science. It enables not only access to populations that may not be routinely included in studies but also promotes partnerships between scientists and the public in pursuing scientific research. Therefore, this method was employed to not only support the scientific endeavor, but also build goodwill by empowering the public to be a part of authentic science that is relevant to their daily lives.
Materials and methods
Participants
Data were obtained through a crowdsourced population study. Crowdsourcing engages people to “create content” (among other things) (Howe 2006). Crowdsourcing in the Genetics of Taste Lab at the Denver Museum of Nature & Science (Museum) consisted of 2 forms of community contribution to the scientific process. First, guests of the Museum could elect to enhance their science education experience by participating in the study as human research subjects. The second form of crowdsourcing entailed training and enabling volunteer citizen scientists to collect data from guests and support data analyses.
The study was conducted between November 2013 and August 2015 during which time a total of 1020 participants took part in some or all data collection activities. All procedures were approved by the Purdue University Institutional Review Board, and the study complied with the Declaration of Helsinki for Medical Research involving Human Subjects. Written consent was obtained for adults (≥18 year olds); children (≤17 year olds) gave both assent and had written approval obtained in person from a legal guardian present at the time of enrollment. Participants volunteered their time.
Participant demographics and data points
Participants were able to enroll in the study in small groups of up to 6 participants with barriers between participants so that answers could not be copied. They were asked not to discuss results with each other during the study. Each participant recorded their answers to questions and ratings on a paper ballot. Participants self-reported age, sex, race, ethnicity, and genetic relatedness to other members in their group. As a control for our heredity analysis, we measured body fat percentage (BF%) by bioelectrical impedance (Tanita, Body Composition Analyzer, model TBF-215; Tanita Corp. of America, Inc.). Finally, additional health and demographic information were obtained by questionnaire.
Fatty acid taste stimuli and sensitivity
Participants were first trained on the stimuli. While wearing nose clips, they first tasted a blank strip as a control to familiarize themselves with the vehicle. This was followed by a strip with the highest linoleic concentration to familiarize themselves with the taste of LA. This approach obviated the need for any sensation descriptor. They were instructed to focus on the difference in taste intensity between the control and the high concentration strip and to use that for ratings in the double-blinded taste tests that followed. Participants having not received descriptor language were asked to record any words that came to mind for them personally to describe the taste while ignoring texture and liking. These descriptors were used to characterize the predominant sensation quality and identify the range of responses. It is not possible to know definitively that all participants perceived the sensation quality or intensity identically, as is generally the case with subjective responses.
Taste tests were then administered double-blind. Participants were instructed to place the clear edible strip embedded with varying concentrations of LA as far back on their tongues as possible. A timer was set, and after 45 s they were asked to record their responses on a visual analog scale (100 mm). Scale anchors ranged from “Extremely Weak” to “Extremely Strong.” Concentration order was randomized. The taste strips minimized visual and textural cues about the taste stimulus that might have influenced responses. The edible strips were made using a solution of pullulan-hydroxypropyl methylcellulose (HPMC) combined with a stable emulsion of 0.5% weight/volume (w/v) LA, 12% w/v gum Arabic, 0.01% w/v ethylenediaminetetraacetic acid (EDTA, an antioxidant), and 0.01% w/v tert-butylhydroquinone (TBHQ, an antioxidant) (Smutzer et al. 2008; Tucker et al. 2015). After drying, the resulting LA concentrations were 0.0% (control/blank), 0.06% (low), 0.15% (medium), and 0.38% (high) v/v by calculation. Strips were made once a month and kept frozen until testing. To eliminate olfactory cues, the subjects wore nose clips during all taste tests.
Statistical analysis
Analyses were restricted to individuals of European (EUR) descent, as no other racial group was large enough to yield adequate power for estimation of heritability and association testing. Complete data were available for 643 EUR individuals for taste ratings at all 4 levels of LA concentration, sex, age, and BF%. Of this group, 289 individuals represent 106 families ranging in size from 2 to 9 (median family size = 2, quartile 3 for family size = 3). Because analyses were conducted per LA concentration level, these counts represent a minimum sample size as complete data for each covariate-adjusted taste response were used in the final analysis with sample sizes as given in Tables 1 and 2 for the number of families, number of related individuals in total, and the number of unrelated individuals.
Table 1.
Mean response differences across concentrations
| Hypothesis | Number of families/ related/unrelated | P-value |
|---|---|---|
| LA 0% = LA 0.06% | 145/598/339 | 0.00360* |
| LA 0% = LA 0.15% | 146/603/340 | 5.06E−26* |
| LA 0% = LA 0.38% | 149/612/331 | 5.37E−23* |
| LA 0.06% = LA 0.15% | 148/609/337 | 4.46E−13* |
| LA 0.15% = LA 0.38% | 151/619/331 | 8.64E−29* |
Mean response differences were assessed for each concentration combination.
*Significant at α < 0.05.
Table 2.
Heritability estimates for oleogustus
| Traita | Number of families/related/unrelated | Heritabilityb | Covariatesc | P-valued |
|---|---|---|---|---|
| LA 0.06%–LA 0% | 145/598/339 | 15% (0.122) | None | 0.093 |
| LA 0.15%–LA 0% | 146/603/340 | 8% (0.102) | None | 0.209 |
| LA 0.38%–LA 0% | 149/612/331 | 19% (0.114) | Sex (0.02) | 0.043* |
| BF% | 147/603/338 | 31% (0.112) | Age, sex (0.365) | 0.0021* |
Heritability estimates and significance were assessed for each concentration adjusting for baseline. The trait BF% is representative of a positive control of the statistical test for an estimation of heritability.
aLA measures are square root transformed.
bNarrow-sense heritability estimates (percentage) shown with the standard error of estimate in parentheses.
cCovariates are shown with the proportion of variance due to that factor in parentheses.
d P-value represented as a test of the null hypothesis (H0: ).
*Significant at α < 0.05.
The variance component model used requires an assumption of multivariate normality (Almasy and Blangero 1998). Hence, to ensure that our conclusions would be valid, we first analyzed the distribution of the responses. The responses at all concentration levels had right skewed distributions; the square root transformation is a common procedure for reducing right skewness and was applied. All analyses are reported based on the square root scale unless otherwise specified; responses were scaled as a difference from the control/blank, and the pedigree structure reported by the participants was used. Backward selection was utilized to determine adjustment of covariates (sex, age, and BF%) with α = 0.05. Following covariate selection, the full likelihood model is given as follows. Let the response for the LA concentration level under study be denoted by with model for a single individual given as , where is the population mean, is a vector of covariates for the individual, is the vector of regression coefficients for the covariates, is the genetic component of , and represents residual error. Responses are modeled as a multivariate normal distribution for each family with covariance matrix , where is the matrix of expected kinship coefficients, is the variance of after subtracting the population mean and covariate effects, is narrow-sense heritability after covariate adjustment, is the identify matrix, and is the proportion of variance due to residual error beyond the effects of covariates. A likelihood ratio test was applied to test whether the mean difference in response from control/blank was significantly different from zero—this corresponds to a test of and uses a Chi-squared distribution with 1 degree of freedom. Maximum likelihood estimation was used for narrow-sense heritability, and a test of the null hypothesis of h2 = 0 was conducted using a likelihood ratio statistic. Here, significance is based on a 0.5:0.5 mixture of a Chi-squared distribution with 1 degree of freedom and a point mass at 0. Analyses were conducted in the SOLAR [Sequential Oligogenic Linkage Analysis Routines] software package (Almasy and Blangero 1998). Throughout, statistical significance was assessed at a type 1 error of 0.05 as this was an initial/exploratory study.
Results
Taste sensitivity analysis
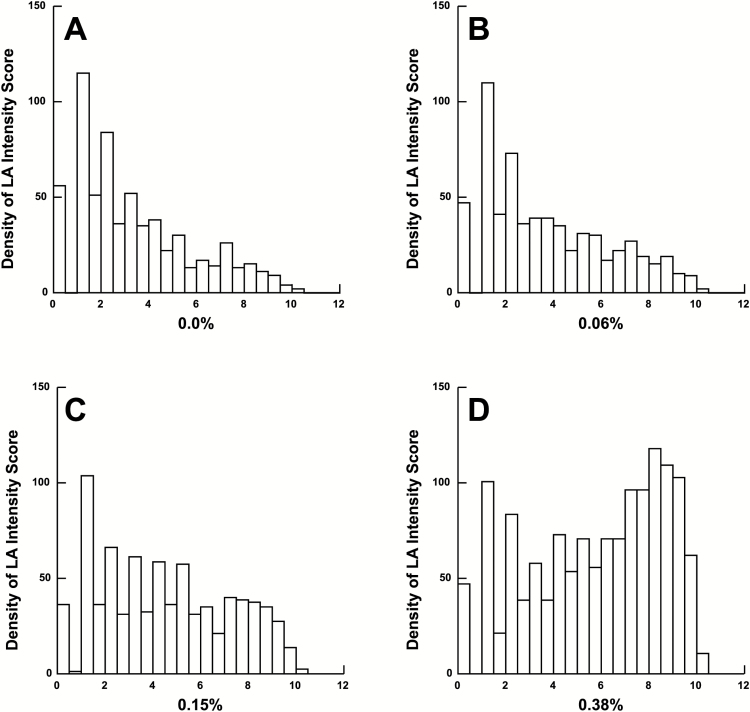
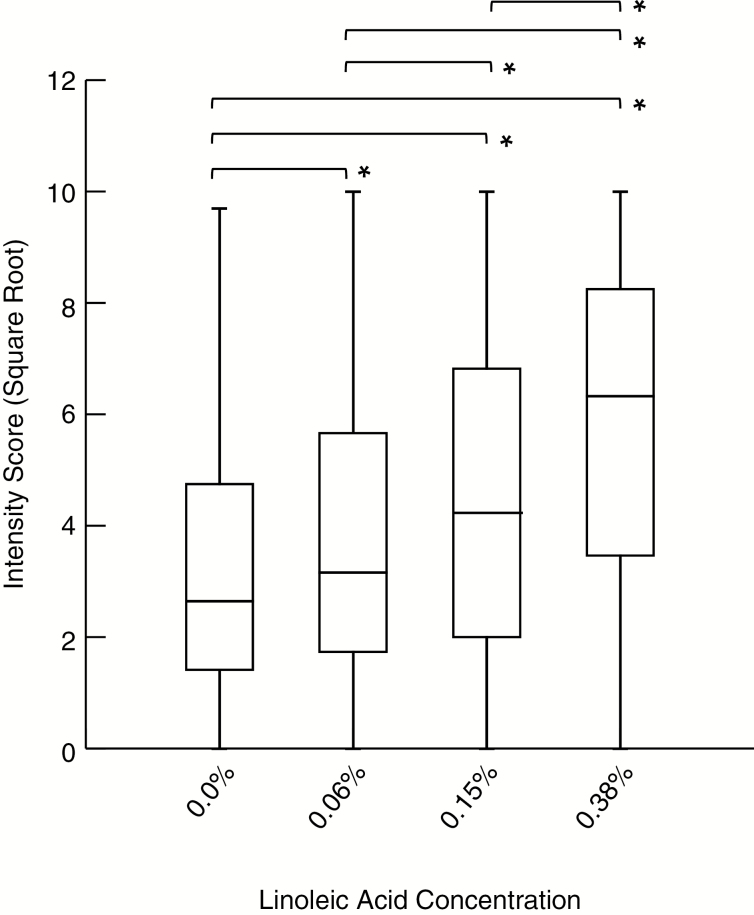
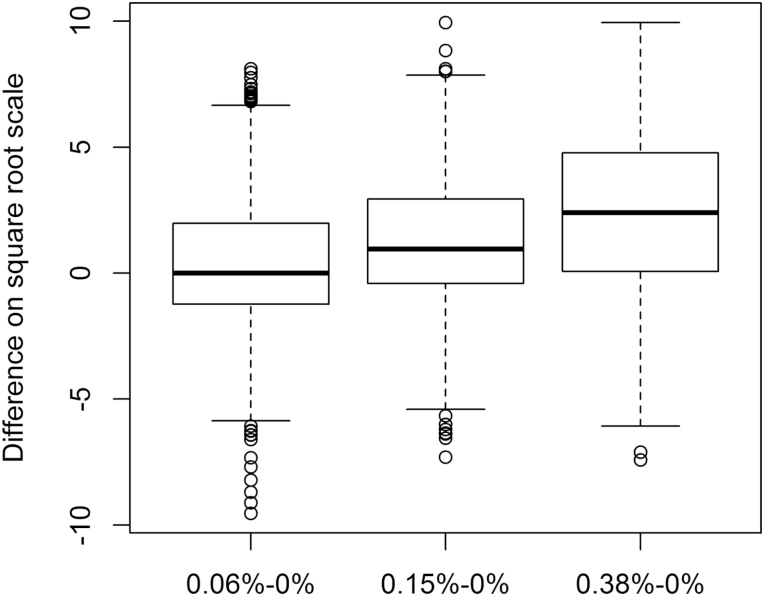
Intensity scores for the control strip (0.0%) and the 3 concentrations of LA (0.06%, 0.15%, and 0.38%) transformed by their square root are presented in Figure 1. The mean response differed for consecutive levels of LA and from each concentration level from baseline (Figure 2 and Table 1). Finally, some people respond to the blank vehicle. To account for this response, we examined the difference from each concentration to the next when subtracting the response to baseline. We find that the distribution of the difference from the baseline taste response (0.0%) increases as the concentration of the LA increases, revealing participants understood and were capable of performing the sensory assessment specifically for the taste of LA (Figure 3).
Figure 1.
Histogram of LA intensity scores. Histograms of the raw scores (transformed by square root) for each concentration of LA. The X-axis (range from 0 to 12) represents the square root of the raw taste intensity score at the indicated concentration of LA. The Y-axis plots the number of participants at each intensity score. (A) Control strip (0.0% LA); (B) Low concentration (0.06% LA); (C) Medium concentration (0.15% LA); and (D) High concentration (0.38% LA).
Figure 2.
Box plot of LA intensity scores. Box and whisker plot showing quartiles of raw intensity scores (transformed by square root). The median is represented by the middle line within each box with the second quartile the lower segment of the box and the third quartile the upper segment of the box. The whiskers of the plot represent the lower quartile (bottom whisker) and the upper quartile (top whisker). There were significant differences observed between each concentration tested as represented by *.
Figure 3.
Significant differences in taste response from baseline. Box and whisker plot of matched data. Box and whisker plot showing quartiles of raw intensity scores (transformed by square root). The median is represented by the middle line within each box with the second quartile the lower segment of the box and the third quartile the upper segment of the box. The whiskers of the plot represent the lower quartile (bottom whisker) and the upper quartile (top whisker). There were significant differences observed between each concentration tested when accounting for baseline response to the control (0.0%LA). Heritability analyses correlate similarity in the response between relatives with the degree of their relationship.
Hereditary analysis
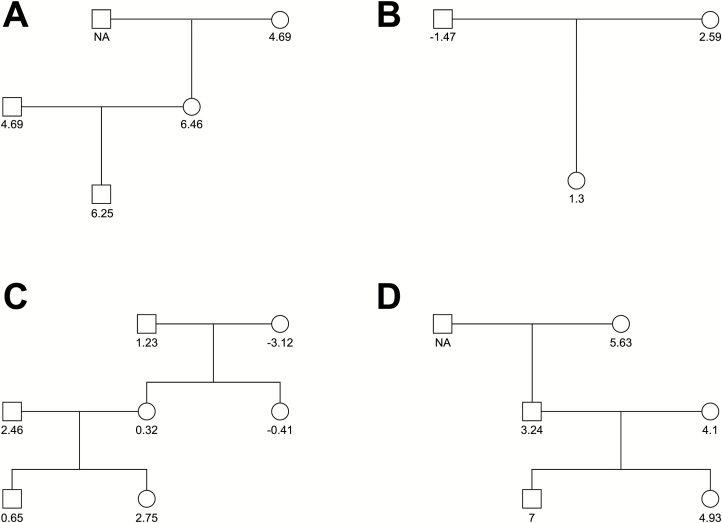
Example pedigrees are presented in Figure 4 representing small and large collected families. Genetically related family members are plotted along with their difference in taste response at LA 0.38% from baseline (0.0%). Estimated heritability was statistically significant only at the highest concentration (Table 1) with 19% of the variability being heritable, after adjustment of biological sex as a significant covariate (P = 0.043).
Figure 4.
Example pedigrees. Example pedigrees from the dataset are plotted along with their difference in taste response at LA 0.38% from the baseline level of LA 0%. Squares represent male relatives, circles represent female relatives. The score below each family member represents their difference in their taste score from the high concentration of LA (0.38%) from the control (0.0%) NA indicates the relative was not present for testing.
Discussion
Given that the concept of fat taste has only recently gained acceptance, most work has focused on determining its validity rather than the heritable and environmental determinants of its expression. Reviews of the animal literature reveal a probable heritable component of fat preference and intake, but there is marked species variability (Reed 2010; Reed and Knaapila 2010). The suitability of rodents as models for human oleogustus is uncertain as rodents and humans differ in lingual lipase secretion (Kawai and Fushiki 2003; Kulkarni and Mattes 2013) and nonesterified fatty acid preference (Mattes 2010). There is a body of evidence indicating an association between a heritable component for fat preference and intake in humans that ranges from no association to r = 0.48 (Reed 2010). However, to our knowledge, the present data are the only estimate of the heritability of fat taste, or more specifically, LA intensity ratings in humans. These data reveal that 19% of the variance in LA taste intensity rating compared with baseline is heritable at the highest concentration tested.
High variability decreases levels of heritability. Therefore, we attribute the lack of a consistent level of heritability at each concentration to the higher variability associated with ratings for the 2 lower LA concentrations compared with the high concentration. It is also plausible that different receptors are recruited at varying concentrations (Abdoul-Azize et al. 2014) and that heritability may vary with receptor type activity. For example, CD36 is rapidly down-regulated in mice with exposure to high fat stimuli whereas FFAR4 (GPR120) is more stable (Martin et al. 2011). Thus, CD36 may be more active at low/medium fatty acid concentrations (Ozdener et al. 2014) and FFAR4 (GPR120) at higher concentrations. Additionally, we cannot exclude the possibility that the quality changes with concentration, and the heritability is stronger for the quality associated with the high concentration. It is clear that the quality changes with fatty acid chain length where it is sour for short-chain fatty acids and shifts to oleogustus for long-chain fatty acids (Running et al. 2015; Running and Mattes 2016). Finally, our heritability estimate is modest (19%). This was not unexpected and may be attributable to the crowdsourcing methodology and use of a convenience sample. As heritability is the proportion of total variation in a characteristic that is attributed to genetics, the larger the experimental and environmental variation, the more noise introduced in the analysis and the smaller the resulting heritability estimate. So, while the heredity pattern was not consistent across all concentrations tested, we do not believe this to reflect a false positive at the highest level of LA tested.
In addition to these considerations, it is of note that similarly conservative findings have been reported for heritability estimates of other tastes. For example, current best estimates from studies of humans hold that approximately 30% of the variance in sweet taste intensity is genetically determined, and this holds equally for nutritive (glucose, fructose) and low-calorie sweeteners (aspartame, neohesperidine dihydrochalcone) (Hwang et al. 2015). The similarity of estimates across these sweeteners suggests a common set of genes underlie the sensation of sweetness despite evidence that there is more than 1 sweet taste transduction mechanism (Damak et al. 2003). Shared environmental influences on sweet intensity ratings are weak (Hwang et al. 2015). Salt (NaCl) detection and recognition thresholds do not appear to have a heritable component, but there is a substantive environmental influence (Beauchamp et al. 1985; Wise et al. 2007). Up to 50% of the variance in sour (citric acid) recognition thresholds may have a genetic basis (Wise et al. 2007). In contrast, no heritable component has been reported for hydrochloric acid detection threshold (Kaplan et al. 1967). Whether this discrepancy is due to the stimulus (e.g., weak versus strong acid), transduction mechanism, or taste dimension (detection versus recognition threshold) is not known. The heritable component for intensity ratings of selected bitter compounds ranges from very high (phenylthiocarbamide (71%) and propylthiouracil (72%) of phenotypic variance) to moderate (sucrose octaacetate (28%); quinine (34%), caffeine (30%)) (Hansen et al. 2006; Knaapila et al. 2012). Environmental contributions are weak, ranging from 7% to 22% of the variance (Hansen et al. 2006). Correlations between intensity ratings for these bitter compounds (r = 0.12–0.56) are consistent with different transduction mechanisms (Hansen et al. 2006; Knaapila et al. 2012). Based on twin studies, the genetic basis for detection thresholds for quinine range from 11% (Krondl et al. 1983) to 85% (Smith and Davies 1973). It is known that there is variation in sensory perception to monosodium glutamate, but the heritable component has not been determined (Reed and Knaapila 2010). Thus, the present findings on oleogustus fall within the range of other tested taste qualities.
Several caveats warrant mention. First, as previously mentioned, variability in responses reduces the proportion of variance that may be identified from other sources (e.g., genetics, environment). Thus, the current estimate is likely to be conservative. Related to this, the degree of experimental control in a crowdsourcing approach to data collection is not comparable to that possible under tightly controlled laboratory studies. This will add to the variance of the responses but would not be expected to bias findings towards observing a heritable effect for LA taste intensity. Therefore, the data provided are regarded as suggestive, rather than confirmatory, of a genetic contribution to oleogustus and its magnitude. Second, in order to account for any genetic contribution to ratings of the vehicle used to deliver the LA stimuli, we analyzed the data using the difference between ratings for each concentration and the control strip (0% LA). Third, given the large array of dietary fatty acids that vary in degree of saturation and chain length, it is possible, if not likely, that values for 1 are not valid indicators for all. Preliminary evidence indicates fatty acids varying in chain length differ in perceived quality and intensity (Running et al. 2015), and therefore may differ in the gene(s) responsible for detection of fat taste. Finally, the present methodologic approach of crowdsourcing did not permit assessment of the reliability of the sensory ratings; however, the one study that has performed repeated measures of fat taste intensity observed responses to be significantly correlated (Mattes 2009). In addition, prior controlled studies of oleogustus reveal that, similar to work with other taste qualities, there is a reduction of threshold with repeated testing over several trials followed by a stable level of performance (Running et al 2013; Tucker and Mattes 2013; Tucker et al. 2014).
Conclusion
Here, we report data providing an initial estimate of a genetic contribution for the oral fat sensation, oleogustus. First, we show that intensity ratings grow monotonically with gradations of LA concentration. Second, we obtained evidence supporting a heritable contribution (~19%) for the intensity rating of the highest LA concentration versus a control.
Funding
This work was supported by the National Center for Research Resources, National Institutes of Health [1R25RR025066] and the University of Colorado Denver, CLAS Research Innovation Seed Program, and US Department of Agriculture Hatch Grant [208684].
Acknowledgments
The authors wish to thank current and previous volunteer citizen scientists, interns, and staff members in both the Genetics of Taste Lab and on the Expedition Health core team for their support of the crowdsourcing research model. Special thanks to Laura Hancock, MD/PhD candidate in the Human Medical Genetics and Genomics Program at the University of Colorado Anschutz Medical Campus, for developing a qRT-PCR protocol suitable for our citizen scientists. Members of the Genetics of Taste Lab Citizen Science corps that contributed to this work include (in alphabetical order): Michael Archer, Sunanda Babu, Emma Boxer, Wendy Covert, Vanessa Dorrance, Katelyn Engel, Michael Hamby, Laura Harmacek, Joyce Hutchens, Sally Huynh, Rachel Jones, Torry Knodell, Wim Leenhouts, Stephania Lukjan, Kathy Lutchi, Joshua Mak, Dani Meyers, Anjelica Miranda, Jericho Oviedo, Brian Reinhart, Wilbur Reusser, Valerie Schowinsky, Claire Simon, Sean Stahle, Sonnie Talley, Rudy Torres, Weston Truman, Johanna Van De Wege, Tyler Wilson, and Diane Woltkamp.
References
- Abdoul-Azize S, Selvakumar S, Sadou H, Besnard P, Khan NA. 2014. Ca2+ signaling in taste bud cells and spontaneous preference for fat: unresolved roles of CD36 and GPR120. Biochimie. 96:8–13. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp G, Bertino M, Engleman K. 1985. Sensory basis for human salt consumption. In: Horan M, Blaustein M, Dunbar J, Kachadorian T, Kaplan N, Simopoulos A, editors. NIH workshop on nutrition and hypertension proceedings from a symposium; 1984 March 12–14; Bethesda, MD, New York: Biomedical Information Corporation. [Google Scholar]
- Chevrot M, Passilly-Degrace P, Ancel D, Bernard A, Enderli G, Gomes M, Robin I, Issanchou S, Vergès B, Nicklaus S et al. 2014. Obesity interferes with the orosensory detection of long-chain fatty acids in humans. Am J Clin Nutr. 99:975–983. [DOI] [PubMed] [Google Scholar]
- Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Varadarajan V, Zou S, Jiang P, Ninomiya Y, Margolskee RF. 2003. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science. 301:850–853. [DOI] [PubMed] [Google Scholar]
- DiPatrizio NV. 2014. Is fat taste ready for primetime? Physiol Behav. 136:145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson TA, Khan NA. 2014. Cell signaling mechanisms of oro-gustatory detection of dietary fat: advances and challenges. Prog Lipid Res. 53:82–92. [DOI] [PubMed] [Google Scholar]
- Greenberg D, Smith GP. 1996. The controls of fat intake. Psychosom Med. 58:559–569. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Reed DR, Wright MJ, Martin NG, Breslin PA. 2006. Heritability and genetic covariation of sensitivity to PROP, SOA, quinine HCI, and caffeine. Chem Senses. 31:404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. 2006. The rise of crowdsourcing (Wired Magazine) [Online, cited 2016 July 22. https://www.wired.com/2006/06/crowds/].
- Hwang LD, Zhu G, Breslin PA, Reed DR, Martin NG, Wright MJ. 2015. A common genetic influence on human intensity ratings of sugars and high-potency sweeteners. Twin Res Hum Genet. 18:361–367. [DOI] [PubMed] [Google Scholar]
- Kaplan AR, Fischer R, Karras A, Griffin F, Powell W, Marsters RW, Glanville EV. 1967. Taste thresholds in twins and siblings. Acta Genet Med Gemellol (Roma). 16:229–243. [DOI] [PubMed] [Google Scholar]
- Kawai T, Fushiki T. 2003. Importance of lipolysis in oral cavity for orosensory detection of fat. Am J Physiol Regul Integr Comp Physiol. 285:R447–R454. [DOI] [PubMed] [Google Scholar]
- Keast RS, Costanzo A. 2015. Is fat the sixth primary? Evidence and implications. Flavour. 4:5. [Google Scholar]
- Knaapila A, Hwang LD, Lysenko A, Duke FF, Fesi B, Khoshnevisan A, James RS, Wysocki CJ, Rhyu M, Tordoff MG et al. 2012. Genetic analysis of chemosensory traits in human twins. Chem Senses. 37:869–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krondl M, Coleman P, Wade J, Milner J. 1983. A twin study examining the genetic influence on food selection. Hum Nutr Appl Nutr. 37 A:189–198. [PubMed] [Google Scholar]
- Kulkarni B, Mattes R. 2013. Evidence for presence of nonesterified fatty acids as potential gustatory signaling molecules in humans. Chem Senses. 38:119–127. [DOI] [PubMed] [Google Scholar]
- Lawrence GD. 2013. Dietary fats and health: dietary recommendations in the context of scientific evidence. Am Soc Nutr Adv Nutr. 4:294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Passilly-Degrace P, Gaillard D, Merlin JF, Chevrot M, Besnard P. 2011. The lipid-sensor candidates CD36 and GPR120 are differentially regulated by dietary lipids in mouse taste buds: impact on spontaneous fat preference. PLoS One. 6:e24014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes RD. 2009. Oral detection of short-, medium-, and long-chain free fatty acids in humans. Chem Senses. 34:145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes RD. 2010. Fat taste in humans: is it a primary? In: Montmayeur JP, le Coutre J, editors. Fat detection: taste, texture, and post ingestive effects. Boca Raton: CRC Press/Taylor & Francis; p. 167–193. [Google Scholar]
- Ozdener MH, Subramaniam S, Sundaresan S, Sery O, Hashimoto T, Asakawa Y, Besnard P, Abumrad NA, Khan NA. 2014. CD36- and GPR120-mediated Ca²⁺ signaling in human taste bud cells mediates differential responses to fatty acids and is altered in obese mice. Gastroenterology. 146:995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passilly-Degrace P, Chevrot M, Bernard A, Ancel D, Martin C, Besnard P. 2014. Is the taste of fat regulated? Biochimie. 96:3–7. [DOI] [PubMed] [Google Scholar]
- Ramirez I. 1993. Role of olfaction in starch and oil preference. Am J Physiol. 265:R1404–R1409. [DOI] [PubMed] [Google Scholar]
- Reed DR. 2010. Heritable variation in fat preference. In: Montmayeur JP, le Coutre J, editors. Fat detection: taste, texture, and post ingestive effects. Boca Raton: CRC Press/Taylor & Francis; p. 395–415. [PubMed] [Google Scholar]
- Reed DR, Knaapila A. 2010. Genetics of taste and smell: poisons and pleasures. Prog Mol Biol Transl Sci. 94:213–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running CA, Mattes RD, Tucker RM. 2013. Fat taste in humans: sources of within- and between- subject variability. Prog Lipid Res. 52:438–445. [DOI] [PubMed] [Google Scholar]
- Running CA, Craig BA, Mattes RD. 2015. Oleogustus: the unique taste of fat. Chem Senses. 40: 507–516. [DOI] [PubMed] [Google Scholar]
- Running CA, Mattes RD. 2016. A review of the evidence supporting the taste of non-esterified fatty acids in humans. J. Am. Oil Chem. Soc. 93: 1325. [Google Scholar]
- Schwab U, Lauritzen L, Tholstrup T, Haldorsson TI, Riserus U, Uusitupa M, Becker W. 2014. Effect of the amount and type of dietary fat on cardiometabolic risk factors and risk of developing type 2 diabetes, cardiovascular diseases, and cancer: a systematic review. Food & Nutr Res. 58:25–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Davies PD. 1973. Quinine taste thresholds: a family study and a twin study. Ann Hum Genet. 37: 227–232. [DOI] [PubMed] [Google Scholar]
- Smutzer G, Lam S, Hastings L, Desai H, Abarintos RA, Sobel M, Sayed N. 2008. A test for measuring gustatory function. Laryngoscope. 118:1411–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JE, Feinle-Bisset C, Golding M, Delahunty C, Clifton PM, Keast RS. 2010. Oral sensitivity to fatty acids, food consumption and BMI in human subjects. Br J Nutr. 104:145–152. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Nurse RJ. 1997. Fat perception is related to PROP taster status. Physiol Behav. 61:949–954. [DOI] [PubMed] [Google Scholar]
- Tucker RM, Mattes RD. 2013. Influences of repeated testing on nonesterified fatty acid taste. Chem Senses. 38:325–332. [DOI] [PubMed] [Google Scholar]
- Tucker RM, Mattes RD, Running CA. 2014. Mechanisms and effects of “fat taste” in humans. Biofactors. 40:313–326. [DOI] [PubMed] [Google Scholar]
- Tucker RM, Nuessle TM, Garneau NL, Smutzer G, Mattes RD. 2015. No difference in perceived intensity of linoleic acid in the oral cavity between obese and nonobese individuals. Chem Senses. 40:557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise PM, Hansen JL, Reed DR, Breslin PA. 2007. Twin study of the heritability of recognition thresholds for sour and salty taste. Chem Senses. 32:749–754. [DOI] [PMC free article] [PubMed] [Google Scholar]