SUMMARY
Single-cell RNA sequencing has generated catalogs of transcriptionally defined neuronal subtypes of the brain. However, the cellular processes that contribute to neuronal subtype specification and transcriptional heterogeneity remain unclear. By comparing the gene expression profiles of single layer 6 corticothalamic neurons in somatosensory cortex, we show that transcriptional subtypes primarily reflect axonal projection pattern, laminar position within the cortex, and neuronal activity state. Pseudotemporal ordering of 1,023 cellular responses to sensory manipulation demonstrates that changes in expression of activity-induced genes both reinforced cell-type identity and contributed to increased transcriptional heterogeneity within each cell type. This is due to cell-type biased choices of transcriptional states following manipulation of neuronal activity. These results reveal that axonal projection pattern, laminar position, and activity state define significant axes of variation that contribute both to the transcriptional identity of individual neurons and to the transcriptional heterogeneity within each neuronal subtype.
Graphical abstract
INTRODUCTION
Single-cell RNA sequencing (scRNA-seq) has revealed previously hidden levels of complexity in cell types and states within tissues (Junker and van Oudenaarden, 2014; Liu and Trapnell, 2016). The neocortex is a brain region dependent on a wide variety of neuronal cell types for its function (Custo Greig et al., 2013; Molyneaux et al., 2015; Zeng and Sanes, 2017). Cortical neurons are also highly dynamic, exhibiting, for example, activity-dependent changes in gene expression (Flavell and Greenberg, 2008; Lyons and West, 2011; West and Greenberg, 2011). Although recent studies have yielded insights into the diversity of cortical cell types (Darmanis et al., 2015; Hevner et al., 2003; Lake et al., 2016; Pollen et al., 2014; Sorensen et al., 2015; Sugino et al., 2006; Tasic et al., 2016; Zeisel et al., 2015), the sources of transcriptional variation both within and across cell types remain poorly understood (Dueck et al., 2016; Sanes and Masland, 2015; Wagner et al., 2016; Zeng and Sanes, 2017).
We compared expression profiles of layer 6 corticothalamic neurons (L6CThNs), a heterogeneous population of cortical projection neurons defined by anatomical, functional, and gene expression studies, making them ideally suited for investigating relationships between transcriptional subtypes and other cellular properties (Bourassa and Deschênes, 1995; Bourassa et al., 1995; Briggs et al., 2016; Katz, 1987; Killackey and Sherman, 2003; Kwegyir-Afful and Simons, 2009; Shima et al., 2016; Sorensen et al., 2015; Tasic et al., 2016; Zhang and Deschênes, 1997). By combining scRNA-seq with an enrichment strategy that preserved axonal target information, we identified two transcriptionally distinct L6CThN subtypes whose transcriptional profiles reflected their long-range projection targets and laminar position within layer 6 (L6). These two L6CThN subtypes also exhibited divergent signatures of neuronal activity both at baseline and following manipulation of sensory input. Subtype biases in the choice of response following sensory manipulation increased transcriptional heterogeneity within each type and reinforced the transcriptional differences between the two L6CThN subtypes. These results demonstrate that scRNA-seq resolves relationships between gene expression and features such as axonal projection pattern, spatial organization, and cell state and identifies the independent contributions of multiple biological signals that together determine transcriptional heterogeneity within and across neuronal populations.
RESULTS
Transcriptional Profiling of L6CThNs Reveals Subtypes that Reflect Axonal Projection Bias
Studies of primary sensory cortex demonstrate that L6CThNs are heterogeneous (Bourassa and Deschênes, 1995; Bourassa et al., 1995; Briggs et al., 2016; Katz, 1987; Killackey and Sherman, 2003; Kwegyir-Afful and Simons, 2009; Shima et al., 2016; Tasic et al., 2016; Zhang and Deschênes, 1997). In rat barrel cortex (BC), L6CThNs in upper L6 project to the ventral posterior medial nucleus (VPM) of the thalamus, while L6CThNs in lower L6 project primarily to both VPM and the posterior medial nucleus (POm; Bourassa et al., 1995; Killackey and Sherman, 2003). To distinguish between these two projection patterns, we first validated Cre recombinase expression as a reliable marker for L6CThNs in BC of Neurotensin receptor 1-Cre mice (Figures 1A–1E; Ntsr1-Cre, Gensat 220; Bortone et al., 2014; Gong et al., 2007; Guo et al., 2017; Kim et al., 2014). Next, we showed that a subset of Ntsr1-Cre-expressing, VPM-projecting L6CThNs in lower L6 also projects to POm (Figures 1F–1J).
Figure 1. L6CThNs Distinguished by Their Axonal Projections Have Distinct Gene Expression Profiles.
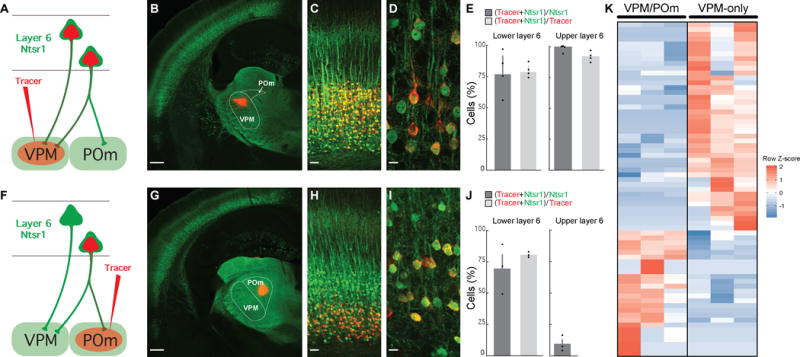
(A and F) Labeling schemes for L6CThNs projecting to the ventral posterior medial nucleus (VPM, A) and to VPM and the posterior medial nucleus (POm, F).
(B) Retrograde tracer injection (red) into VPM in an Ntsr1-Cre;YFP mouse.
(C and D) Colocalization of tracer and YFP in low-magnification (C) and high-magnification (D) images of layer 6 (L6) of barrel cortex (BC).
(E) Quantification of the colocalization (n = 4 mice; error bars: SEM).
(G) Injection of retrograde tracer (red) in POm of an Ntsr1-Cre;YFP mouse.
(H and I) Colocalization of tracer in CThNs in lower L6 in low-magnification (H) and high-magnification (I) images of BC.
(J) Quantification of the colocalization (n = 3 mice; error bars: SEM).
(K) Matrix showing the 69 genes differentially expressed between pools of VPM/POm and VPM-only L6CThNs (three replicates).
Scale bars, 500 μm (B and G), 50 μm (C and H), and 10 μm (D and I). See also Figure S1, Table S1, and Data S2.
To determine whether Ntsr1-Cre-expressing L6CThNs projecting to VPM only or to both VPM and POm are distinguished by their gene expression profiles, we labeled the two subclasses in adolescent mice as described above and collected enriched populations of each subclass for bulk RNA sequencing (Figures S1A and S1B). We identified 69 differentially expressed genes between the two populations (Figure 1K; Data S2; Table S1; Cuffdiff2; 10% false discovery rate [FDR]), demonstrating that L6CThNs distinguished by their long-range axonal projection patterns are differentiated by their gene expression profiles.
Because this bulk analysis was predicated on prior knowledge of existing morphological subclasses and may have obscured underlying transcriptional subtypes within each projection class, we next evaluated the gene expression landscape of single L6CThNs using an unbiased classification approach. We sorted and collected individual, labeled L6CThNs (Figure S1C) from two replicate mice. Individual cell lysates were subjected to a modified Smart-Seq2 library preparation and scRNA-seq analysis. 346 single L6CThNs passed quality control filters (Figures S1D–S1I; Data S3). We confirmed the fidelity of our enrichment by assessing each cell for neuronal and non-neuronal markers (Figure S1J).
To identify transcriptional subtypes of L6CThNs, we selected high-variance genes common to both replicates (Figure S2A; Supplemental Experimental Procedures). The resulting 261 genes were enriched for genes contributing to transcriptional variation across neurons and depleted for genes associated with technical variation between replicates (Table S2; Brennecke et al., 2013; Hicks et al., 2015). Weights on the first three significant principal components (PCs) of a principal-component analysis (PCA) across all cells using this gene set (Figure S2B; permutation parallel analysis; p < 0.001; Chung and Storey, 2015) were used for a t-distributed stochastic neighbor embedding (t-SNE) dimensionality reduction followed by k-means clustering (Figure 2A). Single-cell transcriptional profiles of the 346 L6CThNs clustered into at least two distinct subtypes. We bootstrapped this analysis over 1,000 iterations to confirm that the clustering solution was independent of the non-deterministic variation in t-SNE results with different random seeds (Figure S2C). Fitting these data to three or more subtypes (k ≥ 3) resulted in lower silhouette scores (Figure S2C). We next compared our classification approach to several recently described single-cell clustering utilities (Kiselev et al., 2017; Lin et al., 2017) and found a high-degree of agreement (Figure S2D; SC3, 93.77% agreement; CIDR, 90% agreement). These results indicate that independent, unbiased clustering approaches based on genes with higher than expected variance across the Ntsr1-Cre-expressing L6CThN population identify two major subtypes.
Figure 2. Unbiased Clustering of Single L6CThN Transcriptomes Defines Two Subtypes with Strong Axonal Projection Bias.
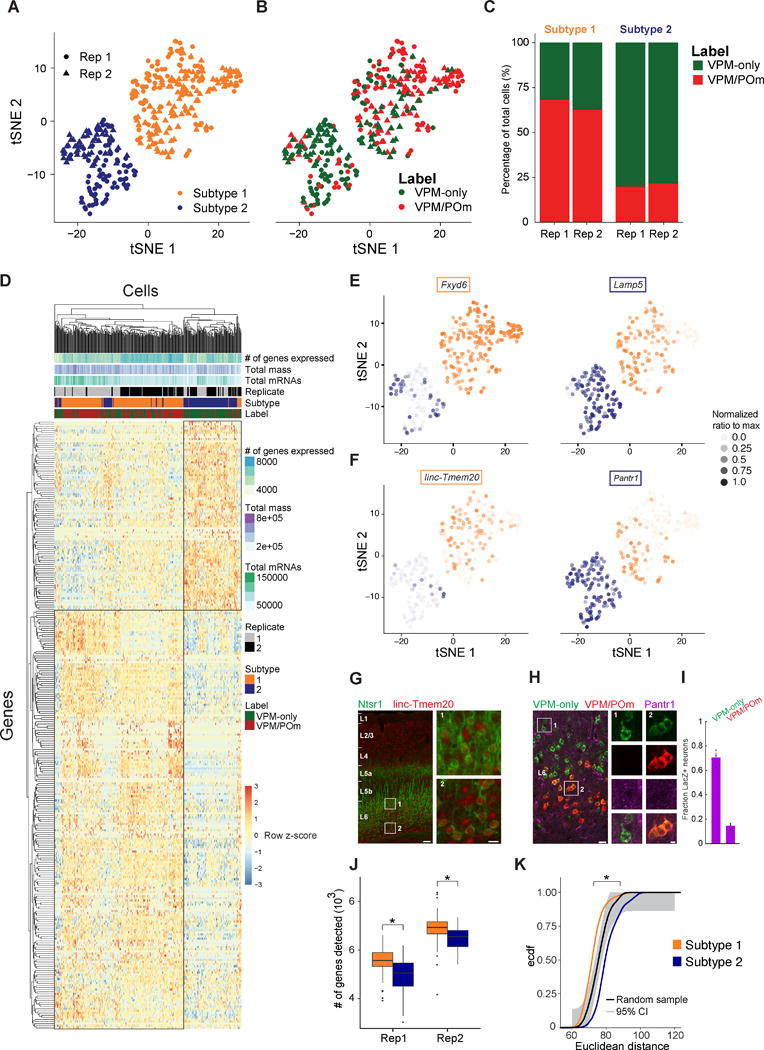
(A) t-SNE plot showing two subtypes of L6CThNs classified via unsupervised clustering.
(B) t-SNE plot as in (A) with each L6CThN color-coded by axonal projection label.
(C) Fraction of VPM-only (green) and VPM/POm (red) L6CThNs in each transcriptionally defined subtype for each replicate.
(D) Hierarchical clustering of the 346 L6CThNs (x axis) and the 286 genes differentially expressed between the two subtypes (y axis) (0.1% FDR).
(E and F) t-SNE plots showing the normalized expression levels of two differentially expressed protein-coding and long-noncoding RNAs enriched in subtype 1 (E, Fxyd6, left; F, linc-Tmem20, left) and subtype 2 (E, Lamp5, right; F, Pantr1, right).
(G) Low-magnification image of linc-Tmem20 (red) in barrel cortex (BC) of an Ntsr1-Cre;tdTomato (green) mouse combining in situ hybridization (linc-Tmem20) and immunohistochemistry (tdTomato). Insets show higher expression of linc-Tmem20 in L6CThNs in lower layer 6 (L6; inset 2) relative to upper L6 (inset 1).
(H) LacZ expression in BC of a heterozygous Pantr1-LacZ mouse following tracer injections in VPM (green) and in POm (red). Insets show LacZ puncta in VPM-only L6CThNs (column 1, green) and not in VPM/POm L6CThNs (column 2, red and green).
(I) Fraction of VPM-only and VPM/POm L6CThNs expressing LacZ (n = 3 mice).
(J) Median number of genes detected across all cells for each subtype by replicate pair (replicate 1: subtype 1 5,582 ± 526.3 [SD], subtype 2 5,080 ± 650.0 [SD], p < 2.169 × 10−10, Mann-Whitney test; replicate 2: subtype 1 6,950 ± 545.4 [SD], subtype 2 6,569 ± 478.7 [SD], p < 7.071 × 10−7, Mann-Whitney test).
(K) Cumulative probability distribution of the pairwise Euclidean distances among cells in subtypes 1 (gold) and 2 (blue; p < 2.2 × 10−16, Welch’s two-sample t test). The black line represents the pairwise distances among a random sample of 100 cells drawn from the 346 cells. Ninety-five percent confidence interval is shown in light gray (Dvoretzky-Kiefer-Wolfowitz inequality).
Scale bars, 100 and 20 μm (G) and 20 and 5 μm (H). See also Figures S1–S4, Tables S2 and S3, and Data S3.
To determine the relationship between transcriptional identity and morphological subtypes, we next compared the distribution of VPM-only and VPM/POm projection labels across the two subtypes (Figures 2B and 2C). The majority of neurons in subtype 2 were labeled VPM only (79% [103 of 130 cells], p < 3.768 × 10−17, hypergeometric test), whereas most in subtype 1 were VPM/POm (65% [141 of 216 cells], p < 3.768 × 10−17, hypergeometric test), a distribution significantly different from that expected by chance. Together our results indicate that each transcriptionally defined subtype of L6CThN is enriched for neurons targeting specific sets of thalamic nuclei.
Transcriptional Differences between Subtypes of L6CThNs
To assess transcriptional differences between the two L6CThN subtypes, all expressed genes were subjected to the Monocle2 differential test (Supplemental Experimental Procedures). We identified 286 genes that were significantly differentially expressed between the two subtypes (Figure 2D; Monocle likelihood ratio test, 0.1% FDR; Data S3; Table S3; mean RNA copies per cell 58.12), only 6 of which overlapped with the 69 differentially expressed genes observed in our bulk RNA-seq analysis despite the high correlation between our bulk RNA-seq and scRNA-seq data (Figures S2E–S2G). These results, in conjunction with the incomplete label segregation across L6CThN subtypes, suggest that this discrepancy is due primarily to sample heterogeneity arising from retrograde label inefficiencies. Importantly, parameters such as total mapped fragments (mass), total estimated mRNAs per cell, number of genes detectably expressed per cell, and replicate did not result in biased clustering across the differentially expressed gene list (Figure 2D), suggesting a minimal influence of technical variation on the list of differentially expressed genes. The significant differential expression of genes, including Fxyd6 and Lamp5, between the two subtypes was consistent with expression patterns in the Allen Mouse Brain Atlas (Figure 2E; http://mouse.brain-map.org, Fxyd6-RP_051017_01_E10-coronal, Lamp5-RP_050725_01_B03-coronal; Lein et al., 2007). The two subtypes also shared transcriptional similarities with highlighted genes expressed in two recently defined subtypes of L6CThNs in primary visual cortex (Figure S3A; Tasic et al., 2016). When we pooled our data across the two types, we also found a close correspondence between our data and a recent study which assessed the gene expression of small pools of Ntsr1-Cre-expressing L6CThNs using bulk RNA-seq (Figures S3B and S3C; Shima et al., 2016).
Several long non-coding RNAs (lncRNAs) were specifically enriched in each subtype. For example, linc-Tmem20 (Molyneaux et al., 2015) was significantly enriched in subtype 1 (Figure 2F) and was preferentially expressed in lower L6 (Figure 2G), while Pantr1 was identified as the gene with the greatest predictive power for neurons in subtype 2 (Figure 2F; area under the curve [AUC] = 0.876, power = 0.752, receiver-operating characteristic [ROC] analysis). A mouse line in which LacZ was knocked into the Pantr1 locus (Sauvageau et al., 2013) confirmed greater LacZ expression in VPM-only L6CThNs relative to VPM/POm L6CThNs (Figures 2H and 2I). Furthermore, in contrast to our analysis of L6CThNs by anatomical labeling, we found that the total number of genes with detectable expression in each transcriptional subtype was significantly greater for subtype 1 relative to subtype 2, a finding consistent across replicates (Figure 2J), and that the mean pairwise Euclidean distance between cells within each subtype was also greater in subtype 2 than in subtype 1, indicating greater cell-cell variation within subtype 2 (Figure 2K; subtype 1, μ = 72.16, σ = 5.80; subtype 2, μ = 80.59, σ = 7.07; p < 2.2 × 10−16, Welch’s two-sample t test). Importantly, we found only two genes, Lypd1 and Calm2, with a significant combinatorial effect of subtype and label, suggesting that label does not distinguish subpopulations within each subtype. Together, these data identify two Ntsr1-Cre-expressing L6CThN subtypes, each biased for projection target.
To identify cellular processes that differentiate the two transcriptional subtypes of L6CThNs, we queried the differentially expressed gene list for enrichment of annotated gene sets from public databases (Figures S4A–S4D). Significant gene sets included Gene Ontology and Reactome terms related to general features of neurons such as “Neuronal part” and “Synaptic transmission” (p < 0.01, hypergeometric test, Benjamini-Hochberg corrected), highlighting the limited resolution of currently available public databases for generating biological insights among neuronal subtypes. To identify more informative biological processes that shape differences in L6CThNs, we compared the expression of voltage-gated ion channels, neurotransmitter receptors and neuropeptides, a number of which were differentially expressed (Figures S4E–S4H). For example, Adcyap1 (PACAP) and a gene encoding a peptide for processing PACAP, Pam, were preferentially expressed in subtype 2 (Figures S4H and S4I). Interestingly, receptors for PACAP are found in primary sensory thalamic nuclei (Joo et al., 2004) and modulate thalamocortical interactions (Sun et al., 2003), consistent with the projection bias of subtype 2 L6CThNs. Thus, a focused analysis identifies gene expression differences that reflect relevant functional features.
Distinct Cellular Processes Are Coordinately Regulated within L6CThN Subtypes
To identify cellular processes that contribute to the heterogeneity of gene expression across all L6CThNs analyzed, we performed a weighted gene co-expression network analysis (WGCNA) on all genes expressed in the 346 L6CThNs (Langfelder and Horvath, 2008), yielding 22 modules of co-regulated genes (Figures 3A and S5A). The eigenvalues of three modules were significantly correlated with subtype 1 (black, turquoise, and cyan) and four with subtype 2 (red, purple, blue and midnight blue; Figures 3B and S5B; p < 0.01, Pearson’s product moment correlation test). Correlation coefficients and confidence measures were all weakened when cell labels were used instead (Figures 3B and S5C). These seven modules were also significantly correlated with the first PC of the PCA on the high-variance gene set (Figures 3C and S5H), suggesting that subtype identity explains a significant amount of variation in gene expression across these neurons. Five of these modules were enriched for genes identified as significantly differentially expressed between subtype 1 and 2 (Figures 3D and S5D). No module was correlated with replicate or other potentially confounding technical parameters (Figures S5F and S5G), confirming that the variations are driven by biological differences among neurons rather than technical variation. These data demonstrate that the greatest source of variation in the transcriptomes of L6CThNs is the difference between subtypes and reveal several discrete modules of gene expression contributing to this difference.
Figure 3. Coordinately Regulated Gene Sets Contribute to the Transcriptional Identities of L6CThNs.
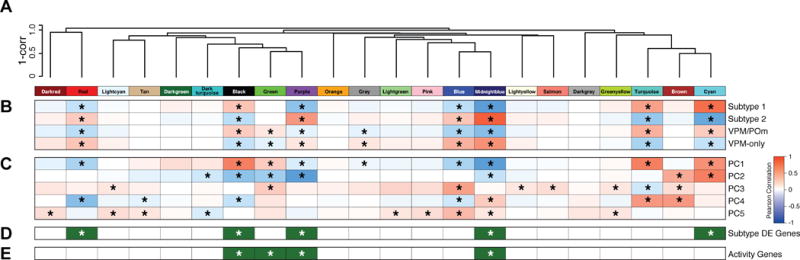
(A) WGCNA on variance-stabilized gene expression estimates identifies modules of coordinately regulated genes grouped using hierarchical clustering of module eigengenes.
(B) Pearson correlation of each module eigengene with both transcriptional subtype and label. Significance (asterisk) was determined using the Pearson’s product moment test (p < 0.01, Benjamini-Hochberg corrected).
(C) Pearson correlation of each module eigengene with component rotations for PCs 1–5.
(D and E) Enrichment of the 286 genes differentially expressed between L6CThN subtypes (D) and genes associated with neuronal activity (E) within each module (hypergeometric test, p < 0.01, Benjamini-Hochberg corrected).
Projection-Dependent and Position-Dependent Gene Expression Differences Contribute to the Transcriptional Identity of L6 Neurons
Because axonal projection pattern and sublaminar position are confounded among L6CThNs (Figures 1A–1J; Zhang and Deschênes, 1997), gene expression differences may represent differences in sublaminar position within L6 rather than axonal projection pattern per se. Because L6CThNs represent approximately half of the neurons in L6 (Kim et al., 2014), we tested whether a gene’s expression reflects axonal projection pattern, in which case its expression should be restricted to L6CThNs either in upper or lower L6, or laminar position, in which case its expression should be restricted to neurons in either upper or lower L6, regardless of their projection pattern.
To select target genes to evaluate, we performed a PCA on the mean-centered expression estimates of the high-variance genes across all 346 L6CThNs and used the rotations from this analysis to project all expressed genes into this PCA space to rank-order candidate genes (Figures S6A–S6D). We quantified the expression of target gene mRNAs in individual tdTomato-positive, NeuN-positive L6CThNs and in tdTomato-negative, NeuN-positive neurons in BC of Ntsr1-Cre;tdTomato mice and fit these data to a generalized additive model to test the independent contributions of laminar position and expression of tdTomato (Figures 4A–4F). We found that the expression of Lamp5, Serpini1, and Gabra5 was dependent on the combined effect of neuronal subtype and position (Figures 4B, 4E, and 4F). In contrast, Pantr1 varied with laminar position within L6 in both L6CThNs and non-L6CThNs (Figure 4C). Our findings reveal that information about position within L6 and long-range axonal projection pattern is contained in the gene expression differences between the two transcriptional subtypes of L6CThNs.
Figure 4. Variation in the Transcriptional Profiles of L6CThNs Is Defined by Subtype-Specific Genes, Genes Reflecting Laminar Location, and Genes Induced by Neuronal Activity.
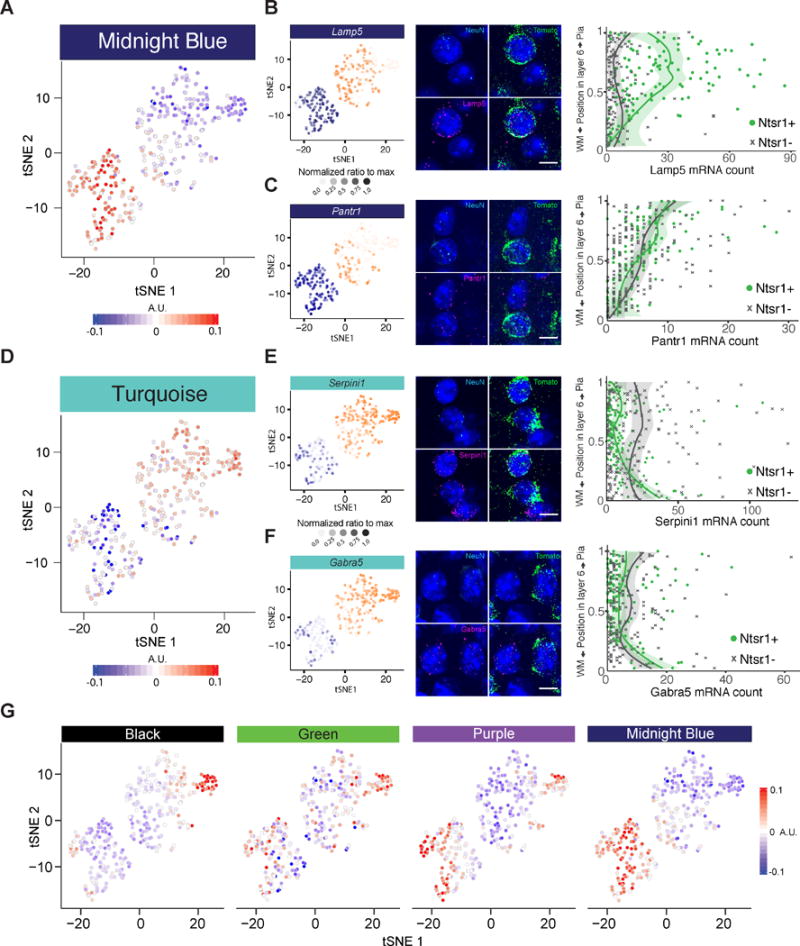
(A and D) t-SNE plots showing the eigenvalue for each cell for the two WGCNA modules most correlated with PC1 (A, midnight blue; D, turquoise).
(B, C, E, and F) t-SNE plots (left) showing the normalized gene expression in each cell for representative genes with significant weights on PC1. (B) and (C) belong to the midnight blue module and (E) and (F) to the turquoise module. Single-molecule fluorescence in situ hybridization (smFISH; middle) of mRNAs detected for each gene of interest (magenta), tdTomato (green), NeuN (cyan), and DAPI (blue) in L6 of Ntsr1-Cre;tdTomato mice. Quantitative gene expression analysis (right) showing the number of mRNAs expressed per neuron as a function of normalized vertical position in layer 6 (L6) and neuronal cell type (L6CThNs: Ntsr1;tdTomato-positive;NeuN-positive neurons in green; non-L6CThNs: Ntsr1;tdTomato-negative, NeuN-positive neurons in gray). Curves represent LOESS fits to individual data points, grouped by cell type; shaded areas correspond to 95% confidence intervals. Statistics: (B, Lamp5) “Subtype specific,” p < 7.3231 × 10−19; “CThN+ position specific,” p < 1.175 × 10−17; “CThN− position specific,” p < 0.0035; (C, Pantr1) “Subtype specific,” p < 0.9931; “CThN+ position specific,” p < 9.243 × 10−11; “CThN− position specific,” p < 1.021 × 10−14; (E, Serpini1) “Subtype specific,” p < 1.606 × 10−6; “CThN+ position specific,” p < 1.045 × 10−6; ”CThN− position specific,” p < 0.3342; (F, Gabra5) ”Subtype specific,” p < 0.1020; “CThN+ position specific,” p < 1.994 × 10−3; “CThN− position specific,” p < 0.09873. Scale bars, 10 μm.
(G) Module eigengenes for the four modules with significant enrichment for genes associated with neuronal activity.
See also Figure S6.
Neural Activity Significantly Contributes to Transcriptionally Defined Subtype Identity
Because neural activity strongly influences gene expression (Flavell and Greenberg, 2008; Lyons and West, 2011; West and Greenberg, 2011), we hypothesized that activity state influences the transcriptional profiles of L6CThNs and contributes to their transcriptional identity. To test whether any modules reflect activity state, we assessed enrichment of a curated set of genes induced by neural activity (Table S4; Cho et al., 2016; Kim et al., 2010; Lacar et al., 2016; Mardinly et al., 2016). Three of the seven modules correlated with transcriptional subtype identity (black, purple, and midnight blue) were significantly enriched for genes induced by neuronal activity (Figures 3E and S5E; p < 0.01, hypergeometric test). All four modules enriched for activity-associated genes (black, green, purple, and midnight blue) were also significantly correlated with PC1 and PC2 of the PCA on the high-variance gene set (Figures 3 and S5), suggesting that differences in neuronal activity state explain a significant amount of transcriptional variation across L6CThNs. Together, our results show that long-range axonal projection pattern, laminar position within L6, and the activity state of each neuron are all reflected in the transcriptional profiles of individual L6CThNs and are principal contributors to the identity of the two L6CThN subtypes.
Among the four activity-associated modules (Figure 4G; black, green, purple, and midnight blue), black was specifically correlated with subtype 1 and purple and midnight blue with subtype 2, suggesting subtype-specific engagement of activity-induced genes in the steady state. To assess whether these signatures of neuronal activity represent a fundamental aspect of subtype identity, we re-evaluated our classification workflow after regressing out the eigenvalues for each activity-associated module and observed a reduction in the separation of the two L6CThN subtypes (Figure S6F), suggesting that steady-state differences in neuronal activity genes are a defining characteristic of these two L6CThN subtypes. We also evaluated the enrichment of activity genes along the first two PCs of a gene-centric PCA of the mean-centered expression estimates of high-variance genes across all 346 L6CThNs (Figures S6A–S6D). Both PC1 and PC2 were significantly enriched for the activity-induced gene list (p < 0.01, Kolmogorov-Smirnov test, pre-ranked gene set enrichment analysis [GSEA]). Expression levels of the most heavily weighted PC1 genes varied predominantly between subtypes (Figures S6F and S6G), while the expression levels of heavily weighted PC2 genes additionally varied within each subtype similar to classical activity-induced genes such as Fos and Bdnf (Figures S6H–S6K). These data indicate that neuronal activity accounts for significant variation in the gene expression of L6CThNs, contributing to subtype identity as well as transcriptional heterogeneity both within and between L6CThN subtypes.
Modulation of Neuronal Activity Influences the Transcriptional State and Identity of L6CThNs
Our data demonstrate that the gene expression profiles of L6CThNs reflect an integration of multiple sources of variation dominated by axonal projection pattern, sublaminar position in L6 and neuronal activity. Thus, alterations in the molecular cascades engaged by different patterns of neural activity may modulate the transcriptional identity of L6CThNs. To test this hypothesis, we removed whiskers unilaterally in a chessboard pattern to engage activity-dependent plasticity mechanisms in BC of mice labeled as in our baseline experiments (Figure 5A; Fox, 2008; Wallace and Fox, 1999). At 1 (day 1) and 7 (day 7) days following this sensory manipulation, we collected and sequenced single L6CThNs from BC both contralateral and ipsilateral to the manipulation. After preprocessing and quality control, we obtained transcriptional profiles for 133 day 1 (two replicates) and 550 day 7 (two replicates) L6CThNs, for a total of 1,023 sequenced L6CThNs when combined with L6CThNs collected at baseline (day 0; Figures S7A–S7D; Data S3 and S4).
Figure 5. Sustained Modulation of Gene Expression after Sensory Manipulation in L6CThNs.
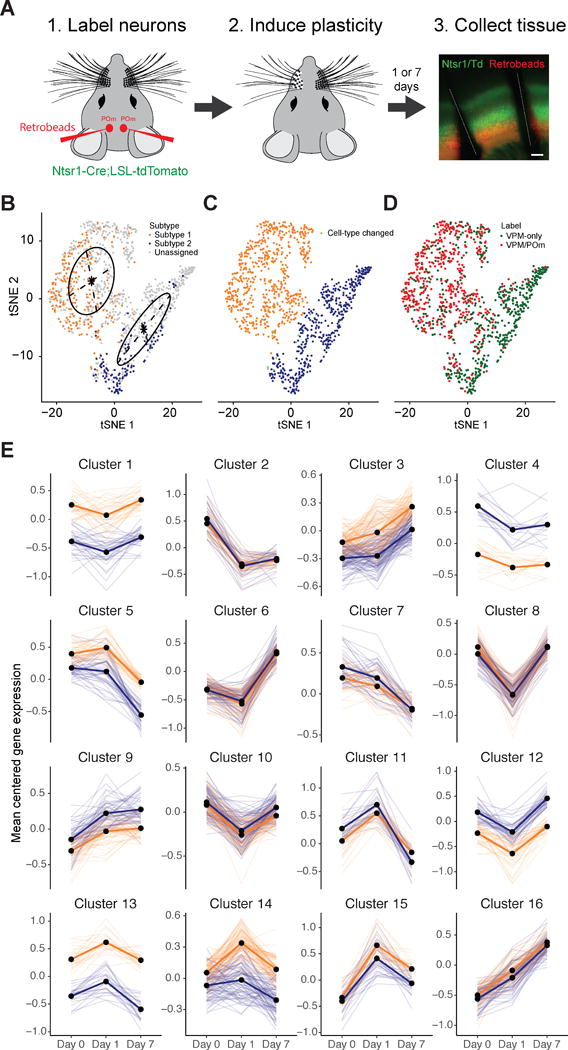
(A) Experimental design. Scale bar, 200 μm.
(B) t-SNE plot of all 1,023 neurons obtained from baseline (day 0), 1, and 7 days following sensory manipulation using the 286 differentially expressed genes identified between L6CThN subtypes at day 0. Day 0 neurons are colored by transcriptional subtype; day 1 and day 7 neurons are colored light gray. t-SNE positions were fit to a Gaussian mixture model (black lines) to classify day 1 and 7 neurons as subtype 1 or 2.
(C) t-SNE plot colored by transcriptional subtype as assigned in (B). Ten of 340 day 0 neurons (2.9%; green) were assigned to a different subtype than in Figure 2A.
(D) t-SNE plot colored by projection label.
(E) k-Means clustering analysis of mean-centered gene expression, aggregated by day and transcriptional subtype (subtype 1, gold; subtype 2, blue) for genes with significant differential expression after sensory manipulation. Semi-transparent lines represent individual genes; bold lines represent cluster centroids.
To assign each L6CThN to its transcriptional subtype, we performed a PCA using the 286 genes differentially expressed between L6CThN subtypes under baseline conditions and visualized the 1,023 L6CThNs using a t-SNE analysis (Figure 5B). Neurons were clustered using a Gaussian mixture model (Fraley and Raftery, 2002; Fraley et al., 2012), which largely recapitulated our original subtype assignments (Figure 5C). The significant axonal projection bias was maintained: subtype 1 neurons were predominantly labeled VPM/POm (70.7% [408 of 585 cells], p < 2.97 × 10−71, hypergeometric test) and subtype 2 VPM only (84.2% [369 of 438 cells], p < 2.97 × 10−71, hypergeometric test), confirming the robustness of our subtype assignment (Figure 5D).
We identified 1,134 genes with significant differential expression across the three time points, independent of baseline differences in expression between the two transcriptional subtypes (Figure 5E; Table S5; q ≤ 0.001, Monocle2 test). A k-means clustering analysis of mean expression profiles identified 16 clusters of genes with different temporal expression following deprivation but failed to identify any clusters with significant subtype-specific effects. However, transcriptional changes induced by altered sensory input are unlikely to be synchronous across all neurons collected at each time point. To describe the cellular responses without the confounding effects of neurons in diverse states intermixed at each time point, we established a pseudotemporal ordering for the 1,023 L6CThNs derived from the 1,134 genes with significant differential expression across time points (Figure 6A). Briefly, using the Monocle2 DDRTree algorithm, cells were arranged in an embedded graph representation in a reduced dimensional space such that cells with similar expression profiles across the 1,134 target genes were positioned next to each other, and a traversal through the graph revealed the sequence of transcriptional changes to altered sensory input. As expected, the distribution of L6CThNs along pseudotime generally followed the temporal order of collection, although neurons from each time point were found throughout pseudotime, confirming that the transcriptional response to sensory manipulation is asynchronous across the population (Figures 6A and 6B). L6CThNs, whether classified transcriptionally (Figure S7F) or by retrograde label (Figure S7G), were distributed throughout the pseudotemporal reconstruction. We observed no technical parameterization that uniformly biased all members of a batch or cell type to one lineage, indicating that these data represent biological divergence in response to sensory manipulation (Figures S7F–S7H). Interestingly, we observed no bias in the distribution of L6CThNs across pseudotime when the neurons were grouped by hemisphere ipsilateral or contralateral to whisker removal, suggesting that longer term transcriptional responses in L6CThNs from both hemispheres are similar (Figure S7E).
Figure 6. Pseudotemporal Reconstruction of Transcriptional Responses to Sensory Manipulation in L6CThNs.
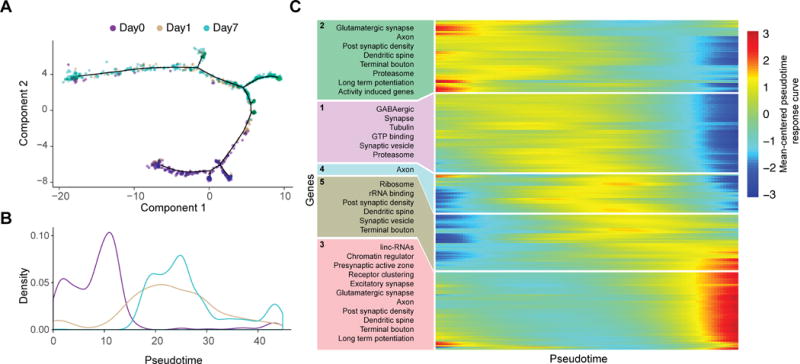
(A) Discriminative dimensionality reduction projection of 1,023 L6CThNs using genes identified as significantly differentially expressed after sensory manipulation. Neurons are colored by day following manipulation.
(B) Density distribution of L6CThNs across pseudotime, grouped by day following manipulation.
(C) Heatmap of normalized response curves for the 1,507 genes with significant differential expression across pseudotime and significantly enriched gene sets identified for each cluster (p < 1.0 × 10−2, hypergeometric test).
We identified 1,507 genes that were differentially expressed across pseudotime at a higher stringency than used in our aggregate analysis (Table S6; q ≤ 0.0000001, Monocle2), indicating that pseudotemporal ordering identified a significantly greater fraction of the transcriptome as modulated by sensory manipulation. To identify cellular processes modulated along pseudotime, the normalized response curves of the differentially expressed genes were clustered and queried for enrichment of annotated gene sets from public databases and the curated list of activity-induced genes (Figure 6C). The cluster with the earliest changes in expression (cluster 2) corresponded to significant downregulation of the activity-associated gene list (p < 5.65 × 10−7, hypergeometric test). In contrast, the cluster representing the late response (cluster 3) corresponded to upregulation of genes associated with chromatin remodeling and lncRNAs, suggesting a slower epigenetic response to sensory manipulation. Interestingly, genes associated with long-term potentiation (LTP) from cluster 2 (Calm1, Calm2, Gria1, Gria2, Plcb1, Plcb4, Ppp1ca, and Ppp3r1) were downregulated early in the response, whereas LTP-associated genes in cluster 3 (Crebbp, Adcy1, Grin1, Prkcb, Ppp3ca, and Ppp3cb) were upregulated toward the end of pseudotime, suggesting that non-overlapping subsets of genes in this gene set are regulated at distinct phases of the L6CThN response. We also identified 75 transcription factors (TFs) with significant differential regulation including activity-associated TFs (i.e., Arc, Fos, Ier2, Junb, Mef2c, and Nr4a1; Flavell and Greenberg, 2008; Lyons and West, 2011), which were expressed early and downregulated over pseudotime. Several TFs involved in neural development (i.e., Neurod6, Fezf2, Mef2c, and Foxp2; Molyneaux et al., 2007) were transiently expressed, suggesting a regulatory relationship between activity-dependent plasticity and neural development.
Activity-Induced Changes in Gene Expression Enhance the Distinction between Subtypes
Because a transcriptional signature of activity significantly contributes to L6CThN subtype identity under baseline conditions, and altered sensory input results in dramatic transcriptional responses in L6CThNs, the response to sensory manipulation may alter the transcriptional relationship among neurons of the same subtype as well as the distinction between the two subtypes. To test these hypotheses, we assessed the distribution of pairwise Euclidean distances of the variance stabilized gene expression estimates across all expressed genes for L6CThNs within each subtype for each day following sensory manipulation. In both subtypes, the response to altered sensory input resulted in an increased mean distance among cells across days (Figure 7A; p < 2.2 × 10−16, Welch’s two-sample t test), and a significant increase in the variance of the intra-subtype distances across days (p < 3.59 × 10−5, F test) for all adjacent time points except for subtype 2 day 1 versus day 7, indicating that the cell-to-cell variation within both L6CThN subtypes increases in response to altered sensory input. Second, we found that the mean inter-subtype pairwise Euclidean distance between subtypes 1 and 2 significantly increased from day 0 to day 1 as well as from day 1 to day 7 (Figure 7B; p < 2.2 × 10−16, Welch’s two-sample t test) as did the variance of inter-subtype distances (p < 4.67 × 10−8, F test). These results demonstrate that modulation of neuronal activity increases both the transcriptional heterogeneity within each L6CThN subtype and the relative transcriptional differences between L6CThN subtypes, confirming a dependent relationship between activity state and transcriptional identity.
Figure 7. Sensory Manipulation Induces Distinct Cellular Responses in L6CThNs Biased with Respect to Transcriptional Subtype.
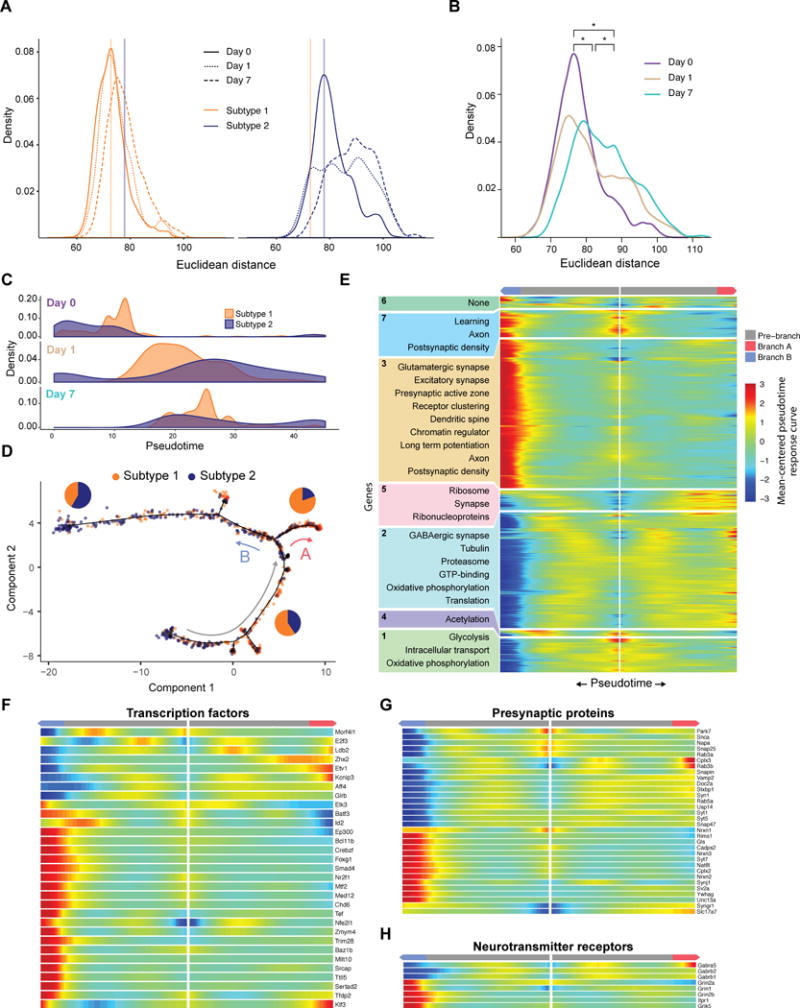
(A) Distribution of the pairwise Euclidean distances within each subtype (subtype 1: left, gold; subtype 2: right, blue), using variance-stabilized expression estimates for all expressed genes.
(B) Distribution of pairwise inter-subtype Euclidean distances between transcriptionally defined L6CThN subtypes across all expressed genes plotted for each day following sensory manipulation. The significant divergence between subtypes across time points is indicated by a positive shift in the distances after manipulation.
(C) Density distributions of L6CThNs at each day plotted across pseudotime for the two L6CThN subtypes.
(D) Discriminative dimensionality reduction projection of 1,023 L6CThNs shown in Figure 6A, colored by transcriptional subtype. Red and blue arrows indicate the major cellular response branchpoint following sensory manipulation. Gray arrow indicates the direction of response progression in the root state. Pie charts depict the proportion of each subtype for each branch.
(E) BEAM analyses of gene sets with significant differential expression dependent on either major branchpoint.
(F–H) BEAM heatmap for branch-dependent transcription factors not detected in the aggregate pseudotime response (F), presynaptic proteins (G), and ligand-gated neurotransmitter receptors (H).
Subtype-Biased Responses Contribute to Transcriptional Heterogeneity and Subtype Identity
These results raise the possibility that subtype-specific responses to sensory manipulation drive the increase in transcriptional heterogeneity and enhance the distinction between L6CThN subtypes. We found that the two subtypes were differentially distributed along pseudotime at each time point assessed (Figure 7C). Pseudotemporal ordering identified a major branchpoint that exhibited significant biases for L6CThN subtype (branch A and branch B; Figure 7D). Although neither subtype 1 nor subtype 2 was significantly enriched in the root state (gray arrow; subtype 1, p < 0.0715; subtype 2, p < 0.91; hypergeometric test), indicating that they share a similar early response, branch A and branch B exhibited significant subtype bias: branch A (red arrow) was biased for subtype 1 (VPM/ POm; 80.4% [148 of 184 neurons], p < 7.03 × 10−14, hypergeometric test) and branch B (blue arrow) for subtype 2 (VPM only; 58.4% [202 of 346 neurons], p < 2.04 × 10−13, hypergeometric test). A subsequent branchpoint also showed subtype-specific biases (Figure S7F). Biases along branches were similar when L6CThNs were classified by retrograde label (Figure S7G). These results indicate that although either cell type may engage the processes represented by each branch, the cellular decisions to engage a particular response are biased with respect to L6CThN subtypes.
Using the Monocle2 branch expression analysis modeling (BEAM) test, we identified 1,392 genes with significant branch-dependent differential expression (Table S7; q ≤ 0.0001, Monocle2 BEAM test); 926 of these overlapped with the 1,507 genes with pseudotime-dependent expression, suggesting that discrete cellular responses independently contribute to the aggregate transcriptional response to altered sensory manipulation. The branch-dependent genes were organized into seven clusters using hierarchical clustering of the Monocle2 branched model fits (Figure 7E). Hypergeometric testing showed that neurons that progress along branch A (VPM/POm enriched) were enriched for genes associated with the proteasome complex, a process involved in synaptic remodeling (Ehlers, 2003), while branch B cells (VPM-only enriched) were enriched for genes related to the postsynaptic density and LTP. Seventy-nine TFs were expressed in a branch-dependent manner, including 31 TFs not identified in our initial analysis of the aggregate response (Figure 7F), suggesting that much of the aggregate L6CThN response induced by altered sensory input is confounded across these two alternative cellular responses. The remaining 27 pseudotime-dependent TFs may regulate a uniform response independent of these two subtype-biased responses. Thirty-two presynaptic genes were regulated in a branch-specific manner (Figure 7G). Neurons committing to a branch A response enhanced expression of GABA receptor subunits, while neurons along branch B induced ionotropic glutamate receptors (Figure 7H). These subtype biases in transcriptional responses induced by sensory manipulation likely underlie the overall effect we observed on L6CThN identity, enhancing the distinctions between subtypes 1 and 2 and significantly increasing both the inter- and intra-subtype heterogeneity of L6CThNs.
DISCUSSION
Gene expression differences among L6CThNs showed that variations in axonal projection pattern, laminar position, and neuronal activity state all significantly contribute to transcriptional identity. Manipulating the activity states of L6CThNs further showed that each subtype was biased for particular responses to sensory manipulation. These subtype-biased transcriptional responses not only increased transcriptional heterogeneity within each subtype but also enhanced the transcriptional differences between the two subtypes. Together, these data identify the most significant influences on the transcriptional identity of individual cortical projection neurons, and show how cellular responses affect the population-level variation and classification of neuronal subtypes.
Although projection target and transcriptional identity may be decoupled for a minority of neurons, the incomplete segregation of retrograde label across subtypes likely represents mislabeling of a subset of L6CThNs because of both the proximity of VPM and POm and the difficulty of retrogradely labeling all neurons projecting to a target. These data reinforce the importance of measuring response variables at single-cell resolution, as they may not be uniform in retrogradely or genetically labeled populations. Profiling a greater number of neurons or selecting the profiled neurons differently may reveal additional subtypes of L6CThNs (Bourassa and Deschênes, 1995; Bourassa et al., 1995; Shima et al., 2016). Alternatively, such studies may reveal that subtle variations in axonal projection pattern seen in some anatomical studies are not apparent in the neurons’ expression profiles.
Interestingly, the gene expression pooled across the two BC L6CThN subtypes identified here is consistent with previously observed aggregate gene expression signatures of Ntsr1-Cre-expressing neurons (Figures S3B and S3C; Shima et al., 2016). The two subtypes also share some transcriptional similarity with two recently defined subtypes in primary visual cortex (Figure S3A; Tasic et al., 2016). These comparisons suggest a conserved relationship between transcriptional identity and axonal projection target bias across cortical regions. Future studies are needed to identify sources of cellular variation across different cortical areas. Although recent studies have further clarified roles for L6CThNs as a whole (Guo et al., 2017; Hasse and Briggs, 2017; Mease et al., 2014; Olsen et al., 2012; Wang et al., 2016), the distinct contributions of L6CThN subtypes to sensory processing remain unresolved.
Pseudotemporal ordering of states induced in L6CThN transcriptomes by altered sensory input revealed that L6CThNs engage at least two molecularly distinct responses in a subtype-biased, but not subtype-specific, manner. Although the distinct transcriptional responses were dominated by neurons collected at days 1 and 7, day 0 neurons were found throughout pseudotime, suggesting that individual cortical neurons engage these plasticity responses in the baseline state. Previous studies showed that averaged responses induced by neural activity measured over hours compared across brain regions or between inhibitory and excitatory neurons exhibit common early transcriptional responses leading to cell-type specific late responses (Spiegel et al., 2014; Whitney et al., 2014). Our single-cell RNA sequencing data reveal that the decision to engage a particular response to experience-dependent plasticity on longer timescales is not intrinsically linked to subtype identity.
We demonstrate a non-dissociable relationship between neuronal identity and neuronal activity as the differential response to sensory manipulation resulted in a significant enhancement of the distinction between the subtypes. Because single L6CThNs have the potential to engage either transcriptional response regardless of subtype, our data suggest that extrinsic factors, such as distinct activation patterns generated by differences in each subtype’s synaptic inputs, induce neurons from a given subtype preferentially toward a similar response. Furthermore, expression of genes that strongly contributed to subtype identity, such as Lamp5, was altered in response to sensory manipulation. These data indicate that factors that change cell state such as plasticity or injury may affect our ability to accurately define stable subtypes.
Our results indicate not only that the gene expression profiles of cortical neurons reflect specific functional features of these cells but that cell-to-cell variation across individual neurons itself is a principal feature of subtype identity (Dueck et al., 2016). Subtype 2 L6CThNs were more transcriptionally heterogeneous than subtype 1 neurons under steady-state conditions in part because of baseline differences in gene expression associated with neural activity. In addition, the intra-cell type variation across subtype 2 at day 1 and day 7 was greater than the inter-cell type variation at the same time points, suggesting that changes in cell-to-cell variation, rather than subtype-specific differences, dominate the transcriptional responses to experience dependent plasticity. Together, these data indicate that the contribution of neuronal activity to gene expression differs across neuronal subtypes and that the transcriptional variation due to differences in neuronal activity state plays a central role in defining the identity of cortical projection neurons.
EXPERIMENTAL PROCEDURES
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Mice
All procedures were approved by the Johns Hopkins Animal Care and Use Committee. Mice used for RNA-seq ranged from postnatal day 23 (P23) to P28; mice used for immunohistochemistry and in situ hybridization ranged from P23 to P32. Both males and females were used, and the gender for each replicate is reported in Supplemental Experimental Procedures.
Statistical Methods
Statistical testing (likelihood ratio test) and curve fitting (LOESS) was performed in R/Bioconductor for comparison of the distributions of single mRNA molecules (RNAscope; ACDbio). Log2 expression estimates (with a pseudocount of 1) of high-variance genes were used as input for PCA analysis and t-SNE clustering of individual cells (Krijthe, 2015; Macosko et al., 2015).
After cluster assignment, differential expression testing was performed using the Monocle2 VGAM model comparison test (Trapnell et al., 2014; 0.1% FDR, Monocle2 test, Benjamini-Hochberg corrected). Gene-centric PCA was performed on a mean-centered matrix of variance-stabilized expression estimates for high-variance genes across all cells, and the resulting rotations were used to project all expressed genes into the same PCA space to identify their weights. These weights were used to rank-order all expressed genes for input into a pre-ranked GSEA (Subramanian et al., 2005). Gene sets for the GSEA were derived from the Monocle2 differential gene tests described above or a curated list of neuronal activity genes (Cho et al., 2016; Kim et al., 2010; Lacar et al., 2016; Mardinly et al., 2016) using an adjusted p value cutoff of <0.01.
To identify modules of correlated gene expression, we used the Weighted Gene Correlation Network Analysis (WGCNA) package (Langfelder and Horvath, 2008). Module eigenvalues were correlated with cellular traits using the Pearson product moment correlation test. Module gene membership was determined in a similar manner. To test the effect of each module on the segregation of L6CThN cell types, each module eigenvalue was separately regressed out of the expression matrix using limma, and subsequent values were used as input for a t-SNE using identical parameters to the original assay.
Pseudotemporal ordering was performed using the prescribed Monocle2 workflow on all 1,023 L6CThNs. To reconstruct a trajectory that reflected cellular progression in response to altered sensory input, we performed a differential test to identify genes whose expression changed as a function of collection day, independent of baseline differences between cell types. These 1,134 genes were used as a filtering set for the DDRTree dimensionality reduction. All significant gene lists were tested for gene set enrichment using the hypergeometric test.
Supplementary Material
Acknowledgments
We thank Hao Zhang and the Johns Hopkins Bloomberg School of Public Health Flow Cytometry and Cell Sorting Core Facility. Sequencing service was provided by the Johns Hopkins Genetic Resources Core Facility. We thank Dr. Joshua R. Sanes, Harvard University, for the anti-LacZ antibody and Dr. Jesse Gray, Harvard Medical School, for the curated list of neuronal activity-induced genes. This work was supported by a Klingenstein-Simons Fellowship (to S.P.B.), a Johns Hopkins Science of Learning grant (to S.P.B. and L.A.G.), a National Science Foundation grant (IOS-1656592 to S.P.B. and L.A.G.), an NIH training grant (T32 GM07814 to J.D.R.), a Boerhinger-Ingelheim Fonds Fellowship (to M.C.), and a National Institute of Neurological Disorders and Stroke (NINDS) grant (NS050274).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, seven figures, eight tables, and four data files and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.12.046.
AUTHOR CONTRIBUTIONS
M.C., S.P.B., and L.A.G. conceived the experiments. M.C., J.D.R., and G.H.C. performed all experiments. M.C., S.P.B., and L.A.G. analyzed the data and wrote the paper with input from all authors.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
The accession number for the sequencing data reported in this paper is GEO: GSE107632. Source code and software tools are available upon request.
References
- Bortone DS, Olsen SR, Scanziani M. Translaminar inhibitory cells recruited by layer 6 corticothalamic neurons suppress visual cortex. Neuron. 2014;82:474–485. doi: 10.1016/j.neuron.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourassa J, Deschênes M. Corticothalamic projections from the primary visual cortex in rats: a single fiber study using biocytin as an anterograde tracer. Neuroscience. 1995;66:253–263. doi: 10.1016/0306-4522(95)00009-8. [DOI] [PubMed] [Google Scholar]
- Bourassa J, Pinault D, Deschênes M. Corticothalamic projections from the cortical barrel field to the somatosensory thalamus in rats: a single-fibre study using biocytin as an anterograde tracer. Eur J Neurosci. 1995;7:19–30. doi: 10.1111/j.1460-9568.1995.tb01016.x. [DOI] [PubMed] [Google Scholar]
- Brennecke P, Anders S, Kim JK, Ko1odziejczyk AA, Zhang X, Proserpio V, Baying B, Benes V, Teichmann SA, Marioni JC, Heisler MG. Accounting for technical noise in single-cell RNA-seq experiments. Nat Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- Briggs F, Kiley CW, Callaway EM, Usrey WM. Morphological substrates for parallel streams of corticogeniculate feedback originating in both V1 and V2 of the macaque monkey. Neuron. 2016;90:388–399. doi: 10.1016/j.neuron.2016.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Huang BS, Gray JM. RNA sequencing from neural ensembles activated during fear conditioning in the mouse temporal association cortex. Sci Rep. 2016;6:31753. doi: 10.1038/srep31753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung NC, Storey JD. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics. 2015;31:545–554. doi: 10.1093/bioinformatics/btu674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Custo Greig LF, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck H, Eberwine J, Kim J. Variation is function: are single cell differences functionally important? Testing the hypothesis that single cell variation is required for aggregate function BioEssays. 2016;38:172–180. doi: 10.1002/bies.201500124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox K. Barrel Cortex. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- Fraley C, Raftery AE. Model-based clustering, discriminant analysis and density estimation. J Am Stat Assoc. 2002;97:611–631. [Google Scholar]
- Fraley C, Raftery AE, Murphy TB, Scrucca L. mclust Version 4 for R: Normal mixture modeling for model-based clustering, classification, and density estimation. Department of Statistics, University of Washington; 2012. (Technical Report No. 597). [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, Heintz N, Gerfen CR. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Clause AR, Barth-Maron A, Polley DB. A corticothalamic circuit for dynamic switching between feature detection and discrimination. Neuron. 2017;95:180–194.e5. doi: 10.1016/j.neuron.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasse JM, Briggs F. Corticogeniculate feedback sharpens the temporal precision and spatial resolution of visual signals in the ferret. Proc Natl Acad Sci U S A. 2017;114:E6222–E6230. doi: 10.1073/pnas.1704524114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Daza RA, Rubenstein JL, Stunnenberg H, Olavarria JF, Englund C. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–151. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- Hicks S, Teng M, Irizarry RA. On the widespread and critical impact of systematic bias and batch effects in single-cell RNA-seq data. bioRxiv. 2015 https://doi.org/10.1101/025528.
- Joo KM, Chung YH, Kim MK, Nam RH, Lee BL, Lee KH, Cha CI. Distribution of vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide receptors (VPAC1, VPAC2, and PAC1 receptor) in the rat brain. J Comp Neurol. 2004;476:388–413. doi: 10.1002/cne.20231. [DOI] [PubMed] [Google Scholar]
- Junker JP, van Oudenaarden A. Every cell is special: genome-wide studies add a new dimension to single-cell biology. Cell. 2014;157:8–11. doi: 10.1016/j.cell.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Katz LC. Local circuitry of identified projection neurons in cat visual cortex brain slices. J Neurosci. 1987;7:1223–1249. doi: 10.1523/JNEUROSCI.07-04-01223.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killackey HP, Sherman SM. Corticothalamic projections from the rat primary somatosensory cortex. J Neurosci. 2003;23:7381–7384. doi: 10.1523/JNEUROSCI.23-19-07381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Matney CJ, Blankenship A, Hestrin S, Brown SP. Layer 6 corticothalamic neurons activate a cortical output layer, layer 5a. J Neurosci. 2014;34:9656–9664. doi: 10.1523/JNEUROSCI.1325-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M. SC3: consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijthe J. Rtsne: T-distributed stochastic neighbor embedding using Barnes-Hus implementation. 2015 R package version 0.10. https://CRAN.R-project.org/package=Rtsne.
- Kwegyir-Afful EE, Simons DJ. Subthreshold receptive field properties distinguish different classes of corticothalamic neurons in the somatosensory system. J Neurosci. 2009;29:964–972. doi: 10.1523/JNEUROSCI.3924-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacar B, Linker SB, Jaeger BN, Krishnaswami S, Barron J, Kelder M, Parylak S, Paquola A, Venepally P, Novotny M, et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun. 2016;7:11022. doi: 10.1038/ncomms11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung HL, Chen S, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Lin P, Troup M, Ho JW. CIDR: ultrafast and accurate clustering through imputation for single-cell RNA-seq data. Genome Biol. 2017;18:59. doi: 10.1186/s13059-017-1188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Res. 2016;5:182. doi: 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons MR, West AE. Mechanisms of specificity in neuronal activity-regulated gene transcription. Prog Neurobiol. 2011;94:259–295. doi: 10.1016/j.pneurobio.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardinly AR, Spiegel I, Patrizi A, Centofante E, Bazinet JE, Tzeng CP, Mandel-Brehm C, Harmin DA, Adesnik H, Fagiolini M, Greenberg ME. Sensory experience regulates cortical inhibition by inducing IGF1 in VIP neurons. Nature. 2016;531:371–375. doi: 10.1038/nature17187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mease RA, Krieger P, Groh A. Cortical control of adaptation and sensory relay mode in the thalamus. Proc Natl Acad Sci USA. 2014;111:6798–6803. doi: 10.1073/pnas.1318665111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Goff LA, Brettler AC, Chen HH, Hrvatin S, Rinn JL, Arlotta P. DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron. 2015;85:275–288. doi: 10.1016/j.neuron.2014.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen SR, Bortone DS, Adesnik H, Scanziani M. Gain control by layer six in cortical circuits of vision. Nature. 2012;483:47–52. doi: 10.1038/nature10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol. 2014;32:1053–1058. doi: 10.1038/nbt.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez-Gomez DB, Hacisuleyman E, Li E, Spence M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima Y, Sugino K, Hempel CM, Shima M, Taneja P, Bullis JB, Mehta S, Lois C, Nelson SB. A Mammalian enhancer trap resource for discovering and manipulating neuronal cell types. eLife. 2016;5:e13503. doi: 10.7554/eLife.13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen SA, Bernard A, Menon V, Royall JJ, Glattfelder KJ, Desta T, Hirokawa K, Mortrud M, Miller JA, Zeng H, et al. Correlated gene expression and target specificity demonstrate excitatory projection neuron diversity. Cereb Cortex. 2015;25:433–449. doi: 10.1093/cercor/bht243. [DOI] [PubMed] [Google Scholar]
- Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ, Nelson SB. Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat Neurosci. 2006;9:99–107. doi: 10.1038/nn1618. [DOI] [PubMed] [Google Scholar]
- Sun QQ, Prince DA, Huguenard JR. Vasoactive intestinal polypeptide and pituitary adenylate cyclase-activating polypeptide activate hyperpolarization-activated cationic current and depolarize thalamocortical neurons in vitro. J Neurosci. 2003;23:2751–2758. doi: 10.1523/JNEUROSCI.23-07-02751.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Menon V, Nguyen TN, Kim TK, Jarsky T, Yao Z, Levi B, Gray LT, Sorensen SA, Dolbeare T, et al. Adult mouse cortical cell taxonomy revealed by single cell transcriptomics. Nat Neurosci. 2016;19:335–346. doi: 10.1038/nn.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Regev A, Yosef N. Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol. 2016;34:1145–1160. doi: 10.1038/nbt.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace H, Fox K. The effect of vibrissa deprivation pattern on the form of plasticity induced in rat barrel cortex. Somatosens Mot Res. 1999;16:122–138. doi: 10.1080/08990229970564. [DOI] [PubMed] [Google Scholar]
- Wang W, Andolina IM, Lu Y, Jones HE, Sillito AM. Focal gain control of thalamic visual receptive fields by layer 6 corticothalamic feedback. Cereb Cortex, Published online December. 2016;17:2017. doi: 10.1093/cercor/bhw376. https://doi.org/10.1093/cercor/bhw376. [DOI] [PubMed] [Google Scholar]
- West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb Perspect Biol. 2011;3:3. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney O, Pfenning AR, Howard JT, Blatti CA, Liu F, Ward JM, Wang R, Audet JN, Kellis M, Mukherjee S, et al. Core and region-enriched networks of behaviorally regulated genes and the singing genome. Science. 2014;346:1256780. doi: 10.1126/science.1256780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zeng H, Sanes JR. Neuronal cell-type classification: challenges, opportunities and the path forward. Nat Rev Neurosci. 2017;18:530–546. doi: 10.1038/nrn.2017.85. [DOI] [PubMed] [Google Scholar]
- Zhang ZW, Deschênes M. Intracortical axonal projections of lamina VI cells of the primary somatosensory cortex in the rat: a single-cell labeling study. J Neurosci. 1997;17:6365–6379. doi: 10.1523/JNEUROSCI.17-16-06365.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


