Abstract
The present study aimed to identify whether microRNA (miRNA/miR)-34a regulates the proliferation and apoptosis of gastric cancer cells by targeting silent information regulator 1 (SIRT1). The expression of miR-34a and SIRT1 and cell viability was investigated in gastric cancer cells. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was applied to determine miR-34a expression in gastric adenocarcinoma, normal pericarcinomatous tissues, human normal gastric mucosa epithelial cell line GES and various gastric cancer cell strains. A bioinformatics method was then used to predict the target gene of miR-34a. A human miR-34a over expression lentiviral vector system was constructed and then used for transfection of the gastric cancer cell line SCG-7901 to determine the expression of SIRT1 mRNA and SIRT1 protein using RT-qPCR and western blot analysis. The MTT method and flow cytometry was used to measure cell proliferation and apoptosis. The relative expression of miR-34a in gastric cancer tissues was significantly decreased compared with that in normal tissues (P<0.01). miR-34a expression was also significantly decreased in low differentiated N2, N3 gastric cancer tissues (P<0.01). However, tumor size and filtration degree were not significantly associated with miR-34a expression. The relative expression of miR-34a was decreased in gastric cancer cells, especially in the SGC-7901 cell line (P<0.01) compared with the GES group. The relative expression of SIRT1 protein was decreased in the miR-34a group compared with the negative control (P<0.01). The rate of proliferation was significantly decreased, whereas the rate of apoptosis was significantly increased in the miR-34a group compared with the NC group (P<0.01). Therefore, the present results suggested that miRNA-34a serves a pivotal role in gastric cancer as a cancer suppressor gene by targeting SIRT1 to regulate the proliferation and apoptosis of gastric cancer cells.
Keywords: microRNA, microRNA-34a, silent information regulator 1, gastric cancer, proliferation, apoptosis
Introduction
Gastric cancer is the second leading cause of cancer-associated mortality worldwide due to its high incidence. Various inherited and environmental factors, including genetic characteristics, infectious agents and dietary habits serve an essential role in the development of gastric cancer (1). Furthermore, signaling pathways, including the epidermal growth receptor, vascular endothelial growth factor, phosphoinositide 3-kinase/protein kinase b/mechanistic target of rampamycin (PI3K/AKT/mTOR) and hepatocyte growth factor/hepatocyte growth factor receptor signaling pathways have been demonstrated as being associated with gastric cancer in a molecular classification study (2). Surgical resection and chemo-radiation are the most efficient strategies for treating gastric cancer (3). However, due to the lack of convenient, noninvasive biomarkers for routine population screening, diagnosis of early stage gastric cancer is challenging in the majority of patients (3). In addition, peritoneal dissemination and local metastases occur in the late stages of gastric cancer (3). A previous study suggested that microRNAs (miRNAs or miRs) are important regulators of the oncogenesis pathway and may be used as clinical biomarkers (4). The emergence of miRNAs serving as biomarkers has provided a novel method of diagnosing cancer and determining the prognosis for various types of cancer, including gastric cancer (3).
The mechanism of epigenetic regulation and control does not include sequence alteration of the coding gene and is important in post-translational regulation as an independent process (5). It is important in tumor formation and development, especially with the abnormal expression and dysfunction of miRNA, which is one of the components of the p53 tumor suppressor network (6,7). miR-34a is a member of the miRNA family, which has tumor suppressor properties, promotes apoptosis and cell arrest, and has an aging function (8). miR-34a has gradually drawn considerable attention due to its tumor suppressor effect. It serves an essential role as a tumor suppressor gene in many types of tumors through impacting various potential tumor associated genes, including sirtuin 1, SNAIL and zinc finger E-box-binding homeobox 1 (9,10). The present study aimed to compare the difference in expression of miR-34a between gastric adenocarcinoma and paired paricarcinomatous, various gastric cancer cells and gastric epithelial cells in order to determine the expression of miR-34a in human gastric cancer and whether miR-34a targets its downstream gene to affect the biological function of gastric cancer. A previous study demonstrated that miR-34a targets receptor tyrosine kinases and platelet-derived growth factor receptor to inhibit gastric cancer via the PI3K/Akt pathway (11). Therefore, in the present study sirtuin 1 (SIRT1) was predicted as the potential tumor associated target gene of miR-34a, as bioinformatics technology indicated that it was associated with cell proliferation and apoptosis. SIRT1 is a nicotinamide adenine dinucleotide (NAD)-dependent deacetylase that regulates cellular processes, including energy metabolism, cell-cycle progression, apoptosis, aging and migration (7). Therefore, SIRT1-3′-untranslated region (UTR) recombinant plasmid and luciferase reporter vector were constructed in the present study to verify the prediction and the impact of miR-34a on cell proliferation and apoptosis of gastric cancer cells by targeting SIRT1 was subsequently investigated.
Materials and methods
Gastric cancer and pericarcinomatous tissue specimens
Gastric cancer and pericarcinomatous tissue specimens were obtained from the First Affiliated Hospital of Bengbu Medical College (Bengbu, China). Gastric cancer samples were obtained from surgical cases immediately following diagnosis by gastroscope and pathology biopsy between January 2015 and December 2015. None of the patients were treated via radiotherapy and chemotherapy prior to surgery. Cancer tissues were derived from tumor lesions and pericarcinomatous tissue specimens were taken from normal stomach tissue >5 cm away from the tumor edge. All specimens were stored in liquid nitrogen in eppendorf tubes filled with RNA preserving fluid (RNAStore; cat no. DP408-02; Tiangen Biotech., Co., Ltd., Beijing, China) following sampling. Clinical data were recorded for pathology research and subsequent statistical analysis. A total of 38 patients with confirmed gastric adenocarcinoma following surgical diagnosis were selected for the present study. There were 27 male patients and 11 female patients, and the age range was 32–79 years. Gastric cancer stages were identified according to the 7th edition of the Tumor Node Metastasis Staging System issued by International Union Against Cancer in 2010 (12), which was used for subsequent analysis between clinical pathology parameters and miR-34a expression. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Bengbu Medical College and written informed consent was obtained from all participants.
Cell strains, vectors and reagents
Human normal gastric epithelial cell line GES-1, human gastric cancer strain AGS, SGC-7901, MKN-45, BGC-823 and 293 cells were purchased from Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). Escherichia coli strain DH5α (BeNa Culture Collection) were cultured in LB medium at 37°C for 24 h (BeNa Culture Collection) and stored at −20°C in the laboratory. The TRIzoI RNA extraction kit was purchased from Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA) and the reverse transcription kit M-MLV was obtained from Promega Corporation (Madison, WI, USA). A SYBR premix ex taq kit was purchased from Axygen Scientific, Inc. (Union City, CA, USA), and the upstream and downstream primers of the targeting gene and internal reference genes were purchased from Guangzhou Ribo Bio Co., Ltd. (Guangzhou, China). Over-expression hsa-miR-34a vector, GV272 luciferase report empty vector and GV259, pHelper 1.0, pHelper 2.0 lentiviral empty vector were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). The dual-luciferase reporter assay system and plasmid extraction kit were purchased from Promega Corporation and the X-tremegene HP transfection reagents were purchased from Roche Applied Science (Penzberg, Germany). Lipofectamine 2000 was purchased from Invitrogen; Thermo Fisher Scientific, Inc. XbaI, XhoI and BamHI restriction enzymes and T4DNA ligase were purchased from New England Biolabs, Inc. (Ipswich, MA, USA). The In-Fusion™ PCR Cloning kit was purchased from Clontech Laboratories, Inc. (Mountainview, CA, USA). The agarose gel recovery kit was purchased from Takara Bio, Inc. (Otsu, Japan). Rabbit anti-human SIRT1 antibody was obtained from Cell Signaling Technology, Inc. (Danvers, MA, USA), GAPDH antibody (cat no. AF0006), SIRT1 antibody (cat no. AF1267), horseradish peroxide conjugated goat-anti-rabbit Immunoglobulin g (H+L) secondary antibody (cat no. A0208) and the ECL kit were purchased from Beyotime Institute of Biotechnology (Haimen, China). The MTT kit was purchased from Beijing Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China) and the cell apoptosis kit was purchased from eBioscience (Thermo Fisher Scientific, Inc.).
miR-34a expression in gastric cancer and normal tissue detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
TRIzoI from an RNA extraction kit was used to isolate RNA from the gastric cancer and normal tissues according to the manufacturer's protocol. A Nanodrop 2000 spectrophotometer (Nanodrop; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA) was used to identify the total RNA concentration. The M-MLV kit was used to synthesize cDNA through RT and U6 was adopted as the internal reference according to the manufacturer's protocol. Upstream and downstream primer sequences were as follows: miR-34a, forward 5′-TGGCAGTGTCTTAGCTGGTTGT-3′ and reverse 5′-CATTGGTGTCGTTGTGCTCT-3′; and U6, forward 5′-CTCGCTTCGGCAGCACATATA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′, and 2X SYBR premix ex taq was used for qPCR. qPCR thermocycling conditions were set as 98°C for 5 min, 96°C for 30 sec, 62°C for 30 sec and 73°C for 1.5 min for 35 cycles and 4°C for storage. The experiment was performed in triplicate and PCR quantification was performed using the 2−ΔΔCq method (13).
miR-34a target gene prediction
miR-34a target gene predication was performed using the miRWalk database (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html) and the overlap of the miRanda (http://34.236.212.39/microrna/home.do), TargetScan (http://www.targetscan.org/vert_71/) and PICTAR (http://pictar.mdc-berlin.de/) online analysis results was selected for further analysis.
SIRT1 gene and 3′-UTR luciferase reporter recombinant plasmid construction
The wild type 3′-UTR gene sequence and mutation type SIRT1 gene sequence was downloaded from Genbank (https://www.ncbi.nlm.nih.gov/nuccore/215982795/?Report=genbank) by inputting accession number NM_012238. Double enzyme digestion of the target gene SIRT1 and GV272 luciferease reporter vector was performed using XbaI/XbaI. The exchanging reaction was performed in ddH2O and the In-Fusion exalter enzyme system at 25°C for 30 min and 42°C for 15 min. The GV272 self-ligation empty vector was used as a negative control and the GV272 vector binding to GAPDH was used as a positive control. Following transfection of the vectors into the competent DH5α monoclonal colony, the cells were enzyme digested and purified followed by PCR amplification and 10% agarose gel electrophoresis. The Taq DNA polymerase (cat no. D7205) was purchased from Beyotime Institute of Biotechnology (Haimen, China). Additionally, the primers used in PCR were as follows: miR-34a, forward, 5′CTTGAACTCCTGGGGCCTGAAG3′ and reverse, 5′GCCAAAGAAACACTCACAGCT3′; and SIRT1, forward, 5′TAGCCTTGTCAGATAAGGAAGGA3′ and reverse, 5′ACAGCTTCACAGTCAACTTTGT3′. The amplification conditions consisted of 2 min at 95°C, followed by 40 cycles of 95°C for 15 sec and 60°C for 30 sec and storage at 4°C. The agarose gel electrophoresis was stained using ethidium bromide and visualized under UV light at 254 nm. Fragment sequencing was conducted by Beijing SBS Genetech Co., Ltd (Beijing, China).
Cell co-transfection and luciferase activity verification
Using trypsin, 293 cells growing in log-phase were harvested and 1×105 cells were transfected. Transfection was performed in triplicate in 96-well plates. Plasmids were transfected using the X-tremeGene HP DNA transfection reagent and 100 µl opti-MEM (cat no. 31985-070, Shanghai Bai Gen Biological Technology Co., Ltd., Shanghai, China) at a ratio of 1 µg plasmid to 2 µl X-tremeGene HP reagent. Following incubation at 37°C for 6 h, the medium was replaced with fresh culture medium containing 10% bovine serum. Following incubation for 24 h, the Dual-Luciferase® Reporter assay system (cat no. E1910; ‘Promega Corporation) was used to verify the transfection efficiency according to the manufacturer's protocols. Experimental groups are presented in Table I.
Table I.
Cell groups co-transfected with recombinant vectors.
| Group | Transfection vectors |
|---|---|
| Group 1 | 3′-UTR empty plasmid + miRNA empty plasmid |
| Group 2 | 3′-UTR empty plasmid + miR-34a over-expression plasmid |
| Group 3 | SIRT1-3′-UTR recombinant plasmid + miRNA empty plasmid |
| Group 4 | SIRT1-3′-UTR recombinant plasmid + miR-34a over-expression plasmid |
| Group 5 | SIRT1-3′-UTR mutant plasmid + miRNA empty plasmid |
| Group 6 | SIRT1-3′-UTR mutant plasmid + miR-34a over-expression plasmid |
Negative control group: Group 1. SIRT1, silent information regulator 1; UTR, untranslated regions; miRNA, microRNA.
hsa-miR-34a over-expressing lentiviral vector and cell transfection construction
The hsa-miR-34a over-expression lentiviral vector system was constructed via combination ligation (hsa-miR-34a, 5′-UGGCAGUGUCUUAGCUGGUUGU-3′; negative control, 5′-TTCTCCGAACGTGTCACGTCTC-3′). The pHelper 1.0, pHelper 2.0 lentiviral vectors and has-miR-34a plasmids were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). SGC-7901 cells (5×106) were transfected with lentivirus using X-tremegene HP (Roche Diagnostic, Basel, Switzerland) and incubated in opti-MEM medium at 37°C with 5% CO2 for 6 h. The cells were grouped into three groups: Group A transfected with 10 µl lentiviral stock solution; Group B transfected with 1 µl lentivirus and Group C transfected with 0.1 µl lentivirus. The three concentration of lentivirus were set to avoid the 100% positive and 100% negative of lentiviral transfection. Following transfection and incubation for 3 days, the cells were employed for the subsequent experiment. The CON group was defined as the non-transfected group; the NC group was defined as the negative control group transfected with empty lentiviruses; and the miR-34a group was the group transfected with miR-34a overexpression vectors. TCID50 (Tissue Culture Infective Dose) was used to calculate the lentiviral titer (14).
Determination of the expression of SIRT1 mRNA by RT-qPCR
Total RNA from the cells transfected with miR-34a plasmids was extracted using a TRIzoI RNA extraction kit, and cDNA was synthesized by reverse transcription with GoTaq® DNA Polymerase (cat no. M3005, Promega Corporation, Madison, WI, USA) and amplified by qPCR using the M-MLV RT kit (cat no. 28025-013; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The temperature protocol was 93°C for 12 min for reverse transcription. A probe with FAM fluorophore at the 5′ end and TAMARA fluorophore at the 3′ end from Shanghai Shinegene Molecular Biotechnology (Shanghai, China) was employed for detection. The primer sequences used were as follows: SIRT1, forward 5′-GACTTCAGGTCAAGGGAT-3′ and reverse 5′-CGTGTCTATGTTCTGGGTA-3′; and GAPDH, forward 5′-TGACTTCAACAGCGACACCCA-3′ and reverse 5′-CACCCTGTTGCTGTAGCCAAA-3′. The PCR reaction was set as 96°C for 5 min, 95°C for 30 sec, 66°C for 30 sec and 76°C for 1 min for 32 cycles and 4°C for storage. The expression of SIRT1 mRNA was relatively quantified using the 2−ΔΔCq method (13). This experiment was performed in triplicate and there were three wells per row for each experiment.
SIRT1 protein expression determined by western blot analysis
Cells were lysed using radioimmunoprecipitation buffer (cat no. 211-40; AmyJet Scientific Co., Ltd., Wuhan, China) and total protein concentration was verified using the BCA method. A total of 2 µg per lane protein was used for the SDS-PAGE assay with 10% gel electrophoresis. Proteins were then transferred to PVDF membranes and blocked with 5% milk at room temperature for 1 h. The PVDF membrane was incubated with rabbit anti-human SIRT1 primary antibody (cat no. AF0282; 1:10,000; Beyotime Institute of Biotechnology) for 24 h at 4°C. GAPDH was used as the reference protein stated above. The secondary antibody (1:10,000) was added to the membrane and incubated for 1 h with agitation at room temperature. An ECL kit was used to visualize the blots and the gel image was recorded by a Chemiluminescent imager (Thermo Fisher Scientific, Inc.). Version 1.5.2 ImageJ analytical software (National Institutes of Health, Bethesda, MD, USA) was used for quantification. Each sample was tested three times independently.
Cell proliferation determined using an MTT assay
The inoculated SGC-7901 cells (5×105) were plated on 96-well plates following trypsin digestion and cell counting, and 5 wells in a row were used for each sample. Cells were incubated at 37°C with 5% CO2 for 5 days. MTT stain was performed according to the manufacturer's protocol. Formazan was dissolved by DMSO. Then, a microplate reader was used to measure the OD490 value of solution.
Cell apoptosis profile detection by flow cytometry
SGC-7901 cells (5×105) were digested with trypsin and samples were plated in 96-well plates in triplicate. 5 µl Annexin V-APC (Wuhan Bot Biological Technology Co., Ltd., Wuhan, China) was added to 100 µl cell suspension and incubated at room temperature in the dark for 15 min. The cell apoptosis profile of SGC-7901 was detected by flow cytometry using a flow cytometer (FACScan; BD Biosciences). Version 2.5 WinMDI from Purdue University Cytometry Laboratories (West Lafayette, USA) was employed for data analysis.
Statistical analysis
SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was used for statistical analysis. Data are expressed as the mean ± standard deviation and non-normal distribution data was shown by boxplot. A Student t-test was adopted for paired comparison and one-way ANOVA analysis of variance followed by Tukey's test were performed for comparisons of multiple groups. According to the analysis of variance homogeneity by Bartlett's test, the corresponding t- and P-values were selected on the basis of P<0.05 indicating a statistically significant difference.
Results
Expression of miR-34a in gastric cancer tissues
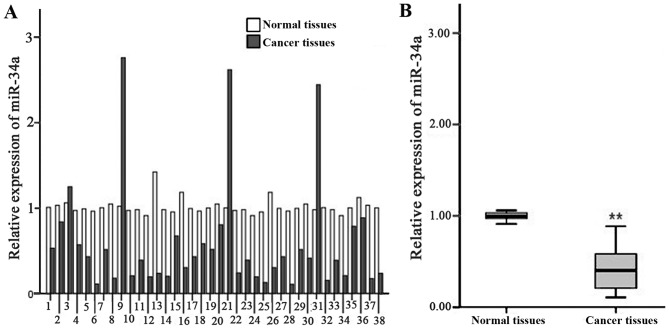
The results of RT-qPCR analysis indicated that there was a significant decrease in the relative expression of miR-34a in gastric cancer tissue compared with that in paired pericarcinomatous tissue (0.59±0.65 vs. 1.01±0.09; t=4.007; P<0.01; Fig. 1). T-test analysis on the pathology parameters and relative expression of miR-34a suggested that the expression of miR-34a was significantly decreased in low differentiated gastric cancer cells N2, N3 compared with that of high differentiated and middle differentiated gastric cells N0, N1 (P<0.01). There was no distinct association between tumor size and infiltration degree with the expression of miR-34a (Table II).
Figure 1.
Relative expression of miR-34a in gastric cancer and pericarcinomatous normal tissues determined using reverse transcription-quantitative polymerase reaction. (A) Relative expression of miR-34a in gastric cancer tissues and normal tissues from 38 cases of gastric cancer. (B) Overall analysis of miR-34a relative expression in normal and cancer tissues. **P<0.01 vs. normal tissues. miR-34a, microRNA-34a.
Table II.
Association between pathological parameters and miR-34a expression level.
| Pathology parameters | Cases (n) | Relative expression of miR-34a in gastric cancer cells | P-value |
|---|---|---|---|
| Tumor size | 0.643 | ||
| ≥5 cm | 11 | 0.67±0.77 | |
| <5 cm | 27 | 0.56±0.61 | |
| Degree of differentiation | 0.004a | ||
| High, middle differentiation | 15 | 1.05±0.84 | |
| Low differentiation | 23 | 0.29±0.14 | |
| Infiltration degree | 0.804 | ||
| T1, T2 | 13 | 0.55±0.58 | |
| T3, T4 | 25 | 0.61±0.69 | |
| Lymphatic metastasis | 0.007a | ||
| N0, N1 | 16 | 0.98±0.85 | |
| N2, N3 | 22 | 0.31±0.18 |
Expression of miR-34a was determined by 2−ΔΔCq relative quantification following RT-qPCR. Statistical analysis was performed using Student's t-test.
P<0.01. miR-34a, microRNA-34a.
Relative expression of miR-34a in human gastric cancer cells and GES
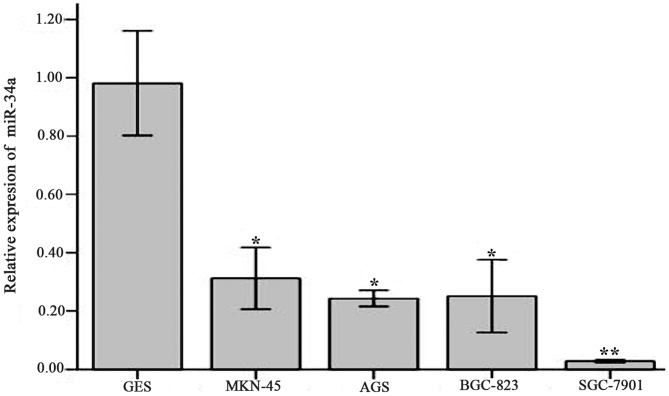
The results of RT-qPCR indicated that the relative expression of miR-34a in human gastric cell strains AGS (0.24±0.01; P<0.05), SGC-7901 (0.03±0.00; P<0.01), MKN-45 (0.31±0.04; P<0.05) and BGC-823 (0.25±0.05; P<0.05) was significantly decreased compared with GES cells (0.98±0.07; Fig. 2). These differences were significant with respective t-values of t=17.522, 22.879, 13.836 and 14.393. The biggest decrease in the relative expression of miR-34a compared with GES cells was in the gastric cancer cell line SGC-7901 (P<0.01). Therefore, this cell line was selected for the subsequent lentivirus infection and cell viability tests.
Figure 2.
Relative expression of miR-34a in various cell strains compared with GES. The relative expression was determined using reverse transcription-quantitative polymerase chain reaction. miR-34a relative expression in cell strains GES, MNK-45, AGS, BGC-823, SGC-7901 was 0.98±0.07, 0.31±0.04, 0.24±0.01, 0.25±0.05 and 0.03±0.00, respectively. Values are expressed as the mean ± standard deviation and experiments were performed in triplicate. *P<0.05, **P<0.01 vs. GES. miR-34a, microRNA-34a; GES, human normal gastric mucosa epithelial cells.
miR-34a target gene prediction
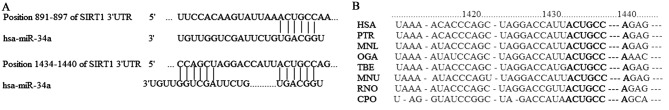
The results of the bioinformatic analysis indicated that there were two miR-34a binding sites in the SIRT1-3′-UTR region from 891–897 bp and 1,434-1,440 bp, which are associated with tumor proliferation and apoptosis. The free energy of binding was −20.1 and −37.5 kcal/mol respectively (data not shown). Analysis of species homology implied that the base sequence of SIRT1-3′-UTR region from 1,434-1,440 bp was highly conserved among different species (Fig. 3). Therefore, SIRT1 was chosen for subsequent experiments in the present study.
Figure 3.
Results of miR-34a targeting gene prediction. (A) Binding sites for miR-34a within the SIRT1-3′-UTR extend from 891–897 and 1434–1440 bp. (B) Seeding sequence of SIRT1-3′-UTR and species homology analysis derived from miRanda, TargetScan and PICTAR databases. miR-34a, microRNA-34a; SIRT1, sirtuin 1; UTR, untranslated region.
Construction of SIRT-1 3′-UTR region luciferase reporter vector
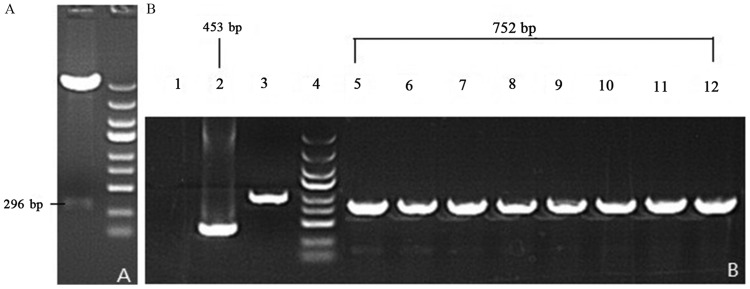
The agarose gel electrophoresis indicated that the base pairs of the recombinant SIRT1-3′-UTR luciferase reporter vector plasmid were 752 bp and the base pairs of converter of negative control were 453 bp following double restriction enzyme digestion (Fig. 4).
Figure 4.
Gel images of PCR products of recombinant vectors. Following PCR, the double enzyme restriction digestion and agarose gel electrophoresis was performed to determine the fragment size. (A) Enzyme-digested SIRT1-3′-UTR fragments. (B) Lane 1, ddH2O; lane 2, negative control (empty vector self-ligation); lane 3, positive control (GAPDH); lane 4, MW scale (molecular weight of marker protein); lanes 5–12, recombinant SIRT1-3′-UTR luciferase reporter vector plasmid. PCR, polymerase chain reaction; miR-34a, microRNA-34a; SIRT1, sirtuin 1; UTR, untranslated region.
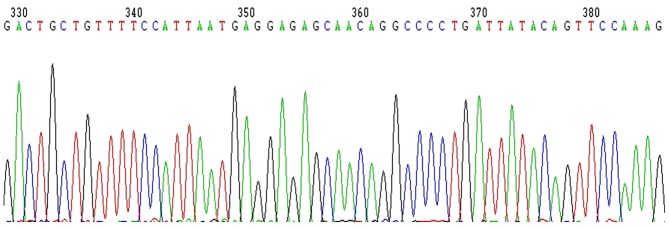
Therefore, 752 bp-453 bp=299 bp and the segment size of SIRT1-3′-UTR is 293 bp following deduction of the 6 bp enzyme digestion sites (Fig. 4). This result was consistent with the GenBank search result without site mutation (Fig. 5).
Figure 5.
Sequencing results of recombinant vectors with sirtuin 1–3′-untranslated region.
Co-transfection efficiency and luciferase activity evaluation
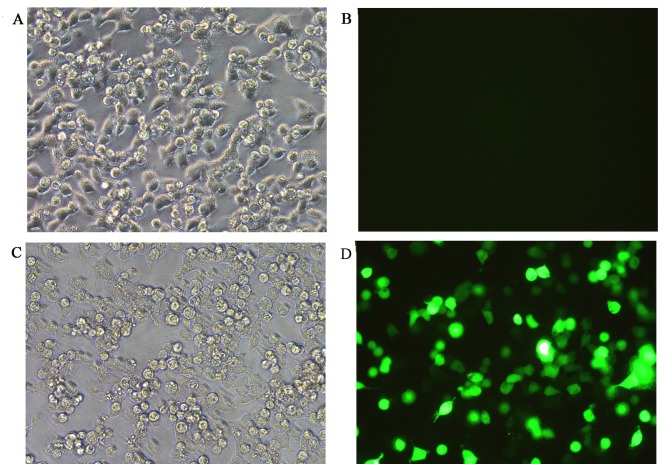
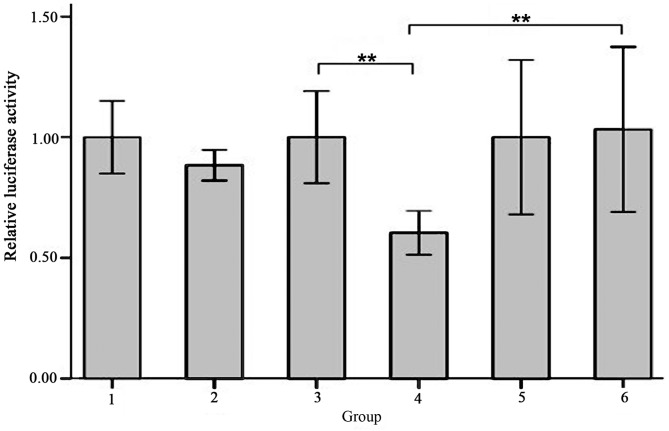
None of the co-transfection plasmids contained green fluorescent protein (GFP) in order to avoid the effects of luciferase activity, whereas the reference group, which used an empty plasmid was marked with GFP. The green fluorescence expression profile of all experiments including the reference group was then evaluated. Successful co-transfection was defined as 80% of GFP expression rate, as indicated by fluorescence microscopy (Fig. 6). The results of the luciferase activity test indicated that there was a significantly decreased fluorescence value for the cells co-transfected with SIRT1-3′-UTR recombinant plasmid and miR-34a over-expression plasmid (Group 4) compared with cells co-transfected with SIRT1-3′-UTR recombinant plasmid and miRNA empty plasmids (Group 3; 0.60±0.04 vs. 1.00±0.08, t=−8.062, P=0.001) and the cells co-transfected with SIRT1-3′-UTR mutant plasmid and miR-34a over-expression plasmid (Group 6; 0.60±0.04 vs. 1.03±0.14, t=−5.211; P=0.006; Fig. 7). The differences between the other groups were not significant (Fig. 7).
Figure 6.
Co-transfection evaluation via fluorescence microscopy. (A) Bright field image of randomly selected experimental group. (B) Green fluorescence image of the randomly selected experimental group. (C) Bright field image of the reference group marked with GFP. (D) Green fluorescence image of the reference group marked with GFP. Magnification, ×200 GFP, green fluorescent protein.
Figure 7.
Relative luciferase activity of each group. Group 1, 3′-UTR empty plasmid + miRNA empty plasmid; group 2, 3′-UTR empty plasmid + miR-34a over-expression plasmid; group 3, SIRT1-3′-UTR recombinant plasmid + miRNA empty plasmid; group 4, SIRT1-3′-UTR recombinant plasmid + miR-34a overexpression plasmid; group 5, SIRT1-3′-UTR mutant plasmid + miRNA empty plasmid; group 6, SIRT1-3′-UTR mutant plasmid + miR-34a over-expression plasmid. Values are expressed as the mean ± standard deviation for three experiments. **P<0.01. miR-34a, microRNA-34a; SIRT1, sirtuin 1; UTR, untranslated region.
Titer measurement of hsa-miR-34a over-expressing lentivirus
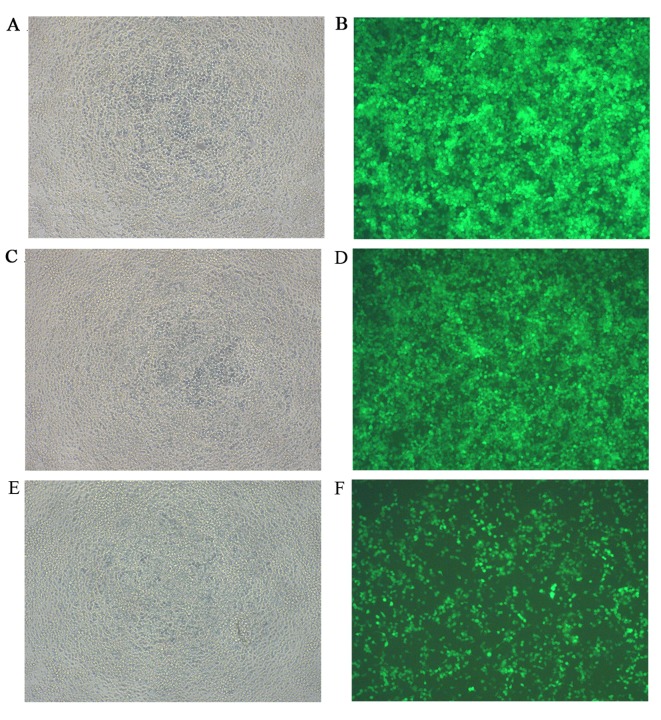
Transfection was performed with 293 cells using miR-34a over-expressing lentiviral vector marked with GFP. The decrease in the number of fluorescent cells indicated an increase in virus dilution fold (Fig. 8). According to the equation virus titer (Tu/ml)=(fluorescent cell number × virus dilution fold)/0.1, the titer of the miR-34a over-expressing lentiviral system was calculated and the result was 4×108 Tu/ml. The recombinant lentiviruses were then transfected into SGC-7901 cells in the experimental group and cultured for 3 days. Subsequent experiments were performed when culture plates were filled with transfected cells.
Figure 8.
Titer measurement of lentivirus expressing hsa-miR-34a. (A) Bright field image of group with 10 µl lentivirus. (B) Green fluorescent image of group with 10 µl lentivirus. (C) Bright field image of group with 1 µl lentivirus. (D) Green fluorescent image of group with 1 µl lentivirus. (E) Bright field image of group with 0.1 µl lentivirus. (F) Green fluorescent image of group with 0.1 µl lentivirus. Magnification, ×100. Virus titer was calculated using the following formula: (Tu/ml)=(fluorescent cell number × virus dilution fold)/0.1. Virus titer of hsa-miR-34a was 4×108 Tu/ml. miR-34a, microRNA-34a.
Effect of miR-34a on SIRT1 mRNA and protein expression
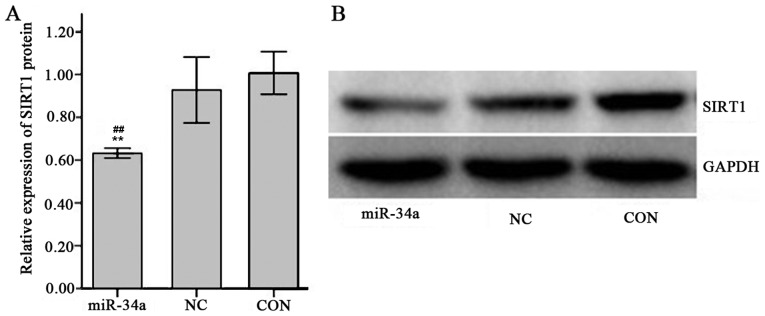
The expression of SIRT1 mRNA in SGC-7901 cells in the miR-34a group was decreased compared with the NC group, however this result was not significant (0.98±0.11 vs. 1.01±0.01; Fig. 9). The expression of SIRT1 protein was significantly decreased in the miR-34a group compared with the NC group (0.63±0.01 vs. 0.93±0.06, t=−8.17, P<0.01; Fig. 10).
Figure 9.
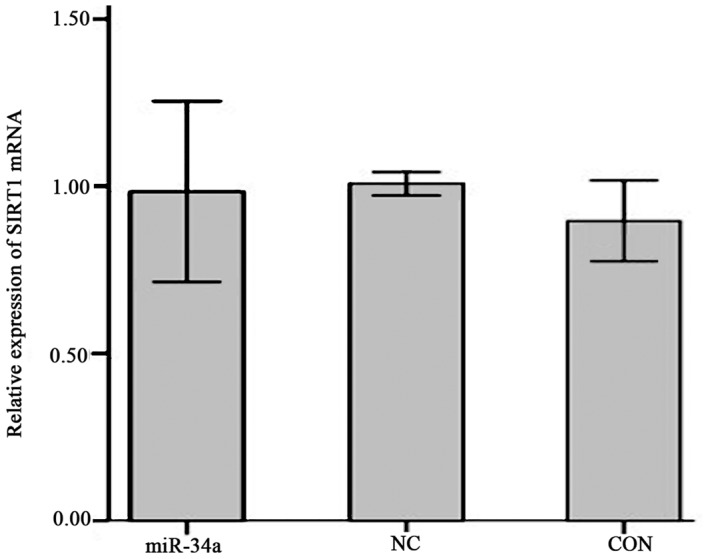
Relative expression of SIRT1 mRNA. The groups were as follows: miR-34a, infected hsa-miR-34a over-expression viral group; CON, uninfected non-viral group; NC, infected negative control viral group. Values are expressed as the mean ± standard deviation. The experiment was performed in triplicate. miR-34a, microRNA-34a; SIRT1, sirtuin 1.
Figure 10.
Relative expression of SIRT1 protein determined by western blot analysis. (A) Quantified western blot results. (B) Images of western blot analysis. The groups were as follows: miR-34a, infected hsa-miR-34a over-expression viral group; CON, uninfected non-viral group; NC, infected negative control viral group. Values are expressed as the mean ± standard deviation for three experiments. **P<0.01 vs. NC group, ##P<0.01 vs. CON group. miR-34a, microRNA-34a; SIRT1, sirtuin 1.
Inhibition of SGC-7901 cell proliferation by miR-34a
The MTT method was used to determine the cell proliferation of all experimental groups over 5 days. The results indicated that SGC-7901 cell proliferation was significantly decreased in the miR-34a group compared with the NC group between days 3 and 5 (day 3, 1.39±0.19 vs. 1.84±0.10, t=4.567; day 4, 1.82±0.16 vs. 2.46±0.11, t=7.265; day 5, 2.37±0.04 vs. 3.14±0.38, t=4.500; all P<0.01; Fig. 11).
Figure 11.
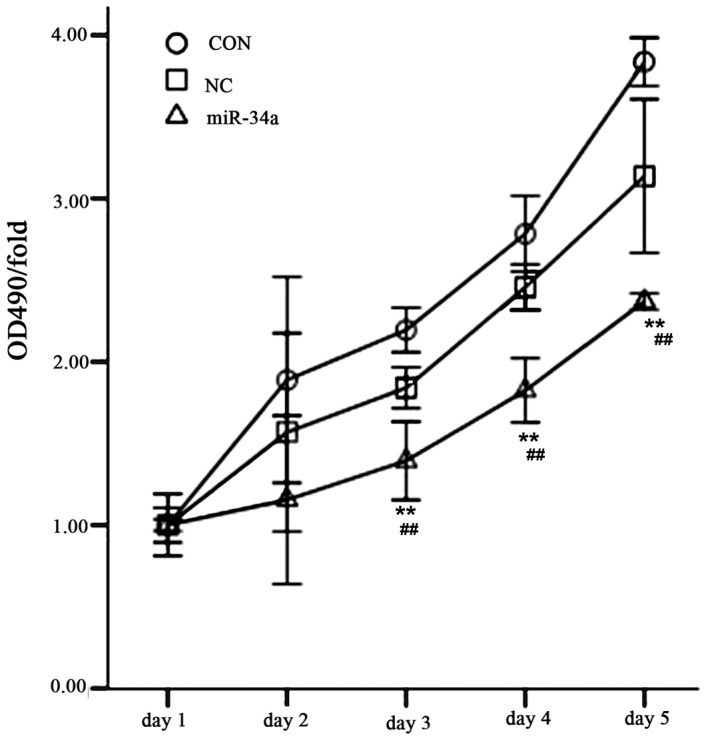
OD490/fold of transfected SGC-7901 cells. The groups were as follows: miR-34a, transfected hsa-miR-34a over-expression viral group; CON, untransfected non-viral group; NC, transfected negative control viral group. Values are expressed as the mean ± standard deviation for three experiments **P<0.01 vs. NC group, ##P<0.01 vs. CON group. OD, optical density; miR-34a, microRNA-34a.
miR-34a promotes SGC-7901 cell apoptosis
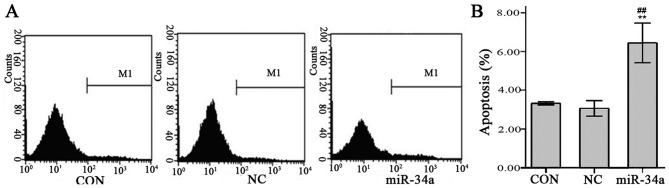
Results of flow cytometry indicated that the SGC-7901 cell apoptosis rate was significantly increased in the miR-34a group compared with the NC group (6.44±0.41 vs. 3.06±0.16, t=−13.226, P<0.01, Fig. 12).
Figure 12.
SGC-7901 cell apoptosis rate in the different groups. At 24 h following transfection, apoptosis was analyzed by flow cytometry. (A) Flow cytometry analysis. (B) Percentage of cell apoptosis in each group. Values are expressed as the mean ± standard deviation for three experiments. **P<0.01 vs. NC group, ##P<0.01 vs. CON group. miR-34a, microRNA-34a; CON, uninfected non-viral group; NC, infected negative control viral group.
Discussion
miR-34a is an endogenous, non-coding regulatory small RNA containing 22 base pairs (15). Its coding gene is located on chromosome 1p36.22 and is associated with regulation of cell growth, proliferation and metabolism. Previous studies have demonstrated that there is abnormal expression of miR-34a due to the lack of 1p36 in certain tumors and metabolic-associated diseases (16,17). Furthermore, abnormal expression of miR-34a caused by an abnormal methylation promoter region and mutation of p53 widely exists in various tumor-associated diseases (18,19). Therefore, miR-34a is studied due to its vital role in tumor development. It has been previously demonstrated that there is a low expression level of miR-34a in gastric cancer cells (20). To further investigate the association between miR-34a and gastric cancer, the results of the present study indicated that the expression of miR-34a was significantly lower in gastric cancer tissue compared with pericarcinomatous tissue and the expression level was associated with tumor differentiation degree and lymphatic metastasis. The significant decrease in the expression of miR-34a in various gastric cancer cell strains implied that there was an association between miR-34a and gastric cancer development. The 3′-UTR region of miR-34a is able to identify and bind to the 3′-UTR of its targeting sequence, which is able to induce the degradation of the target gene and inhibit transcription (15). Therefore, the expression of the downstream protein is inhibited, which leads to various biological effects, including DNA damage repair, cell proliferation and apoptosis (21,22). Bioinformatics was used to overlap the search results from different data banks to ensure that target gene prediction was correct in the present study. Furthermore, the seed sequences were analyzed and screened according to the high tumor correlation biological function, high thermodynamic stability of binding and high conservatism of the predicted target gene. The miR-34a specific targeting gene SIRT1 was subsequently selected as the focus of the present study.
SIRT1 is a member of the Sirtuin protein family, which is associated with cell stress reaction regulation, DNA repair, chromatin recombination, metabolism, aging and apoptosis due to the acetyl enzyme function of co-enzyme NAD+ to SIRT1 (23,24). The function of SIRT1 in cancer is unclear. It has previously been indicated that SIRT1 blocks certain cancer pathways and serves a role as a cancer suppressor (25). However, it has also been demonstrated that SIRT1 undergoes de-acetylation to transcription factors, including the p53 ‘gene guardian’, which weakens its mediation of cell proliferation inhibition, cell cycle arrest and apoptosis process (26). In addition, SIRT1 has an oncogenic function via the direct inhibition of pro-apoptotic proteins, including Bax and caspase (27). Yamakuchi previously demonstrated that human HCT116 cells exhibited p53/miR-34a/SIRT1 positive feedback regulation (20). miR-34a is the downstream activating receptor, which serves an important role in elevating the acetylation level of p53 by inhibiting SIRT1 expression to recover the activity of miR-34a and subsequently promoting apoptosis and the anti-tumor function of p53 (20). However, the de-acetylation function of SIRT1 decreases the activity of p53 due to the interruption of positive feedback loop regulation when the expression of miR-34a is decreased or exogenously inhibited (20). Previous studies have suggested that the association between miR-34a and SIRT1 was the key regulatory mechanism of damaged DNA double-strand break repair (28). It has also been demonstrated that the expression of SIRT1 is increased in gastric cancer (29). In order to determine whether miR-34a targets SIRT1 to regulate and control cell proliferation and apoptosis, a SIRT1-3′-UTR luciferase reporter plasmid was successfully constructed in the present study and it was demonstrated that miR-34a specifically binds to the SIRT1-3′-UTR. The results of the luciferase reporter assay demonstrated that SIRT1 was the target gene of miR-34a. Therefore, an miR-34a over-expression lentiviral vector system was constructed and then transfected into human gastric cancer SGC-7901 cells to determine the regulatory role served by miR-34a on SIRT1 using RT-qPCR. Western blot analysis was then used to determine the expression of SIRT1 protein. The results indicated that miR-34a inhibits SIRT1 protein expression. Cell proliferation was inhibited and apoptosis was promoted in the group with cells transfected with miR-34a, therefore it is suggested that miR-34a serves a role in gastric cancer inhibition via its targeting of SIRT1.
In conclusion, the expression of miR-34a was decreased in human gastric cancer tissues and cells compared with normal tissues. The expression level was associated with gastric cancer differentiation degree and lymphatic metastasis, suggesting that the development of gastric cancer was associated with miR-34a expression disorder and dysfunction. The results also demonstrated that miR-34a specifically binds to the SIRT1-3′-UTR to inhibit SIRT1 protein expression. Therefore, miR-34a serves a role in cell proliferation inhibition and apoptosis promotion of gastric cancer, which suggests that miR-34a is a tumor suppressor. The level of SIRT1 mRNA was not significantly decreased in the miR-34a group and this may be due to the more than 16 miRNAs that regulate SIRT1 expression and activity (30). Therefore, further study is required to investigate the pathway network and concrete downstream regulation of miR-34a. The aim of the present study was to determine the role of miR-34a in gastric cancer and the results indicated its tumor suppression mechanism via the regulation of the expression of SIRT1. Therefore, the present study identified miR-34a as a potential target for the diagnosis and treatment of gastric cancer.
The current first line therapy for gastric cancer is combination chemotherapy, which consists of a multi-agent chemotherapy regimen combining fluoropyrimidines and platinum derivatives (31). In addition, irinotecan, 5-fluorouracil and leucovorin are acceptable alternatives to the platinum-based ECX (epirubicin, cisplatin and capecitabine) regimen (31). Second line therapy includes cytotoxic agents and biological agents such as Ramucirumab, Lapatinib, Gefitinib and Everolimus (32). Gastric cancer treatment is now shifting from disease-specific novel drug development to biomarker-oriented investigations (19). Biomarker-oriented investigations may improve the prognosis of patients with gastric cancer.
Acknowledgements
The present study was supported by grants from the Anhui Natural Science Foundation of China (grant no. KJ2015A177) and The Natural Science Foundation of China (grant nos. 81372899 and 81172087).
References
- 1.Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10:643–655. doi: 10.1038/nrclinonc.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu HH, Lin W, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med. 2014;16:e1. doi: 10.1017/erm.2013.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan H, Lu H, Wang X, Jin H. MicroRNAs as Potential Biomarkers in Cancer: Opportunities and Challenges. Biomed Res Int. 2015;2015:125094. doi: 10.1155/2015/125094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costa FF. Epigenomics in cancer management. Cancer Manag Res. 2010;2:255–265. doi: 10.2147/CMAR.S7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–392. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Yan C, Xin M, Han L, Zhang Y, Sun M. Sirtuin 1 (Sirt1) overexpression in BaF3 cells contributes to cell proliferation promotion, apoptosis resistance and pro-inflammatory cytokine production. Med Sci Monit. 2017;23:1477–1482. doi: 10.12659/MSM.900754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmid G, Notaro S, Reimer D, Abdel-Azim S, Duggan-Peer M, Holly J, Fiegl H, Rössler J, Wiedemair A, Concin N, et al. Expression and promotor hypermethylation of miR-34a in the various histological subtypes of ovarian cancer. BMC Cancer. 2016;16:102. doi: 10.1186/s12885-016-2135-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 10.Li XJ, Ren ZJ, Tang JH. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014;5:e1327. doi: 10.1038/cddis.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng Y, Guo JJ, Liu YM, Wu XL. MicroRNA-34A inhibits the growth, invasion and metastasis of gastric cancer by targeting PDGFR and MET expression. Biosci Rep. 2014;34:e00112. doi: 10.1042/BSR20140020. pii. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Edge SB, Compton CC. The american joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.Pourianfar HR, Javadi A, Grollo L. A colorimetric-based accurate method for the determination of enterovirus 71 titer. Indian J Virol. 2012;23:303–310. doi: 10.1007/s13337-012-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, et al. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet. 2011;7:e1002363. doi: 10.1371/journal.pgen.1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, Campbell J, Bhimavarapu A, Luikenhuis S, de Cabo R, Fuchs C, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji Q, Hao X, Meng Y, Zhang M, Desano J, Fan D, Xu L. Restoration of tumor suppressor miR-34 inhibits human p53-mutant gastric cancer tumorspheres. BMC Cancer. 2008;8:266. doi: 10.1186/1471-2407-8-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Leeuwen IM, Higgins M, Campbell J, McCarthy AR, Sachweh MC, Navarro AM, Laín S. Modulation of p53 C-terminal acetylation by mdm2, p14ARF, and cytoplasmic SirT2. Mol Cancer Ther. 2013;12:471–480. doi: 10.1158/1535-7163.MCT-12-0904. [DOI] [PubMed] [Google Scholar]
- 20.Yamakuchi M, Ferlito M, Lowenstein CJ. miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci USA. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu M, Lu L, Mao B, Lü X, Wu XS, LI L, Liu DP. Mutual inhibition between miR-34a and SIRT1 contributes to regulation of DNA double-strand break repair. Chin Sci Bulletin. 2013;58:979–985. doi: 10.1007/s11434-012-5599-8. [DOI] [Google Scholar]
- 22.Ma W, Xiao GG, Mao J, Lu Y, Song B, Wang L, Fan S, Fan P, Hou Z, Li J, et al. Dysregulation of the miR-34a-SIRT1 axis inhibits breast cancer stemness. Oncotarget. 2015;6:10432–10444. doi: 10.18632/oncotarget.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price NL, Gomes AP, Ling AJY, Duarte FV, Martin-Montalvo A, North BJ, Agarwal B, Ye L, Ramadori G, Teodoro JS, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tulino R, Benjamin AC, Jolinon N, Smith DL, Chini EN, Carnemolla A, Bates GP. SIRT1 activity is linked to its brain region-specific phosphorylation and is impaired in huntington's disease mice. PLoS One. 2016;11:e0145425. doi: 10.1371/journal.pone.0150682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mvunta DH, Miyamoto T, Asaka R, Yamada Y, Ando H, Higuchi S, Ida K, Kashima H, Shiozawa T. SIRT1 regulates the chemoresistance and invasiveness of ovarian carcinoma cells. Transl Oncol. 2017;10:621–631. doi: 10.1016/j.tranon.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch CJ, Shah ZH, Allison SJ, Ahmed SU, Ford J, Warnock LJ, Li H, Serrano M, Milner J. SIRT1 undergoes alternative splicing in a novel auto-regulatory loop with p53. PLoS One. 2010;5:e13502. doi: 10.1371/journal.pone.0013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riquelme I, Saavedra K, Espinoza JA, Weber H, García P, Nervi B, Garrido M, Corvalán AH, Roa JC, Bizama C. Molecular classification of gastric cancer: Towards a pathway-driven targeted therapy. Oncotarget. 2015;6:24750–24779. doi: 10.18632/oncotarget.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng L, Yuan Z, Li Y, Ling H, Izumi V, Fang B, Fukasawa K, Koomen J, Chen J, Seto E. Ubiquitinated sirtuin 1 (SIRT1) function is modulated during DNA damage-induced cell death and survival. J Biol Chem. 2015;290:8904–8912. doi: 10.1074/jbc.M114.612796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bilici A. Treatment options in patients with metastatic gastric cancer: Current status and future perspectives. World J Gastroenterol. 2014;20:3905–3915. doi: 10.3748/wjg.v20.i14.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, Wang Y, Xiong Y, Chen XC, Ma ML, Cai R, Gao Y, Sun YM, Yang GS, Pang WJ. Sirt1 AS lncRNA interacts with its mRNA to inhibit muscle formation by attenuating function of miR-34a. Sci Rep. 2016;6:21865. doi: 10.1038/srep21865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoyagi K, Kouhuji K, Kizaki J, Isobe T, Hashimoto K, Shirouzu K. Molecular targeting to treat gastric cancer. World J Gastroenterol. 2014;20:13741–13755. doi: 10.3748/wjg.v20.i38.13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Vita F, Di Martino N, Fabozzi A, Laterza MM, Ventriglia J, Savastano B, Petrillo A, Gambardella V, Sforza V, Marano L, et al. Clinical management of advanced gastric cancer: The role of new molecular drugs. World J Gastroenterol. 2014;20:14537–14558. doi: 10.3748/wjg.v20.i40.14537. [DOI] [PMC free article] [PubMed] [Google Scholar]