Abstract
Increasing evidence has demonstrated that microRNA (miRNA) serve an important role in the tumorigenesis of various types of cancer, such as renal cell carcinoma (RCC). The expression of miR-211-5p has been detected in RCC tissue by microarray profiling. However, studies regarding miR-211-5p and RCC remain rare. In the present study, the expression of miR-211-5p in RCC tissues and cell lines was revealed to be downregulated by reverse transcription-quantitative polymerase chain reaction analyses. The present results also revealed that the upregulation or downregulation of miR-211-5p inhibited or promoted, respectively, RCC cell proliferation, migration and invasion. In addition, the upregulation or downregulation of miR-211-5p induced or inhibited, respectively, RCC cell apoptosis. However, the present study only identified that downregulation of miR-211-5p promoted 786O and ACHN cell viability. The above results suggest that miR-211-5p may be a tumor suppressor in the tumorigenesis of RCC and may be a potential therapeutic target for RCC in the future. Further research should focus on the underlying mechanism of miR-211-5p in RCC and on investigating the possible use of miR-211-5p as a biomarker for RCC.
Keywords: microRNA, miR-211-5p, renal cell carcinoma, tumor suppressor
Introduction
Renal cell carcinoma (RCC) is the most common type of malignant kidney cancer and occurs primarily on the renal tubular epithelial system, excluding metastatic neoplasms. In addition, due to the aberrant expression of genes in renal tubular epithelial cells, they gradually transform into RCC cells (1). In a study conducted in 2014, ~3.9% of new cancer cases were RCC, with a median age of 64 years at diagnosis in the USA alone (2). Due to the lack of typical clinical symptoms and an early diagnostic marker for RCC, metastasis has already developed in ~40% of patients when RCC is diagnosed (3). Currently, surgery remains to be the most effective treatment strategy for RCC and an abundance of postoperative adjuvant therapies have been approved for treatment of RCC; however, the most effective treatment is unknown (2,4). Therefore, it is necessary to identify a highly specific RCC-associated maker for early diagnosis of RCC.
MicroRNA (miRNA) are a family of short non-coding RNA, with a length of 19–25 nucleotides. miRNA guide the RNA-induced silencing complex to miRNA target sites (3′ untranslated region of mRNA) and then regulate gene expression at the post-translational level, leading to the inhibition of translation or mRNA degradation (5). With increasing research, more and more studies have demonstrated that miRNA may serve an important role in the occurrence and development of various cancer types (6–8), such as RCC (9–11).
miR-211-5p was predicted using computational methods using conservation with mouse and Fugu rubripes sequences (12). The sequence maps to human chromosome 15 and miR-211-5p has been demonstrated to serve an important role in several cancer types, including colorectal cancer (CRC) (13), gastric cancer (14), non-small cell lung cancer (15) and hepatocellular carcinoma (16). The aim of the present study was to reveal the expression and function of miR-211-5p in RCC.
Materials and methods
Ethics statement
All patients signed the informed consent forms prior to initiation of the present study. The present study was approved by the Ethical Review Committee of the Peking University Shenzhen Hospital (Shenzhen, China) and complied with the Declaration of Helsinki.
Specimens and cell lines
RCC tissues and paired adjacent normal tissues were obtained from patients undergoing surgery at the Peking University Shenzhen Hospital from the 17th of March 2013 to the 30th of December 2015. The patients had not received any anticancer treatment prior to surgery. A total of 24 patients were enrolled in the present study (18 males and 6 females), with a mean age of 51 years. Once RCC tissues and paired adjacent normal tissues were completely removed, all tissues were immediately collected and stored at −80°C until the RNA was later extracted. The clinical and pathological characteristics of the patients are presented in Table I.
Table I.
Clinicopathological characteristics of patients with renal cell carcinoma.
| Characteristic | Number of cases |
|---|---|
| Mean age, range (years) | 51 (27–72) |
| Sex | |
| Male | 18 |
| Female | 6 |
| Histological type | |
| Clear cell | 20 |
| Papillary | 4 |
| Fuhrman grade | |
| I | 15 |
| II | 7 |
| III | 1 |
| IV | 1 |
| American Joint Committee on Cancer clinical stage | |
| I | 15 |
| II | 8 |
| III+IV | 1 |
RCC cell lines (ACHN, 786O, Caki-1 and 769P) and the human embryo kidney cell line (293T) were acquired from the Guangdong and Shenzhen Key Laboratory of Male Reproductive Medicine and Genetics (Shenzhen, China). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), 1% antibiotics (100 U/ml penicillin and 100 mg/ml streptomycin; Gibco; Thermo Fisher Scientific, Inc.) and 1% glutamine. The cells were incubated in a 5% CO2 humidified incubator at 37°C.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
According to the manufacturer's protocol, total RNA from tissues and cells was isolated using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and purified using an RNeasy Maxi kit (Qiagen GmbH, Hilden, Germany). Following the measurement of the RNA concentration using a NanoDrop 2000c (Thermo Fisher Scientific, Inc.), an miScript II RT kit (Qiagen GmbH) was used to synthetize cDNA with reverse transcriptase, according to manufacturer's protocol. The temperature protocol was as follows: 37°C for 60 min, 95°C for 5 min and kept at 4°C. Subsequently, the expression of miR-211-5p was detected with an miScript SYBR®Green PCR kit (Qiagen GmbH) by qPCR on a Roche light cycler 480 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland). The 10 µl reaction mixture consisted of 5 µl 2X QuantiTect SYBR Green PCR Master mix, 3.7 µl RNase-free water, 1 µl cDNA template, 0.4 µl specific miRNA primer and 10X miScript Universal Primer. The forward primer of miR-211-5p was 5′-TTCCCTTTGUCATCCTTCGCCT-3′ and the reverse primer was a universal primer, which was provided in the miScript SYBR®Green PCR kit. U6 was chosen as an internal control. The U6 primer sequences were as follows: Forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-ACGCTTCACGAATTTGCGT-3′. The thermocycling conditios were as follows: 95°C for 2 min, 95°C for 10 sec, 55°C for 30 sec and 72°C for 30 sec, for 40 cycles. The expression levels of miR-211-5p in tissues and cells were analyzed using the 2−ΔΔCq method (17).
Cell transfection
The expression of miR-211-5p in 786O and ACHN cells was upregulated/downregulated by transfection of the synthesized miR-211-5p mimic/inhibitor (Shanghai GenePharma Co., Ltd., Shanghai, China) using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.) and Opti-MEM® I Reduced Serum Medium (Gibco; Thermo Fisher Scientific, Inc.), following the manufacturer's protocol. The concentration of the miRNA mimic and inhibitors used for transfection are presented in Table II. The efficiency of transfection was measured by RT-qPCR as aforementioned. The sequences of the miRNA and primers used are presented in Table III.
Table II.
Concentration of the miRNA mimic/inhibitors used for the transfection.
| Plate | siRNA | Final volume | Lipo2000 |
|---|---|---|---|
| 96-well | 0.5 µl (5 pmol) | 100 µl | 0.25 µl |
| 24-well | 1 µl (20 pmol) | 500 µl | 1 µl |
| 12-well | 2 µl (40 pmol) | 1 ml | 2 µl |
| 6-well | 5 µl (100 pmol) | 2 ml | 5 µl |
siRNA, small interfering RNA.
Table III.
Sequences of primers and miRNA.
| Primer/miRNA | Direction | Sequence (5′-3′) |
|---|---|---|
| miR-211-5p | F | TTCCCTTTGUCATCCTTCGCCT |
| R | Universal primers (miScript SYBR Green polymerase chain reaction kit) | |
| U6 | F | CTCGCTTCGGCAGCACA |
| R | ACGCTTCACGAATTTGCGT | |
| miR-211-5p mimic | F | UUCCCUUUGUCAUCCUUCGCCU |
| R | GCGAAGGAUGACAAAGGGAAUU | |
| miR-211-5p inhibitor | – | AGGCGAAGGAUGACAAAGGGAA |
| NC | F | UUCUCCGAACGUGUCACGUTT |
| R | ACGUGACACGUUCGGAGAATT | |
| Inhibitor NC | – | CAGUACUUUUGUGUAGUACAA |
miRNA, microRNA; NC, negative control; F, forward; R, reverse.
MTT assay
The cell activity of the 786O and ACHN cells was determined using an MTT assay. Cells (~5,000 cells/well) were seeded into 96-well plates and transfected with miR-211-5p mimic, inhibitor, negative control (NC) and inhibitor NC using Lipofectamine® 2000. A total of 96 h later, 20 µl MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added into each well of the 96-well plate. Following 4 h at 37°C, 100 µl dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to each well. Subsequently, the 96-well plate was shocked in a reciprocating decolorization shaking table (TSB-108; Qilinbeier, Jiangsu, China) for 10 min in the dark at room temperature. Finally, the optical density (OD) value of each well was measured by an ELISA microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at a wavelength of 595 nm (with 620 nm as the reference wavelength).
Cell Counting Kit-8 (CCK-8) assay
The proliferation ability of the 786O and ACHN cells was analyzed using a CCK-8 assay (Beyotime Institute of Biotechnology, Shanghai, China). Cells (~5,000 cells/well) were seeded into a 96-well plate and transfected according to the aforementioned protocol. Following transfection, 10 µl CCK-8 solution was added to each well and culture was continued for 30 min in the dark at room temperature. The OD value of each well was then measured using an ELISA microplate reader at a wavelength of 450 nm (with 620 nm as the reference wavelength) at 0, 24, 48 and 72 h following transfection.
Transwell assay
The migration and invasion ability of the 786O and ACHN cells in vitro was examined by Transwell assays. Transwell chambers (BD Biosciences, Franklin Lakes, NJ, USA) with Matrigel were used to assess invasion ability, while the Transwell chambers without Matrigel were used to assess migration ability. A total of 24 h after transfection, ~2×104 cells were added into each upper chamber with DMEM and DMEM supplemented with 10% FBS was added to the lower chambers. The chambers were incubated for 48 h in a 5% CO2 incubator at 37°C. The cells in the lower chamber were fixed and stained successively by 4% paraformaldehyde and crystal violet each for 20 min at room temperature. Finally, a light microscope was used to observe the cells in the bottom of chamber at a magnification of ×100.
Wound scratch assay
The migration ability of the 786O and ACHN cells in vitro was also examined by wound scratch assays. In total, ~1×106 cells were cultured in each well of a 6-well plate. After 24 h, cells were transfected with miR-211-5p mimic, inhibitor, NC and inhibitor NC using Lipofectamine® 2000. A vertical line was scratched in the cells using a sterile 1-ml pipette tip. The images of the scratches were respectively captured by a digital camera system at 0, 12 and 24 h.
Flow cytometry assay
The apoptosis rate of the 786O and ACHN cells in vitro was analyzed by a flow cytometry assay. In total, ~1×106 cells were incubated in each well of the 6-well plate and were transfected following the manufacturer's instructions. All cells were harvested after 48 h and washed twice with cold PBS. Following this, 100 µl 1X binding buffer (Invirtogen; Thermo Fisher Scientific, Inc.) was used to resuspend the cells. Next, 5 µl Annexin V-fluorescein isothiocyanate (Invitrogen; Thermo Fisher Scientific, Inc.) and 5 µl propidium iodide (Invitrogen; Thermo Fisher Scientific, Inc.) were added to the experimental group and stained for 15 min in the dark at room temperature. Finally, the apoptosis rate was analyzed by flow cytometry (EPICS Xl-4; Beckman Coulter, Inc., Brea, CA, USA) after adding 400 µl 1X binding buffer to each tube. All assays were repeated at least three times. FlowJo 7.6.1 software was utilized for the analysis of results (FlowJo LLC, Ashland, OR, USA).
Statistical analyses
Data were presented as the mean ± standard error of the mean. Significance of differential expression was analyzed by using the Student's t-test, while paired t-tests were used to compare the expression levels of miR-211-5p in matched tumor/normal tissues. One-way analysis of variance followed by Bonferroni's post hoc tests were used to compare the expression level of miR-211-5p in RCC cell lines. SPSS 23.0 (IBM Corp., Armonk, NY, USA) was used for all statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression level of miR-211-5p is downregulated in RCC tissues and cell lines
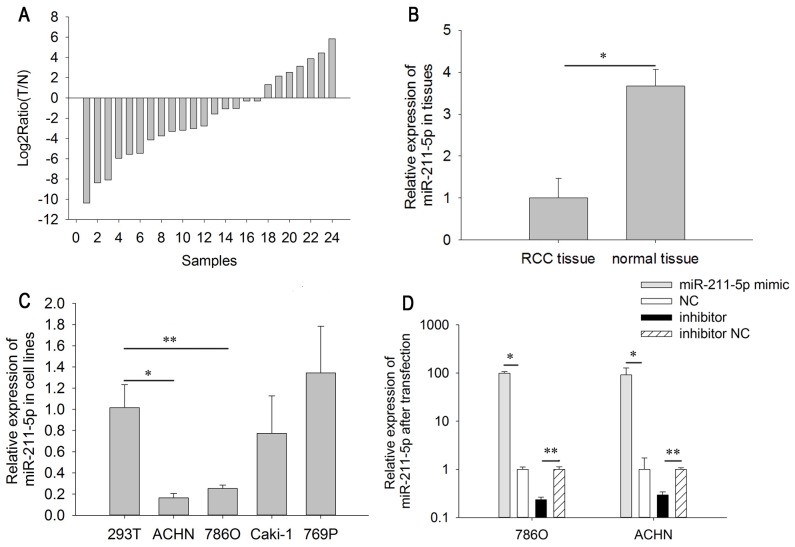
The relative expression level of miR-211-5p in RCC tissues is demonstrated in the Fig. 1A. As indicated in Fig. 1B, the expression level of miR-211-5p in RCC tissues (1.000±0.466) was significantly lower than that in adjacent normal tissues (3.664±0.407) (P<0.05). The results of miR-211-5p expression in the cell lines demonstrated that the relative expression of miR-211-5p was significantly lower in 786O (0.252±0.032, P<0.01) and ACHN cells (0.253±0.032, P<0.001) than in 293T cells (1.000±0.217). However, there was no significant difference between the expression level in Caki-1 (0.772±0.355, P=0.401) and 769P cells (1.341±0.441, P=0.366) compared with that in 293T cells (Fig. 1C). The above results suggest that miR-211-5p may act as a tumor suppressor in RCC.
Figure 1.
Relative expression of miR-211-5p in 24 paired tissues and cell lines. (A) Log2 ratios of miR-211-5p expression in 24 paired Ts and adjacent Ns. (B) The relative expression of miR-211-5p in T and N. (C) The relative expression of miR-211-5p in RCC cell lines. (D) The relative expression of miR-211-5p following transfection in 786O and ACHN cells. *P<0.05 and **P<0.01 as indicated. T, RCC tissues; N, normal tissue; RCC, renal cell carcinoma; NC, negative control.
Cell transfection efficiency validation
RT-qPCR was performed to detect the transfection efficiency of miR-211-5p mimic or inhibitor, compared with NC or inhibitor NC. The results demonstrated that the expression levels of miR-211-5p were 0.234±0.032 times higher (786O cells, P=0.003) and 0.293±0.048 times higher (ACHN cells, P=0.005) in cells transfected with miR-211-5p inhibitor compared with the levels in those transfected with inhibitor NC after 24 h. The expression levels of miR-211-5p were 99.310±7.915 times higher (786O cells, P=0.021) and 91.321±36.326 times higher (ACHN cells, P=0.018) in cells transfected with miR-211-5p mimic compared with the levels in those transfected with NC after 24 h (Fig. 1D).
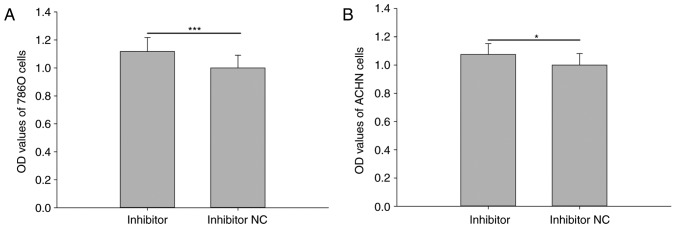
Downregulation of miR-211-5p promotes 786O and ACHN cell viability
Cell viability was analyzed using an MTT assay. The results revealed that the relative viability of 786O cells transfected with miR-211-5p inhibitor was significantly increased compared with that observed in cells transfected with inhibitor NC (1.117±0.100 vs. 1.000±0.090, respectively; P<0.001; Fig. 2A). The relative viability of ACHN cells was also significantly increased in cells transfected with miR-211-5p inhibitor compared with that observed in cells transfected with inhibitor NC (1.075±0.077 vs. 1.000±0.082, respectively; P<0.05; Fig. 2B). However, there was no significant difference observed between the mimic group and NC group for the viability of 786O and ACHN cells (P>0.05; data not shown).
Figure 2.
Cell viability assay following transfection with miR-211-5p inhibitor or inhibitor NC. Results of cell viability assay in (A) 786O and (B) ACHN cells following transfection with miR-211-5p inhibitor or inhibitor NC. *P<0.05 and ***P<0.001 as indicated. NC, negative control; OD, optical density.
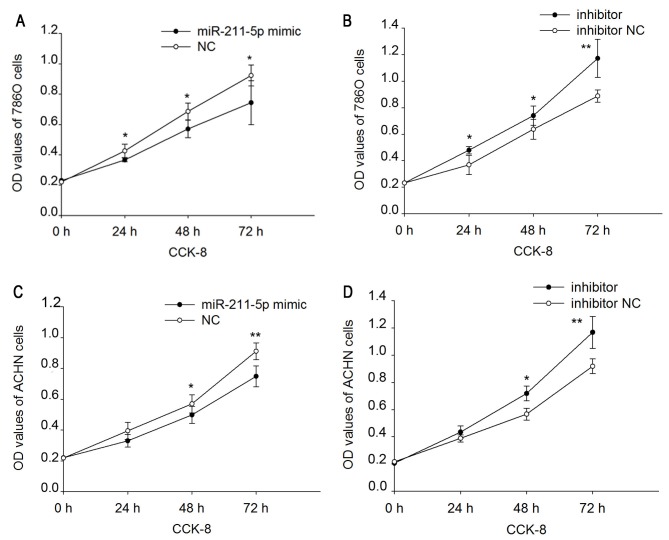
Upregulation/downregulation of miR-211-5p inhibits/promotes 786O and ACHN cell proliferation
A CCK-8 assay was performed to assess the proliferation ability of 786O and ACHN cells (Fig. 3). The results demonstrated that the upregulation/downregulation of miR-211-5p inhibited/promoted the proliferation of the 786O and ACHN cells. Proliferation was downregulated by 14.030 (P<0.05), 16.802 (P<0.05) and 19.416% (P<0.05) (Fig. 3A) in 786O cells, and 16.385 (P=0.150), 12.627 (P<0.05) and 17.886% (P<0.01) (Fig. 3C) in ACHN cells following transfection with miR-211-5p mimic at 24, 48 and 72 h, respectively, compared with that observed in cells transfected with NC. Proliferation was upregulated by 29.957 (P<0.05), 16.118 (P<0.05) and 31.945% (P<0.01) (Fig. 3B) in 786O cells, and 11.691 (P=0.095), 26.840 (P<0.05) and 27.187% (P<0.01) (Fig. 3D) in ACHN cells following transfection with miR-211-5p inhibitor at 24, 48 and 72 h, respectively, compared with that observed in cells transfected with inhibitor NC.
Figure 3.
Cell proliferation assay of 786O and ACHN cells. A CCK-8 assay was used to measure cell proliferation in 786O cells transfected with (A) miR-211-5p mimic or NC and (B) miR-211-5p inhibitor or inhibitor NC. Cell proliferation was also measured in ACHN cells transfected with (C) miR-211-5p mimic or NC and (D) miR-211-5p inhibitor or inhibitor NC. *P<0.05 and **P<0.01 vs. miR-211-5p mimic or inhibitor NC. CCK-8, Cell Counting Kit-8; NC, negative control; OD, optical density.
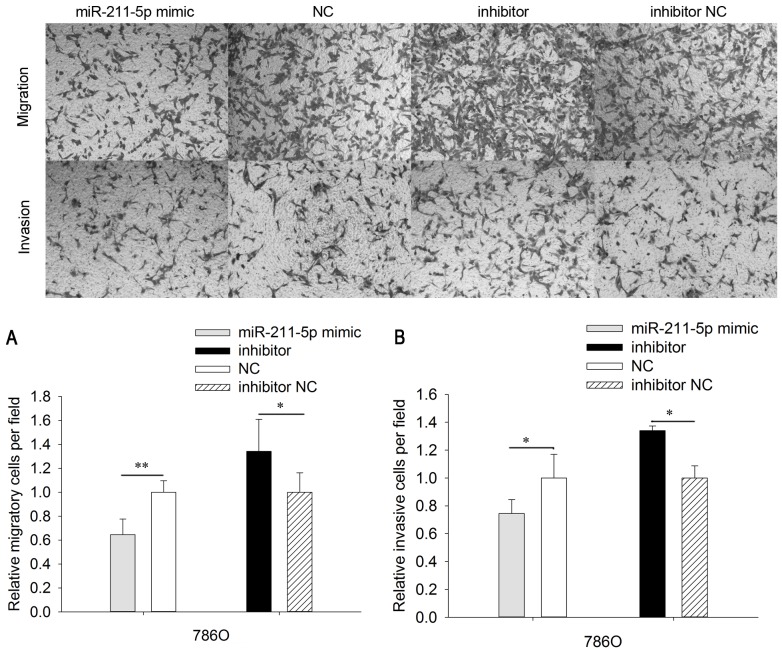
Upregulation/downregulation of miR-211-5p inhibits/promotes 786O and ACHN cell mobility
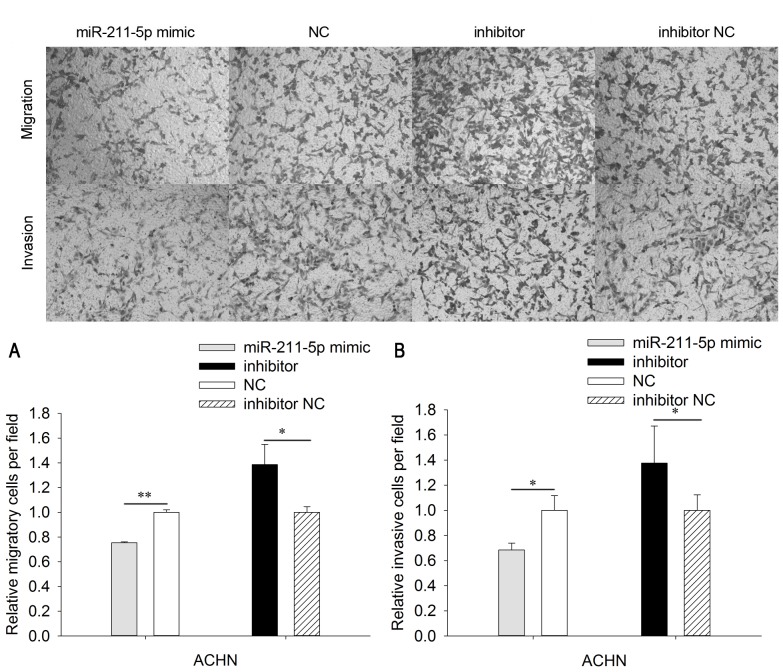
To analyze the mobility of 786O and ACHN cells, wound scratch and Transwell assays were designed (Figs. 4 and 5). The results of Transwell assays demonstrated that the migratory ability of 786O cells was significantly downregulated by 35.351% (P<0.01) in the miR-211-5p mimic group compared with that observed in the NC group (Fig. 4). Furthermore, the migratory ability of 786O cells was significantly upregulated by 34.207% (P<0.05) in the miR-211-5p inhibitor group compared with the level observed in the inhibitor NC group. In ACHN cells, the migratory ability was significantly downregulated by 24.582% (P<0.01) in the miR-211-5p mimic group compared with that observed in the NC group. The migratory ability was significantly upregulated by 38.620% (P<0.05) in ACHN cells of the miR-211-5p inhibitor group compared with the level observed in the inhibitor NC group (Fig. 5).
Figure 4.
Migration and invasion assays of 786O cells. The cell (A) migratory and (B) invasion abilities of 786O cells transfected with miR-211-5p mimic or NC and miR-211-5p inhibitor or inhibitor NC were measured using Transwell assays (magnification, ×100). *P<0.05 and **P<0.01 as indicated. NC, negative control.
Figure 5.
Migration and invasion assays of ACHN cells. The cell (A) migratory and (B) invasion abilities of ACHN cells transfected with miR-211-5p mimic or NC and miR-211-5p inhibitor or inhibitor NC were measured using Transwell assays (magnification, 100×). *P<0.05 and **P<0.01 as indicated. NC, negative control.
In order to determine the invasive ability of cells, Transwell chambers with Matrigel were utilized. The invasive ability of 786O cells was significantly downregulated by 35.351% (P<0.05) in the miR-211-5p mimic group compared with that observed in the NC group. The invasive ability of 786O cells was significantly upregulated by 34.207% (P<0.05) in the miR-211-5p inhibitor group compared with that observed in the inhibitor NC group (Fig. 4). In addition, the invasive ability of ACHN cells was significantly downregulated by 31.625% (P<0.05) in the miR-211-5p mimic group compared with that observed in the NC group, while the invasive ability of ACHN cells was significantly upregulated by 37.628% (P<0.05) in the miR-211-5p inhibitor group compared with that observed in the inhibitor NC group (Fig. 5).
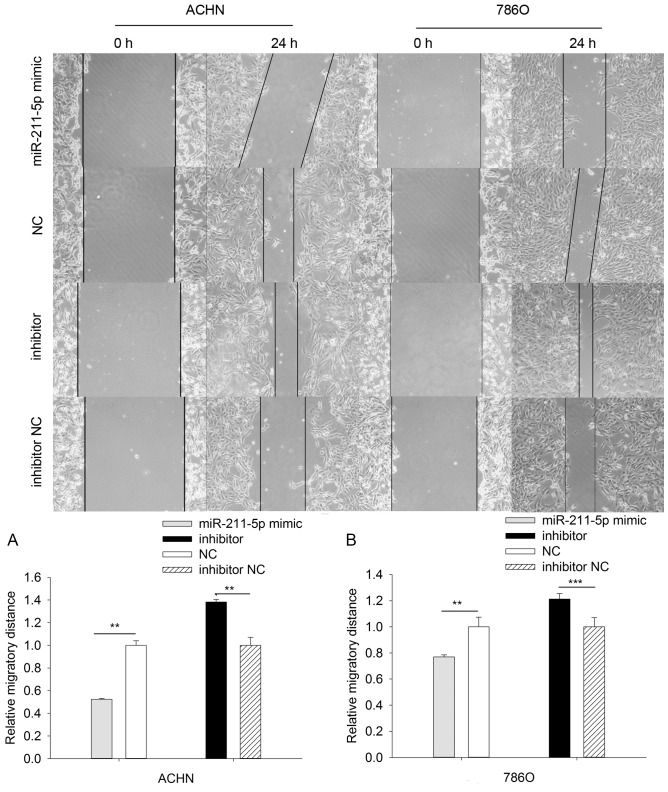
The results of the scratch wound assay demonstrated that the migratory ability of 786O cells was significantly downregulated by 23.131% (P<0.01) in the miR-211-5p mimic group compared with that observed in the NC group, while the migratory ability was significantly upregulated by 21.287% (P<0.001) in the miR-211-5p inhibitor group compared with the observed in the inhibitor NC group. The results in ACHN cells were similar to those in the 786O cells, which demonstrated that the migratory ability was significantly downregulated by 47.671% (P<0.01) in the miR-211-5p mimic group compared with that observed in the NC group, while the migratory ability was significantly upregulated by 38.300% (P<0.01) in the miR-211-5p inhibitor group compared with the observed in the inhibitor NC group (Fig. 6).
Figure 6.
Wound scratch assays to determine the migratory ability of (A) ACHN and (B) 786O cells transfected with miR-211-5p mimic or NC and miR-211-5p inhibitor or inhibitor NC. **P<0.01 and ***P<0.001 as indicated. NC, negative control.
Upregulation/downregulation of miR-211-5p induces/inhibits 786O and ACHN cell apoptosis
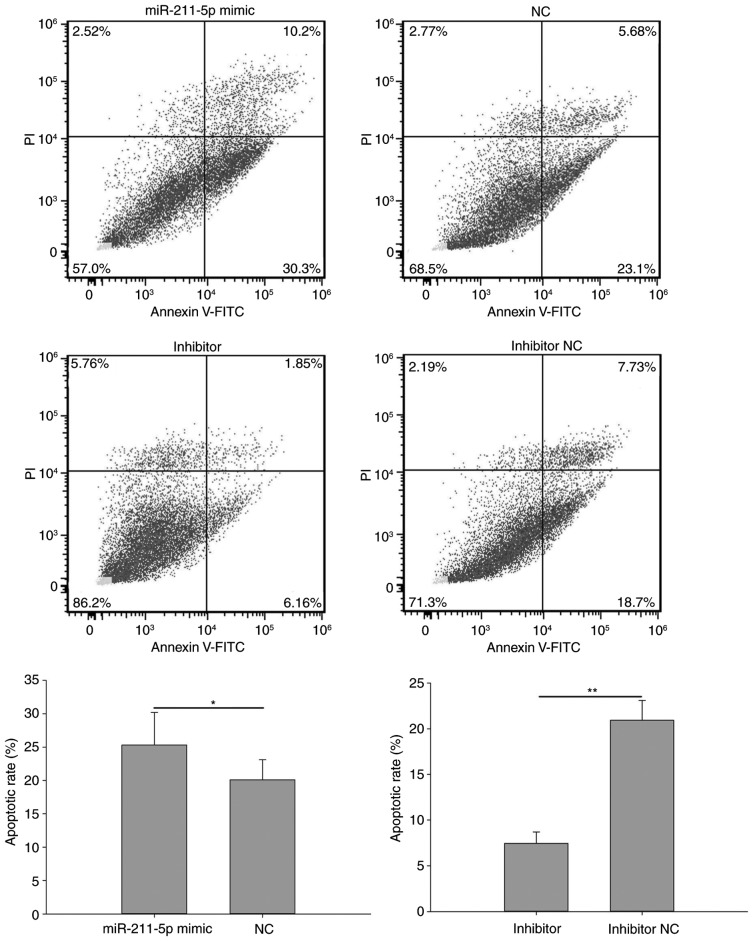
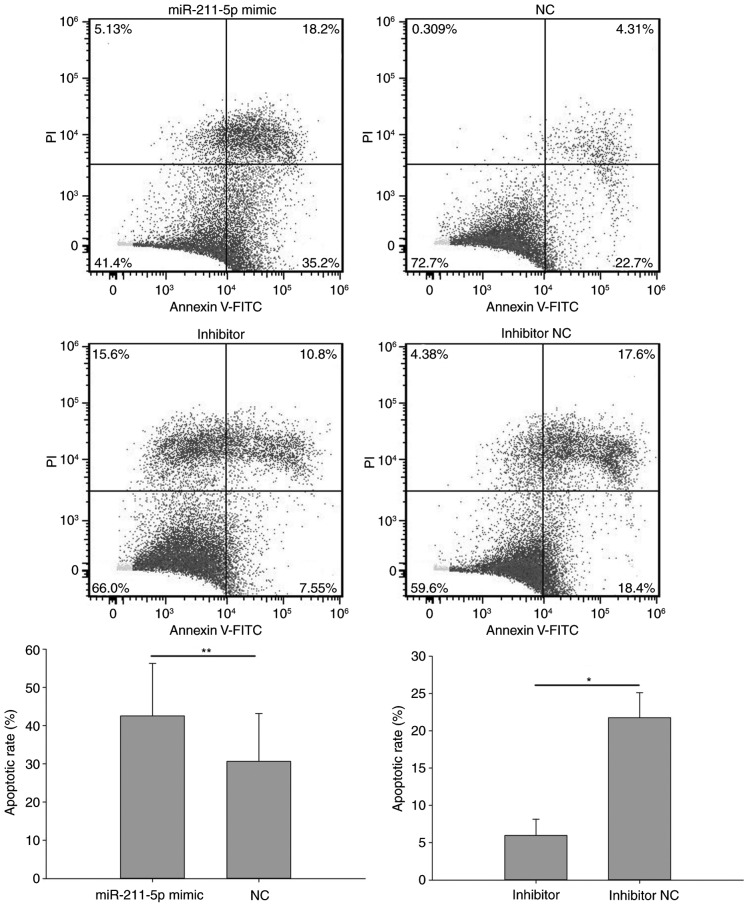
Flow cytometry was performed to analyze the apoptotic rate of cells (Figs. 7 and 8). The results demonstrated that the apoptotic rate of 786O cells transfected with miR-211-5p mimic was significantly increased compared with that observed in the NC group (25.333±4.854 vs. 20.100±3.000%, respectively; P<0.05; Fig. 7). A similar outcome was also observed in ACHN cells, with an apoptotic rate of 42.567±13.723 vs. 30.633±12.548% (P<0.01) in the miR-211-5p mimic and NC groups, respectively (Fig. 8). Additionally, the apoptotic rate of 786O cells transfected with miR-211-5p inhibitor was significantly decreased compared with that observed in the inhibitor NC group (7.453±1.243 vs. 20.933±2.155%, respectively; P<0.01; Fig. 7). Similarly, the apoptotic rate of ACHN cells was 5.963±2.170 and 21.767±3.350% (P<0.05) in the inhibitor and inhibitor NC groups, respectively (Fig. 8). These results suggest that the upregulation/downregulation of miR-211-5p induced/inhibited 786O and ACHN cell apoptosis.
Figure 7.
Flow cytometry results of the apoptosis rates in 786O cells transfected with miR-211-5p mimic or NC and miR-211-5p inhibitor or inhibitor NC. *P<0.05 and **P<0.01 as indicated. NC, negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Figure 8.
Flow cytometry results of the apoptosis rates in ACHN cells transfected with miR-211-5p mimic or NC and miR-211-5p inhibitor or inhibitor NC. *P<0.05 and **P<0.01 as indicated. NC, negative control; PI, propidium iodide; FITC, fluorescein isothiocyanate.
Discussion
It is well known that miRNA are important post-transcriptional regulators and that they are involved in various physiological and pathological processes, including cell differentiation, cell proliferation and tumorigenesis (18). miRNA have been demonstrated to serve as oncogenes when they are overexpressed or tumor suppressor genes when they downregulated (19). Previous studies have indicated that miR-211-5p was downregulated in hepatocellular carcinoma, gastric cancer and ovarian cancer (14,16,20). However, miR-211-5p was overexpressed in non-small cell lung cancer (15). Previous microarray analyses indicated that miR-211-5p was significantly downregulated in RCC tissue compared with adjacent normal tissues (21). The results of the present study also indicated that the expression of miR-211-5p was downregulated in human RCC tissues and cell lines, according to RT-qPCR.
Furthermore, the present study also demonstrated that miR-211-5p has a negative impact in RCC cell lines, particularly on migration and invasion. This result was similar to that demonstrated in a study by Wang et al (22). In addition, the present study indicated that upregulation/downregulation of miR-211-5p induced/inhibited 786O and ACHN cell apoptosis. This phenomenon was also demonstrated in ovarian cancer (20). It has also been suggested that miR-211 acted as a tumor suppressor in epithelial ovarian cancer (EOC) and inhibited cell proliferation by regulating cyclin-dependent kinase 6 in EOC cells (20). In addition, long non-coding (lnc)RNA ucoo2kmd.1 was highly expressed in CRC tissues compared with adjacent normal tissues and regulated cluster of differentiation 44 as a molecular decoy for miR-211-3p (13). However, a study by Xu et al (23) revealed that miR-211-3p served as an oncogene in CRC and promoted cell proliferation by targeting lncRNA tumor suppressor candidate 7 in CRC cell lines. Furthermore, a case-control study discovered that miR-211 was associated with poor prognosis in CRC, while no statistically significant differences between clinicopathological factors and miR-211 expression level were identified in the CRC group (24).
In addition to cancer, miR-211 also serves an important role in the initiation and development of other diseases. A study by Sun et al (25) indicated that miR-133, miR-135, miR-204 and miR-211 acted as negative regulators and inhibited differentiation of osteoprogenitors by attenuating the essential transcription factor Runt-related transcription factor 2 (RUNX2). Furthermore, Atlasi et al (26) demonstrated that miR-211 acted as an endogenous attenuator of this transcription factor and the effect in calcium deposition did not correlate with the effect in RUNX2, which demonstrated that miR-211 could exert its effects on calcification through the Wnt signaling pathway. In addition, a study by Panizo et al (27) concluded that miR-29b, miR-133b and miR-211 have direct roles in vascular smooth muscle calcification induced by high phosphorus, and may be novel therapeutic targets in the management of vascular calcification.
In conclusion, the present study demonstrated that miR-211-5p was downregulated in RCC tissues and cell lines. The present study also indicated that miR-211-5p serves important roles in cellular functions, including proliferation, migration, invasion and apoptosis. Thus, this study suggests that miR-211-5p acts as a tumor suppressor in RCC. Further research should focus on the underlying mechanism of miR-211-5p in RCC and on investigating the possible use of miR-211-5p as a biomarker for RCC.
Acknowledgements
The authors thank reviewers for helpful comments on the manuscript and all the patients for consenting to provide tissue samples.
Funding
This study was supported by grants from the National Natural Science Foundation of China (grant no. 81101922), Science and Technology Development Fund Project of Shenzhen (grant nos. JCYJ20150403091443329 and JCYJ20170307111334308), the fund of ‘San-ming’ Project of Medicine in Shenzhen (grant no. SZSM201612066) and the fund of Guangdong Key Medical Subject.
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YL and TW conceived and designed the experiments. JQ, XP, TH, CL and PC performed the experiments, analyzed the data and drafted the paper. ZZ and SY conceived the experiments. All authors have read and approved this manuscript.
Ethics approval and consent to participate
All patients signed the informed consent forms prior to initiation of the present study. The present study was approved by the Ethical Review Committee of the Peking University Shenzhen Hospital (Shenzhen, China) and complied with the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chow WH, Dong LM, Devesa SS. Epidemiology and risk factors for kidney cancer. Nat Rev Urol. 2010;7:245–257. doi: 10.1038/nrurol.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Motzer RJ, Jonasch E, Agarwal N, Beard C, Bhayani S, Bolger GB, Chang SS, Choueiri TK, Costello BA, Derweesh IH, et al. Kidney cancer, version 3.2015. J Natl Compr Canc Netw. 2015;13:151–159. doi: 10.6004/jnccn.2015.0022. [DOI] [PubMed] [Google Scholar]
- 3.Flanigan RC, Campbell SC, Clark JI, Picken MM. Metastatic renal cell carcinoma. Curr Treat Options Oncol. 2003;4:385–390. doi: 10.1007/s11864-003-0039-2. [DOI] [PubMed] [Google Scholar]
- 4.Afriansyah A, Hamid AR, Mochtar CA, Umbas R. Targeted therapy for metastatic renal cell carcinoma. Acta Med Indones. 2016;48:335–347. [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Shui Y, Yu X, Duan R, Bao Q, Wu J, Yuan H, Ma C. miR-130b-3p inhibits cell invasion and migration by targeting the Notch ligand Delta-like 1 in breast carcinoma. Gene. 2017;609:80–87. doi: 10.1016/j.gene.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Chang C, Liu T, Huang Y, Qin W, Yang H, Chen J. MicroRNA-134-3p is a novel potential inhibitor of human ovarian cancer stem cells by targeting RAB27A. Gene. 2017;605:99–107. doi: 10.1016/j.gene.2016.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Chen T, Ren H, Thakur A, Yang T, Li Y, Zhang S, Wang T, Chen M. miR-382 inhibits tumor progression by targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother. 2017;86:248–253. doi: 10.1016/j.biopha.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Zhou W, Bi X, Gao G, Sun L. miRNA-133b and miRNA-135a induce apoptosis via the JAK2/STAT3 signaling pathway in human renal carcinoma cells. Biomed Pharmacother. 2016;84:722–729. doi: 10.1016/j.biopha.2016.09.074. [DOI] [PubMed] [Google Scholar]
- 10.Jin L, Li Y, Liu J, Yang S, Gui Y, Mao X, Nie G, Lai Y. Tumor suppressor miR-149-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Mol Med Rep. 2016;13:5386–5392. doi: 10.3892/mmr.2016.5205. [DOI] [PubMed] [Google Scholar]
- 11.Kurozumi A, Kato M, Goto Y, Matsushita R, Nishikawa R, Okato A, Fukumoto I, Ichikawa T, Seki N. Regulation of the collagen cross-linking enzymes LOXL2 and PLOD2 by tumor-suppressive microRNA-26a/b in renal cell carcinoma. Int J Oncol. 2016;48:1837–1846. doi: 10.3892/ijo.2016.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 13.Wu X, He X, Li S, Xu X, Chen X, Zhu H. Long Non-Coding RNA ucoo2kmd.1 regulates CD44-dependent cell growth by competing for miR-211-3p in colorectal cancer. PLoS One. 2016;11:e0151287. doi: 10.1371/journal.pone.0151287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang CY, Hua L, Sun J, Yao KH, Chen JT, Zhang JJ, Hu JH. MiR-211 inhibits cell proliferation and invasion of gastric cancer by down-regulating SOX4. Int J Clin Exp Pathol. 2015;8:14013–14020. [PMC free article] [PubMed] [Google Scholar]
- 15.Ye L, Wang H, Liu B. miR-211 promotes non-small cell lung cancer proliferation by targeting SRCIN1. Tumour Biol. 2016;37:1151–1157. doi: 10.1007/s13277-015-3835-y. [DOI] [PubMed] [Google Scholar]
- 16.Jiang G, Cui Y, Yu X, Wu Z, Ding G, Cao L. miR-211 suppresses hepatocellular carcinoma by downregulating SATB2. Oncotarget. 2015;6:9457–9466. doi: 10.18632/oncotarget.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 19.Shenouda SK, Alahari SK. MicroRNA function in cancer: Oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28:369–378. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 20.Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer. 2015;14:57. doi: 10.1186/s12943-015-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng T, Wang L, Li Y, Huang C, Zeng L, Yang J. Differential microRNA expression in renal cell carcinoma. Oncol Lett. 2013;6:769–776. doi: 10.3892/ol.2013.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K, Jin W, Jin P, Fei X, Wang X, Chen X. miR-211-5p suppresses metastatic behavior by targeting SNAI1 in renal cancer. Mol Cancer Res. 2017;15:448–456. doi: 10.1158/1541-7786.MCR-16-0288. [DOI] [PubMed] [Google Scholar]
- 23.Xu J, Zhang R, Zhao J. The novel long noncoding RNA TUSC7 inhibits proliferation by sponging MiR-211 in colorectal cancer. Cell Physiol Biochem. 2017;41:635–644. doi: 10.1159/000457938. [DOI] [PubMed] [Google Scholar]
- 24.Sümbül AT, Göğebakan B, Bayram S, Batmacı CY, Öztuzcu S. MicroRNA 211 expression is upregulated and associated with poor prognosis in colorectal cancer: A case-control study. Tumour Biol. 2015;36:9703–9709. doi: 10.1007/s13277-015-3708-4. [DOI] [PubMed] [Google Scholar]
- 25.Sun Y, Byon CH, Yuan K, Chen J, Mao X, Heath JM, Javed A, Zhang K, Anderson PG, Chen Y. Smooth muscle cell-specific runx2 deficiency inhibits vascular calcification. Circ Res. 2012;111:543–552. doi: 10.1161/CIRCRESAHA.112.267237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atlasi Y, Noori R, Gaspar C, Franken P, Sacchetti A, Rafati H, Mahmoudi T, Decraene C, Calin GA, Merrill BJ, Fodde R. Wnt signaling regulates the lineage differentiation potential of mouse embryonic stem cells through Tcf3 down-regulation. PLoS Genet. 2013;9:e1003424. doi: 10.1371/journal.pgen.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panizo S, Naves-Díaz M, Carrillo-López N, Martínez-Arias L, Fernández-Martín JL, Ruiz-Torres MP, Cannata-Andía JB, Rodríguez I. MicroRNAs 29b, 133b and 211 regulate vascular smooth muscle calcification mediated by high phosphorus. J Am Soc Nephrol. 2016;27:824–834. doi: 10.1681/ASN.2014050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.