Abstract
Although there have been notable improvements in treatments against cancer, further research is required. In colon cancer, nearly all patients eventually experience drug resistance and stop responding to the approved drugs, making treatment difficult. Steroid receptor coactivator (SRC) is an oncogenic nuclear receptor coactivator that serves an important role in drug resistance. The present study generated a doxorubicin-resistant colon cancer cell line, in which the upregulation/activation of SRC was responsible for drug resistance, which in turn activated AKT. Overexpression of receptor tyrosine kinase-like epidermal growth factor receptor and insulin-like growth factor 1 receptor also induced SRC expression. It was observed that doxorubicin resistance in colon cancer also induced epithelial to mesenchymal transition, a decrease in expression of epithelial marker E-cadherin and an increase in the expression of mesenchymal markers, including N-cadherin and vimentin. Additionally, the present study indicated that SRC acts as a common signaling node, and inhibiting SRC in combination with doxorubicin treatment in doxorubicin-resistant cells aids in reversing the resistance. Thus, the present study suggests that activation of SRC is responsible for doxorubicin resistance in colon cancer. However, further research is required to understand the complete mechanism of how drug resistance occurs and how it may be tackled to treat patients.
Keywords: colon cancer, steroid receptor coactivator, drug, resistance, doxorubicin, epithelial to mesenchymal transition
Introduction
Colon cancer, a frequently diagnosed cancer type worldwide, is a disease in which malignant tumors form in the tissues of the colon and it is one of the leading causes of cancer-related mortality worldwide (1). Currently, the two main options for treatment of colon cancer are chemotherapy and surgery, and the treatment option is dependent upon the tumor location, size and stage of cancer, and overall characteristics of the patient affected (2,3). Chemotherapy may be given at any stage and is generally administered following surgery; however, in some cases it is also given prior to surgery in order to reduce the tumor size (4). The overall survival of patients with colon cancer has increased over the past decade due to improvements in medical sciences and chemotherapeutic regimens (5,6). Despite this, almost all patients with colon cancer develop drug resistance, which decreases the efficacy of the drugs and ultimately leads to failure of chemotherapy (7,8).
Decreases in the effect of drugs, including antibiotics or chemotherapeutic agents, such as doxorubicin, is termed drug resistance (9,10). Treating drug resistance, particularly multidrug resistance, is one of the major obstacles to successful chemotherapy (11). Furthermore, the majority of cancer-related mortalities occur due to failure of chemotherapy that occurs due to the generation of drug resistance during the process of chemotherapy and cancer progression (12). Evidence suggests that cancer cell resistance to chemotherapy is due to a transition from an epithelial to mesenchymal phenotype (13). Other research has demonstrated an association of epithelial to mesenchymal transition (EMT) with acquired resistance to cancer therapy, as well as cancer metastasis (14). As an outcome of these findings, if the epithelial phenotype is restored, it increases the sensitivity of tumor cells to chemotherapy (15). EMT of tumor cells not only causes increased metastasis, but also contributes to drug resistance (16). Intricate links between cells with an EMT-like phenotype and drug resistance in tumors have also been proposed (17). A diverse array of cytokines and growth factors may contribute to the regulation of the process of drug resistance development (16,18). The development of drug resistance may also be regulated by a higher apoptotic threshold, aerobic glycolysis, regions of hypoxia and elevated activity of drug efflux transporters (19). Furthermore, a previous report has indicated that drug resistance emergence may occur as a consequence of EMT (16). Evidence for an eminent role of steroid receptor coactivator (SRC) in invasion and in other tumor progression-related events, such as EMT, also exists (19). There are several receptors, as well as non-receptor tyrosine kinases (RTKs), which are modulated by SRC and are responsible for persistence and robustness of RTK signaling (20). It is important to study the mechanism underlying drug resistance in cancer and the ways by which drug resistance may be reversed.
Over the past few years, multiple mechanisms responsible for drug resistance have been proposed by different groups, which may be broadly divided into two types: Cellular and non-cellular resistance mechanisms (21,22). Cellular mechanisms include enzymes, targets of drugs and inside transport systems of cancer cells, whereas non-cellular mechanisms include extracellular factors, such as the microenvironment of the tumor and vascular accessibility (23–25). The present study focused on the generation of a drug-resistant cell line and the ways by which the chemoresistance may be reversed (26). The present study was able to successfully demonstrate that, instead of targeting multiple signaling pathways that are activated in drug resistance, it is possible to target a common signaling node that aids in the reversal of drug resistance.
Materials and methods
Drugs, reagents and chemicals
Doxorubicin, saracatinib and triciribine were purchased from Selleck Chemicals (Shanghai, China). RPMI-1640, radioimmunoprecipitation assay (RIPA) buffer, Hanks buffer, MTT and Bradford reagent were obtained from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany). Probes, SuperScript™ One-Cycle cDNA kit cDNA kits for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and the following primers were obtained from Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA): E-cadherinP-F, 5′-GGGGTACCTGTCTCTCTACAAAAAGGCA-3′ and E-cadherinP-R, 5′-GGAAGATCTGGGCTGGAGCGGGCTGGAGT-3′; epithelial cell adhesion molecule (EpCAM)P-F, 5′-CGCAGCTCAGGAAGAATGTG-3′ and EpCAMP-R, 5′-TGAAGTACACTGGCATTGACG-3′; vimentinP-F, 5′-GGCTCAGATTCAGGGGAACAGC-3′ and vimentinP-R, 5′-CAGGTTGTGCAGGTTGTTCTA-3′; N-cadherinP-F, 5′-CACTGCTCAGGACCCAGAT-3′ and N-cadherinP-R, 5′-TAAGCCGAGTGATGGTCC-3′; GAPDHP-F, 5′-GGTGTGAACGGATTTGGCCGTATTG-3′ and GAPDHP-R, 5′-CCGTTGAATTTGCCGTGAGTGGAGT-3′. Fetal bovine serum (FBS), Opti-MEM (cat. no. 11058021) and Anti-Anti were procured from Gibco (Thermo Fisher Scientific, Inc.). FuGENE 6 (cat. no. PRE2691) was obtained from Promega Corporation (Madison, MI, USA). Phosphorylated (p)-SRC 416 (10% SDS-PAGE; cat. no. 2101; dilution 1:1,000), AKT (10% SDS-PAGE; cat. no. 9272; dilution 1:1,000), phosphatase and tensin homolog (PTEN; 10% SDS-PAGE; cat. no. 9188; dilution 1:1,000), epidermal growth factor receptor (EGFR; 6% SDS-PAGE; cat. no. 4267; dilution 1:1,000), p-SRC 416 (10% SDS-PAGE; cat. no. 2101; dilution 1:1,000), insulin-like growth factor 1 receptor (IGF-1R; 8% SDS-PAGE; cat. no. 9750; dilution 1:1,000), p-AKT ser473 (10% SDS-PAGE; cat. no. 4060; dilution 1:1,000), SRC (10% SDS-PAGE; cat. no. 2109; dilution 1:1,000), E-cadherin (6% SDS-PAGE; cat. no. 14472; dilution 1:1,000), EpCAM (12% SDS-PAGE; cat. no. 2929; dilution 1:1,000), N-cadherin (8% SDS-PAGE; cat. no. 13116; dilution 1:1,000), vimentin (10% SDS-PAGE; cat. no. 5741; dilution 1:1,000), β-actin (cat. no. 4970; dilution 1:2,000), anti-rabbit secondary antibody (cat. no. 93702; dilution 1:2,500), anti-mouse secondary antibody (cat. no. 14709; dilution 1:2,500), small interfering (si)RNA control (cat. no. 6568; dilution 100 nM), siRNA EGFR (cat. no. 6480; dilution 200 nM) and siRNA insulin-like growth factor receptor (IGFR; cat. no. 6610; dilution 200 nM) were obtained from Cell Signaling Technology Inc. (Danvers, MA, USA). shRNA control (cat. no. sc-108060; dilution 1 µg), shRNA EGFR (cat. no. sc-29301; dilution 1 µg) and shRNA IGFR (cat. no. sc-35638; dilution 1 µg) were obtained from Santa Cruz Biotechnology Inc. (Dallas, TX, USA).
Cell line and culture conditions
The LS180 cell line was purchased from American Type Culture Collection (Manassas, VA, USA), cultured in RPMI-1640 supplemented with 10% FBS and 1% antibiotic Anti-Anti and grown in an incubator at 37°C with 5% CO2 and 95% humidity. For generation of doxorubicin resistance, LS180 cells were continuously treated with the drug (from 10 nM up to 30 µM) for a period of 6 months in an incubator at 37°C containing 5% CO2 and 95% humidity. Prior to two days of subsequent experiments, resistant cells were grown in media without the addition of any drug. For overexpression/knockdown, transient transfection was performed. Cells were grown in six-well plates at a density of 1×106 cells in transfection media (Opti-MEM). The transfection mixture contains 35 µl of FuGENE 6 and siRNA (1:300) or shRNA (1:200) was prepared and incubated for 30 min prior to addition to the cells. After 24 h of transfection, cells were washed with PBS and lysates were prepared for western blot analysis.
Cell viability assay
The parental and doxorubicin-resistant LS180 cell lines were seeded at a density 1.5×104 cells/well and allowed to grow for 24 h to attain morphology and stationary state. After 24 h, both parental and doxorubicin-resistant cells were treated with doxorubicin at a concentration of 2, 5, 10, 20, 40, 80 and 100 µM in an incubator containing 5% CO2 at at 37°C and 95% humidity for 12 and 24 h. MTT was added at a concentration of 2.5 mg/ml to each well before 4 h of termination. Finally, the formazan crystals were dissolved in dimethyl sulfoxide and absorbance was measured at 570 nm using a synergy MX plate reader (BioTek Instruments, Inc., Winooski, VT, USA).
Western blotting
Parental and resistant LS180 cells were seeded in 60-mm dishes at a density of 1.5×106 cells for 24 h. After 24 h, the cells were treated with different inhibitors (1 µM doxorubicin, 1 µM saracatinib and 1 µM triciribine). These cells were lysed in RIPA buffer and protein estimation was performed using the Bradford method. Proteins (70 µg) were separated on 10% SDS-PAGE and transferred onto a nitrocellulose membrane at 100 V for 2 h. Membranes were blocked in 5% fat-free skimmed milk for 1 h at room temperature to avoid non-specific binding. After 1 h, primary antibodies were added and incubated overnight at 4°C. Protein blots were washed with Tris-buffered saline-Tween-20 twice for 5 min each. Subsequently, horseradish peroxidase-conjugated secondary antibody was added at room temperature for 1 h, followed by washing thrice with blocking buffer for 5 min each. Finally, using an enhanced chemiluminescence kit (GE Healthcare, Chicago, IL, USA), the bands of proteins were analyzed on an X-ray film. Quantification of all western blots were was performed by normalized by β-actin and using Image J software (v.1.48, National Institutes of Health, Bethesda, MD, USA).
Colony formation assay
Doxorubicin-resistant LS180 cells were seeded in a six-well plate and allowed to grow for 24 h. After 24 h, these cells were treated with 1 µM doxorubicin and 1 µM saracatinib alone and in combination in an incubator at 37°C with 5% CO2 and 95% humidity for 48 h. Following this, cells were trypsinized and replated in a six-well plate at a density of 500 cells/well and allowed to grow for 21 days. On experiment termination, cells were first washed thrice with PBS and then fixed in 4% paraformaldehyde for 10–15 min at room temperature. Crystal violet (0.06%) at room temperature was used to stain live cells at 25°C for 10 min and images were captured using a light microscope at a magnification of ×30 (Olympus Corporation, Tokyo, Japan).
Three-dimensional (3D) sphere formation assay
Using MammoCult medium (Stemcell Technologies, Inc., Vancouver, BC, USA), single Doxo LS180 cells were seeded in ultra-low attachment plates at a density of 1×105 cells/well and allowed to grow for 7 days. Following this, the cells were treated with doxorubicin (1 µM) and saracatinib (1 µM) alone and in combination for 48 h. Subsequent to treatment, the primary spheres were dissociated by pipetting and single cells were replated in ultra-low attachment 6-well plates at a density of 5×104 cells/well. Secondary spheres were counted using a light microscope at a magnification of ×30 after 21 days of incubation.
mRNA quantification
Parental and doxorubicin-resistant cells were cultured and total RNA was isolated using TRIzol reagent (cat. no. T9424 obtained from Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). RNA was purified using an RNeasy mini kit (Qiagen China Co., Ltd., Shanghai, China). Purified RNA was first used to generate cDNA using an M-MLV RT kit (Promega Corporation, Madison, WI, USA) according to the instructions of manufacturer and then qPCR analysis was conducted using a TaqMan universal PCR master mix (Roche Applied Science, Penzberg, Germany). Reverse transcription was performed according to the following thermocycling conditions: Denaturation at 94°C for 30 sec and annealing and elongation at 72°C for 1 min, followed use of the aforementioned primers on an ABI PRISM sequencing detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.) The relative fold change of differential inducible expression of the genes vs. control group was quantified by using 2−∆∆Cq method (27).
Statistical analysis
GraphPad Instat3 software (GraphPad software Inc., La Jolla, CA, USA) was used for statistical analysis. All experiments were performed three times and values of each experiment were demonstrated as the mean ± standard deviation. For comparison of each experiment, one-way analysis of variance was performed followed by Bonferroni's test. P<0.05 was considered to indicate a statistically significant difference.
Results
Development of a chemoresistant cell line
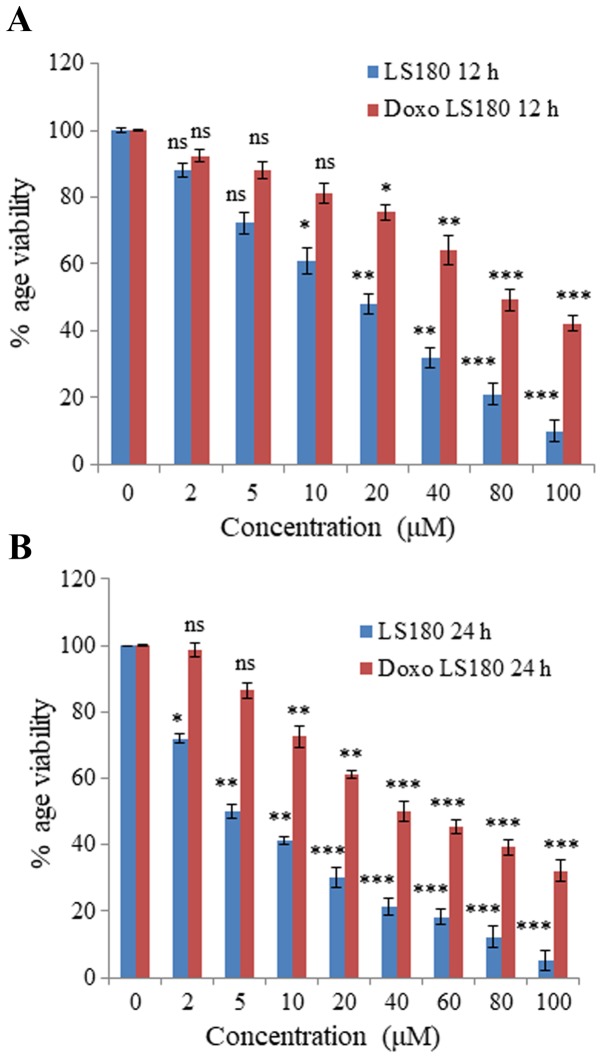
Colon cancer cell line LS180 was selected to generate resistance towards doxorubicin. Following attainment of 80% confluency, cells were treated with doxorubicin beginning with a low dose of 50 nM and treatment was retained continuously until the growth of cells began to decrease and they demonstrated morphological changes. The resistance of these cells was obtained in 6 months and was confirmed by an MTT assay. The IC50 of doxorubicin-resistant cells was 80 µM whereas the parental cancer cell line was 18 µM at 12 h (Fig. 1A). Notably, IC50 of doxorubicin in parental cell line was 5 µM, whereas in resistant cells the IC50 was increased up to 40 µM at 24 h (Fig. 1B).
Figure 1.
Generation of chemoresistant LS180 cells. Colon cancer cell line LS180 was selected and chemoresistance against Doxorubicin was generated by treating the cells with increasing concentrations of Doxorubicin over a period of 6–8 months. Percentage viability of parental and resistant cells following (A) 12 and (B) 24 h of Doxo treatment. The IC50 of parental cells was 5 µM, which was increased up to 40 µM, confirming the generation of resistance, as determined by an MTT assay. Data is presented as the mean ± standard deviation of three different experiments. Statistical comparisons were made using Bonferroni's method. *P<0.05, **P<0.01 and ***P<0.001 vs. 0 µM groups. Doxo, doxorubicin; Doxo LS180, doxorubicin-resistant LS180 cells.
Doxorubicin resistance induces an EMT-like phenotype
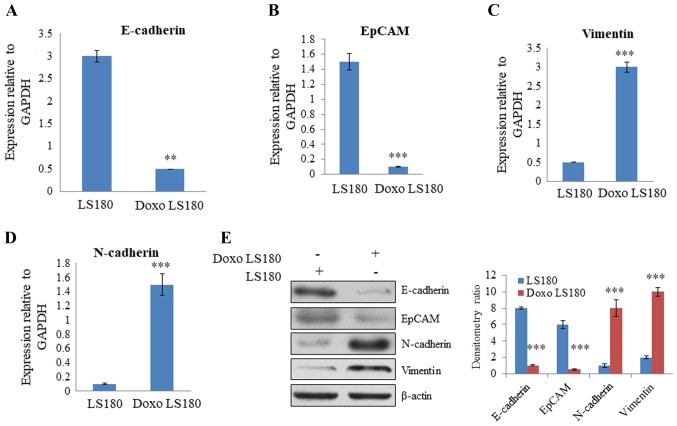
For generation of a stable doxorubicin-resistant cell line, LS180 cells were continuously treated with doxorubicin for 6 months. After 6 months, there was a marked difference in the morphology of parental and resistant cells. Resistant cells were mesenchymal in shape (analyzed by microscopy; data not shown). RT-qPCR analysis of EMT-related genes, including N-cadherin, vimentin, EpCAM and E-cadherin, was performed to further analyze the difference between parental and resistant cells. The results revealed that there was a significant decrease in the expression of E-cadherin and EpCAM, two epithelial markers, in resistant cells compared with the level in parental cells (Fig. 2A and B). Furthermore, there was a significant increase in the expression level of mesenchymal markers, vimentin and N-cadherin, in the doxorubicin-resistant cells compared with the levels observed in the parental cells (Fig. 2C and D). Western blotting demonstrated that E-cadherin and EpCAM were expressed at a higher level in parental cells than in resistant cells. However, the expression of mesenchymal markers, N-cadherin and vimentin, were significantly increased in cells resistant to doxorubicin, and the epithelial markers, E-cadherin and EpCAM, demonstrated reduced expression levels compared with the levels in parental cells (Fig. 2E). Together these results indicate that doxorubicin leads to resistance in colon cancer and induces an EMT-like phenotype.
Figure 2.
Induction of EMT in LS180 cells by Doxo resistance. Quantification of epithelial markers (A) E-cadherin and (B) EpCAM by RT-qPCR using GAPDH as the normalizing marker. Quantification of mesenchymal markers (C) vimentin and (D) N-cadherin by RT-qPCR using GAPDH as the normalizing marker. (E) Assessment of EMT phenotype at the protein by western blotting demonstrated decreased expression of epithelial markers and increased expression of mesenchymal markers in Doxo-resistant cells. Data is presented as the mean ± standard deviation of three different experiments. Statistical comparisons were made using Bonferroni's method. **P<0.01 and ***P<0.001 vs. LS180 cells. EMT, epithelial to mesenchymal transition; Doxo, doxorubicin; Doxo LS180, doxorubicin-resistant LS180 cells; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; EpCAM, epithelial cell adhesion molecule.
SRC is activated in doxorubicin-resistant cells
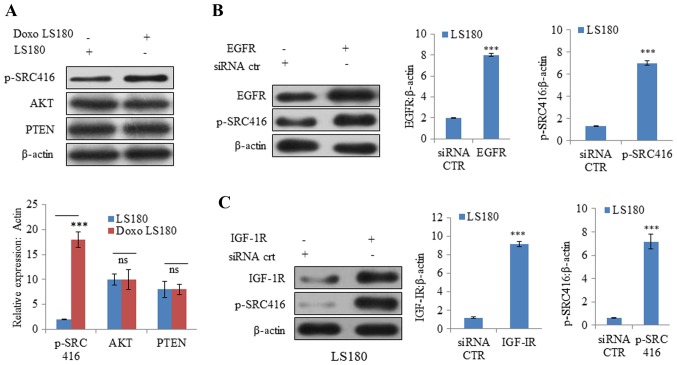
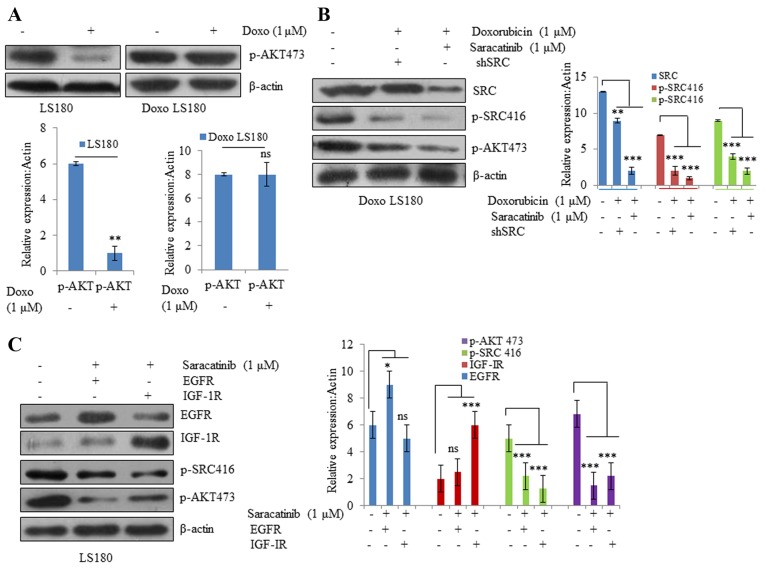
Cancer cells are known to be evolved that undergo reprogramming and become drug-resistant (28). In resistant LS180 cells, SRC was observed to be activated (indicated by phosphorylation at position 416) compared with the parental cell line (Fig. 3A). However, there was no notable change in the protein expression levels of PTEN and AKT, which further confirmed that resistance was due to doxorubicin and not due to the presence of any pre-existing phosphoinositide 3-kinase mutation (Fig. 3A).
Figure 3.
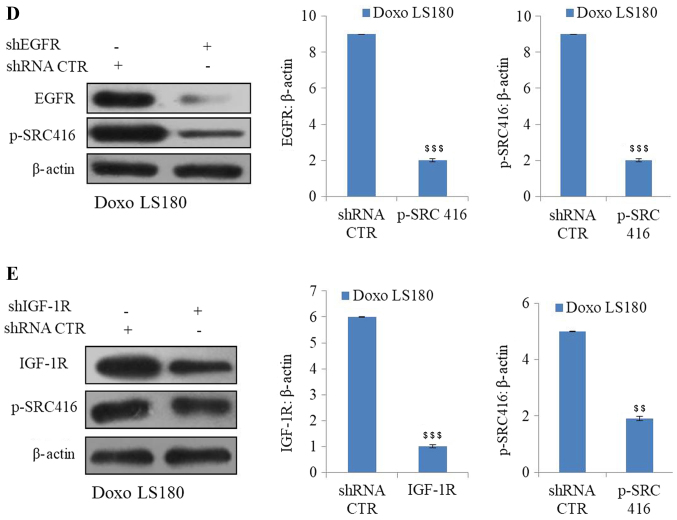
SRC is activated by Doxo resistance. (A) Doxo resistance induces activation of oncogenic protein SRC independent of AKT and PTEN, as demonstrated by increased phosphorylation of SRC at Y416 with no notable effect on AKT and PTEN. Overexpression of receptor tyrosine kinases (B) EGFR activates SRC activation in Doxo LS180 and (C) IGF-1R also resulted SRC activation in LS180 cells. Knockdown of tyrosine kinases (D) EGFR and (E) IGF-1R in Doxo-resistant cells led to decreased phosphorylation of SRC. Data is presented as the mean ± standard deviation of three different experiments. Statistical comparisons were made using Bonferroni's method ***P<0.001 vs. siRNA CTR; $$P<0.01 and $$$P<0.001 vs. shRNA CTR. SRC, steroid receptor coactivator; Doxo, doxorubicin; Doxo LS180, doxorubicin-resistant LS180 cells; PTEN, phosphatase and tensin homolog; EGFR, epidermal growth factor receptor; IGF-1R, insulin-like growth factor 1 receptor; p, phosphorylated; siRNA, small interfering RNA; sh, short hairpin; CTR, control.
Subsequently, the roles of other RTKs, including EGFR and IGF-1R, were examined in SRC activation. Overexpression of EGFR and IGF-1R resulted in increased phosphorylation at position 416, indicating SRC activation (Fig. 3B and C). Furthermore, knockdown of EGFR and IGF-1R in doxorubicin-resistant cells resulted in SRC inactivation (decreased phosphorylation of SRC at position 416; Fig. 3D and E).
Activation of RTKs by SRC induces doxorubicin resistance
To understand the mechanism of doxorubicin-induced SRC activation in resistant cells, SRC activity was inhibited by using a small molecule SRC inhibitor, saracatinib, as well as SRC siRNA. It was observed that AKT phosphorylation was significantly inhibited by doxorubicin in the parental cell line, whereas doxorubicin-resistant cells were resistant towards doxorubicin-mediated inhibition of AKT (Fig. 4A). However, when resistant cells were treated with SRC inhibitor saracatinib and SRC shRNA in combination with doxorubicin, there was a significant decrease in the phosphorylation of AKT compared with non-treated cells (Fig. 4B). In EGFR and IGF-1R overexpressing cells, SRC inhibition led to significant inhibition of AKT compared with non-treated cells (Fig. 4C). These results suggest that different RTKs, along with their downstream targets that are activated due to drug resistance, may be effectively blocked by inhibiting SRC.
Figure 4.
SRC activation drives Doxo resistance. (A) Parental LS180 cells responded to Doxo and there was a significant decrease in phosphorylation of AKT; whereas in the resistant cells, there was no significant difference in the p-AKT levels following treatment with Doxo. (B) SRC inhibition by saracatinib or SRC shRNA resensitized the resistant cells towards Doxo and there was a decrease in the expression levels of p-AKT and p-SRC. (C) SRC inhibition resulted in inhibition of AKT even in the presence of overexpressed tyrosine kinases EGFR and IGF-1R. Data are presented as the mean ± standard deviation of three different experiments. Statistical comparisons were made using Bonferroni's method. *P<0.05, **P<0.01 and ***P<0.001 vs. LS180 and Doxo LS180 without Doxo treatment. SRC, steroid receptor coactivator; Doxo, doxorubicin; Doxo LS180, doxorubicin-resistant LS180 cells; p, phosphorylated; EGFR, epidermal growth factor receptor; IGF-1R, insulin-like growth factor 1 receptor, ns, not significant.
Inhibiting SRC in vitro reverses doxorubicin resistance
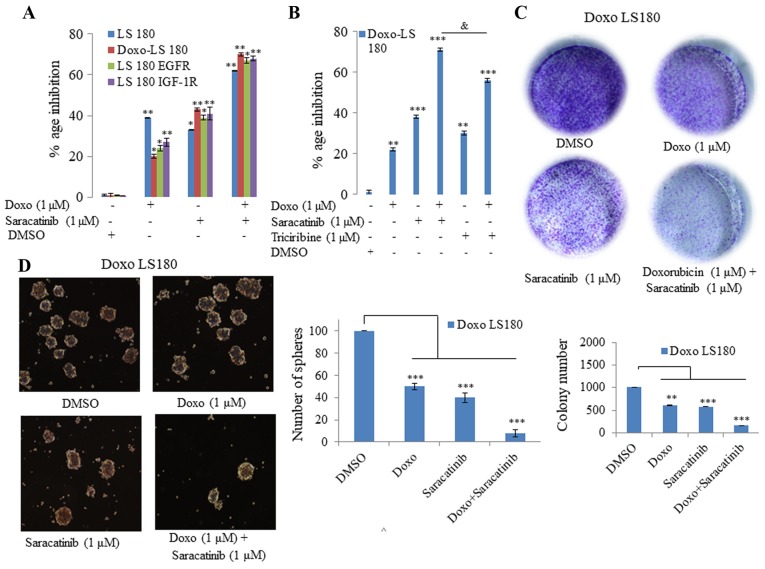
The present study demonstrated that SRC was driving doxorubicin resistance and served as a common node of various signaling pathways. Therefore, it was hypothesized that targeting SRC may be effective in reverting doxorubicin resistance. In order to test this hypothesis, the orally available SRC inhibitor, saracatinib, was utilized. As demonstrated in Fig. 5, it was observed that saracatinib not only sensitized the doxorubicin-resistant cells to doxorubicin, but also induced inhibition of cell growth in EGFR- and IGF-1R-overexpressing cells compared with doxorubicin (Fig. 5A). Additionally, 3D tumor spheroid formation (Fig. 5D) and colony forming potential (Fig. 5C) of doxorubicin-resistant cells was inhibited when the cells were treated with doxorubicin in combination with saracatinib.
Figure 5.
SRC inhibition reverses Doxo resistance in vitro. (A) Saracatinib in combination with Doxo was more effective in all four models tested (LS180, Doxo LS180, LS180 EGFR and LS180 IGF-1R) than when administered alone, as determined by an MTT assay. (B) MTT assay determined that Doxo in combination with the SRC inhibitor was more effective than a combination of Doxo and AKT inhibitor, triciribine. (C) Combination of SRC inhibitor, saracatinib, with Doxo inhibited the clonogenic potential effectively in Doxo LS180 cells. (D) Doxo in combination with SRC inhibitor, saracatinib, efficiently blocked the proliferation of Doxo LS180 cells, as observed by inhibition of mammospheres. Scale bar, 100 µM. Data are presented as the mean ± standard deviation of three different experiments. Statistical comparisons were made using Bonferroni's method. *P<0.05, **P<0.01 and ***P<0.001 vs. LS180 and Doxo LS180 without Doxo treatment. &P<0.05 as indicated. SRC, steroid receptor coactivator; Doxo, doxorubicin; Doxo LS180, doxorubicin-resistant LS180 cells; EGFR, epidermal growth factor receptor; IGF-1R, insulin-like growth factor 1 receptor; DMSO, dimethyl sulfoxide.
Subsequently, the present study investigated whether inhibition of AKT with its inhibitor, triciribine, induced the same effect in combination with doxorubicin in doxorubicin-resistant cells as that observed following SRC inhibition. However, it was observed that AKT inhibition in combination with doxorubicin was not as effective as SRC inhibition (Fig. 5B). Therefore, saracitinib demonstrated a greater effect at re-sensitizing the cells towards doxorubicin than the AKT inhibitor, triciribine.
Discussion
In colon cancer, which is one of the leading causes of cancer-related mortality worldwide, drug resistance remains one of the major challenges to be tackled (29). However, there are currently some effective strategic approaches including the combination of ATP-binding cassette transporter and EGFR inhibitors with conventional drugs, against cancer that have been effective against drug resistance in colon cancer (30). There are various targeted therapies used for treatment of colon cancer, such as bevacizumab, which is a Food and Drug Agency-approved first-line treatment, as well as a second-line therapy for colon cancer approved in 2017 (31). Regorafenib was approved in 2012 for metastatic colon cancer, and ramucirumab along with ziv-aflibercept is the second-line treatment in metastatic colorectal cancer (32). Doxorubicin, a potent anticancer drug that is widely used to fight various cancer types, including colon cancer, is cost effective compared with other anticancer drugs (33). The present study generated a drug-resistant cell line against doxorubicin by treating the cell line with the drug for ~6 months. Drug resistance induced EMT in the colon cancer cell line, LS180, as indicated by a decrease in the expression of epithelial marker, E-cadherin, in resistant cells as compared with the levels observed in parental cells. There was a visible increase in the expression of mesenchymal markers, including vimentin and N-cadherin, in the drug-resistant cells compared with the expression in parental cells at the mRNA and protein levels, as determined by RT-qPCR and western blotting, respectively.
SRC, which acts as a signaling node between cell surface receptors and cytoplasmic pathways, is also activated in drug-resistant cells (34). In fact, SRC activation appears to be the predominant factor responsible for drug resistance, as it results in the activation of the downstream AKT pathway (35). SRC is also activated due to overexpression of RTKs, including EGFR and IGF-1R (36,37). In the present study, it was demonstrated that SRC inhibition not only resulted in reversal of drug resistance, but also led to inactivation of AKT in resistant cells. SRC was also inhibited upon inhibition of EGFR and IGF-1R. The present results led to the conclusion that different RTKs, along with their downstream targets that are activated due to drug resistance, may be effectively blocked by SRC inhibition. Furthermore, in vitro inhibition of SRC in combination with doxorubicin also leads to inhibition of AKT in EGFR- and IGF-1R-overexpressing cells. The present study revealed that 3D tumor spheroid formation and colony forming potential of doxorubicin-resistant cells was inhibited when cells were treated with doxorubicin in combination with SRC inhibitor, saracatinib. The present study also indicated that inhibition of AKT in combination with doxorubicin was not as effective as treatment with SRC inhibitor, saracatinib. Notably, previous studies have suggested that paclitaxel and cisplatin inhibit SRC tyrosine kinase, which enhances toxicity to human ovarian cancer cells as well as mouse ovarian cancer cells (38,39).
In conclusion, the present study demonstrated that SRC is activated in doxorubicin resistance, as well as overexpression of RTKs, including EGFR and IGF-1R, which lead to further activation of downstream pathways, such as the AKT signaling pathway. Apoptosis assays in doxorubicin-resistant LS180 and parental LS180 cell lines are intended to be performed in future studies. The combinatorial treatment of doxorubicin along with inhibition of SRC not only helps in the inactivation of the AKT pathway, but also in the reversal of resistance.
References
- 1.Haggar FA, Boushey RP. Colorectal cancer epidemiology: Incidence, mortality, survival, and risk factors. Clin Colon Rectal Surg. 2009;22:191–197. doi: 10.1055/s-0029-1242458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sheth KR, Clary BM. Management of hepatic metastases from colorectal cancer. Clin Colon Rectal Surg. 2005;18:215–223. doi: 10.1055/s-2005-916282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoyagi T, Terracina KP, Raza A, Takabe K. Current treatment options for colon cancer peritoneal carcinomatosis. World J Gastroenterol. 2014;20:12493–12500. doi: 10.3748/wjg.v20.i35.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nordlinger B, Sorbye H, Glimelius B, Poston GJ, Schlag PM, Rougier P, Bechstein WO, Primrose JN, Walpole ET, Finch-Jones M, et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): A randomised controlled trial. Lancet. 2008;371:1007–1016. doi: 10.1016/S0140-6736(08)60455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colon Cancer Laparoscopic or Open Resection Study Group. Buunen M, Veldkamp R, Hop WC, Kuhry E, Jeekel J, Haglind E, Påhlman L, Cuesta MA, Msika S, et al. Survival after laparoscopic surgery versus open surgery for colon cancer: Long-term outcome of a randomised clinical trial. Lancet Oncol. 2009;10:44–52. doi: 10.1016/S1470-2045(08)70310-3. [DOI] [PubMed] [Google Scholar]
- 6.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: An evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- 7.Diaz LA, Jr, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, Allen B, Bozic I, Reiter JG, Nowak MA, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 9.Nikaido H. Prevention of drug access to bacterial targets: Permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 10.Ozben T. Mechanisms and strategies to overcome multiple drug resistance in cancer. FEBS Lett. 2006;580:2903–2909. doi: 10.1016/j.febslet.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Ghannoum MA, Rice LB. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim AY, Kwak JH, Je NK, Lee YH, Jung YS. Epithelial-mesenchymal transition is associated with acquired resistance to 5-fluorocuracil in HT-29 colon cancer cells. Toxicol Res. 2015;31:151–156. doi: 10.5487/TR.2015.31.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosanò L, Cianfrocca R, Spinella F, Di Castro V, Nicotra MR, Lucidi A, Ferrandina G, Natali PG, Bagnato A. Acquisition of chemoresistance and EMT phenotype is linked with activation of the endothelin A receptor pathway in ovarian carcinoma cells. Clin Cancer Res. 2011;17:2350–2360. doi: 10.1158/1078-0432.CCR-10-2325. [DOI] [PubMed] [Google Scholar]
- 15.Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A, Rimm DL, Wong H, Rodriguez A, Herschkowitz JI, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene. 2010;29:4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiscox S, Jiang WG, Obermeier K, Taylor K, Morgan L, Burmi R, Barrow D, Nicholson RI. Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of beta-catenin phosphorylation. Int J Cancer. 2006;118:290–301. doi: 10.1002/ijc.21355. [DOI] [PubMed] [Google Scholar]
- 18.Jones VS, Huang RY, Chen LP, Chen ZS, Fu L, Huang RP. Cytokines in cancer drug resistance: Cues to new therapeutic strategies. Biochim Biophys Acta. 2016;1865:255–265. doi: 10.1016/j.bbcan.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Sui H, Zhu L, Deng W, Li Q. Epithelial-mesenchymal transition and drug resistance: Role, molecular mechanisms, and therapeutic strategies. Oncol Res Treat. 2014;37:584–589. doi: 10.1159/000367802. [DOI] [PubMed] [Google Scholar]
- 20.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 21.Guarino M. Src signaling in cancer invasion. J Cell Physiol. 2010;223:14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 22.Alfarouk KO, Stock CM, Taylor S, Walsh M, Muddathir AK, Verduzco D, Bashir AH, Mohammed OY, Elhassan GO, Harguindey S, et al. Resistance to cancer chemotherapy: Failure in drug response from ADME to P-gp. Cancer Cell Int. 2015;15:71. doi: 10.1186/s12935-015-0221-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu C, Krishnan J, Xu XY. Intrinsic and induced drug resistance mechanisms: In silico investigations at the cellular and tissue scales. Integr Biol (Camb) 2015;7:1044–1060. doi: 10.1039/C5IB00088B. [DOI] [PubMed] [Google Scholar]
- 24.Florea AM, Büsselberg D. Cisplatin as an anti-tumor drug: Cellular mechanisms of activity, drug resistance and induced side effects. Cancers (Basel) 2011;3:1351–1371. doi: 10.3390/cancers3011351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imming P, Sinning C, Meyer A. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 26.Trédan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99:1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Jang M, Kim SS, Lee J. Cancer cell metabolism: Implications for therapeutic targets. Exp Mol Med. 2013;45:e45. doi: 10.1038/emm.2013.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDermott M, Eustace AJ, Busschots S, Breen L, Crown J, Clynes M, O'Donovan N, Stordal B. In vitro development of chemotherapy and targeted therapy drug-resistant cancer cell lines: A practical guide with case studies. Front Oncol. 2014;4:40. doi: 10.3389/fonc.2014.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma X, Yu H. Global burden of cancer. Yale J Biol Med. 2006;79:85–94. [PMC free article] [PubMed] [Google Scholar]
- 31.Tartari F, Santoni M, Pistelli M, Berardi R. Healthcare cost of HER2-positive and negative breast tumors in the United States (2012–2035) Cancer treatment reviews. 2017;60:12–17. doi: 10.1016/j.ctrv.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Giantonio BJ, Catalano PJ, Meropol NJ, O'Dwyer PJ, Mitchell EP, Alberts SR, Schwartz MA, Benson AB., III Eastern Cooperative Oncology Group Study E3200: Bevacizumab in combination with oxaliplatin, fluorouracil, and leucovorin (FOLFOX4) for previously treated metastatic colorectal cancer: Results from the Eastern Cooperative Oncology Group Study E3200. J Clin Oncol. 2007;25:1539–1544. doi: 10.1200/JCO.2006.09.6305. [DOI] [PubMed] [Google Scholar]
- 33.Sonowal H, Pal PB, Wen JJ, Awasthi S, Ramana KV, Srivastava SK. Aldose reductase inhibitor increases doxorubicin-sensitivity of colon cancer cells and decreases cardiotoxicity. Sci Rep. 2017;7:3182. doi: 10.1038/s41598-017-03284-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martino A, San KK, Daniel N, Ezekiel U. Chemoresistance-induced epithelial-mesenchymal transition of a colorectal cancer cell line. FASEB J. 2015;29(Suppl 1):S721.17. [Google Scholar]
- 35.Sen B, Johnson FM. Regulation of SRC family kinases in human cancers. J Signal Transduct. 2011;2011:865819. doi: 10.1155/2011/865819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–7968. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 37.Furcht CM, Buonato JM, Lazzara MJ. EGFR-activated Src family kinases maintain GAB1-SHP2 complexes distal from EGFR. Sci Signal. 2015;8:ra46. doi: 10.1126/scisignal.2005697. [DOI] [PubMed] [Google Scholar]
- 38.Pengetnze Y, Steed M, Roby KF, Terranova PF, Taylor CC. Src tyrosine kinase promotes survival and resistance to chemotherapeutics in a mouse ovarian cancer cell line. Biochem Biophys Res Commun. 2003;309:377–383. doi: 10.1016/j.bbrc.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 39.Chen T, Pengetnze Y, Taylor CC. Src inhibition enhances paclitaxel cytotoxicity in ovarian cancer cells by caspase-9-independent activation of caspase-3. Mol Cancer Ther. 2005;4:217–224. [PubMed] [Google Scholar]