Abstract
Objective
The aim of this study was to identify variants associated with familial late-onset Alzheimer disease (AD) using whole-genome sequencing.
Methods
Several families with an autosomal dominant inheritance pattern of AD were analyzed by whole-genome sequencing. Variants were prioritized for rare, likely pathogenic variants in genes already known to be associated with AD and confirmed by Sanger sequencing using standard protocols.
Results
We identified 2 rare ABCA7 variants (rs143718918 and rs538591288) with varying penetrance in 2 independent German AD families, respectively. The single nucleotide variant (SNV) rs143718918 causes a missense mutation, and the deletion rs538591288 causes a frameshift mutation of ABCA7. Both variants have previously been reported in larger cohorts but with incomplete segregation information. ABCA7 is one of more than 20 AD risk loci that have so far been identified by genome-wide association studies, and both common and rare variants of ABCA7 have previously been described in different populations with higher frequencies in AD cases than in controls and varying penetrance. Furthermore, ABCA7 is known to be involved in several AD-relevant pathways.
Conclusions
We conclude that both SNVs might contribute to the development of AD in the examined family members. Together with previous findings, our data confirm ABCA7 as one of the most relevant AD risk genes.
Several genome-wide association studies (GWASs) have identified ABCA7 (ATP-binding cassette transporter A7) as a risk factor for sporadic late-onset Alzheimer disease (AD).1–3 ABCA7 encodes a protein with major function in lipid transport.4 The protein is involved in AD pathology, as it was demonstrated to play a role in formation, clearance, and aggregation of amyloid beta, the etiologic agent in AD.5,6 Recently, multiple rare loss-of-function variants in ABCA7 associated with AD risk and possible causal variants in familial cases and pedigrees have been identified through sequencing efforts.7–11 Alterations in ABCA7 have not only been observed in European but also in African American12 and Asian13,14 populations either by GWASs or targeting sequencing with varying minor allele frequencies (MAFs). In addition, a protective ABCA7 variant has also been described, emphasizing the role of this gene in AD.15 We now present the data of 2 rare variants of ABCA7 in 2 German families.
Methods
Standard protocol approvals, registrations, and patient consents
All individuals provided written informed consent before their participation in this study for the clinical evaluation and genetic analysis of leukocyte DNA. Clinical phenotyping, whole-genome sequencing (WGS), and genetic analysis were approved by the Central Ethics Committee of the Bavarian Medical Association and the Ethics Review Panel of the University of Luxembourg.
Patient information
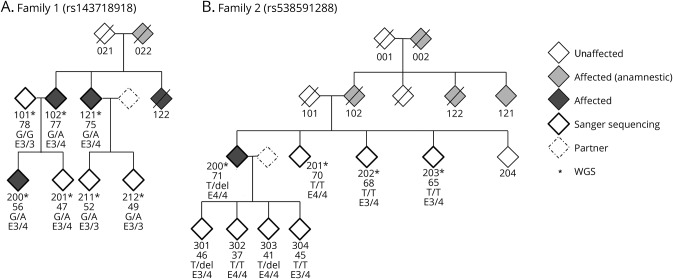
Two families with an autosomal dominant inheritance pattern of AD were analyzed, and a pedigree chart is shown in figure 1. The 3 patients with AD sequenced in family 1 had reported ages at onset of <56, 70–75, and 71–77 years. The APOE status for all 3 patients was ε3/4. Family members 021, 022, and 122 died at the age of 47, 56, and 75 years, respectively. The patient with AD in family 2 had an age at onset of 66 years; the APOE status was ε4/4. Family members 101 and 102 died at the age of 74 and 73, respectively. For 001, 002, and 122, the age at death is unknown. Blood samples were taken from 7 (family 1) and 8 (family 2) family members, respectively, and DNA was extracted from leukocytes using standard procedures.
Figure 1. Pedigree charts.
The age at examination of each individual sequenced in this study is given beneath the identifier number. Individuals diagnosed with AD are indicated as affected (dark gray), individuals with AD-like symptoms reported by their family members are indicated in light gray. (A) Pedigree of family 1. The genotypes are wild type (G/G) or the alteration (G/A) that causes the ABCA7 missense mutation. (B) Pedigree of family 2. The genotypes are wild type (T/T) or the alteration (T/del) that causes the ABCA7 frameshift mutation.
WGS and analysis
WGS was performed by Complete Genomics Inc. (CG, Mountain View, CA) using their proprietary paired-end, nanoarray-based sequencing-by-ligation technology.16 Sequencing, quality control, mapping, and variant calling for the sequencing data were performed by CG as part of their sequencing service using the Standard Sequencing Service pipeline version 2.0. Sequencing reads were mapped against NCBI Build 37. For further analysis, only single nucleotide variants (SNVs) and small insertions, deletions, and block substitutions up to a size of about 50 nt (indels) were used.
Variant prioritization
Variants were annotated by ANNOVAR17 (version 2015 March 12) using the NCBI RefSeq release 60 and the Ensembl release 74 genome. As input for our family WGS analysis pipeline,18 we first combined all variants from all genomes of every sequenced family member into the union of variants using CG analysis software (CGATOOLS, version 1.5) listvariant tool and the CG “var” files of all individuals per family as input. We used CGATOOLS testvariant to test each sample for the presence of each allele at every variant position from the union set of variants. We removed variants that were not called as high-quality calls (VQHIGH) in at least 1 individual. For both families, we used ISCA version 0.1.919 to search for shared haplotype blocks between pairs of samples and determined the number of shared alleles per block. For family 1, we filtered for haplotype blocks that shared 1 allele between the cases that was not shared with the unaffected individual 101 (figure 1A). For family 2, we excluded blocks where any pair of unaffected siblings shared 2 alleles (figure 1B). For each family, we applied an autosomal dominant inheritance model and filtered for exonic variants excluding synonymous variants and for variants in essential splice sites (±2 nucleotides from the exon boundary).
Variants within regions that are known to show very high mutation rates, like in mucins and olfactory receptors, were excluded (commonly mutated region).20 We filtered for rare variants having an MAF of less than 5% in the European American population of the 1000 Genomes Project, the European NHLBI ESP exomes, and the Non-Finnish European population from the ExAC project as well as in the control data set CG69 provided by CG. We annotated the remaining variants for pathogenicity by considering either loss-of-function variants (indels, stop-gain, stop-loss, and splice-site variants) or missense mutations predicted to be deleterious by SIFT, PolyPhen-2_HDIV, LRT, and MutationTaster or mutated at highly conserved positions (GERP_RS>3). All annotations were derived from dbNSFP3.0a.21 We further used a list of AD candidate genes that was collected from various GWAS in the dbGAP, the Alzgene database,22 and the Genotator tool23 to prioritize variants.
Population stratification
We performed population stratification by using EIGENSTRAT24 with default parameters. First, we merged our data with the 1000 Genomes data. We chose only the autosomal SNVs concordant with hapmap25 that were biallelic and not in linkage disequilibrium (LD) with each other by using PLINK (version 1.9)26 with the parameters—indep 50 5 2, MAF of at least 0.1, and minimum call rate of 0.99 to perform the population stratification. To identify the ethnicity of samples in the current study, the first and the second principal components were visualized.
Genetic and linkage analysis
For linkage analysis, high-quality SNV positions (complete call rate over all individuals from VQHIGH status in CG var files) were extracted from the WGS data. Variants with high LD were removed using PLINK 1.926; further thinning of variants was performed using mapthin.27 A set of 2,000 variants per chromosome along with the identified variants segregating with the disease through the pedigree were used to check for genotype errors and mendelian inconsistencies using MERLIN28 and were subsequently removed if they were identified as errors. The remaining variants were used for linkage analysis and their genomic positions were linearly interpolated based on the hapmap genetic map (2011-01_phaseII_B37). MERLIN was used to perform both haplotyping and multipoint parametric linkage analysis with a rare autosomal dominant disease model with a disease frequency of 0.0001 and penetrance of 0.0001, 1.0, and 1.0. Haplotyping results were visualized using HaploPainter.29 Using the R package “paramlink,”30 we calculated the power of each pedigree given as the maximal LOD scores for each family under an autosomal dominant inheritance model and 10,000 simulated markers. Relationship detection between all individuals was performed using software GRAB.20
Validation by sanger sequencing
The presence of both variants identified by WGS were validated and replicated by Sanger sequencing in each family member of both pedigrees using standard protocols with the following oligonucleotide sets: ABCA7_delT_1055908_FWD: 3′-TTGTCCACCCTTGACTCTGTGC-5′; ABCA7_delT_1055908_REV: 3′-CTTGAGACTGTCCTGAGCATCC-5′; ABCA7_rs143718918_FWD: 3′-ACAGGTCCATCTTGAGTGGC-5′; ABCA7_rs143718918_REV: 3′-GAGACCAGCCCCACATCC-5′.
Results
We used WGS to identify the genetic cause of AD in 10 families with an autosomal dominant pattern of inheritance. Among these families, variants in ABCA7 were identified in 2 families (family 1 and family 2). In family 1, we sequenced the genomes of 7 family members; 3 of them were diagnosed with AD (figure 1A). In the second family (family 2), we sequenced genomes of 4 family members, 1 affected index patient with AD (age at diagnosis 66 years) and 3 unaffected siblings (figure 1B). Relationship estimation20 confirmed all relationships in both families, given the original pedigree information. All families were self-reported of German ethnicity. European ancestry could be confirmed using EIGENSTRAT24 analysis (figure e-1, links.lww.com/NXG/A38).
WGS fully called on average 97% of genome and 98% of exonic regions. Seventy-seven percent of the genome and 86% of the exome were covered with at least 30X. We detected on average over all samples from both families 3,415,106 SNVs and 577,534 indels and substitutions per genome (table e-1, links.lww.com/NXG/A39).
In total, 7,516,717 and 6,139,540 variants (SNVs and indels) different from the reference genome were identified in at least 1 family member for family 1 and 2, respectively. Disease-associated variants were searched using an autosomal dominant inheritance model. In family 2, only variants present in the affected and not present in the unaffected individuals were considered. After strict quality, mode of inheritance, shared haplotype and MAF < 0.05 filtering, 51,269 and 56,962 variants remained (table e-2, links.lww.com/NXG/A39). After annotation using RefSeq, we screened for exonic splice-site affecting variants. After excluding variants within commonly mutated and brain-expressed genes, we prioritized the remaining variants according to their predicted pathogenicity and conservation. In total, only 11 (family 1, tables e-3 and e-5) and 8 (family 2, tables e-4 and e-6) variants were found in AD-related genes (table e-7) and were therefore considered to be relevant to AD. Strict variant filtering revealed for each family rare ABCA7 variants, rs143718918 (family 1) and rs538591288 (family 2) as best candidate variants.
European (Non-Finnish) population allele frequencies for rs143718918 and for rs538591288 add up to 0.0021 and 0.0016, respectively. Both variants are very rare (MAF < 0.01) in the European population according to the ExAC31 and 1000 Genomes (tables e-3 and e-4, links.lww.com/NXG/A39).
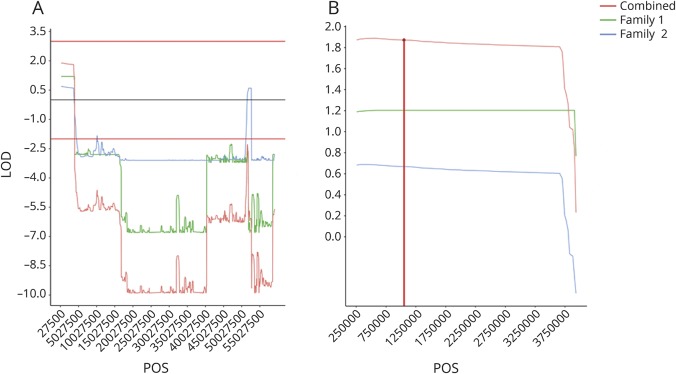
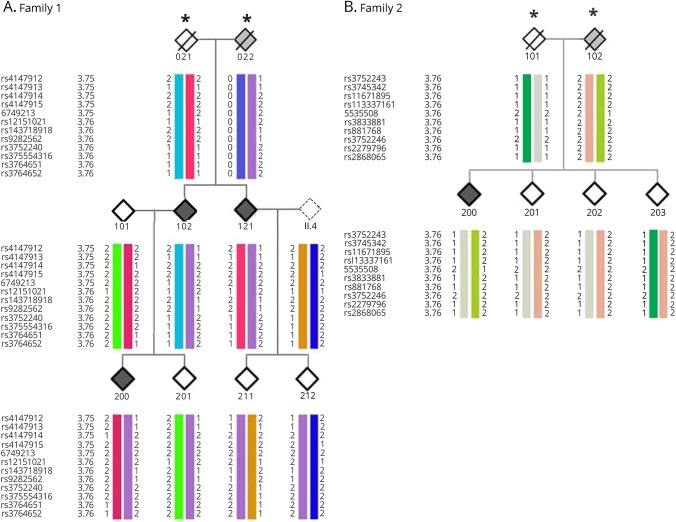
In addition, the performed linkage (figure 2, A and B) and haplotype block analysis (figure 3, A and B) show cosegregation and association of both variants with AD in both German families. We confirmed the presence of both variants in the initially screened and the additional family members by Sanger Sequencing (figure 1).
Figure 2. Linkage analysis of chromosome 19.
(A) The maximum LOD score (1.8) over the whole chromosome is seen in the region containing ABCA7. (B) Linkage analysis of the ABCA7 region on chromosome 19. The maximal LOD score (1.8) could be found on chromosome 19 in the region from 257,507 to 3,909,104 suggesting linkage, the region in red spans the gene ABCA7. Of the combined LOD score of 1.8 in the region spanning ABCA7, family 1 and family 2 contributed LOD scores of 1.2 and 0.6, respectively.
Figure 3. Segregating haplotype blocks.
The affected, unaffected, and disease status of unknown individuals are filled in black, white, and gray, respectively. An asterisk indicates the individuals who were not sequenced and their haplotypes were inferred. (A) The disease haplotype is indicated in purple. (B) The disease haplotype is indicated in light green. In both families, cosegregation of the disease haplotype including the corresponding ABCA7 variant can be seen in all affected individuals.
The SNV rs143718918 identified in family 1 causes a missense mutation of ABCA7 (c.2693G>A) that affects the ABC1 domain of the protein (p.R880Q). This variant was previously identified in patients with AD and controls of a larger Belgian cohort in a French as well as in an European cohort with early-onset patients and in patients with AD of a Caucasian cohort.7,32,33 We identified the SNV in all sequenced family members except for 1 healthy member (figure 1A). Three family members (201, 211, and 212) also carrying the risk variant were not affected and/or did not report cognitive deficits at the time of the last consultation, but were considerably younger than the affected family members and therefore possibly presymptomatic at the time of examination. As such, genetic counseling and clinical follow-up examinations will be conducted.
The second variant (rs538591288) identified in family 2 causes a frameshift deletion in exon 31 of ABCA7 (c.4208delT; p.L1403fs). This variant was also previously identified in patients with AD and controls of a larger Belgian cohort as well as in a French and in a European Cohort with early-onset AD patients.7,32,33 Of interest, in one of these studies, additional Italian relatives with EOAD carrying the deletion were reported.32 Furthermore, 2 groups have recently shown that p.L1403fs variant carriers had decreased ABCA7 protein levels but unchanged mRNA levels.32,34
We have identified the SNV (rs538591288) in 3 family members, including 2 children of the index patient (301 and 303, figure 1B), which were not diagnosed with AD but due to young age possibly presymptomatic at the time of examination. Of interest, both so far unaffected carriers reported already having occasional memory problems.
Discussion
We conducted a whole-genome sequencing (WGS) study to search for SNVs cosegregating with Alzheimer disease (AD) cases in German families. Of interest, we identified 2 rare variants of ABCA7 possibly contributing to AD pathogenesis in 2 families, respectively. ABCA7 is one of more than 20 AD risk loci that have so far been identified by GWASs and sequencing studies. ABCA7 is also involved in AD-relevant pathways (lipid metabolism, microglial phagocytosis, and altered amyloid-beta processing) and abundantly expressed in the brain.
We identified the rs143718918 to cosegregate with AD in family 1. The SNV causes a missense mutation of ABCA7 in exon 19 (c.2693G>A) that affects the ABC1 domain of the protein (p.R880Q) and is probably damaging. This variant has previously been identified by GWASs in Caucasians with late-onset AD.8 Furthermore, several studies reported the presence of this variant in AD and in control subjects of (1) a Belgian cohort,7 (2) a French EOAD cohort,33 and (3) an EOAD cohort including samples of diverse origin.32 Overall, the variant was present with higher frequency in AD cases compared with controls.
The rs538591288 cosegregated with AD in family 2 and causes a frameshift mutation in exon 31 of ABCA7 (c.4208delT; p.L1403fs). Initially, this SNV has been reported in an Icelandic cohort9 and was later also identified with higher frequency in cases than controls of German, Swedish, Italian,32 French,33 and Belgian7 cohorts. Mutation carriers express lower levels of full-length ABCA7 protein with unchanged mRNA expression levels.32,34 However, it has been reported that by in-frame exon skipping of the premature termination codon bearing exon 31, the transcript escapes nonsense-mediated mRNA decay.7,32 Exon skipping leads to the production of a shorter version of ABCA7 protein, which might partly compensate for the reduced full-length protein levels and might cause incomplete penetrance of rs538591288.
It has to be mentioned that we cannot exclude that other variations might cause additive effects on the development of AD in both families. Because of the previously shown involvement of ABCA7 in AD, the presented variants represent the most promising candidates. Together, our results support the notion that rare variants of ABCA7 exert considerable risk to the development of AD.
Acknowledgment
The authors are grateful to the probands and their families for contributing to this study. They thank the ISB Family Genomics Group for their support and project management of WGS data. They also thank Katrin Williams for excellent technical assistance.
Glossary
- AD
Alzheimer disease
- GWAS
genome-wide association study
- MAF
minor allele frequency
- WGS
whole-genome sequencing
- SNV
single nucleotide variant
- LD
linkage disequilibrium
Author contributions
Study concept and design: P. May, J.G. Schneider, and M. Riemenschneider. Acquisition and analysis of the data: all authors. Collection, analysis, and interpretation the data: P. May, S. Pichler, D. Hartl, D.R. Bobbili, and C. Spaniol. Drafting the manuscript and/or figure: all authors.
Study funding
Financial support was provided by the University of Luxembourg (UL)–Institute for Systems Biology (ISB) Strategic Partnership by “le plan Technologies de la Santé par le Gouvernement du Grand-Duché de Luxembourg” through the Luxembourg Centre for Systems Biomedicine and the UL to P. May, from the Saarland University (Homfor 2014 to D. Hartl), the German Federal Ministry of Education and Research (BMBF) National Genome Research Network (NGFN) grant No. 01GS08125 to M. Riemenschneider, and through the Helmholtz Alliance for Mental Health in an Aging Society (HELMA) Grant No. Ha-15 to M. Riemenschneider. Computational results presented in this article were carried out using the HPC facility of the UL (hpc.uni.lu).
Disclosure
P. May, S. Pichler, D. Hartl, and D.R. Bobbili report no disclosures. M. Mayhaus has been employed by Evotec AG, Hamburg, Germany. C. Spaniol reports no disclosures. A. Kurz has served on the scientific advisory board of MSD Germany; has served on the editorial board of Der Nervenarzt; and has received research support from the Federal Ministry of Health, the Federal Ministry of Education and Research, and the German Academic Exchange Service. R. Balling has served on the scientific advisory board of the Novo Nordisk Foundation; has received funding for travel and/or speaker honoraria from Bayer; has served on the editorial board of Annual Review Nutrition; and has held stock, stock options, and/or board of director's compensation from Megeno SARL and ITTM. J.G. Schneider has received research support from the FNR. M. Riemenschneider has received research support from the BMBF, NGFN, and the Helmholtz Alliance for Mental Health in an Aging Society. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Hollingworth P, Harold D, Sims R, et al. . Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. . Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013;45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Logue MW. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol 2011;68:1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Kimura Y, Nagata K, Yamamoto A, Matsuo M, Ueda K. ABC proteins: key molecules for lipid homeostasis. Med Mol Morphol 2005;38:2–12. [DOI] [PubMed] [Google Scholar]
- 5.Sakae N, Liu CC, Shinohara M, et al. . ABCA7 deficiency accelerates amyloid-β generation and Alzheimer's neuronal pathology. J Neurosci 2016;36:3848–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Karl T, Garner B. Understanding the function of ABCA7 in Alzheimer's disease. Biochem Soc Trans 2015;43:920–923. [DOI] [PubMed] [Google Scholar]
- 7.Cuyvers E, De Roeck A, Van den Bossche T, et al. . Mutations in ABCA7 in a Belgian cohort of Alzheimer's disease patients: a targeted resequencing study. Lancet Neurol 2015;14:814–822. [DOI] [PubMed] [Google Scholar]
- 8.Vardarajan BN, Ghani M, Kahn A, et al. . Rare coding mutations identified by sequencing of Alzheimer disease genome-wide association studies loci. Ann Neurol 2015;78:487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinberg S, Stefansson H, Jonsson T, et al. . Loss-of-function variants in ABCA7 confer risk of Alzheimer's disease. Nat Genet 2015;47:445–447. [DOI] [PubMed] [Google Scholar]
- 10.Bellenguez C, Charbonnier C, Grenier-Boley B, et al. . Contribution to Alzheimer's disease risk of rare variants in TREM2, SORL1, and ABCA7 in 1779 cases and 1273 controls. Neurobiol Aging 2017;59:220.e1–220.e9. [DOI] [PubMed] [Google Scholar]
- 11.Kunkle BW, Carney RM, Kohli MA, et al. . Targeted sequencing of ABCA7 identifies splicing, stop-gain and intronic risk variants for Alzheimer disease. Neurosci Lett 2017;649:124–129. [DOI] [PubMed] [Google Scholar]
- 12.Reitz C, Jun G, Naj A, et al. . Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E ϵ4, and the risk of late-onset Alzheimer disease in african americans. JAMA 2013;309:1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang P, Sun YM, Liu ZJ, et al. . Association study of ABCA7 and NPC1 polymorphisms with Alzheimer's disease in Chinese Han ethnic population. Psychiatr Genet 2013;23:268. [DOI] [PubMed] [Google Scholar]
- 14.Chung SJ, Lee JH, Kim SY, et al. . Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer Dis Assoc Disord 2012;27:1. [DOI] [PubMed] [Google Scholar]
- 15.Sassi C, Nalls MA, Ridge PG, et al. . Neurobiology of aging ABCA7 p.G215S as potential protective factor for Alzheimer' s disease. Neurobiol Aging 2016;46:235.e1–235.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drmanac R, Sparks AB, Callow MJ, et al. . Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 2010;327:78–81. [DOI] [PubMed] [Google Scholar]
- 17.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert J, Siekierska A, Langlois M, et al. . Mutations in STX1B, encoding a presynaptic protein, cause fever-associated epilepsy syndromes. Nat Genet 2014;46:1327–1332. [DOI] [PubMed] [Google Scholar]
- 19.Roach JC, Glusman G, Smit AF, et al. . Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 2010;328:636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Glusman G, Huff C, Caballero J, Roach JC. Accurate and robust prediction of genetic relationship from whole-genome sequences. PLoS One 2014;9:e85437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Jian X, Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat 2011;32:894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 2007;39:17–23. [DOI] [PubMed] [Google Scholar]
- 23.Wall DP, Pivovarov R, Tong M, et al. . Genotator: a disease-agnostic tool for genetic annotation of disease. BMC Med Genomics 2010;3:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006;38:904–909. [DOI] [PubMed] [Google Scholar]
- 25.International HapMap Consortium. The international HapMap project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 26.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2014;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howey R, Cordell HJ. MapThin: thinning your map files for linkage analyses! 2011. Available at: staff.ncl.ac.uk/richard.howey/mapthin/mapthin.pdf. Accessed July 28, 2017.
- 28.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 2002;30:97–101. [DOI] [PubMed] [Google Scholar]
- 29.Thiele H, Nürnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics 2005;21:1730–1732. [DOI] [PubMed] [Google Scholar]
- 30.Egeland T, Pinto N, Vigeland MD. A general approach to power calculation for relationship testing. Forensic Sci Int Genet 2014;9:186–190. [DOI] [PubMed] [Google Scholar]
- 31.Lek M, Karczewski KJ, Minikel EV, et al. . Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Roeck A, Van den Bossche T, van der Zee J, et al. . Deleterious ABCA7 mutations and transcript rescue mechanisms in early onset Alzheimer's disease. Acta Neuropathol 2017;134:475–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Guennec K, Nicolas G, Quenez O, et al. . ABCA7 rare variants and Alzheimer disease risk. Neurology 2016;86:2134–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allen M, Lincoln SJ, Corda M, et al. . ABCA7 loss-of-function variants, expression, and neurologic disease risk. Neurol Genet 2017;3:e126. [DOI] [PMC free article] [PubMed] [Google Scholar]