Abstract
Post-translational modification of lysine residues via reversible acylation occurs on proteins from diverse pathways, functions, and organisms. While nuclear protein acylation reflects the competing activities of enzymatic acyltransferases and deacylases, mitochondrial acylation appears to be driven mostly via a non-enzymatic mechanism. Three protein deacylases, SIRT3, SIRT4 and SIRT5 reside in the mitochondria and remove these modifications from targeted proteins in an NAD+-dependent manner. Recent proteomic surveys of mitochondrial protein acylation have identified the sites of protein acetylation, succinylation, glutarylation and malonylation and their regulation by SIRT3 and SIRT5. Here, we review recent advances in this rapidly moving field, their biological significance and their implications for mitochondrial function, metabolic regulation and disease pathogenesis.
Introduction
Lysine acylation was first identified on the amino terminal tails of histone proteins. The simplest form of acylation, the addition of an acetate group, is called acetylation and was first discovered on histone tail lysine residues (reviewed in (Verdin and Ott 2015)). Histone acetylation is under the control of competing enzymatic activities: histone acetyltransferases and histone deacetylases (Verdin and Ott 2015);(Choudhary et al. 2014)). During the past 20 years, the scope of protein acetylation has dramatically expanded beyond histones to every subcellular compartment and most cellular processes: a recent proteomic survey identified more than 3,000 distinct sites of protein acetylation ((Choudhary et al. 2009)). During the same period, a growing number of acetyltransferases and protein deacetylases have been identified ((Choudhary et al. 2014)). More recently, the field has come to the realization that acetylation is only one of many acyl modifications that include butyrylation, propionylation, succinylation, glutarylation, malonylation, and others. These modifications are found on proteins across all kingdoms of life and regulate diverse cellular processes including transcription and metabolism.
Mitochondrial sirtuins: SIRT3, SIRT4 and SIRT5 are protein deacylases
An unexpected aspect of the broader investigation of acetylation was the early realization that the NAD+-dependent protein deacetylase, SIRT3, was a mitochondrial protein ((Schwer et al. 2002), (Onyango et al. 2002)) suggesting that acetylation was also present in mitochondria. This was directly demonstrated by the identification of the first mitochondrial acetylated protein and target of SIRT3, acetyl-CoA synthetase 2 ((Schwer et al. 2006),(Hallows, Lee, and Denu 2006)), and by the demonstration that mitochondrial protein acetylation represents a significant fraction of the cellular acetylproteome ((Kim et al. 2006)).
Early experiments using western blotting analysis and antisera specific for acetyl-lysine showed globally increased mitochondrial protein acetylation in mice lacking SIRT3 ((Lombard et al. 2007)), indicating that SIRT3 was the major mitochondrial protein deacetylase. Interestingly, mice lacking the homologous proteins SIRT4 and SIRT5 showed no such increased protein acetylation, suggesting that these proteins targeted low-abundance or a limited subset of acetylated proteins or, alternatively, targeted other lysine modifications ((Lombard et al. 2007)). Since no deacetylase activity could be found associated with purified, recombinant SIRT4 or SIRT5, a search for novel possible targets was initiated. This work resulted in the identification of novel modifications targeted by SIRT5: succinylation, malonylation and glutarylation as described in details below ((Du et al. 2011{Zhang, 2011 #16);(Peng et al. 2011);(Tan et al. 2014)}.
To date, SIRT4 remains the least understood member of the mitochondrial sirtuins in terms of its enzymatic activity. Recent evidence indicates that it might function as a weak protein deacetylase on a limited number of substrates ((Laurent et al. 2013)), as a lipoamidase (delipoylation) and de-biotinylase ((Mathias et al. 2014)), and as a demethylglutarylase ((Anderson 2017)).
Mitochondrial protein acetylation is regulated by SIRT3
The first proteomic analysis of lysine acetylation was reported in 2006 ((Kim et al. 2006)) and used a novel pan-acetyl-lysine polyclonal antibody raised against acetyl-lysine-containing peptides to enrich an input pool of trypsin-generated peptides for mass spectrometry, a strategy previously employed in studies targeting other post-translational modifications. This enriched pool of peptides was then separated and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). Using this proteomics approach, 388 sites of acetylation were identified in 195 proteins in the cytosolic and nuclear fractions of HeLa-S3 cells and isolated mouse liver mitochondria. Only 13 of the 195 acetylated proteins they identified were known previously. Unexpectedly, more than half of the acetylation sites and proteins were found on mitochondrial proteins, a surprising observation given that no mitochondrial proteins had been previously reported to be acetylated. Of 277 acetyllysine sites identified in 133 mitochondrial proteins from mouse liver, many were metabolic proteins, including many dehydrogenases. This study showed that lysine acetylation is abundant in the mitochondria and provided the conceptual framework for a distinct and regulated ‘mitochondrial acetylome’.
Using the novel affinity enrichment proteomic approach established by Zhao et al., two groups followed up with more sensitive, more accurate, and higher-resolution mass spectrometry instrumentation. In the first of these reports, Choudhary and colleagues ((Choudhary et al. 2009)) greatly expanded the cellular acetylation landscape by cataloguing 3600 lysine acetylation sites in 1750 proteins. They further fractionated the enriched acetyllysine peptide fraction by isoelectric focusing to increase coverage and employed stable isotope labeling by amino acids in cell culture (SILAC) (Ong et al. 2002) to compare the responses of the acetylome after the addition of two different lysine deacetylase inhibitors. A key finding of this study was that acetylation seems to be functionally important mediator of protein-protein interactions. Based on their results, the cellular ‘acetylome’ affects proteins involved in nearly all cellular functional categories.
The same affinity-enrichment discovery approach was employed to assess acetylation sites in proteins from human liver samples. ((Zhao et al. 2010)). They separated nuclear, mitochondrial and cytosolic fractions, and analyzed the cytosolic and mitochondrial fractions for acetylation. Over 1300 acetylated peptides and 1047 proteins were identified by mass spectrometry, mapping acetylated proteins to metabolic pathways including fatty acid metabolism, glycolysis, and the TCA and urea cycles, in agreement with the previous observations from cell line experiments that mitochondrial metabolism represented an enriched category of acetylated proteins.
Having established that metabolic pathways are modified by acetylation, and that those acetylations can have functional effects, the natural next step was understanding how protein acetylation is regulated through attachment and removal. Although the origin of protein acetylation is now generally accepted as non-enzymatic, the removal is performed by the Sirtuins. Sites of acetylation regulated by sirt3 have been determined by comparing the acetylation profiles of wild-type and SIRT3-deficient mice (Sirt3−/−) ((Schwer et al. 2002),(Onyango et al. 2002)). SIRT3-deficient mice exhibit significant and widespread protein hyperacetylation, unlike mice lacking SIRT4 or SIRT5 ((Lombard et al. 2007)). The Gibson and Verdin labs exploited this opportunity in a global, in depth analysis of acetylation sites starting with purified mitochondria from SIRT3 WT and KO mouse liver ((Rardin, Newman, et al. 2013)). They employed a novel label-free quantification (LFQ) proteomic approach and a mixture of two commercial anti-acetyllysine antibodies ((Schilling et al. 2012)) (see Figure 1a). This analysis led to the identification of 2187 acetylation sites in 483 proteins, of which 283 sites in 136 proteins (or about 13% of all quantifiable sites) were significantly increased (>2-fold) in mice lacking SIRT3. These results were surprising in several ways. First, only a small fraction of all modified acetyllysine sites appeared to be removed by SIRT3. Second, the degree of regulation (i.e. differential acetylation in the presence vs. absence of SIRT3) varied considerably ranging 1.5 to 20-fold. Third, many of the ‘regulated sites’ were concentrated on a subset of the proteins identified; for example, 8 sites in 3-hydroxy-3-methyglutaryl-CoA synthase isoform 2 (HMGCS2) and 16 sites in the trifunctional protein complex. These proteins with many regulated sites mapped to various KEGG pathways, including fatty acid β-oxidation (FAO), the TCA cycle, branched chain amino acid catabolism (BCAA), ketone body metabolism, electron transfer proteins (ETC), the urea cycle and pyruvate metabolism. Using the same SIRT3 KO vs. WT model and proteomic workflow, the same groups subsequently identified 549 acetylated peptides among 147 proteins in skeletal muscle mitochondria ((Jing et al. 2013)), including acetylation of K336 in PDH E1alpha subunit leading to inhibition of pyruvate dehydrogenase activity. Collectively, these results depict SIRT3 coordinately regulating mitochondrial function via multiple pathways
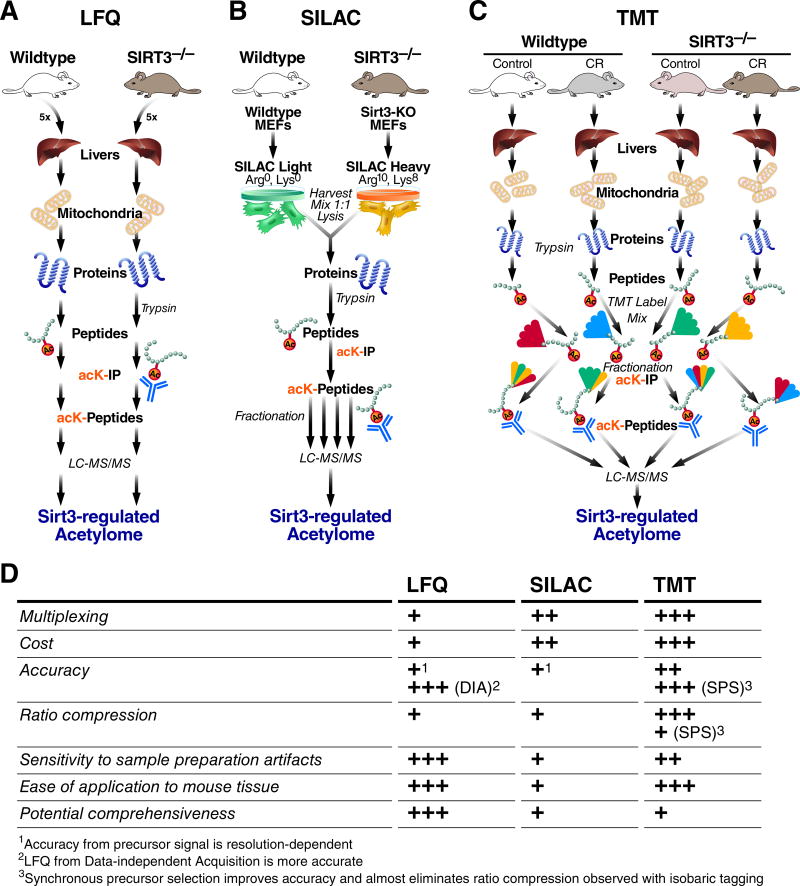
Figure 1. Mass Spectrometry Methods for Relative Quantification of Site-Specific Protein Acylation.
Workflows pictured here reflect the respective studies from the text, and are representative of the most common strategy although slight variations are used. The number of arrows along each workflow indicates where sample mixing occurs, if at all. A, Workflow for Label-Free Quantification (LFQ). B, Workflow for metabolic labeling of MEFs from mutant mice using SILAC. C, Workflow for in vitro isobaric labeling of peptides using TMT reagent. D, Comparison of relative strengths and weaknesses among the methods in (A–C). All strategies use protein isolation, trypsin digestion, immuno-affinity enrichment of the low-abundance peptides containing acetyl-lysine, and LC-MS/MS for identification and quantification. All strategies can be combined with fractionation before LC-MSMS to increase the number of identified acetylation sites, but this greatly complicates LFQ data analysis.
Additional reports examined the SIRT3-regulated acetylome and came to similar conclusions Choudhary and colleagues ((Sol et al. 2012)) isolated mouse embryonic fibroblasts (MEFs) from WT and SIRT3 KO mice and again employed an isotopic metabolic-labeling method in cell cultures or SILAC to differentially label proteins in SIRT3 MEFs (see Fig 1b). They identified over 1500 acetyllysine sites, with 933 sites in common and quantifiable. Of these sites, over 100 showed a >2-fold increase between SIRT3WT and SIRT3KO cells with the majority of these annotated as mitochondrial. The majority of acetylation sites showing a statistically significant increase in the SIRT3KO cells were of mitochondrial origin. Mapping of these putative SIRT3-regulated sites onto enzymes in the TCA cycle and FAO metabolism showed good correlation between the mouse MEFs and human cell cultures (U2OS) and with data from the label-free proteomic study mentioned above ((Rardin, He, et al. 2013)).
In a separate study, the Coons and Denu labs reported that calorie restriction (CR) alters mitochondrial protein acetylation ((Hebert et al. 2013)). They employed a multiplexed chemical labeling strategy that allowed them to quantitatively compare the acetylation levels of proteins form mouse tissues under four conditions simultaneously, i.e., WT and SIRT3 KO mice under ad libitum feeding or CR. They also employed extensive fractionation after a 4-plex tandem mass tagging (TMT) of each individual sample but prior to immuno-affinity purification (see Fig. 1c). With this experimental design, they identified 3285 acetylation sites, 2193 of which could be assigned to 434 mitochondrial proteins. Interestingly, cluster analysis revealed three classes of acetylation sites that could be distinguished by comparing these four regimens or conditions, each with distinct functional and structural features. The first class (Class I acetyl sites) are responsive to SIRT3 expression levels: low under conditions where SIRT3 expression is high, such as CR, and elevated when SIRT3 was absent (SIRT3KO). The acetylation level of class II sites increased under CR only but were sirtuin-insensitive, whereas class III sites showed minimal change under any of the three experimental conditions tested.
The same group recently examined lysine acetylation in skeletal muscle in rats selectively bred for high running capacity and their controls under regimens of both running and resting conditions ((Overmyer et al. 2015)). Key enzymes in oxidative pathways (e.g., oxidative phosphorylation, fatty acid metabolism, and BCAA degradation) underwent rapid deacetylation under running conditions, a process that was more pronounced in the high running capacity animals. This study also examined acetylation in 5 tissues (brain, heart, kidney, liver and skeletal muscle) in WT and SIRT3 KO mouse backgrounds under fasting conditions. Overall, they identified 6286 sites of acetylation, quantified the abundance of 80% of these sites in all five tissues, and found a relatively small set of 87 shared sites, with a much larger number of sites unique to each tissue, i.e., 893 kidney, 451 brain, 389 liver, 306 heart, and 89 muscle. This complicated overlap suggests a common set of core mitochondrial processes shared and perhaps similarly regulated in these diverse tissues such as the citrate cycle and oxidative phosphorylation, but also implies that additional tissue types will reveal as-yet-unexplored patterns of acetylation. Further studies of this type will be important to unravel the complexities of how nutrient stress might play out across these different tissues in the organism, and provide deeper understanding of protein acetylation dynamics that are under SIRT3 regulation.
The acidic acyl modifications, lysine succinylation, malonylation and glutarylation are regulated by SIRT5
a. Lysine succinylation was first identified in E. coli as a new acyl modification carrying a carboxylic acid ((Zhang et al. 2011)). Similar to studies surveying the acetylome landscape, an anti-succinyl-lysine immunoprecipitation was used to find 69 succinylation sites in 14 proteins. Hening Lin’s group solved structure of SIRT5 while attempting to explain its lack of deacetylase activity and found that its substrate binding pocket contains an arginine residue, suggesting that SIRT5 might bind and target an acidic acyl group ((Du et al. 2011)). NAD-dependent desuccinylase activity of SIRT5 was then demonstrated in vitro, as well as indirectly in vivo by showing an increase in protein succinylation in a SIRT5 KO mouse proteins probed with anti-succinyllysine antibodies. Independently, another study confirmed that SIRT5 regulates lysine succinylation and another acid acyl group, malonylation, of mammalian and prokaryotic proteins in vitro and in vivo ((Peng et al. 2011)).
Two additional, independent studies provided evidence for the extensive presence of mitochondrial succinylation and its regulation by SIRT5. Using liver tissue and MEFs from a Sirt5 KO mouse ((Park et al. 2013)), 2565 succinylation sites in 779 proteins were identified. KEGG pathway analysis identified mitochondrial BCAA metabolism and TCA cycle as the top two pathways and a strong enrichment in mitochondrial proteins, although the nuclear and cytosolic compartments were also highly represented, particularly for histones and ribosomal proteins. One of their more contentious findings was the high stoichiometry of succinylation, reported to be over 10% for the majority of sites seen in the Sirt5 KO cells, and only slightly less in WT MEFs (32% of sites). These stoichiometry values were determined indirectly using a previously reported method developed for estimating phosphorylation site occupancy, that requires SILAC ratios from both modified and unmodified peptides along with overall protein values ((Olsen et al. 2010)). This method assumes that the site of interest is only modified by the modification of interest, which is likely true for phosphorylation which modifies amino acids that only contain a limited range of modifications, serine, threonine, and tyrosine, but is almost certainly not true for lysine residues, which accommodate a myriad of modifications.
Different results have obtained when comparing the sites of succinylation and acetylation. Park et al. reported a small overlap (28%) between succinylation and acetylation sites in MEFs ((Park et al. 2013)) while the Gibson and Verdin groups ((Rardin, He, et al. 2013)) found a significant (79%) overlap of lysine acetylation and succinylation in mouse liver mitochondria. The discrepancy between these two studies may be due to differences in source material and growth condition, i.e., in vitro MEFs vs. in vivo mouse liver. However, it should be noted that while overall acetylation and succinylation have a strong overlap in sites in liver mitochondria, the regulated site sets for deacetylation and desuccinylation by SIRT3 and SIRT5 are different with only a minority overlap (24%). Pathway analysis of the liver mitochondria succinylome identified fatty acid oxidation (FAO), BCAA catabolism, TCA cycle, and ATP and ketone body synthesis as enriched pathways – similarly to acetylation - with all four enzymes of the ketogenesis pathway and most enzymes in the FAO pathways carrying one or more SIRT5-regulated succinyllysine site. In agreement with this data, fatty acid oxidation was decreased in SIRT5 KO mice hepatocytes and plasma levels of β-hydroxybutyrate were decreased in SIRT5 KO mice.
b. Lysine Malonylation was identified as a novel lysine modification that could also be removed by SIRT5 in vitro ((Peng et al. 2011)). Using a newly developed and highly selective anti-malonyllysine antibody, the Verdin and Gibson groups found malonylated proteins in multiple tissues ((Nishida et al. 2015)); in the absence of SIRT5, hypermalonylated proteins were more prevalent in kidney and liver, particularly in the cytoplasmic fraction. This observation contrasted with SIRT5-dependent succinylation, which was more prevalent in liver mitochondria and barely detected in a cytoplasmic fraction following mitochondrial depletion ((Rardin, He, et al. 2013)). Label-free mass spectrometry of immunoprecipitated malonyllysine peptides from SIRT5 wt and KO livers identified 1137 unique malonylation sites in 430 total proteins of which 183 sites (13%) were increased, and therefore presumably regulated, in mice lacking SIRT5. Pathway analysis of the SIRT5-regulated proteins showed the cytoplasmic gluconeogenesis and glycolysis pathways as the two most enriched, and the urea cycle as third. Closer examination of this data revealed 8 out of 10 glycolytic enzymes carrying a SIRT5-regulated malonyl-lysine site and study of primary hepatocytes derived from SIRT5 KO versus WT mice showed decreased production of lactic acid and glycolytic flux in mice lacking SIRT5 ((Nishida et al. 2015)).
As three of the proteomic survey studies of SIRT3 and SIRT5 KO mice all used a similar proteomic approach with liver as the source material ((Rardin, He, et al. 2013), (Rardin, Newman, et al. 2013), (Nishida et al. 2015)) these investigators compared the overlap between the three modifications, malonylation with acetylation and succinylation. Interestingly, and in contrast to succinylation, mitochondrial malonylated peptides showed distinctly lower overlap with mitochondrial acetyl-and succinyl-lysines. Coupled with the predominantly cytoplasmic localization of malonylation, this indicates a very different pattern of modification for malonylation than for previously studied lysine acylations.
A separate large-scale analysis of protein malonylation was recently carried out in db/db mice, a type 2 diabetes mouse model ((Du et al. 2015)). Using affinity enrichment and LC-MS/MS, a total of 573 malonylated lysine sites were identified on 268 proteins, with an over-representation of metabolic processes such as oxoacid metabolism, glucose catabolism, and organic acid metabolism. The demonstration that db/db mice show enhanced lysine malonylation suggests that this modification might play a pathogenic role in the metabolic disturbances associated with the db mutation.
Similarly, another study used malonyl-CoA decarboxylase deficiency (MCD−/−) in human fibroblasts to show that the increased level of malonyl-CoA present in these cells resulted in hypermalonylation, mitochondrial dysfunction, and impaired fatty acid oxidation ((Colak et al. 2015)). This study identified a staggering 4943 malonyllysine sites, 461 of which showed an increase of greater than 2-fold abundance in MCD−/− cells. The authors propose that protein hypermalonylation and may be partially responsible for the pathophysiology of malonic aciduria.
c. Lysine glutarylation, Glutarylation of lysine residues was recently reported as yet another modification regulated by SIRT5 ((Tan et al. 2014)). Glutaryl-CoA is produced as an intermediate in the mitochondrial catabolism of amino acids including lysine, during which it presumably reacts with mitochondrial proteins. Lysine -hydroxy-3-methylglutarylation (HMG-K), -methyglutaryl-lysine (MG-K), and 3-methylglutaconyl-lysine (MGc-K) of lysine residues was recently reported as yet another modification regulated by SIRT5 ((Tan et al. 2014)). (Wagner et al., 2017) Collectively, these modifications’ reversal points to a paradigm of side reaction regulation by SIRT5 in which the enzyme promiscuously reverts any negatively charged acylation species capable of fitting into its active site from whichever mitochondrial protein lysine residues it can access.
The Emerging Targets of SIRT4
The third mitochondrial sirtuin, SIRT4, has lagged behind SIRT3 and SIRT5 in the study of its targets and specificities; however, recent work has illuminated many aspects of its function. One study has shown that SIRT4 can regulate the levels of both lipoyl- and biotinyl-lysine modifications in pyruvate dehydrogenase and likely other proteins ((Mathias et al. 2014)). Whether this is the only or the most important activity of SIRT4 is not yet clear, as previous reports have shown that SIRT4 can also function as an ADP-ribosyl transferase and regulate insulin secretion in pancreatic β cell mitochondria ((Haigis et al. 2006)) ((Ahuja et al. 2007)).
Recently, three additional target acylations have been identified for SIRT4. Methylglutaryl-, hydroxymethylglutaryl-, and 3-methylglutaconyl-lysines have all been identified on mitochondrial enzymes, including those involved in leucine catabolism and its production of branched-chain acyl-CoA intermediates (Anderson 2017). Recombinant SIRT4 was sufficient to demethylglutarylate lysine residues, and SIRT4-overexpressing cells contained lower levels of these branched-chain acyl-lysines. As SIRT4KO mice are known to exhibit increased insulin secretion in response to BCAA stimulation ((Haigis et al. 2006),(Ahuja et al. 2007),(Anderson 2017)) the generation of these newly described acyl-lysine species from BCAA catabolites finally provides a molecular mechanism linking SIRT4’s putative target modifications to the observed phenotypes in SIRT4KO models.
The recent publication of a crystal structure for the Xenopus sequence of SIRT4 may reconcile these seemingly conflicting observations (Pannek et al. 2017). The authors described catalytic activities for SIRT4 against a variety of target acylations, with the highest reported for the nonnatural modification 3,3-dimethylsuccinylation. The next highest activities were reported against HMGylation and octanoylation, with biotinylation and lipoylation removed at ~80% and ~40% of the efficiency of octanoylation. Lysine octanoylation is reversed by multiple sirtuins (Feldman, Baeza, and Denu 2013), and the structurally related lipoylation modification required for pyruvate (and other 2-oxoacid) dehydrogenase function may represent a biologically important side reaction for this shared specificity of SIRT4. This crystallographic study also revealed an unusual sensitivity of SIRT4 to inhibition by NADH (Pannek et al. 2017); if HMGylation and other branched-chain acylations are also targeted by SIRT5, perhaps SIRT4’s unique contribution is redox-sensitive regulation of its target modifications.
Other lysine acylations targeted by sirtuins
Evidence is also emerging that most sirtuins may act on multiple acyl substrates, at least in vitro ((Feldman, Baeza, and Denu 2013)). These include propionylation, butyrylation, crotonylation, hexanoylation, octanoylation, decanoylation, dodecanoylation, myristoylation, palmitoylation, and lipoylation. Of note, de-decanoylase (C-10 acyl group) activity is associated with all three mitochondrial sirtuins, and SIRT3 further exhibits significant depalmitoylase activity. This raises the possibility that the mitochondrial sirtuins may regulate currently uncharacterized mitochondrial modifications.
A Menu of Mass Spectrometry Options for Studying Mitochondrial Acylation
The wealth of acylomic studies mapping out mitochondrial acylation over the past several years can be hard to compare from an outside perspective; different technical approaches to acylomic studies offer different strengths and weaknesses, and selecting an appropriate tool for a project is both challenging and essential. Mass Spectrometry-based proteomics methods for quantifying protein acylation directly measure either: (1) relative changes, or (2) less commonly, stoichiometry, which refers to the % of a given modification site occupied by acylation. Stoichiometry can be used to infer relative changes, but the converse is not true. Methods for relative quantification of acylation discussed here include: (1) label-free quantification (LFQ), (2) stable isotope labeling by amino acids in cell culture (SILAC), and (3) multiplexed, isobaric chemical tagging (e.g. TMT). Here (and in Fig. 1D) we provide a discussion of the pros and contras of these methods as applied to acylation profiling, but previous reviews cover them more generally and in greater depth (Bantscheff et al. 2012), (Meyer and Schilling 2017).
Measuring Relative Changes in Acylation Levels
The first strategy, LFQ (Fig. 1A), is easily applied to any mass spectrometry dataset, even in hindsight after data collection, with no additional cost. However, some LFQ strategies are better than others (see (Rardin et al. 2015)). Compared to isotope labeling-based quantification methods, where ratios of acylation between conditions are locked-in at the point of sample mixing, LFQ is more sensitive to slight differences in sample preparation and mass spectrometry data collection. Importantly, LFQ is most easily applied to human or mouse tissue, but has relatively low throughput, because each replicate requires an separate mass spectrometry data collection.
The second strategy, SILAC (Fig 1B), is less sensitive to differences in sample preparation because the ratio of treatment to control is locked-in before much of the sample manipulation, ideally based on cell count before lysis. Therefore, when using SILAC, peptides can be pre-fractionated before mass spectrometry analysis, which increases identifications and quantification quality. Because at least two SILAC samples are mixed, this strategy can have slightly greater throughput compared with LFQ. However, SILAC is not as easily applied to human or mouse tissue, because stable-isotope metabolic labeling of whole animals can be expensive.
The final strategy, isobaric labeling (e.g. TMT, Fig. 1C), of peptides offers several benefits over the other two methods at the greatest reagent cost. The cost is further amplified in studies of PTMs, where a large amount of input material (usually 1–20 mg of protein) must be chemically labeled before enrichment. Cost aside, isobaric labeling has several advantages over the other two methods. TMT allows the greatest multiplexing, up to 10 sample conditions can be quantified from a single mass spectrometry acquisition. This method is also easily applied to animal tissue as the isotopic labeling occurs in vitro after protein digestion. However, TMT requires the newest mass spectrometry, such as the Thermo Orbitrap Fusion, to realize the highest quality data (McAlister et al. 2014).
Among the available methods for quantification of acylation, we propose that despite the advantages of the isobaric labeling such as TMT, the cost is often prohibitive, and LFQ can be the most accurate, comprehensive, low-cost, and applicable method for a variety of experimental designs.
Measuring Acylation Stoichiometry Directly
While differences between samples can illuminate gene functions and the effects of metabolic perturbations, evaluating which of the thousands of acylation sites discovered are most biologically significant demands absolute numbers; a ten-fold increase from 0.1% to 1% occupancy may have no effect on enzyme function, while an increased occupancy from 10% to 100% may completely block an enzyme’s activity. New proteomic methods using differential labeling of all lysine residues with stable isotope-labeled acetic anhydrides (e.g., 2H6- or 13C2-acetic anhydride) may answer this question (table 1) ((Baeza et al. 2014), (Nakayasu et al. 2014)). In an similar approach that incorporates several levels of partial acetylation to provide internal quality control of measurement accuracy, Choudhary and colleagues have come to the conclusion that the stoichiometry of nearly all acetylation sites is low and consistent with a nonenzymatic process for lysine acetylation ((Weinert, Moustafa, et al. 2015), (Weinert et al. 2017b)). However, both methods struggle with peptides containing multiple lysine residues; this is particularly problematic as trypsin, commonly used to cleave protein samples into peptides, fails to cleave at lysine following chemical acetylation.
In 2016, two additional new methods for stoichiometry measurement were reported, both of which enabled the determination of stoichiometry from peptides with multiple lysine residues. Yue Chen’s group demonstrated a creative method with optimized labeling chemistry and a data analysis package that uses both precursor and fragment intensities to determine acetylation stoichiometry (Zhou et al., 2016). Subsequently, the Schilling group published another strategy using fragment-ion quantification from DIA to reduce quantification interferences (Meyer et al., 2016). The latter method also enables, for the first time, measurement of succinylation stoichiometry, but there are not yet published reports of this applied to interesting biological questions.
How does mitochondrial protein acylation occur?
In the nucleus, histone acetyltransferase activity is associated with many transcriptional coactivators such as GCN5, PCAF, p300/CBP, MOF and others. These proteins specifically acetylate individual lysine residues on histone and non-histone proteins associated with transcriptional regulation. Though the bulk of histone acetylation is enzymatically driven by these enzymes, histones have long been known to react with acetyl-CoA in a non-enzymatic manner, particularly at high pH ((Paik et al. 1970)).
Many facts support the hypothesis that the bulk of mitochondrial protein acylation could also be driven by a non-enzymatic process. These facts include: 1. The very large number of identified acylation sites and the apparent lack of specific sequence surrounding the sites of acylation (discussed above); 2. Recent mass spectrometry evidence that most sites, but not all, exist at relatively low stoichiometry in yeast ((Weinert et al. 2014)) and in mice ((Weinert, Moustafa, et al. 2015)), with few residues reaching a stoichiometry of 1% modification; 3. The higher pH of mitochondrial matrix (7.9) which is associated with the deprotonation of the ε-amino groups of lysine and makes them more susceptible to acylation; 4. The high local concentration of several acyl-CoA species within the mitochondrial matrix.
Manipulation of these variables in vitro allowed Wagner and Payne to demonstrate nonenzymatic acylation of mitochondrial proteins on a physiologically relevant time scale ((Wagner and Payne 2013)). They highlighted experimentally the importance of both a basic pH and acyl-CoA levels and showed both progressive and non-selective hyperacetylation of denatured mitochondrial proteins over a period of hours. Another recent study further demonstrated that non-enzymatic acetylation of proteins is semi-selective. Site-specific, nonenzymatic acetylation rates were measured for 90 sites in eight native purified proteins. Lysine reactivity to either acetyl-coA or acetylphosphate (see below) ranged over 3 orders of magnitude with the most highly reactive sites located within clusters of basic residues, whereas low reactivity lysines were engaged in strong attractive electrostatic interactions with acidic residues ((Baeza, Smallegan, and Denu 2015)). Acetyl-phosphate, which drives acetylation under conditions of growth arrest and abundant glucose in bacteria, is an alternative acylation source currently unknown in mammals ((Weinert et al. 2013)); while acetate kinase is found in some eukaryotic microbes ((Ingram-Smith, Martin, and Smith 2006)), the pathway leading to acetylphosphate synthesis is apparently absent from higher organisms. A reactive phosphoacyl species of greater relevance to mammalian systems is 1,3-bisphosphoglycerate, a cytosolic intermediate in glycolysis recently reported to non-enzymatically modify lysine residues - particularly those found in enzyme binding sites - and inhibit enzyme function ((Moellering and Cravatt 2013)). Though neither 1,3-BPG nor acetyl-phosphate are found in the mitochondrial environment, they provide additional evidence for the idea that metabolic intermediate acyl species can be intrinsically reactive with potential physiological consequences, and that acyl groups’ transfer to lysines can readily occur in the absence of lysine acyltransferases.
Recent work has further shown that some acyl-CoA species can modify lysines via a cyclic anhydride intermediate (Wagner et al. 2017). Divalent acyl groups such as succinyl- and glutaryl-CoA can use this alternative non-enzymatic reaction mechanism to achieve higher reaction rates than monovalent acyl species such as acetyl-CoA. Furthermore, anhydride intermediates may be formed by other, less well-documented acyl-CoA metabolites such as the branched-chain glutaric acid derivatives produced during amino acid catabolism.
As an added twist on semi-enzymatic acylation, proximity to thiols such as those provided by cysteine residues apparently increases the rate of acylation for nearby lysines (James et al. 2017). This redox-sensitive mechanism means that certain acetylation sites observed in mitochondria may be regulated by glutathione redox balance and glyoxalase II in addition to mitochondrial sirtuins. This mechanism has been shown to affect both acetylation and succinylation; other acylation modifications are likely to be similarly affected. Furthermore, S-acylation by long-chain fatty acids such as palmitate and myristate has been shown to be important for membrane localization of proteins; these fatty acyl groups could conceivably transfer over to nearby lysine residues using the same mechanism, suggesting as-yet-undescribed long-chain acylations with similar membrane-anchoring functions.
Mitochondrial Acyltransferases: enzymatic and pseudo-enzymatic acylation?
As nuclear and cytosolic acylation are driven by acyltransferases, a considerable amount of work has been poured into finding a mitochondrial acyltransferase to little success; it seems likely that no such general acyltransferase exists. However, the possibility remains that canonical acyltransferases may localize to mitochondria under some conditions, and that enzymes with other functions may moonlight as acyltransferases. Two putative lysine acyltransferases have been identified and characterized and are discussed below.
GCN5L1 is localized to mitochondria and displays significant homology with a prokaryotic acetyltransferase (GCN5) although it lacks a significant fraction of its catalytic domain. Genetic knockdown of GCN5L1 blunts mitochondrial protein acetylation, and its reconstitution in intact mitochondria restores protein acetylation. GCN5L1 interacts with and promotes acetylation respiratory chain target proteins and reverses global SIRT3 effects on mitochondrial protein acetylation, respiration and bioenergetics ((Scott et al. 2012)). Many questions remain unresolved, such as how does GCN5L1 mediate acetylation without an intact catalytic domain? In which tissues and under which conditions is its function most important?
An interesting possibility raised by recent work is that acyltransferases responsible for conjugating acyl-CoA species to non-protein substrates may aid in lysine acylation as a side reaction. In particular, acetyl-CoA acetyltransferase (ACAT1), which synthesizes acetoacetyl-CoA by condensing two molecules of acetyl-CoA as a precursor for ketone body synthesis, may be a driving source of acetylation of the pyruvate dehydrogenase complex component PDHA1 and its activating phosphatase PDP1 by associating with these proteins ((Fan et al. 2014)). Assuming that ACAT1 is capable of holding a reactive acetyl-CoA molecule in place for long enough, increasing the local concentration of reactive thioester, a semi-enzymatic mechanism may account for increased acetylation to PDP1/PDHA1.
Regulation of mitochondrial protein acylation
Even as the mechanistic details of mitochondrial acylation continue to be revealed, many systemic factors affecting it have been discovered. These provide, to date, the best therapeutic opportunities to manipulate the processes of acylation and deacylation in humans. Broadly, dietary inputs and the metabolic demands of various tissues create acyl-CoA concentrations with interact with molecular contexts in mitochondria to drive acylation at differing rates for different targets; gene expression of and NAD+ supply to sirtuins then reverses this process.
Diet
Both over- and under-nutrition create abnormal metabolic states with high levels of acyl-CoAs. During fasting, acetyl-CoA is mobilized from stored energy reserves for fuel, and both glucose and ketone bodies are produced by the liver and to a lesser extent the kidneys. Loss of SIRT3 has been shown to inhibit fatty acid oxidation ((Hirschey et al. 2010)); the same study showed decreased gluconeogenic production of glucose in response to cold challenge and decreased production of the ketone body beta-hydroxybutyrate during fasting. Mitochondrial hyperacetylation thus tends to limit the metabolic adaptations necessary for proper function during fasting; SIRT3 is upregulated during fasting to combat this process (Hirschey et al. 2010), and many mitochondrial proteins are hyperacetylated during fasting (Still et al. 2013).
Calorie restriction (CR) represents a longer-term but less extreme metabolic intervention triggering similar responses to fasting. Metabolic supplier organs such as the liver and kidney demonstrate mitochondrial hyperacetylation under CR, as does the heart (Schwer et al. 2009); brain mitochondrial acetylation was unchanged, however, and skeletal muscle acetylation decreased. Proteomic characterization of the changes in liver reveals a complicated landscape of alterations, but an overall increase in acetylation levels for mitochondrial proteins but not those found in other cellular compartments during CR (Hebert et al. 2013).
In contrast to fasting and calorie restriction, hypernutrition to the point of obesity has been shown to predominantly decrease mitochondrial acetylation in liver (Still et al. 2013). However, prolonged hypernutrition leads to type 2 diabetes, and has been shown to impair skeletal muscle glucose uptake in a manner exacerbated by the loss of SIRT3 (Jing et al. 2013),(Lantier et al. 2015).
Local acyl-CoA concentrations and molecular context
Many mitochondrial metabolic pathways use high-energy coenzyme A thioesters as key intermediates (Figure 3). Acetyl-CoA generated from pyruvate represents the main input to the TCA cycle and succinyl-CoA is another key intermediate in this pathway. Furthermore, amino acid and fat catabolism progress through a diversity of acyl-CoA intermediates: each length of carbon chain for beta oxidation proceeds through no less than four distinct species (saturated, enoyl, 3-hydroxy, 3-keto-acyl-CoA) prior to chain shortening. If the emerging non-enzymatic mechanism for mitochondrial lysine acylation holds, the relative abundance of distinct acyl-CoA species represent the main force driving differences in acylation between tissues.
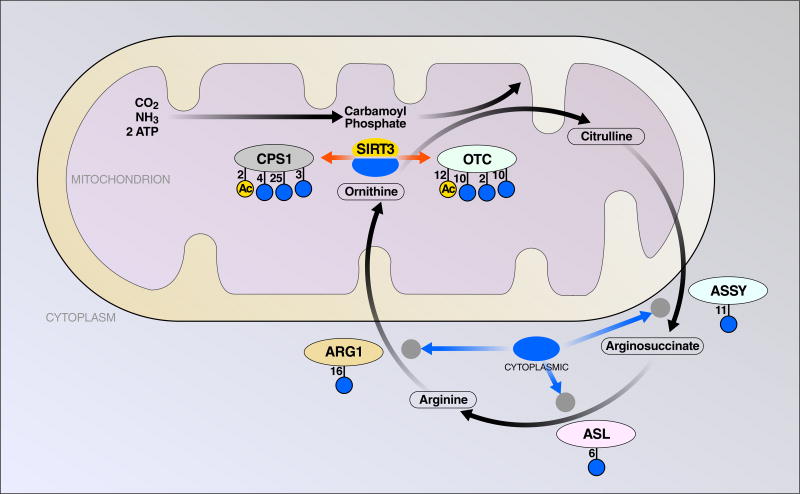
Figure 3. Mitochondrial Acylation and Sirtuin-Mediated Deacylation Affect The Urea Cycle At Multiple Points.
The urea cycle allows cellular disposal of nitrogen from mitochondrially catabolized glutamate via mitochondrial conversion to carbamoyl phosphate, citrulline shuttling to the cytosol, and loss of urea from arginine. Each enzyme in the pathway is depicted with the number of known sirtuin-regulated sites of each acyl-lysine type shown. SIRT5 desuccinylates and deglutarylates CPS1 to allow efficient clearance of mitochondrial ammonia; and SIRT3 deacetylates Ornithine Carbamoyltransfrase (OTC) to maintain its function. Furthermore, SIRT5 expression in the cytosol maintains the conversion of citrulline to argininosuccinate by demalonylating ASSY.
The role of change in mitochondrial matrix acyl-coA concentrations was recently highlighted in another experimental system. Mice with short-chain acyl-CoA dehydrogenase (SCAD) deficiency accumulate high levels of butyryl-CoA and show increased protein lysine butyrylation ((Pougovkina et al. 2014)). Similarly, and from the same study, human SCAD-deficient fibroblasts show increased lysine butyrylation, patients with malonyl-CoA decarboxylase (MCD) deficiency exhibit increased protein malonylation and finally, propionyl-CoA carboxylase (PCC) deficient patient cells show increased lysine propionylation.
Local mitochondrial concentrations of individual acyl-coAs is also likely to be modulated via intercompartmental trafficking. Acyl species are exchanged between mitochondria and the cytosol using carnitine rather than coenzyme A as a cofactor, and use specific proteins to ferry carnitines with and without acyl payloads across the mitochondrial inner membrane. Voltage-dependent anion channels (VDACs) are outer mitochondrial membrane proteins with tissue-dependent expression patterns and are part of the acylcarnitine translocase pathway ((Lee, Kerner, and Hoppel 2011)); their partners on the inner membrane, adenine nucleotide translocases (ANTs), are known to be acetylated and some sites are predicted to be functionally important to channel function (Mielke et al. 2014).
The intramolecular context for mitochondrial proteins’ lysines affects how likely they are to be acylated. For example, denatured proteins are more rapidly acetylated by coincubation with acetyl-CoA (Wagner and Payne 2013), indicating that local structure slows acylation; past studies have shown that sirtuins favor either unstructured regions (Khan and Lewis 2005, Smith et al. 2011) or, in the case of SIRT3, beta sheets (Smith et al. 2011). Another variable likely to influence the level of non-enzymatic modification of specific lysines is their pKa. Lysine residues with altered pKa, and therefore, decreased or increased positive charge, become more or less able to perform nucleophilic attack, respectively. For example, lysine residues near arginine may have decreased pKa and be more likely to have no charge, leaving their free electron pair available for acylation. Excellent reviews have discussed the variables affecting the pKa of individual lysines ((Pace, Grimsley, and Scholtz 2009)).
Regulation of Sirtuin Expression
The mitochondrial sirtuins responsible for controlling mitochondrial protein acylation can be upregulated in stress conditions. SIRT3 is upregulated during fasting (Hirschey et al. 2010) and more generally by PGC-1α (Kong et al. 2010), which allows it to best control the hyperacetylation caused by acetyl-CoA-dependent metabolic processes such as fatty acid oxidation and ketone body metabolism. Similarly, SIRT4 is upregulated as part of the DNA damage response via ATF4 (Jeong et al. 2013). Less is known about SIRT5’s transcriptional regulation, and the process of splicing which distinguishes cytosolic from mitochondrial SIRT5 production is also not well-understood.
NAD+ Availability
As mentioned previously, the only known deacylases in the mitochondrial matrix are sirtuins, which require NAD+ as a cofactor for function. NAD+ has many other roles in mitochondrial metabolism and signaling, most notably as an electron transfer currency for oxidative phosphorylation, the TCA cycle, and fat synthesis and catabolism. Addition of exogenous NAD+, or precursor compounds which are converted to NAD+ in vivo, is currently a hot topic in the field of aging biology due to recent successes in enhancing tissue function and health ((Yoshino et al. 2011), (Canto et al. 2012)).
One potential route for NAD+’s beneficial effects on health may be linked to mitochondrial acylation. Supplementation of NAD+ levels in C elegans ((Mouchiroud et al. 2013)) extends worm lifespan via the induction of the mitochondrial unfolded protein response and upregulation of mitochondrial superoxide dismutase. The authors note that increased MnSOD/SOD2 activity in the affected worms lags behind increased expression by a period of several hours, and postulate that MnSOD acetylation (as discussed in Section 3) may play a key role. As NAD+ supplementation also results in potentially beneficial production of N-methylnicotinamide via sirtuins ((Schmeisser et al. 2013)), evidence such as the link to MnSOD affirms roles for acylation both as a direct effector modulated by NAD+ and in transducing NAD+ into an N-methylnicotinamide signal.
There is also evidence that NAD precursor may work via altering mitochondrial protein acetylation. Hearing loss is induced by intense noise exposure. Administering the NAD precursor nicotinamide riboside prevents noise-induced hearing loss and degeneration of spiral ganglia neurite degeneration. These effects appear to be mediated in the mitochondria because SIRT3-overexpressing mice are resistant to noise-induced hearing loss and SIRT3 deletion abrogates the protective effects of nicotinamide riboside ((Brown et al. 2014))
Mitochondrial acylation from the bottom up: From individual targets to biological effects
Most mitochondrial acylation appears to be functionally inhibitory in enzymes in which it has been tested. This stands in contrast to protein acetylation in other cellular compartments which can be either inhibitory or activating ((Choudhary et al. 2014)). For example, histone acetylation can activate various functions by bromodomain binding and subsequent effector protein recruitment. Biochemistry in the mitochondrial matrix frequently uses small, anionic substrates - amino and carboxylic acids, nucleotide conjugates, superoxide - which interact with positively charged protein patches, frequently lysine residues, in or near enzyme active sites. Neutralization of these positive patches by conjugating negatively charged acyl groups is thus likely to represent a shared mechanism for enzymatic inhibition.
As discussed above, a strong overlap between lysines modified by different acyl groups such as acetylation and succinylation has been noted (Rardin, He, et al. 2013). Below we review a few of the key enzymes and pathways that have been extensively characterized.
Pyruvate dehydrogenase
Pyruvate catabolism in the mitochondria is a key entry point for mitochondrial oxidative metabolism. Flux through the multiprotein pyruvate dehydrogenase complex (PDC) limits TCA cycle activity; its acetyl-CoA product drives the cycle forward. PDC is thus tightly regulated, most notably by the competing activities of various kinases (PDKs) which inactivate the PDC by phosphorylating it and pyruvate dehydrogenase phosphatase (PDP) which reverses this inhibitory phosphorylation. Mammalian pyruvate dehydrogenase is a large, symmetrically constructed complex with five unique protein sequences: DLAT is the lipoylated core protein, with DLD and its adaptor protein E3BP occupying pores in the DLAT structure and outer tethered subunits consisting of PDHA1 and PDHB. Lysine acetylation has been observed on DLD and PDHA1 at multiple residues ((Rardin, Newman, et al. 2013)), but a functional role for acetylation has only been demonstrated for modification of lysine 321 of PDHA1 ((Jing et al. 2013)). Acetylation of PDHA1 K321 recruits PDK leading to PDC phosphorylation and inhibition of its enzymatic activity. In contrast, SIRT3 activates PDC at two different levels. First, SIRT3 deacetylates K321 and second, SIRT3 also deacetylates PDP, enhancing its phosphatase activity on PDC and thereby decreasing PDC phosphorylation. ((Fan et al. 2014)).
Lysine succinylation has been observed on DLAT in addition to DLD and PDHA1, including multiple succinylations controlled by SIRT5 ((Rardin, He, et al. 2013)). However, the best-demonstrated lysine acylation on the PDC is not metabolically-driven acetylation or succinylation but rather lipoylation, which transfers acetyl groups from thiamine pyrophosphate to coenzyme A as a key part of the enzyme complex’s function. Lipoylation occurs at two sites in the E2 N-terminal region (K132 and K259 in the human sequence), and is enzymatically reversed by SIRT4 ((Mathias et al. 2014)). Thus, SIRT3 and SIRT4 regulate the PDC in opposing directions; SIRT3’s clearance of lysine acetylation allows for efficient PDC function, while SIRT4’s removal of enzymatically active cofactors from specific lysine residues inhibits the PDC. The effect of SIRT5-mediated removal of PDC succinylation is not known but would be expected to activate the enzyme similar to SIRT3.
Lipid Metabolism
SIRT3 promotes the efficient utilization of lipids as a primary source of acetyl-CoA during fasting by activating long-chain acyl-CoA dehydrogenase, a key enzyme in the β-oxidation of fatty acids, via deacetylation at K42 ((Hirschey et al. 2010)). Mice lacking SIRT3 accumulate β-oxidation precursors and intermediates, including triglycerides and long-chain fatty acids. These mice also share other characteristics of human disorders of fatty acid oxidation, including cold intolerance and reduced basal ATP levels ((Hirschey et al. 2010)). SIRT3 also deacetylates (at K642 in the human protein or K635 in mice) and activates acetyl-CoA synthetase 2, an enzyme in extrahepatic tissues that activates acetate into acetyl-CoA ((Schwer et al. 2006); (Hallows, Lee, and Denu 2006)). Acetate itself is produced in the liver from acetyl-CoA and can be subsequently used in extrahepatic tissues as a form of energy. SIRT3 therefore facilitates the catabolism of fatty acids in the liver and the peripheral use of lipid-derived acetate and ketone bodies during fasting.
Ketone body synthesis
Increasing evidence support an important role for lysine acylation and sirtuins in regulating ketone body biosynthesis. Ketone bodies are small lipid-derived molecules, which are primarily produced in the liver under fasting conditions and distributed via the bloodstream to metabolically active tissues, such as muscle or brain, and used as an alternative energy source under glucose-sparing conditions. During hepatic ketogenesis, acetyl-CoA derived from fatty acids via mitochondrial β-oxidation goes through four consecutive enzymatic reactions and produces β-hydroxybutyrate as the end product. Interestingly, all the four enzymes involved in this process (Thiolase, HMG-CoA synthase, HMG-CoA lyase, and βOHB dehydrogenase) are heavily modified by lysine acylations including acetylation, succinylation, glutarylation and malonylation ((Rardin, Newman, et al. 2013))((Rardin, He, et al. 2013))((Park et al. 2013))((Tan et al. 2014))((Nishida et al. 2015)). Certain enzymes contain more than a dozen modified sites and almost every enzyme contains at least one site subject to SIRT3 or SIRT5 regulation. Mice lacking SIRT3 or SIRT5 have reduced levels of ketone bodies upon fasting, supporting a positive role for sirtuins in regulating ketone body biosynthesis ((Shimazu et al. 2010))((Rardin, He, et al. 2013)).
The ketone body synthesis enzyme 3-hydroxy-3-methyglutaryl-CoA synthase isoform 2 (HMGCS2) catalyzes the rate limiting and irreversible step of condensation of acetyl-CoA with HMG-CoA. SIRT3 and SIRT5 both positively regulate HMGCS2 activity via respectively deacetylating and desuccinylating it ((Shimazu et al. 2010))((Rardin, He, et al. 2013)). SIRT3 deacetylates HMGCS2 at lysines 310, 447, and 473 ((Shimazu et al. 2010)). Structural simulations show that in silico deacetylation of these three lysines causes conformational changes of HMGCS2 near the catalytic site, suggesting acetylation might interfere with the enzymatic activity via changing its conformation ((Shimazu et al. 2010)). In agreement with this model, mutation of these lysines to arginines, which prevent acetylation, enhance HMGCS2 enzymatic activity ((Shimazu et al. 2010)). SIRT5, on the other hand, desuccinylates HMGCS2 at multiple lysine residues; two of the top regulated sites, K83 and K310, are in close proximity to the substrate binding site of the catalytic pocket ((Rardin, He, et al. 2013)). Structural analysis shows that the substrate binding site for acetyl-CoA and HMG-CoA features a lysinerich highly positively charged patch necessary for binding to the negatively charged phospho-ADP element in coenzyme A. This highly positive microenvironment might be disrupted when the two lysines are modified by negatively charged succinyl groups leading to a reduction in enzymatic activity. SIRT5, by removing these succinyl groups, restores the positively charged microenvironment and facilitates enzymatic function. Substitution of these two lysines with a succinyl-mimicking amino acid (such as the a negatively charged glutamic acid), resulted in a complete loss of enzymatic activity. Moreover, HMGCS2 is also subject to glutarylation and malonylation ((Tan et al. 2014))((Nishida et al. 2015)). Little is known regarding how these two modifications might affect enzymatic activity of HMGCS2.
The health or disease consequences of disrupted ketone body production in the absence of SIRT3 or SIRT5 have not been extensively studied. In addition to its crucial role in intermediary metabolism, ketone bodies also possess signaling activities (Newman & Verdin, 2014). Beta-hydroxybutyrate is an endogenous inhibitor of HDACs and can thereby affect gene expression via chromatin modifications and regulate the response to oxidative stress ((Shimazu et al. 2013)). Once can therefore speculate that a decrease in ketone body production might increase sensitivity to oxidative stress.
Urea Cycle
The urea cycle was the first metabolic pathway found to be regulated by mitochondrial sirtuins ((Nakagawa et al. 2009)). It primarily takes place in the liver and is the main mechanism for the body to clear nitrogen waste resulting from protein turnover. Five enzymes are involved in a series of reactions that occur both in mitochondria and cytosol (Figure 3). The two mitochondrial enzymes, carbamoyl phosphate synthetase 1 (CPS1) and Ornithine transcarbamylase (OTC), which carry out the first two steps of urea cycle reactions, are both regulated by the activities of mitochondrial sirtuins.
CPS1, which catalyzes the initial step of the urea cycle by synthesizing carbamoyl phosphate from ammonia and bicarbonate, is a target of lysine acetylation, succinylation, glutarylation and malonylation ((Nakagawa et al. 2009))((Rardin, Newman, et al. 2013))((Du et al. 2011))((Rardin, He, et al. 2013))((Park et al. 2013))((Tan et al. 2014))((Nishida et al. 2015)). CPS1 is the most heavily modified protein in mitochondria, possibly due to its high abundance and relatively large size. At least 52 lysine sites are found to be acetylated, 44 sites succinylated, 33 sites glutarylated and 29 sites malonylated. SIRT5 positively regulates CPS1 activity by mediating desuccinylation, deglutarylation and possibly by deacetylation across multiple sites ((Du et al. 2011))((Tan et al. 2014))((Nakagawa et al. 2009)). During fasting, calorie restriction, or a high protein diet, CPS1 activity increases to adapt to the increase in amino acid catabolism. Mice lacking SIRT5 fail to upregulate CPS1 activity and show elevated blood ammonia under these conditions ((Nakagawa et al. 2009))((Tan et al. 2014)).
SIRT3 is also an important regulator of the urea cycle. It targets the other mitochondrial enzyme, ornithine transcarboxylase (OTC), by deacetylation of lysine 88 ((Hallows et al. 2011)). Metabolomic analysis of fasted mice lacking SIRT3 shows metabolite alterations in the urea cycle ((Hallows et al. 2011)). Calorie restriction stimulates the urea cycle and up-regulates OTC activity; this up-regulation of OTC activity is lost in the absence of SIRT3, leading to increased orotic acid levels, a known outcome of OTC deficiency ((Hallows et al. 2011)). Interestingly, lysine 88 is also a target of succinylation and glutarylation ((Rardin, He, et al. 2013))((Tan et al. 2014)). Whether SIRT5 also regulates OTC activity by mediating desuccinylation and deglutarylation has not been studied.
The three other enzymes in the urea cycle are located in the cytosol and are also malonylated at multiple sites with demalonylation at some sites by SIRT5 (Nishida et al. 2015). These are argininosuccinate synthase (ASSY), argininosuccinate lyase (ARLY), and arginase (ARGI1); ASSY is demalonylated by SIRT5 at 2 of 11, ARLY at 1 of 6, and ARG1 at 3 of 16 malonyllysine sites. Given that SIRT5 is present in the nucleus and cytoplasm in addition to mitochondria ((Matsushita et al. 2011))((Park et al. 2013))((Nishida et al. 2015)), it will be interesting to test whether SIRT5 modulates the activity of the urea cycle via targeting of both mitochondrial and cytosolic enzymes, or whether one compartment’s effects dominate.
Besides SIRT3 and SIRT5, SIRT4 is also involved in the regulation of the urea cycle activity. Glutamate dehydrogenase (GLUD1) catalyzes the oxidative deamination of glutamate to α-ketoglutarate and ammonia, thereby providing a source of ammonia to enter the urea cycle. SIRT4 inhibits GLUD1 function in pancreatic β cells ((Haigis et al. 2006)), raising the possibility that mitochondrial sirtuins might oppose each other to fine tune the activity of urea cycle via modulating different post-translational modifications on various enzymes in this pathway.
Oxidative Stress
Reactive oxygen species (ROS) such as the superoxide anion (O2−), hydroxyl radical (OH*) and hydrogen peroxide (H2O2) are produced during metabolic activity and both directly damage cellular components and drive signaling cascades throughout the cell ((Merksamer et al. 2013)). As ROS production is chiefly mitochondrial, defenses against oxidative damage in the mitochondrial matrix are particularly important. Mitochondrial protein acylation can suppress the activity of key enzymes that protects again oxidative stress. The mitochondrial SIRT3 deacetylates two of these enzymes: manganese superoxide dismutase (MnSOD) (Tao et al. 2010) and isocitrate dehydrogenase 2 (IDH2)(Someya et al. 2010). SIRT3 also deacetylates key enzymes that produce ROS such as unique subunits of the electron transport chain ((Rahman et al. 2014)) and 2-oxoacid dehydrogenases ((Quinlan et al. 2014)).
Manganese superoxide dismutase (MnSOD) catalyzes the decomposition of superoxide to oxygen and hydrogen peroxide. Both MnSOD and the various mitochondrial superoxide sources are acylated ((Rardin, Newman, et al. 2013), (Rardin, He, et al. 2013);(Tan et al. 2014)); MnSOD also carries glutarylation, succinylation, and acetylation at functionally required lysine residues near its active site, K122 in particular bears all three modifications ((Tao et al. 2010)).
Hydrogen peroxide is detoxified by glutathione peroxidases and peroxiredoxins in the mitochondrial matrix. These enzymes use reversible oxidation of catalytic selenol or thiol groups to transfer electrons to an inter-glutathione disulfide bond, to glutathione peroxidases, or to thioredoxins which are subsequently reduced by oxidizing glutathione. Oxidized glutathione thus underpins all mitochondrial H2O2 detoxification, and is reduced by NADPH-dependent glutathione reductases ((Merksamer et al. 2013)). The NADPH required for this process is generated from NADP+ by the activities of either IDH2 or glutamate dehydrogenase (GLUD1). IDH2 competes with NAD+-dependent IDH3 for isocitrate; tuning the activity of IDH2 thus regulates available NADPH for H2O2 detoxification. IDH2 function is inhibited by acetylation, and its deacetylation by SIRT3 restores function ((Schlicker et al. 2008)). GLUD1 is inhibited by SIRT4 ((Haigis et al. 2006)) and by acetylation which can be reversed by SIRT3 ((Schlicker et al. 2008)).
Mitochondrial acylation from the top down: System-wide effects in disease and in health
A growing body of evidence links disturbances in mitochondrial protein acylation with unique diseases, typically using whole-body genetic knockout mice but also via identification of mutations in human genes. In contrast to the highly specified molecular sites described above, the intermediates linking acylation to these associated disease states are not known. Given the generally low stoichiometry of mitochondrial acylation sites, each disease is likely mediated by multiple presently-unidentified acylations, each slightly but cooperatively impairing mitochondrial function. These system-level confirmations that loss of deacylation alters biology at the organismal level are therefore necessary rebuttals to the contention that low-stoichiometry acylation is biologically insignificant.
Cancer
Given the range of mechanisms giving rise to cancer, it is perhaps unsurprising that SIRT3 can play various roles in tumorigenesis, tumor growth and metastasis depending on context. One potentially key role for mitochondrial acylation in tumorigenesis is in the initiation of the Warburg effect, or the shift to glycolysis as the key energy source of the cell from oxidative metabolism. SIRT3’s attenuation of reactive oxygen species production appears to inhibit a glycolytic shift in potentially cancerous cells ((Finley et al. 2011); (Fan et al. 2014)). SIRT4 might also seem to drive a dependence on glucose via its inhibition of fat oxidation and glutamine catabolism, but its overall growth-inhibiting effect results in decreased metabolism and a sensitization to glucose inhibition rather than a Warburg-type shift (Jeong et al, JBC 2014).
SIRT3 has been reported as pro-apoptotic ((Allison and Milner 2007)) and SIRT3 loss is associated with a tumor-permissive environment via the inhibition of ROS production ((Kim et al. 2010);(Bell et al. 2011);(Finley et al. 2011)). On the other hand, studies have indicated a role in mediating tumor survival ((Alhazzazi et al. 2011);(Cheng et al. 2013)). Additionally, dysfunctional mitochondria in certain types of cancer may be abetted in their effects on cell survival, proliferation, and anabolism by SIRT3’s ability to combat hyperacetylation.
SIRT4 likewise appears to control tumorigenesis ((Jeong et al. 2013), (Csibi et al. 2013)) by inhibiting the high rate of glutamine metabolism associated with cancer growth; ADP-ribosylation of glutamate dehydrogenase limits TCA cycle anaplerosis and NAD[P]H generation ((Haigis et al. 2006)), and SIRT4’s delipoylation activity ((Mathias et al. 2014)) also suggests the potential for inhibitory delipoylation of the downstream oxoglutarate dehydrogenase complex, which functions similarly to pyruvate dehydrogenase (discussed above). SIRT4 control of tumorigenesis is therefore particularly relevant to c-Myc-driven, mitochondrially active, glutamine-dependent tumors, while SIRT3 prevents less mitochondrially active tumors.
Neurological Disease
Given the effects already demonstrated of mitochondrial acylation in allowing tumorigenesis and increasing production of reactive oxygen species, it should come as no surprise that mitochondrial acylation can modulate neuronal function - and, more interestingly, that its removal by sirtuins can help improve health. Recent work on sleep-related cognitive decline found a role in for SIRT3 in maintaining defenses against oxidative damage in the wakefulness-driving nucleus accumbens ((Zhang et al. 2014)). Similarly, SIRT3 function has been shown to attenuate cell death after excitotoxic stress, and mediates the protective effect of exercise against such stress ((Cheng et al. 2016)), possibly via deacetylation of cyclophilin D ((Cheng et al. 2016),(Amigo et al. 2017)). Options for therapeutically assisting sirtuins in maintaining low levels of mitochondrial acylation in the brain are, however, limited - the blood-brain barrier precludes large classes of compounds from reaching neurons. Recently, NAD+ supplementation via its precursors nicotinamide riboside and nicotinamide mononucleotide has been shown to enhance SIRT3 function in tissues such as skeletal muscle ((Canto et al. 2012)); the dosage scheme used in this study was insufficient to induce deacetylation in neurons, but longer-term or higher-dose administration may yet offer the best path towards assisting SIRT3 in neuronal deacetylation.
Age- and noise-associated hearing loss
Caloric restriction extends life span and slows the progression of age-related hearing loss, an age-related disorder associated with oxidative stress. Remarkably, SIRT3 is necessary for the protective effect of calorie restriction on age-related hearing loss supporting a key role for SIRT3-dependent mitochondrial adaptations against aging in mammals (Someya et al. 2010). SIRT3 also protects against hearing loss in response to intense noise exposure (Brown et al. 2014).
As discussed above, intense noise exposure causes hearing loss by inducing degeneration of spiral ganglia neurites that innervate cochlear hair cells. Cochlear NAD(+) levels increased in mice after administering nicotinamide riboside, a precursor to NAD(+) and prevented noise-induced hearing loss. These effects are dependent on SIRT3, since SIRT3-overexpressing mice are resistant to noise-induced hearing loss and SIRT3 deletion abrogates the protective effects of NR.
Metabolic syndrome, obesity, and diabetes
Hypernutrition and its associated disease states, such as obesity and type 2 diabetes, are characterized by an increased availability of the acyl-CoA precursors which are believed to drive the bulk of mitochondrial acylation. As already described, increasing NAD+ by exogenous supplementation can protect against obesity ((Canto et al. 2012)) in part by limiting mitochondrial acylation - but can mitochondrial acylation actually cause the disease symptoms associated with excess calorie intake?
Current research argues both for and against a causative role for mitochondrial protein acylation in metabolic disorders. SIRT3 knockout mice exhibit metabolic changes similar to those observed in the human metabolic syndrome ((Hirschey et al. 2011);(Jing et al. 2013)), and a human SIRT3 polymorphism associated with reduced SIRT3 deacetylase activity is associated with increased risk of metabolic syndrome in humans ((Hirschey et al. 2011). The V208I mutation caused by this polymorphism both increased the KM of SIRT3 for NAD+ and reduced the Vmax of the enzyme towards peptide substrate, reducing overall catalytic efficiency and mimicking the effects of reduced SIRT3 expression at the whole-body level and creating a ~1.4-fold increase in risk for metabolic syndrome ((Hirschey et al. 2011)). However, tissue-specific knockdown of SIRT3 in liver or skeletal muscle does not recapitulate these metabolic alterations, despite similar patterns of hyperacetylation in C57Black6 mice ((Fernandez-Marcos et al. 2012)). The reason for this discrepancy is not clear.
Pulmonary Arterial Hypertension
Idiopathic pulmonary hypertension is characterized by a progressive loss of blood oxygenation due to increased blood pressure and fibrosis of the arteries supplying blood to the lungs. Recent work by the Michelakis’s group has identified loss of SIRT3 function as a potential cause of this disease ((Paulin et al. 2014)) via identification of a SIRT3 polymorphism previously associated with metabolic syndrome ((Hirschey et al. 2011) in a cohort of 162 patients and controls. Increased acetylation of mitochondrial proteins in the affected smooth muscle leads to mitochondrial hyperpolarization, increased proliferation, suppressed apoptosis, and eventually to progressive arterial thickening. In other cell types, loss of SIRT3 function leads to mitochondrial depolarization via impaired oxidative phosphorylation and subsequent NAD+ synthesis. This hypertension study thus adds a new dimension to the connection between acylation and disease; the same polymorphism can lead to pathological gain of function in arterial cells and pathological loss of function in metabolism. Loss of SIRT3 activity thus limits the functional envelope of mitochondria in multiple directions, with the precise failure mechanism for a given disease depending on which of many connected mitochondrial pathways is most taxed and thus most sensitive to hyperacetylation.
Aging
A wide variety of diseases – including those previously discussed in this section – feature age as a major risk factor. Accordingly, sirtuins’ role in protecting against multiple aging-associated diseases places them squarely as “anti-aging” proteins; while sirtuins may not alter the overall rate of organismal aging, their ability to forestall disease states makes their function key to maximizing healthy lifespan. Additionally, aging has consequences for sirtuin function at the organismal level outside the bounds of individual diseases. In particular, age-dependent decreases in NAD+ limit sirtuin function; the NAD-destroying enzyme CD38 was shown to induce mitochondrial dysfunction during aging in part by limiting SIRT3’s ability to control mitochondrial hyperacetylation ((Camacho-Pereira et al. 2016)).
Senescence is an aging-associated defense against tumorigenesis which is characterized by a permanent exit from the cell cycle and by a proinflammatory senescence-associated secretory phenotype (SASP) which, early in life, assists in the clearance of senescent cells and late in life causes pathological alterations to tissue structure and function. Recent work shows that SIRT3 and SIRT5 attenuate the SASP, in part by preventing dysfunctional decreases in NAD+/NADH ratios ((Wiley et al. 2016)). The CD38-induced loss of NAD+ with age thus leads to increases in inflammatory SASP cytokines, which in turn lead to proliferation of CD38-expressing cells; SIRT3 function, by attenuating the resulting loss of mitochondrial function, helps keep this loop in check.
One of the few potential clinical interventions which works against aging across multiple organisms is calorie restriction, which limits dietary intake to upregulate stress resistance pathways and reroute metabolism. SIRT3 is required for many protective aspects of this adaptive shift ((Someya et al. 2010); (Qiu et al. 2010); (Amigo et al. 2017)), such as oxidative stress resistance and attenuated hearing loss. This fits with the larger paradigm of SIRT3 playing a key role in adaptive responses to stress ((Cheng et al. 2016)); SIRT3 thus fights aging by attenuating the development of systemic stressors such as NAD+ loss and by enhancing our ability to survive the resulting insults.
Where Does Acylation Go from Here?
Current work has given us several large, and possibly comprehensive, datasets cataloguing acylation modifications to a vast array of protein targets. In conjunction with this, we have a modest - but growing - list of mitochondrial proteins where the contributions of acylation have been verified to affect enzymatic function, and the beginnings of an understanding of the mechanisms of acylation and its regulation. A remaining key question in the field is to determine the stoichiometry of most acylations. Recent studies have begun to address this key question in bacteria ((Baeza et al. 2014); (Baeza, Smallegan, and Denu 2015); (Weinert, Moustafa, et al. 2015); (Weinert et al. 2017a)), and other systems ((Weinert, Moustafa, et al. 2015); (Weinert, Iesmantavicius, et al. 2015); (Meyer et al. 2016)). With the exception of the SILAC-based indirect stoichiometry method (which overestimates stoichiometry via crosstalk with other lysine modifications), all the chemical labeling methods have concluded that most of the mitochondrial protein acetylation sites are occupied at less than 1% on average. This result raises significant concerns regarding the functional relevance of acetylation in biological systems. Unless these modifications impart new function or activate enzymes, inhibition by acetylation at a level of 1% seems unlikely to be relevant in vivo. On the other hand, mice lacking SIRT3, SIRT4 or SIRT5 exhibit significant biological metabolic phenotypes that parallel the pathways that are shown to be hyperacylated. Given that studies often find pathway-wide acylation, it may be that the cumulative effect of dozens of low-occupancy modifications mediates biological effects. Future work in the field will need to focus on addressing these key issues.
Furthermore, the rapidly expanding set of known acyl modifications should be further expanded into the currently undiscovered products of nucleophilic reactions by various fatty acid and amino acid intermediates found as thioester conjugates to coenzyme A. Past this, one wonders whether other targets will in fact be found to be spontaneous acylated; thiol residues are as capable as deprotonated amines of mediating nucleophilic attack, if not more so, and are found in large numbers in the mitochondrial matrix ((Requejo et al. 2010)). A recent study demonstrated spontaneous acetylation of thiols as a pathway towards lysine modification ((James et al. 2017)). Both technical tools and case studies of biology developed to date through studying lysine acylation will frame future studies into post-translational modifications in the mitochondria caused by reactive metabolites.
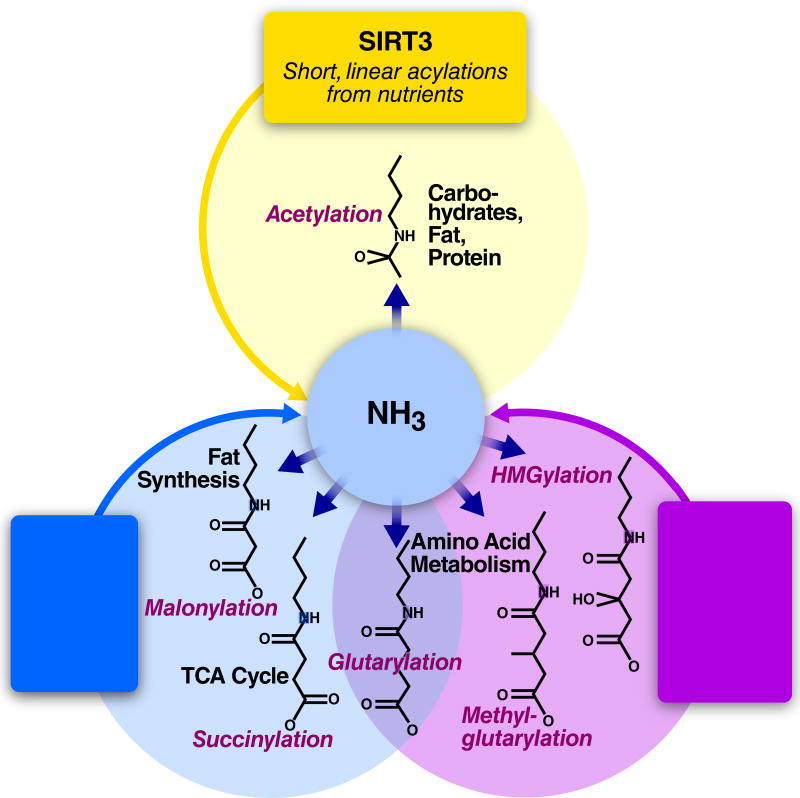
Figure 2. Mitochondrial Acylations are Regulated by SIRT3-5 by Structural Class.
Any unmodified lysine residue can react non-enzymatically with the acyl-CoAs found in the mitochondrial matrix. Which sirtuin reverses the resulting acyllysine is determined by structural features of the product. SIRT3 reverses short-chain modifications such as acetylation. SIRT5 reverses divalent modifications including succinylation, malonylation, and glutarylation. SIRT4 reverses branched-chain modifications such as methylglutarylation. Each sirtuin thus regulates the side products of acyl-CoAs corresponding to different facets of mitochondrial metabolism.
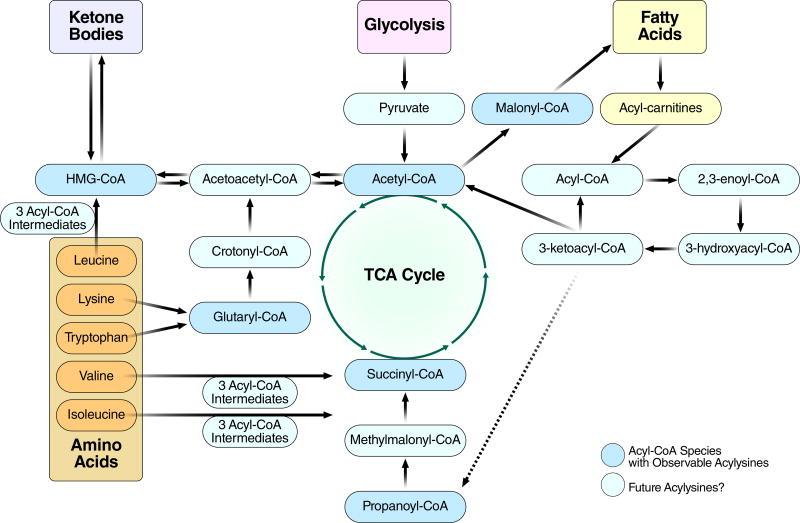
Figure 4. Acyl-CoA Thioesters Are Common Intermediates In Mitochondrial Metabolism.
All major nutrient classes - carbohydrates, fats, proteins, and ketone bodies - are catabolized in part through mitochondrial pathway mediated by acyl-CoAs. Acetylation and succinylation by acetyl-CoA and succinyl-CoA respectively are associated with TCA cycle through all nutrient types, and have been characterized at the proteomic level by mass spectrometry. Similar studies have identified glutarylated, malonylated, and propionylated proteins in mitochondria. Whether the wide variety of lower-abundance acyl-CoA species are in turn transduced into mitochondrial lysine acylation remains to be seen.
Carrico et al. review recent advances in mapping and understanding acylation modifications to mitochondrial proteins, such as acetylation and succinylation, and their regulation by the sirtuins SIRT3-5. Best practices for proteome-wide mass spectrometry are discussed, followed by case studies of how acylation modulates protein function and health.
Acknowledgments
We thank John Carroll for graphics and Gary Howard for editing. This work was supported by an R24 grant from NIDDK (R24 DK085610). CC was supported by a postdoctoral fellowship from the Larry L Hillblom Foundation. JGM was supported by the NIH (T32 AG000266).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282(46):33583–92. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Alhazzazi TY, Kamarajan P, Joo N, Huang JY, Verdin E, D'Silva NJ, Kapila YL. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117(8):1670–8. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6(21):2669–77. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- Amigo I, Menezes-Filho SL, Luevano-Martinez LA, Chausse B, Kowaltowski AJ. Caloric restriction increases brain mitochondrial calcium retention capacity and protects against excitotoxicity. Aging Cell. 2017;16(1):73–81. doi: 10.1111/acel.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, Wagner GR, Thompson JW, Madsen AS, Green MF, Sivley RM, Ilkayeva OR, Stevens RD, Backos DS, Capra JA, Olsen CA, Campbell JE, Muoio DM, Grimsrud PA, Hirschey MD. SIRT4 is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion. 2017 doi: 10.1016/j.cmet.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Dowell JA, Smallegan MJ, Fan J, Amador-Noguez D, Khan Z, Denu JM. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289(31):21326–38. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeza J, Smallegan MJ, Denu JM. Site-specific reactivity of nonenzymatic lysine acetylation. ACS Chem Biol. 2015;10(1):122–8. doi: 10.1021/cb500848p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404(4):939–65. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- Bell EL, Emerling BM, Ricoult SJ, Guarente L. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30(26):2986–96. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Maqsood S, Huang JY, Pan Y, Harkcom W, Li W, Sauve A, Verdin E, Jaffrey SR. Activation of SIRT3 by the NAD(+) precursor nicotinamide riboside protects from noise-induced hearing loss. Cell Metab. 2014;20(6):1059–68. doi: 10.1016/j.cmet.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho-Pereira J, Tarrago MG, Chini CC, Nin V, Escande C, Warner GM, Puranik AS, Schoon RA, Reid JM, Galina A, Chini EN. CD38 Dictates Age-Related NAD Decline and Mitochondrial Dysfunction through an SIRT3-Dependent Mechanism. Cell Metab. 2016;23(6):1127–39. doi: 10.1016/j.cmet.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, Houtkooper RH, Pirinen E, Youn DY, Oosterveer MH, Cen Y, Fernandez-Marcos PJ, Yamamoto H, Andreux PA, Cettour-Rose P, Gademann K, Rinsch C, Schoonjans K, Sauve AA, Auwerx J. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15(6):838–47. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Yang Y, Zhou Y, Maharana C, Lu D, Peng W, Liu Y, Wan R, Marosi K, Misiak M, Bohr VA, Mattson MP. Mitochondrial SIRT3 Mediates Adaptive Responses of Neurons to Exercise and Metabolic and Excitatory Challenges. Cell Metab. 2016;23(1):128–42. doi: 10.1016/j.cmet.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Ren X, Gowda AS, Shan Y, Zhang L, Yuan YS, Patel R, Wu H, Huber-Keener K, Yang JW, Liu D, Spratt TE, Yang JM. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. 2013;4:e731. doi: 10.1038/cddis.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15(8):536–50. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Colak G, Pougovkina O, Dai L, Tan M, Te Brinke H, Huang H, Cheng Z, Park J, Wan X, Liu X, Yue WW, Wanders RJ, Locasale JW, Lombard DB, de Boer VC, Zhao Y. Proteomic and Biochemical Studies of Lysine Malonylation Suggest Its Malonic Aciduria-associated Regulatory Role in Mitochondrial Function and Fatty Acid Oxidation. Mol Cell Proteomics. 2015;14(11):3056–71. doi: 10.1074/mcp.M115.048850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A, Fendt SM, Li C, Poulogiannis G, Choo AY, Chapski DJ, Jeong SM, Dempsey JM, Parkhitko A, Morrison T, Henske EP, Haigis MC, Cantley LC, Stephanopoulos G, Yu J, Blenis J. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153(4):840–54. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, He B, Chen W, Zhang S, Cerione RA, Auwerx J, Hao Q, Lin H. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–9. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Cai T, Li T, Xue P, Zhou B, He X, Wei P, Liu P, Yang F, Wei T. Lysine malonylation is elevated in type 2 diabetic mouse models and enriched in metabolic associated proteins. Mol Cell Proteomics. 2015;14(1):227–36. doi: 10.1074/mcp.M114.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Shan C, Kang HB, Elf S, Xie J, Tucker M, Gu TL, Aguiar M, Lonning S, Chen H, Mohammadi M, Britton LM, Garcia BA, Aleckovic M, Kang Y, Kaluz S, Devi N, Van Meir EG, Hitosugi T, Seo JH, Lonial S, Gaddh M, Arellano M, Khoury HJ, Khuri FR, Boggon TJ, Kang S, Chen J. Tyr phosphorylation of PDP1 toggles recruitment between ACAT1 and SIRT3 to regulate the pyruvate dehydrogenase complex. Mol Cell. 2014;53(4):534–48. doi: 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman JL, Baeza J, Denu JM. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288(43):31350–6. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Marcos PJ, Jeninga EH, Canto C, Harach T, de Boer VC, Andreux P, Moullan N, Pirinen E, Yamamoto H, Houten SM, Schoonjans K, Auwerx J. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Sci Rep. 2012;2:425. doi: 10.1038/srep00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, Clish CB, Pandolfi PP, Haigis MC. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell. 2011;19(3):416–28. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126(5):941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103(27):10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Yu W, Smith BC, Devries MK, Ellinger JJ, Someya S, Shortreed MR, Prolla T, Markley JL, Smith LM, Zhao S, Guan KL, Denu JM. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41(2):139–49. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell. 2013;49(1):186–99. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464(7285):121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B, Stancakova A, Goetzman E, Lam MM, Schwer B, Stevens RD, Muehlbauer MJ, Kakar S, Bass NM, Kuusisto J, Laakso M, Alt FW, Newgard CB, Farese RV, Jr, Kahn CR, Verdin E. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell. 2011;44(2):177–90. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram-Smith C, Martin SR, Smith KS. Acetate kinase: not just a bacterial enzyme. Trends Microbiol. 2006;14(6):249–53. doi: 10.1016/j.tim.2006.04.001. [DOI] [PubMed] [Google Scholar]
- James AM, Hoogewijs K, Logan A, Hall AR, Ding S, Fearnley IM, Murphy MP. Non-enzymatic N-acetylation of Lysine Residues by AcetylCoA Often Occurs via a Proximal S-acetylated Thiol Intermediate Sensitive to Glyoxalase II. Cell Rep. 2017;18(9):2105–2112. doi: 10.1016/j.celrep.2017.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong SM, Xiao C, Finley LW, Lahusen T, Souza AL, Pierce K, Li YH, Wang X, Laurent G, German NJ, Xu X, Li C, Wang RH, Lee J, Csibi A, Cerione R, Blenis J, Clish CB, Kimmelman A, Deng CX, Haigis MC. SIRT4 has tumorsuppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23(4):450–63. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, O'Neill BT, Rardin MJ, Kleinridders A, Ilkeyeva OR, Ussar S, Bain JR, Lee KY, Verdin EM, Newgard CB, Gibson BW, Kahn CR. Sirt3 regulates metabolic flexibility of skeletal muscle through reversible enzymatic deacetylation. Diabetes. 2013;62(10):3404–17. doi: 10.2337/db12-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AN, Lewis PN. Unstructured conformations are a substrate requirement for the Sir2 family of NAD-dependent protein deacetylases. J Biol Chem. 2005;280(43):36073–8. doi: 10.1074/jbc.M508247200. [DOI] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassilopoulos A, Ozden O, Park SH, Singh KK, Abdulkadir SA, Spitz DR, Deng CX, Gius D. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17(1):41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS One. 2010;5(7):e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantier L, Williams AS, Williams IM, Yang KK, Bracy DP, Goelzer M, James FD, Gius D, Wasserman DH. SIRT3 Is Crucial for Maintaining Skeletal Muscle Insulin Action and Protects Against Severe Insulin Resistance in High-Fat-Fed Mice. Diabetes. 2015;64(9):3081–92. doi: 10.2337/db14-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent G, German NJ, Saha AK, de Boer VC, Davies M, Koves TR, Dephoure N, Fischer F, Boanca G, Vaitheesvaran B, Lovitch SB, Sharpe AH, Kurland IJ, Steegborn C, Gygi SP, Muoio DM, Ruderman NB, Haigis MC. SIRT4 coordinates the balance between lipid synthesis and catabolism by repressing malonyl CoA decarboxylase. Mol Cell. 2013;50(5):686–98. doi: 10.1016/j.molcel.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Kerner J, Hoppel CL. Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex. J Biol Chem. 2011;286(29):25655–62. doi: 10.1074/jbc.M111.228692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27(24):8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RA, Greco TM, Oberstein A, Budayeva HG, Chakrabarti R, Rowland EA, Kang Y, Shenk T, Cristea IM. Sirtuin 4 is a lipoamidase regulating pyruvate dehydrogenase complex activity. Cell. 2014;159(7):1615–25. doi: 10.1016/j.cell.2014.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita N, Yonashiro R, Ogata Y, Sugiura A, Nagashima S, Fukuda T, Inatome R, Yanagi S. Distinct regulation of mitochondrial localization and stability of two human Sirt5 isoforms. Genes Cells. 2011;16(2):190–202. doi: 10.1111/j.1365-2443.2010.01475.x. [DOI] [PubMed] [Google Scholar]
- McAlister GC, Nusinow DP, Jedrychowski MP, Wuhr M, Huttlin EL, Erickson BK, Rad R, Haas W, Gygi SP. MultiNotch MS3 enables accurate, sensitive, and multiplexed detection of differential expression across cancer cell line proteomes. Anal Chem. 2014;86(14):7150–8. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merksamer PI, Liu Y, He W, Hirschey MD, Chen D, Verdin E. The sirtuins, oxidative stress and aging: an emerging link. Aging (Albany NY) 2013;5(3):144–50. doi: 10.18632/aging.100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JG, D'Souza AK, Sorensen DJ, Rardin MJ, Wolfe AJ, Gibson BW, Schilling B. Quantification of Lysine Acetylation and Succinylation Stoichiometry in Proteins Using Mass Spectrometric Data-Independent Acquisitions (SWATH) J Am Soc Mass Spectrom. 2016;27(11):1758–1771. doi: 10.1007/s13361-016-1476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JG, Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev Proteomics. 2017;14(5):419–429. doi: 10.1080/14789450.2017.1322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke C, Lefort N, McLean CG, Cordova JM, Langlais PR, Bordner AJ, Te JA, Ozkan SB, Willis WT, Mandarino LJ. Adenine nucleotide translocase is acetylated in vivo in human muscle: Modeling predicts a decreased ADP affinity and altered control of oxidative phosphorylation. Biochemistry. 2014;53(23):3817–29. doi: 10.1021/bi401651e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering RE, Cravatt BF. Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science. 2013;341(6145):549–53. doi: 10.1126/science.1238327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, Guarente L, Auwerx J. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154(2):430–41. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137(3):560–70. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu ES, Wu S, Sydor MA, Shukla AK, Weitz KK, Moore RJ, Hixson KK, Kim JS, Petyuk VA, Monroe ME, Pasa-Tolic L, Qian WJ, Smith RD, Adkins JN, Ansong C. A method to determine lysine acetylation stoichiometries. Int J Proteomics. 2014;2014:730725. doi: 10.1155/2014/730725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Rardin MJ, Carrico C, He W, Sahu AK, Gut P, Najjar R, Fitch M, Hellerstein M, Gibson BW, Verdin E. SIRT5 Regulates both Cytosolic and Mitochondrial Protein Malonylation with Glycolysis as a Major Target. Mol Cell. 2015;59(2):321–32. doi: 10.1016/j.molcel.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, Brunak S, Mann M. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3(104):ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99(21):13653–8. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer KA, Evans CR, Qi NR, Minogue CE, Carson JJ, Chermside-Scabbo CJ, Koch LG, Britton SL, Pagliarini DJ, Coon JJ, Burant CF. Maximal oxidative capacity during exercise is associated with skeletal muscle fuel selection and dynamic changes in mitochondrial protein acetylation. Cell Metab. 2015;21(3):468–78. doi: 10.1016/j.cmet.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem. 2009;284(20):13285–9. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik WK, Pearson D, Lee HW, Kim S. Nonenzymatic acetylation of histones with acetyl-CoA. Biochim Biophys Acta. 1970;213(2):513–22. doi: 10.1016/0005-2787(70)90058-4. [DOI] [PubMed] [Google Scholar]
- Pannek M, Simic Z, Fuszard M, Meleshin M, Rotili D, Mai A, Schutkowski M, Steegborn C. Crystal structures of the mitochondrial deacylase Sirtuin 4 reveal isoform-specific acyl recognition and regulation features. Nat Commun. 2017;8(1):1513. doi: 10.1038/s41467-017-01701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, Zhao Y. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50(6):919–30. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin R, Dromparis P, Sutendra G, Gurtu V, Zervopoulos S, Bowers L, Haromy A, Webster L, Provencher S, Bonnet S, Michelakis ED. Sirtuin 3 deficiency is associated with inhibited mitochondrial function and pulmonary arterial hypertension in rodents and humans. Cell Metab. 2014;20(5):827–39. doi: 10.1016/j.cmet.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Peng C, Lu Z, Xie Z, Cheng Z, Chen Y, Tan M, Luo H, Zhang Y, He W, Yang K, Zwaans BM, Tishkoff D, Ho L, Lombard D, He TC, Dai J, Verdin E, Ye Y, Zhao Y. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10(12) doi: 10.1074/mcp.M111.012658. M111 012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pougovkina O, Te Brinke H, Wanders RJ, Houten SM, de Boer VC. Aberrant protein acylation is a common observation in inborn errors of acyl-CoA metabolism. J Inherit Metab Dis. 2014;37(5):709–14. doi: 10.1007/s10545-014-9684-9. [DOI] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12(6):662–7. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Quinlan CL, Goncalves RL, Hey-Mogensen M, Yadava N, Bunik VI, Brand MD. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. J Biol Chem. 2014;289(12):8312–25. doi: 10.1074/jbc.M113.545301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Nirala NK, Singh A, Zhu LJ, Taguchi K, Bamba T, Fukusaki E, Shaw LM, Lambright DG, Acharya JK, Acharya UR. Drosophila Sirt2/mammalian SIRT3 deacetylates ATP synthase beta and regulates complex V activity. J Cell Biol. 2014;206(2):289–305. doi: 10.1083/jcb.201404118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, Uppala R, Fitch M, Riiff T, Zhu L, Zhou J, Mulhern D, Stevens RD, Ilkayeva OR, Newgard CB, Jacobson MP, Hellerstein M, Goetzman ES, Gibson BW, Verdin E. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18(6):920–33. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, Gibson BW. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110(16):6601–6. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rardin MJ, Schilling B, Cheng LY, MacLean BX, Sorensen DJ, Sahu AK, MacCoss MJ, Vitek O, Gibson BW. MS1 Peptide Ion Intensity Chromatograms in MS2 (SWATH) Data Independent Acquisitions. Improving Post Acquisition Analysis of Proteomic Experiments. Mol Cell Proteomics. 2015;14(9):2405–19. doi: 10.1074/mcp.O115.048181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Requejo R, Hurd TR, Costa NJ, Murphy MP. Cysteine residues exposed on protein surfaces are the dominant intramitochondrial thiol and may protect against oxidative damage. FEBS J. 2010;277(6):1465–80. doi: 10.1111/j.1742-4658.2010.07576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, Maccoss MJ, Gibson BW. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics. 2012;11(5):202–14. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382(3):790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schmeisser K, Mansfeld J, Kuhlow D, Weimer S, Priebe S, Heiland I, Birringer M, Groth M, Segref A, Kanfi Y, Price NL, Schmeisser S, Schuster S, Pfeiffer AF, Guthke R, Platzer M, Hoppe T, Cohen HY, Zarse K, Sinclair DA, Ristow M. Role of sirtuins in lifespan regulation is linked to methylation of nicotinamide. Nat Chem Biol. 2013;9(11):693–700. doi: 10.1038/nchembio.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103(27):10224–9. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8(5):604–6. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158(4):647–57. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott I, Webster BR, Li JH, Sack MN. Identification of a molecular component of the mitochondrial acetyltransferase programme: a novel role for GCN5L1. Biochem J. 2012;443(3):655–61. doi: 10.1042/BJ20120118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobson MP, Verdin E. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12(6):654–61. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Newman J, He W, Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD, Newgard CB, Farese RV, Jr, de Cabo R, Ulrich S, Akassoglou K, Verdin E. Suppression of oxidative stress by beta-hydroxybutyrate, an endogenous histone deacetylase inhibitor. Science. 2013;339(6116):211–4. doi: 10.1126/science.1227166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BC, Settles B, Hallows WC, Craven MW, Denu JM. SIRT3 substrate specificity determined by peptide arrays and machine learning. ACS Chem Biol. 2011;6(2):146–57. doi: 10.1021/cb100218d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sol EM, Wagner SA, Weinert BT, Kumar A, Kim HS, Deng CX, Choudhary C. Proteomic investigations of lysine acetylation identify diverse substrates of mitochondrial deacetylase sirt3. PLoS One. 2012;7(12):e50545. doi: 10.1371/journal.pone.0050545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann JM, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143(5):802–12. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still AJ, Floyd BJ, Hebert AS, Bingman CA, Carson JJ, Gunderson DR, Dolan BK, Grimsrud PA, Dittenhafer-Reed KE, Stapleton DS, Keller MP, Westphall MS, Denu JM, Attie AD, Coon JJ, Pagliarini DJ. Quantification of mitochondrial acetylation dynamics highlights prominent sites of metabolic regulation. J Biol Chem. 2013;288(36):26209–19. doi: 10.1074/jbc.M113.483396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 2014;19(4):605–17. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40(6):893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E, Ott M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16(4):258–64. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- Wagner GR, Payne RM. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288(40):29036–45. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Bhatt DP, O’Connell TM, Thompson JW, Dubois LG, Backos DS, Mitchell GA, Ilkayeva OR, Stevens RD, Grimsrud PA, Hirschey MD. A Class of Reactive Acyl-CoA Species Reveals the Nonenzymatic Origins of Protein Acylation. Cell Metabolism. 2017 doi: 10.1016/j.cmet.2017.03.006. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Iesmantavicius V, Moustafa T, Scholz C, Wagner SA, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2014;10:716. doi: 10.1002/msb.134766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Iesmantavicius V, Moustafa T, Scholz C, Wagner SA, Magnes C, Zechner R, Choudhary C. Acetylation dynamics and stoichiometry in Saccharomyces cerevisiae. Mol Syst Biol. 2015;11(10):833. doi: 10.15252/msb.156513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Iesmantavicius V, Wagner SA, Scholz C, Gummesson B, Beli P, Nystrom T, Choudhary C. Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol Cell. 2013;51(2):265–72. doi: 10.1016/j.molcel.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Weinert BT, Moustafa T, Iesmantavicius V, Zechner R, Choudhary C. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 2015;34(21):2620–32. doi: 10.15252/embj.201591271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Satpathy S, Hansen BK, Lyon D, Jensen LJ, Choudhary C. Accurate quantification of site-specific acetylation stoichiometry reveals the impact of sirtuin deacetylase CobB on the E. coli acetylome. Mol Cell Proteomics. 2017a doi: 10.1074/mcp.M117.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Satpathy S, Hansen BK, Lyon D, Jensen LJ, Choudhary C. Accurate Quantification of Site-specific Acetylation Stoichiometry Reveals the Impact of Sirtuin Deacetylase CobB on the E. coli Acetylome. Mol Cell Proteomics. 2017b;16(5):759–769. doi: 10.1074/mcp.M117.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley CD, Velarde MC, Lecot P, Liu S, Sarnoski EA, Freund A, Shirakawa K, Lim HW, Davis SS, Ramanathan A, Gerencser AA, Verdin E, Campisi J. Mitochondrial Dysfunction Induces Senescence with a Distinct Secretory Phenotype. Cell Metab. 2016;23(2):303–14. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshino J, Mills KF, Yoon MJ, Imai S. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14(4):528–36. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Zhu Y, Zhan G, Fenik P, Panossian L, Wang MM, Reid S, Lai D, Davis JG, Baur JA, Veasey S. Extended wakefulness: compromised metabolics in and degeneration of locus ceruleus neurons. J Neurosci. 2014;34(12):4418–31. doi: 10.1523/JNEUROSCI.5025-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7(1):58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]