Abstract
SNARE-mediated membrane fusion is a ubiquitous process responsible for intracellular vesicle trafficking, including membrane fusion in exocytosis that leads to hormone and neurotransmitter release. The proteins that facilitate this process are highly dynamic and adopt multiple conformations when they interact with other proteins and lipids as they form highly regulated molecular machines that operate on membranes. Solution NMR is an ideal method to capture high-resolution glimpses of the molecular transformations that take place when these proteins come together and work on membranes. Since solution NMR has limitations on the size of proteins and complexes that can be studied, lipid bilayer model membranes cannot be used in these approaches, so the relevant interactions are typically studied in various types of membrane-mimetics that are tractable by solution NMR methods. In this review we therefore first summarize different membrane-mimetic systems that are commonly used or that show promise for solution NMR studies of membrane-interacting proteins. We then summarize recent NMR studies on two SNARE proteins, syntaxin and synaptobrevin, and two related regulatory proteins, complexin and α-synuclein, and their interactions with membrane lipids. These studies provide a structural and dynamical framework for how these proteins might carry out their functions in the vicinity of lipid membranes. The common theme throughout these studies is that membrane interactions have major influences on the structural dynamics of these proteins that cannot be ignored when attempting to explain their functions in contemporary models of SNARE-mediated membrane fusion.
Graphical abstract
1. Brief introduction to the SNARE fusion machinery
Cargo is transferred between different compartments of eukaryotic cells by membrane vesicles that pinch off from the membranes of donor organelles and fuse with the membranes of acceptor organelles. Similarly, hormones or neurotransmitters that are secreted from cells are delivered by secretory vesicles that fuse with the plasma membrane of the secreting cell. These fusion processes are all mediated by specific sets of SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptors) proteins, which in the case of exocytosis are tightly regulated by intracellular calcium. Different SNARE proteins are present on both fusing membranes and have been shown to be sufficient to facilitate fusion between two membranes (Figure 1) [1–2]. All SNAREs contain highly conserved so-called SNARE motifs, which consist of 60-70 amino acids with heptad repeats. When assembled, these heptad repeats form layered hydrophobic contacts, with the exception of the central ionic “0”-layer. The pairing of SNAREs from the two membranes through their SNARE motifs that form a highly stable four-helix bundle is what ultimately drives membrane fusion. The relevant SNAREs in the case of exocytotic release of neurotransmitters at synapses of neurons are synaptobrevin-2 (also called vesicle associated membrane protein-2 or VAMP-2) on the synaptic vesicle and syntaxin-1A and SNAP-25 on the presynaptic cell membrane (Figure 1). The domain structures of these proteins are shown in Figure 2. Synaptobrevin, a v- or R-SNARE, is anchored in the vesicle membrane through its single C-terminal transmembrane (TM) domain, whereas two target SNAREs (t- or Q-SNAREs), syntaxin and SNAP-25, are anchored in the plasma membrane through a single C-terminal TM domain in the case of syntaxin or multiple palmitate chains that are post-translationally added to SNAP-25. The bundle of four parallel α-helices that constitutes the postfusion cis-SNARE complex [3] is believed to be formed by folding (“zippering”) of the four SNARE motifs in a N- to C- terminal direction. SNAP-25 contributes two and the other SNAREs one α-helix each to the final structure. The four helical bundle is stabilized by hydrophobic side-chain interactions in heptad repeats, except in the center where one arginine from synaptobrevin (hence R-SNARE) and three glutamine residues from syntaxin and SNAP-25 (hence Q-SNAREs) form the signature ionic “0”-layer.
Figure 1.
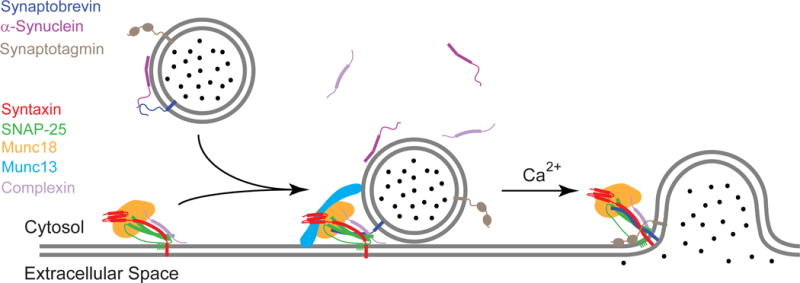
A schematic diagram of neuronal SNARE-mediated membrane fusion. Synaptobrevin on synaptic vesicles can bind to target SNAREs (syntaxin and SNAP-25) on the presynaptic cell membrane. The formation of the trans-SNARE complex (present in intermediate stage of diagram) may be modulated by various synaptic regulatory proteins, such as Munc18, Munc13, complexin, and α-synuclein. Fast and precise neurotransmitter release from the vesicle is triggered by the cytoplasmic rise of Ca2+ and its interaction with synaptotagmin. A four-helix bundle cis-SNARE complex is formed upon completion of fusion (final stage of diagram). Adapted with permission from reference [84].
Figure 2.
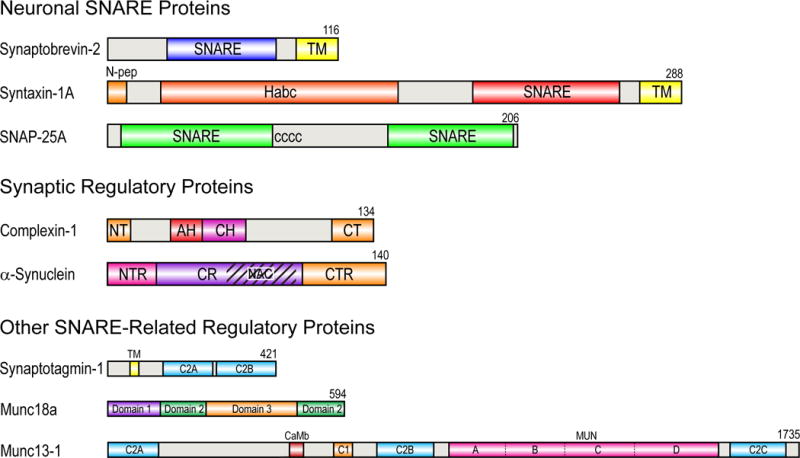
Modular domain structures of neuronal SNAREs and related regulatory proteins. Rat protein sequences and isoforms are shown. The sequence lengths of these proteins are shown to scale, except that the scale of the last three proteins is five times smaller than the scale of the first five proteins. SNARE, SNARE motif; TM, transmembrane domain; N-pep, N-terminal peptide (residues 1-10); Habc, Habc regulatory domain of syntaxin (residues 27-146); CCCC, palmitoylated cysteines; NT, N-terminal (1-12); AH, accessory helix (28-47); CH, central helix (48-70); CT, C-terminal (114-134); NTR, N-terminal region (1-25); CR, central region (26-97); NAC, non-amyloid-β component (61-95); CTR, C-terminal region (98-140); CaMb, calmodulin binding region (459-492); MUN, MUN domain (859-1531).
In addition to the three SNARE proteins, which form the minimal machinery of the neuronal SNARE complex that is sufficient to facilitate membrane fusion in vitro [1], regulated exocytosis in neurons requires a range of additional effector proteins (Figures 1 and 2) [4–6]. Munc18-1 of the Sec1/Munc18 (SM) protein family is essential for proper SNARE assembly and therefore SNARE-mediated membrane fusion in vivo. This soluble protein possesses multiple binding modes with syntaxin, synaptobrevin, as well as the assembled SNARE complex. Specifically, Munc18 regulates the conformation of syntaxin between “open” and “closed” states [7], but it also acts as a chaperone to deliver syntaxin to other binding partners during SNARE complex formation [8–10], and may function as an organizer for precise alignment of SNARE assembly [11–12]. Munc13, which originally was thought to work upstream and contribute to the initial tethering of the two membranes, has been implicated more recently also in the modulation of the Munc18-syntaxin interaction and the organization of the pre-fusion SNARE complex [13–14]. A recent crystal structure of the C1C2BMUN domains of Munc13 reveals a 19.5 nm long multi-helical structure, and the relative positioning of the different domains may lead to cooperative membrane binding and hence may provide points of support for force exertion on the assembly of the cis-SNARE complex [15].
Fast and synchronous SNARE-mediated neuronal excytosis is controlled by Ca2+ via two proteins, synaptotagmin-1 and complexin-1 or -2. It is widely agreed that synaptotagmin is the sensor for Ca2+ in calcium-triggered exocytosis, and the binding of four Ca2+ ions to the two C2 domains of synapotagmin dramatically changes their affinity for membranes with acidic lipids [16–17]. Calcium binding may also modulate the binding of synaptotagmin’s C2 domains to the ternary postfusion SNARE complex [18]. Although multiple interactions between synaptotagmin and different components of the fusion machinery have been observed in vitro, it is still debated how exactly synaptotagmin confers Ca2+ sensitivity to the synaptic fusion process [18–19]. Complexin does not itself bind calcium, but it contributes to the Ca2+ sensitivity of evoked neurotransmitter release. Complexin binds strongly to the postfusion SNARE complex and genetic studies have shown impaired evoked neurotransmitter release in the absence of complexin, although the effect of complexin on spontaneous release can be, depending on conditions, inhibitory or facilitatory [20–21].
There are a number of additional specialized chaperone proteins that affect the cycle of SNARE complex assembly and disassembly. They include cysteine string protein-α and α-synuclein [4]. Both of these proteins are localized on synaptic vesicles, and have been implicated in neurodegenerative diseases. Like synaptotagmin and complexin, these proteins can interact with individual and partially assembled SNAREs as well as with membrane lipids, making their roles in the SNARE-mediated fusion process both interesting and sometimes confusing. Both proteins promote neurodegeneration in a manner that depends on the age of the organism. They affect the misfolding of SNARE intermediates that become more populated later in the life of animal cells, indicating that the chaperone functions of these proteins become more critical in aging cells for reasons that are presently not well understood.
To complete the SNARE assembly and disassembly cycle, postfusion cis-SNARE and misfolded SNARE complexes are disassembled by N-ethylmaleimide sensitive factor (NSF) and soluble NSF attachment proteins (SNAPs) that use ATP as energy source [22–23]. In addition to the already mentioned proteins, which interact with different SNARE proteins specifically or promiscuously at various steps of the fusion process and, therefore, must be in close proximity to the fusion pore, there are more proteins and protein complexes that are important for vesicle tethering to the plasma membrane and recruiting them to the fusion zones [24]. Although very interesting, these proteins will not be discussed in this review.
Just as in other fields of molecular cell biology, most structural insight into how the SNARE fusion machinery might work has come from protein structures of soluble fragments of the relevant proteins, i.e. structures that were solved by x-ray crystallography. Very recently cryo-EM has also contributed some important structures, most notably that of a complex between NSF and the soluble postfusion SNARE complex [23]. Despite the extremely valuable insights that were contributed by these techniques, the importance of partially folded proteins and lipids in the fusion process limits the usefulness of crystallographic or single particle averaging techniques as a means of learning additional important details that are pertinent to understand the molecular interplay that ultimately leads to membrane fusion. NMR, which is uniquely suited to characterize conformationally flexible structural ensembles and study their dynamics in the presence of membrane lipids, has an important role to play to fill these gaps in our current understanding. Both solution and solid-state NMR have made great progress in recent years in the study of structures and dynamics of membrane proteins in native or near-native environments [25]. In the SNARE fusion field significant progress has come recently mostly from solution NMR. Therefore, the purpose of this review is to summarize this progress and to highlight future opportunities in the field. Solution NMR has a well-known limit to the size of systems that can be studied, which is due to severe line-broadening and spectral crowding of solution NMR spectra of large particles caused by slow rotational correlation times. Although instrumental and other technical improvements continue to expand the range of systems that can be studied by these methods, current limits for complete structure determinations of integral membrane proteins by solution NMR are around 40 kDa, and membrane protein complexes in systems over 100 kDa can be studied when only selected components are isotope-labeled. To accommodate this size limit of solution NMR, membrane proteins are commonly studied in membrane-mimetic systems that are discussed in the next section. In the following sections, we highlight important results from recent solution NMR studies of the two SNARE proteins synatpobrevin and syntaxin, as well as complexin and α-synuclein, and we summarize their conformational dynamics and interactions with membrane lipids and other proteins.
2. Membrane-mimetic systems commonly used in solution NMR studies of membrane proteins
2.1. Micelles
Micelles formed by detergent molecules are by far the most widely used membrane mimetics in solution NMR studies of membrane proteins. Empty micelles adopt globular shapes in water [26], with their hydrophobic tails pointing to the center and hydrophilic headgroups orienting towards the surface of the sphere. When membrane proteins are solubilized in micelles, detergent molecules orient their hydrophobic tails towards transmembrane (TM) domains and line their hydrophilic headgroups around the transition regions between TM and water-soluble domains of membrane proteins, likely resulting in prolate or oblate ellipsoids (Figure 3A), depending on the relative sizes of the solvated membrane protein and detergent molecules [27–28]. Micelle-forming detergents are characterized by their aggregation number (N) and critical micelle concentration (CMC). The aggregation number is the average number of detergent molecules in a single micelle, whereas the CMC is the concentration threshold when detergent molecules form a micelle, i.e., the maximum concentration of monomeric detergent molecules. In practice, detergents are labeled as “harsh” or “mild” depending on their tendency to denature or preserve protein structures, respectively. For a series of detergents with the same headgroup, the longer the hydrophobic carbon chain, the smaller is the CMC, i.e., the more stable is the micelle and, ultimately, the lower are detergent costs due to smaller required amounts. These considerations probably reflect the recent trend of choosing diheptanoyl-glycero-phosphocholine (DH7PC) over dihexanoyl-glycero-phosphocholine (DH6PC) in solution NMR studies of a number of membrane proteins [29–30]. However, in many cases, the choice of detergent has mostly been a trial-and-error process. The solubilizing power of a detergent has to be co-optimized with other sample requirements, such as the preservation of the protein’s native fold, the quality of the final spectrum, and the stability of the sample. Dodecylphosphocholine (DPC), with a phosphocholine headgroup and a single 12-carbon aliphatic chain, has been one of the most popular detergents for membrane proteins studied by solution NMR [25]. Lysophospholipids, with either PC or PG headgroups, short-chain phospholipids, lauryldimethylamine oxide (LDAO), n-dodecyl-β-maltoside (DDM), octyl-β-glucoside (β-OG), and sodium dodecyl sulfate (SDS) have also been used successfully.
Figure 3.
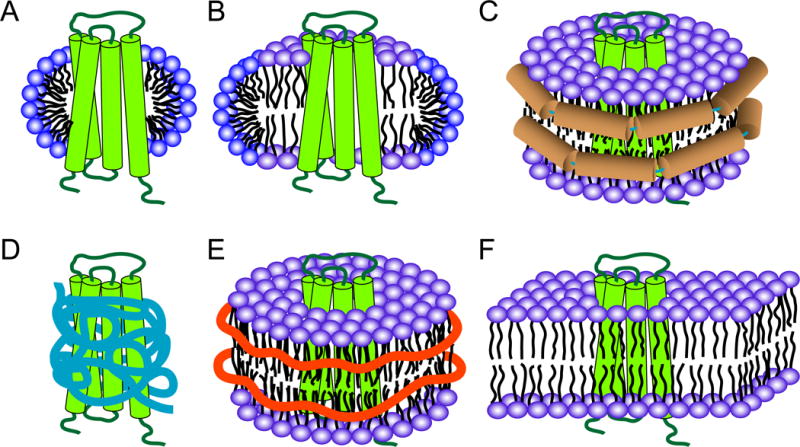
Membrane mimetics for solution NMR studies of membrane proteins. (A) Micelle, (B) bicelle, (C) nanodisc, (D) amphipol, (E) Lipodisq or SMALP, (F) bilayer in a liposome. A hypothetical four-helical bundle protein is shown in green in all cases. Micelles and bicelles are shown in cut-open views in order to show the differences of these two systems. Membrane scaffold protein (MSP) helices in nanodiscs are shown in brown in (C); amphipols are shown in blue in (D); SMA polymers are shown as orange belts in (E).
2.2. Bicelles
Bicelles are formed by a mixture of bilayer-forming lipids and micelle-forming detergents. Common bicelle systems consist of mixtures of long-chain and short-chain phospholipids. In this mixture, long-chain phospholipids supposedly form a patch of planar lipid bilayer with their edge lined by short-chain phospholipids that support a curved surface (Figure 3B). Although such a picture of a bicelle makes intuitive sense, the distribution of both types of lipids in bicelles is in reality much more dynamic. The molar ratio of long-chain to short-chain lipids is defined as the q value, which determines the size of bicelle. For solution NMR studies, “isotropic” bicelles with q values between 0.25–0.5 are preferred to stabilize membrane proteins and provide high-quality spectra. Bicelles with higher q values (>2) have been used to induce alignments for measuring residual dipolar couplings in solution NMR. The most widely used long-chain lipid in bicelles is dimyristoyl-glycerol-phosphocholine (DMPC), with a few recent reports that have used 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) or 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE) instead. The most commonly used short-chain lipid is DH6PC, but other detergents, such as 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO), 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) have also been used. Bicelles provide a more bilayer-like environment than detergent micelles and some proteins have shown a higher stability and higher enzymatic activity in bicelles than in micelles [31]. In addition, further lipids, such as cholesterol, can be incorporated into bicelle samples to study their interactions with membrane proteins [32].
2.3. Nanodiscs
Similar to bicelles, nanodiscs are patches of solubilized phospholipid, such as DMPC, in bilayers. However, instead of detergents, the phospholipids in nanodiscs are surrounded by amphipathic membrane scaffold proteins (MSPs) (Figure 3C). The most widely used scaffold protein MSP1D1 has been genetically engineered from apolipoprotein A-1 [33]. Two copies of this protein are believed to stabilize two monolayers of one nanodisc. An empty MSP1D1 nanodisc has a mass of around 125 kDa and is 10 nm in diameter, which is at the upper limit of being useful for high-field solution NMR spectroscopy. By deleting helical segments from apolipoproteins, hence by shortening the circumference of the nanodisc, Hagn et al have constructed smaller nanodiscs that they used for a de novo solution NMR structure determination of the membrane protein OmpX [34]. A single OmpX-nanodisc complex has total molecular mass of ~107 kDa and contains ~80 DMPC molecules. More recently, circularized nanodiscs, i.e. nanodiscs in which the scaffold proteins were chemically reconnected between the N- and C-termini, have been devised to improve sample stability and homogeneity [35]. More highly resolved NMR spectra were obtained by this approach. Since nanodiscs provide a bilayer-like environment without the presence of a denaturing detergent, they probably provide the most native-like environments for solution NMR studies of integral membrane proteins.
2.4. Amphipols
Amphipols are amphipathic polymers with mixed hydrophobic and hydrophilic side chains. They have been used to stabilize integral membrane proteins since they selectively adsorb to the hydrophobic transmembrane surface of the membrane protein (Figure 3D) [36]. Membrane proteins in amphipols usually display increased thermal stabilities and higher enzymatic activities compared to their counterparts in detergent micelles, probably because amphipols favor the retention of residual native lipids. The most commonly used amphipol, A8-35, self-assembles into globular particles with an average mass of ~40 kDa, a size manageable for solution NMR studies. However, protein samples in amphipols are less homogeneous in comparison to samples in detergents or nanodiscs. There have been no de novo NMR structures solved so far with membrane proteins in amphipols, although a number of studies suggest that it is a promising approach [36–37].
2.5. Lipodisqs or SMALPs
A different type of polymer, styrene-maleic acid (SMA) copolymer, has also been employed to solubilize integral membrane proteins. Lipodisqs or SMA lipid particles (SMALPs) are mixtures of SMA copolymers and lipids that are structurally similar to bicelles or nanodiscs, except that SMA copolymers are employed to stabilize patches of lipid bilayers (Figure 3E) instead of detergents or MSPs [38–39]. The advantage of lipodisqs is that they can be used to extract integral membrane proteins directly from biological membranes and then further purify them without exposure to detergents. Depending on the protein and perhaps also the method of measurement, diameters of membrane proteins embedded in lipodisqs vary between 10 and 24 nm [40]. Applications of lipodisqs to solution NMR studies of membrane proteins are so far still quite limited [38]. A challenge may be their relatively large sizes and their inhomogeneous size distributions.
2.6. Liposomes
Lipid bilayers in liposomes provide the most native environment for membrane proteins (Figure 3F). However, even small unilamellar vesicles (SUVs) with a diameter of ~50 nm and a size of ~17 MDa, are far outside the upper size limit for optimal detection of membrane-embedded residues by solution NMR. On the other hand, liposomes can successfully be employed to study the interaction of amphipathic proteins with membrane surfaces. Measurements of signal attenuation, paramagnetic relaxation enhancement, relaxation dispersion, and transferred nuclear Overhauser effects (NOEs) can be very helpful to observe specific protein-lipid interactions and to detect the dynamics and properties of membrane-associated invisible states. Another obvious advantage of liposomes is that different types of lipids can be easily incorporated during sample preparation, permitting studies on their effects on functionally important lipid interactions.
3. Dynamic NMR structures of SNAREs and related proteins in membrane lipids
Despite the importance of membranes in SNARE-mediated fusion, most studies on the structures of these proteins have focused on soluble domains of these proteins in aqueous buffers, although there are some notable exceptions [41]. In these studies it is frequently overlooked that SNAREs and related proteins ultimately function on membranes and that their structures and interactions may be influenced by the presence of the membrane lipids, especially when they are near the fusion pore where the local lipid concentration is very high. In this section, we summarize recent solution NMR studies of four proteins that are closely involved in SNARE-mediated membrane fusion.
3.1. Synaptobrevin
Synaptobrevin (VAMP) resides on the synaptic vesicle membrane where it is anchored with a single C-terminal TM domain. The crystal structure of the soluble SNARE complex revealed that synaptobrevin forms a ~56-residue α-helix as part of the coiled-coil four helical bundle of the complex [3]. A more recent structure of the membrane-embedded SNARE complex with its TM domains in the detergent n-nonyl β-D-glucopyranoside has revealed a continuous helical bundle through the TM domain [41]. It is believed that folding into a helix and a concomitant force transduction through the juxtamembrane region, i.e., a stretch of about 10 residues that connect the soluble SNARE motif with the TM domain, is important for overcoming the energy barrier of membrane fusion. In aqueous solution, the soluble domains of synaptobrevin appear to be mostly unstructured as analyzed by solution NMR, CD, and FTIR techniques (Figure 4A) [42–44].
Figure 4.
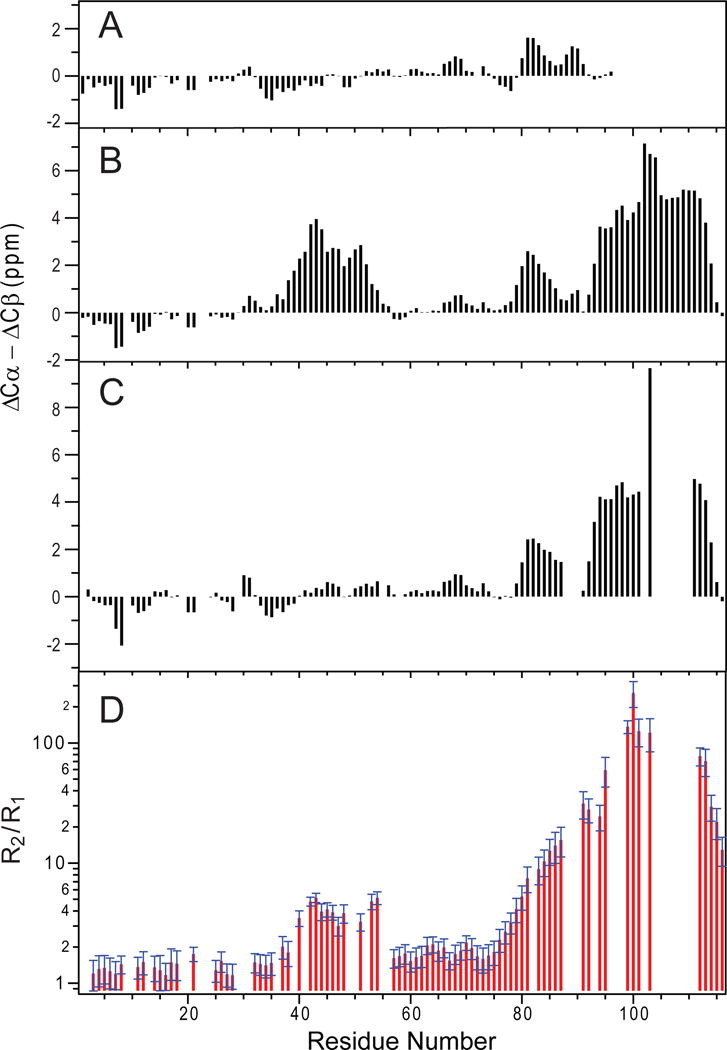
Structural and dynamical changes of synpatobrevin in the presence of membrane lipids. (A–C) Three-bond averaged secondary chemical shift differences versus residue numbers, (A) synaptobrevin (1-96) in aqueous solution; (B) synaptobrevin (1-116) in DPC; (C) synaptobrevin (1-116) in DMPC/DHPC bicelles (q = 0.33). (D) Ratio of 15N R2/R1 relaxation rates for synaptobrevin (1-116) in DMPC/DHPC bicelles (q = 0.33). Adapted with permission from references [44, 46].
Full-length rat synaptobrevin-2 (residues 1-116) reveals a significant amount of ordered structure in DPC micelles [44]. Based on secondary chemical shifts deduced from fully assigned sequential backbone solution NMR spectra, two additional helical segments were identified in addition to the TM helix (Figure 4B). The solution NMR structure of synaptobrevin shows three well-ordered helices: helix I (residues 36-54), helix II (residues 77-88), and the TM helix (residues 93-115), all connected by flexible unstructured regions (Figure 5A). It is surprising that residues 36-54 form helix I before synaptobrevin interacts with target SNARE proteins since these residues are part of the SNARE motif (residues 28-84) and were observed as unstructured in previous NMR studies in aqueous solution. Because of their higher propensity to form a helix near membrane surfaces, it is likely that these residues form a nucleation site for trans-SNARE complex formation, whereas the less structured C-terminal portion of the SNARE motif may serve as a “stop-folding” signal. Importantly, helix I stops right at the critical “0”-layer residue Arg56, underscoring the functional significance of the “0”-layer in SNARE assembly. Helix II is formed in the juxtamembrane region of synaptobrevin, consistent with an earlier NMR study of the soluble fragment in aqueous solution. The short 4-residue flexible linker (Trp-Trp-Lys-Asn) between the helix II and the TM helix is likely important in converting the trans-SNARE into the cis-SNARE complex. The flexibility of this segment is needed to tilt and accommodate the folding of the trans-SNARE complex between the two approaching membranes, and its shortness is needed to allow for an efficient coupling between SNARE complex folding and merging the two membranes. When a soluble construct of synaptobrevin (residues 1-96) is transferred from an aqueous to a DPC environment, similar conformational preferences of the different segments are observed. It is clear that the presence of a membrane mimetic influences the conformation of this protein and that the formation of helix I results from its interaction with a membrane interface.
Figure 5.
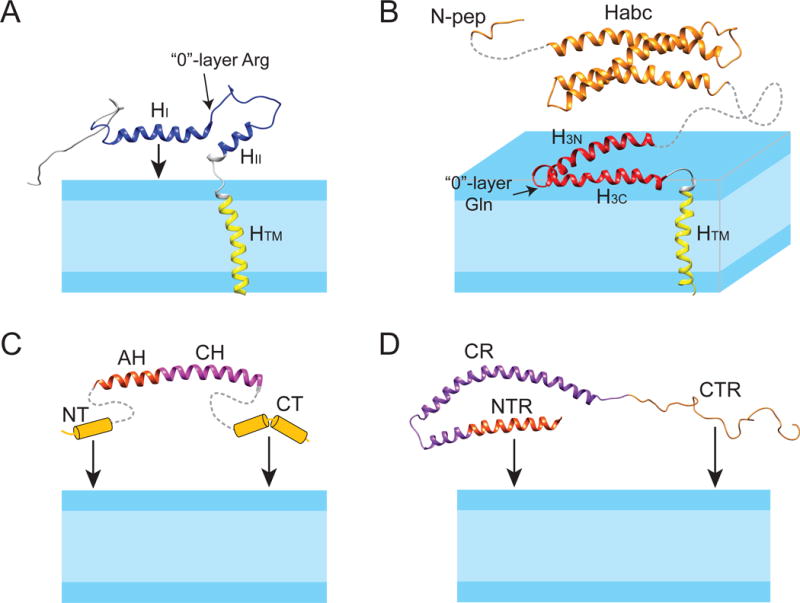
Structure models of synaptobrevin, syntaxin, complexin, and α-synuclein in lipid bilayer membranes. (A) Solution NMR structure of synaptobrevin in DPC (PDB code: 1KOG); (B) full-length syntaxin: solution NMR structure of syntaxin (residues 183-288) in DPC (PDB code: 2M8R) is linked with the NMR structure of the soluble Habc domain (residues 27-146) (1BR0) and the N-peptide (residues 1-12) (3C98); (C) complexin: the structure of the AH and CH helices, taken from the crystal structure of the complexin/SNARE co-complex (1KIL), is linked with the presumed helix-prone N- and C-termini that can tether complexin to the membrane; (D) solution NMR structure of α-synuclein in SDS micelles (1XQ8). The NTR and CTR can tether α-synuclein to the membrane. The protein domains are colored as in Figure 2.
Rizo and co-workers have confirmed that the N-terminal half of the SNARE motif of synaptobrevin adopts an α-helical structure in DPC micelles [45]. However, they also found that the same region remains unstructured in POPC/DOPS nanodiscs or in three types of vesicles with different lipid compositions since their NMR spectra are very similar to that of synaptobrevin (1-96) in aqueous buffer. Transverse relaxation measurements revealed no apparent enhancement of relaxation or decrease in signal intensity for residues 1-74 encompassing the SNARE motif. These authors concluded that the helical content of synaptobrevin in DPC was induced by the micelle environment and that it would not associate with a membrane interface in a more native bilayer environment.
A combined solution NMR and EPR study of synaptobrevin in micelles, bicelles, and vesicles reconciled the two seemingly contradicting results [46]. When synaptobrevin was solubilized in DMPC/DH6PC bicelles, the N-terminal half of the SNARE motif was no longer a stable helix as in DPC micelles since no characteristic α-helical secondary chemical shifts were observed by solution NMR (Figure 4C). Nevertheless, measurements of 15N R1 and R2 relaxation rates (Figure 4D) indicated that this region undergoes conformational exchange on a micro-second to milli-second timescale and additional NMR relaxation dispersion experiments narrowed this exchange rate down to a relatively fast process (about 105 s−1). Interestingly, this chemical exchange process persists in vesicles, albeit to a lesser extent. Site-directed spin-label EPR spectroscopy was employed to further analyze this fast-exchange process. Two motional components were discovered for all labeled synaptobrevin sites in lipid bilayer vesicles, large q=2 bicelles, small q=0.4 bicelles, and DPC micelles. The fast motional component was consistent with that of an unstructured aqueous segment and the slower motional component was consistent with a structured amphipathic segment associated with the membrane-solution interface. In DPC micelles, the structured/interfacial component occupied more than 80% of the population, whereas in POPC/POPS vesicles, the unstructured/mobile component was populated to more than 65%. The combined NMR and EPR data therefore indicate that membrane association and partial helix formation of the SNARE motif of synaptobrevin occurs in every membrane-mimetic environment examined, but does so to a greatly reduced extent as the interface becomes more planar. This dependence on the curvature of the interface explains the differences observed by NMR among DPC, bicelles, nanodiscs, and bilayers. Since membrane fusion depends on the interaction of synaptobrevin with the plasma membrane t-SNARE complex in a region of high protein and lipid content with possibly changing curvature, it is possible that the dynamic exchange between these two conformations could be functionally important. The membrane-associated state of synaptobrevin may well function to nucleate SNARE complex assembly, whereas the acceptor t-SNAREs more likely first encounter synaptobrevin in the aqueous state. In addition, since the curvature of the interface is expected to change in fusion intermediates, the aqueous-membrane partitioning of synaptobrevin may change at different stages of membrane fusion and thereby influence the kinetics of SNARE complex assembly and hence the fusion rate.
3.2. Syntaxin
Syntaxin is an integral membrane protein with a single C-terminal TM helix residing in the plasma membrane. In SNARE-mediated membrane fusion, syntaxin and SNAP-25, i.e. the neuronal target membrane SNAREs (t-SNAREs), form the acceptor SNARE complex (or t-SNARE complex) on the plasma membrane. Similar to synaptobrevin, syntaxin has a ~70-residue SNARE motif that is continuous with the TM helix as visualized in the crystal structure of the post-fusion membrane-embedded SNARE complex [41]. In addition to the SNARE motif and TM domain, syntaxin has two regulatory domains: an autonomously folded three helical bundle Habc domain and a short N-terminal peptide (N-peptide) (Figure 2). Although neither of these two domains is required for fast fusion in vitro, the interactions of these domains with the SNARE motif in the presence of Munc18, Munc13, and other effector proteins are required for regulated SNARE-mediated fusion in vivo. In a crystal structure of a complex of soluble syntaxin and Munc18, the Habc domain and the SNARE motif form a “closed” conformation [47]. Since this “closed” conformation cannot facilitate SNARE assembly, it has been proposed that Munc18 and Munc13 control synaptic vesicle fusion by regulating the dynamic equilibrium between the “closed” and “open” conformations of syntaxin [48]. Interestingly, a recent EPR study found that this equilibrium is shifted towards the “open” state on the surface of a membrane compared to the same complex in solution [49]. In addition, the membrane-bound form of syntaxin can self-aggregate, and the effect of syntaxin clustering on membrane fusion has been explored and debated in the literature [50].
Early solution NMR studies on syntaxin-1a have focused on its soluble domains in aqueous buffers. The three helix bundle structure of the Habc domain was solved first by solution NMR (Figure 5B) [51] and was later confirmed by X-ray crystallography [52]. NMR studies on a construct encompassing the whole cytoplasmic region of syntaxin revealed that its Habc domain still formed a three-helix bundle, but the remainder of the protein, including the N-peptide, the SNARE motif and the long linker between the Habc and SNARE motif were either mostly unstructured or not observed, probably due to unfavorable conformational exchange [53]. In this isolated form, syntaxin appears to be present mostly in a “closed” conformation with Munc18 binding preventing the opening of this conformation.
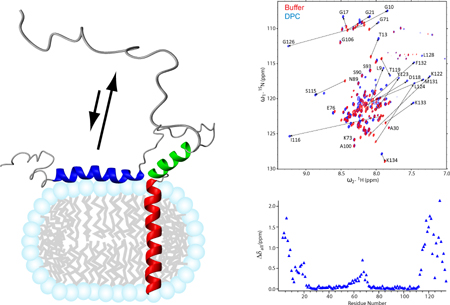
Full-length syntaxin with its TM domain can be solubilized in DPC micelles. In this monomeric form, the Habc domain still appears to be α-helical as indicated by the characteristic secondary chemical shifts observed by solution NMR [54]. Surprisingly, the SNARE motif exhibits a much more well-ordered α-helical structure in DPC compared to its soluble counterpart. A 1D-TRACT experiment for measurement of rotational correlation times could not be fitted to single exponential decays (Figure 6A), revealing the existence of distinctly different correlation times in different regions of this construct, likely reflecting the open/closed equilibrium of the Habc and SNARE domains. These results suggest that syntaxin has a higher propensity to be in the open conformation in membrane mimetic DPC micelles than it does in aqueous buffer.
Figure 6.
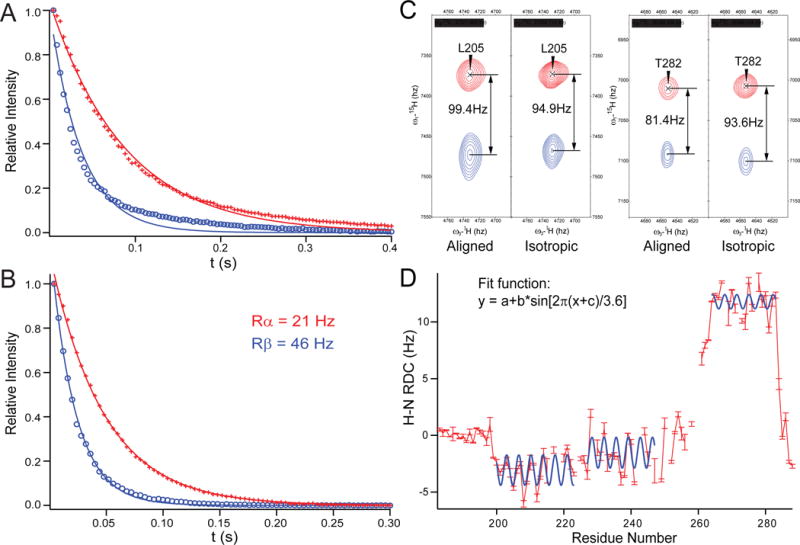
Syntaxin (183-288) is well structured in DPC micelles. 1D-TRACT NMR experiments of (A) the full-length syntaxin (1-288), and (B) syntaxin (183-288) in DPC micelles. The best-fit single-exponentials to the Rα (red) and Rβ (blue) components are displayed as solid lines. (C) Two pairs of resonances showing different alignments in a 50% negatively charged acrylamide copolymer gel [85] and a final polymer concentration of 4%. (D) Observed H-N RDC values (red bars) versus residue numbers, with three stretches of helical segments (200-224, 227-247, and 264-283) fitted to dipolar waves, i.e., sinusoids of periodicity of ~3.6 residues [86]. Adapted with permission from reference [54].
To determine the structure of the SNARE motif and TM domain in a membrane environment, a construct encompassing only the SNARE motif, the TM domain, and their connecting linker region has been employed. This construct shows a homogeneous rotational correlation time profile in the 1D-TRACT experiment that could be fitted to single exponential decays (Figure 6B) with an average size of 35-45 kDa for the protein-detergent complex as determined by NMR relaxation measurements. This size estimate is consistent with the size estimated from size-exclusion chromatography. It signifies that syntaxin exists as a monomer in DPC. It is also important to note that syntaxin is fully functional in DPC micelles since an acceptor SNARE complex that was formed under these conditions can rapidly and efficiently react with synaptobrevin to form the SDS-resistent SNARE complex [54]. Based on an extensive collection of NMR constraints, including NOE, paramagnetic relaxation enhancement (PRE), residual dipolar coupling (RDC) (Figure 6C and 6D), J-coupling, and chemical shift-based dihedral angles, a high-resolution structure of syntaxin(183-288) in DPC micelles has been determined (Figure 5B). This solution structure is characterized by three well-ordered long α-helices: H3N (residues 197-224), H3C (residues 227-251), and HTM (residues 261-282). When positioned in a hypothetical DPC micelle, the first two helices lie on the micelle-water interface, whereas the helical axis of HTM forms a 70° angle against the plane defined by H3N and H3C. The linker region between the H3C and HTM is unstructured. Both H3N and H3C are located within the SNARE motif, connected by a turn at residues Ser225 and Gln226. The Gln226 residue is the single Gln residue in the heptad repeat of syntaxin that forms the “0”-layer in the SNARE complex. This result is reminiscent of the nucleation helix in synaptobrevin, which also stops two residues before its “0”-layer residue Arg. Thus, both synaptobrevin and syntaxin have significant propensities for helix formation in their open prefusion states, and the observation that these helices terminate right before the “0”-layer supports the notion that the “0”-layer may act as a proofreading site for SNARE complex formation (Figure 5A and 5B) [55].
The association of the SNARE motif with the interface of DPC micelles that was observed by NMR also happens in lipid bilayers. By performing fluorescence interference contrast (FLIC) microscopy on fluorescently labeled syntaxin, the SNARE motif was also found to be associated with the membrane interface [54]. In addition, interaction of the SNARE motif with the membrane appears to be critical for efficient SNARE assembly. After assembly with other SNAREs the SNARE motif of syntaxin moves away from the membrane surface, likely by changing the conformation of the linker region.
Solution NMR has also been employed to study the interaction of SNAP-25 and Munc18 with DPC-bound syntaxin. Whereas the addition of SNAP-25 does not result in any significant chemical shift perturbations, the addition of Munc18 induces significant chemical shift changes [54]. Interestingly, Munc18 appears to interact only with the H3N helix, signifying the functional importance of the segmented structure of membrane-associated syntaxin. Therefore, Munc18 likely binds preferentially to the H3N helix in SNARE-mediated exocytosis. This also points to a mechanism, in which a partially zippered SNARE complex is arrested at the “0”-layer, where it could be triggered by additional effectors to complete zippering and fusion.
3.3. Complexin
Complexin is a highly charged cytosolic protein that is essential for Ca2+-triggered exocytosis. It is expressed at presynaptic sites and was found to be colocalized with syntaxin and SNAP-25 [56]. Biochemical studies have shown that complexin binds strongly to the assembled cis-SNARE complex, but with only limited binding to individual monomeric SNAREs. Solution NMR studies have identified a stretch of α-helical residues which were later named the “accessory” and “central” helices owing to their relative position and association with the cis-SNARE complex in a crystal structure of a soluble complexin/SNARE co-complex (Figure 5C). The precise functions of the central and accessory helices of complexin have been hotly debated, with a number of contradicting models that were each supported by different sets of electrophysiological, biochemical, and biophysical data [20].
In a solution NMR study of full-length C. elegans complexin (residues 1-143), a subset of resonances exhibit reduced intensities in the presence of POPC/POPS liposomes [57]. Since these residues were largely disordered in solution, the reduced intensities indicate residues that directly interact with the lipid bilayer. Complexin residues preceding residue 92, i.e. the entire accessory and central helices, were largely unaffected, whereas residues at the C-terminus exhibited a decrease in signal intensity that depended on the amount of added liposomes. Additionally, this binding depended on the curvature of added membranes. When SUVs with a ~30 nm diameter were added at 1 mM total lipid, attenuation was stronger than when large unilamellar vesicles (LUVs) with a ~120 nm diameter were added at 13.5 mM total lipid concentration [58]. In addition to the C-terminal (residues 128-143), an amphipathic motif (residues 110-124), upstream of the C-terminal domain, displayed an even higher preference for binding to highly curved than to moderately curved lipid bilayers. Therefore, these two domains likely target complexin to the more curved synaptic vesicle rather than to the flatter plasma membrane. These results were used to explain in vivo observations of synaptic activities at neuromuscular junctions with various complexin C-terminal mutants [57].
Dramatic changes of the NMR spectra were observed when DPC micelles were added to rat complexin (residues 1-134) in solution (Figure 7A) [59]. Residues in the N- and C-terminal regions (residues 1-22 and 114-134, respectively) displayed large chemical shift changes (Figure 7B). In contrast, residues in the accessory and central helices displayed almost no chemical shift changes, except for a few residues at the C-terminal end of the central helix (residues 63-70) that showed modest changes, probably indicating an extension of the α-helix in this region. It appears that protein-detergent interactions induce α-helices in the N- and C-terminal regions of complexin. The lipid-induced helix formation in the C-terminal region was also observed in C. elegans complexin (residues 91-143) [60]. When rat complexin was bound to nanodiscs or liposomes, resonances of the N- and C-terminal residues revealed dramatic reductions in NMR peak intensities (Figure 7C), indicative of lengthened rotational correlation times for these residues that directly interacted with the bilayers [59]. Regions between these N- and C-terminal regions, including the accessory and central helices, showed only small or no intensity changes. The most dramatic intensity attenuation was observed for the N-terminal residues 1-12 and the C-terminal residues 114-134, indicating that these residues were simultaneously bound to lipid bilayers (Figure 5C). Site-directed spin-labeling EPR data confirmed the NMR findings, indicating that lipid binding of both terminal regions depends on membrane curvature with increasing affinity to more highly curved membranes. In addition, these two regions bound to lipid bilayers independently, which would be expected given the long disordered regions connecting the two terminal domains.
Figure 7.
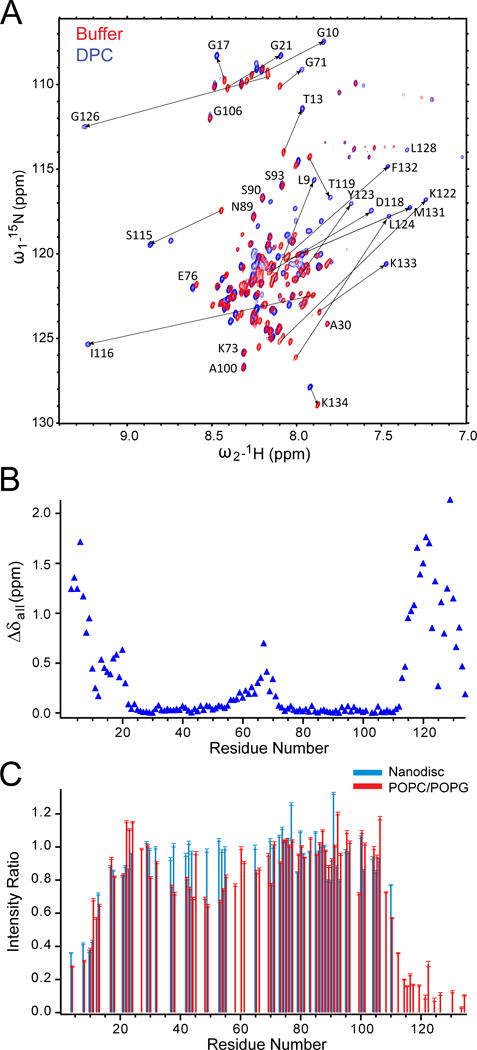
The mechanisms of complexin binding to the t-SNARE complex and its inhibitory, so-called “clamping” activity on membrane fusion have been debated extensively. Early NMR studies showed a direct binding of the central helix of complexin to the soluble cis-SNARE complex [61], a result that was later confirmed when the crystal structure of the soluble complexin/cis-SNARE co-complex was solved [62]. Neither the accessory nor the central helix of complexin bind to individual SNARE proteins and all previous structural models for the complexin-SNARE interaction depict complexin interacting with an assembled or partially assembled SNARE complex comprising all three SNAREs. By taking advantage of the monomeric form of syntaxin in DPC micelles, a binary complex of syntaxin:SNAP-25 in a strictly 1:1 ratio can be assembled [63]. When complexin is added to this 1:1 binary t-SNARE complex, direct binding is observed. A dose-dependent attenuation of the NMR signals indicates that binding of complexin to the binary complex is initiated from the C-terminal half of the central helix and then gradually extends towards both ends of complexin [59]. Importantly, the binding of complexin to the binary t-SNARE complex additionally promotes binding of the N and C-terminal regions of complexin to the lipid bilayer. EPR spectroscopic data show that the presence of the 1:1 binary t-SNARE complex increases the partition coefficient of complexin to the membrane 19-fold.
These recent data clearly demonstrate that complexin associates with the binary t-SNARE complex in the absence of v-SNAREs and potentially before the initiation of v- and t-SNARE assembly, providing a plausible starting point for the clamping activity of complexin. Simultaneous membrane and t-SNARE binding of complexin inhibits the docking of v-SNARE vesicles and weakens the affinity of synaptobrevin for the acceptor SNARE complex. These observations support a model, in which complexin acts to lower the frequency of spontaneous fusion events in neuronal exocytosis by binding to the acceptor t-SNARE complex and thereby delaying assembly of the ternary SNARE complex. Perhaps additional factors, such as Munc18 or Munc13 can re-arrange the complexin/t-SNARE complex. Upon Ca2+ binding, synaptotagmin may then interact either with this complex or with the fusing membranes or both to trigger the final steps of v-SNARE assembly, providing an attractive mechanistic model for the regulation of exocytotic membrane fusion by calcium.
3.4. α-synuclein
The presynaptic protein α-synuclein has attracted much attention because of its pathological connection to Parkinson’s disease and other neurodegenerative disorders. The abnormal aggregation of α-synuclein leads to amyloid formation, a hallmark of Parkinson’s disease. At the presynaptic terminal, α-synuclein is abundantly associated with synaptic vesicles; however, its biological function in its native non-aggregated form is still not clear. In vivo studies suggested that α-synuclein acts as a special chaperone for synaptobrevin and that its C-terminal domain interacts with the N-terminal domain of synaptobrevin [64]. In cysteine string protein-α knockout mice, the full-length, but not the C-terminally truncated, α-synuclein promotes SNARE complex assembly [64]. However, another study found no direct interaction between α-synuclein and SNARE proteins and found that α-synuclein has an indirect inhibitory effect on SNARE-mediated membrane fusion [65]. These authors suggested that α-synuclein sequesters arachidonic acid and thereby inhibits the stimulatory action of this polyunsaturated fatty acid on synaptic vesicle membrane fusion.
The sequence of α-synuclein encompasses 140 residues with seven copies of an unusual 11-residue imperfect repeat in the N-terminal and central regions, followed by a hydrophilic C-terminal tail. The repeat units share a characteristic lipid-binding motif that is also present in apolipoprotein and that has long been proposed as curvature-sensing lipid-binding motif. Solution NMR studies on α-synuclein confirmed that the N-terminal repeat domain is indeed detergent- and lipid-binding, especially when the liposomes contain the negatively charged lipid PS. Ordered helical structures can be induced in a disordered α-synuclein sample in buffer by the addition of detergent or liposomes. Based on the observed secondary chemical shifts and NOE patterns, most of the 98 N-terminal residues adopt helical structures, with a notable interruption of the helix around residues 43 and 44. However, the 42 most C-terminal residues remain unstructured after addition of detergent micelles [66–67]. The solution NMR structure of α-synuclein has been determined in SDS micelles (Figure 5D) [68]. Residues 3-37 and 45-92 form curved α-helices in an antiparallel arrangement and are connected by a well-ordered linker. This structure is followed by an unstructured and highly mobile tail (residues 98-140). The curved helices exhibit average radii of curvature of 77 and 41 Å, respectively. These radii are considerably larger than the radius of a globular SDS micelle (~23 Å). Apparently, the curvature preference of these helices leads to a deformation of the micelle and has been suggested as evidence that α-synuclein has the ability to sense curvature and thereby target membranes with specific curvatures. NMR dynamics measurements singled out a region encompassing residues 30-37, which includes a known mutant (A30P) of familial Parkinson’s disease. These residues exhibit large amplitude dynamics on all timescales, reduced helical content, and their side-chains do not contact the micelle surface. The reduced stability of this region may promote the formation of non-helical structures and their initial aggregation leading to pathological forms of α-synuclein [68].
When bound to negatively charged SUVs, α-synuclein shows a pattern of gradual attenuation of NMR resonances along the protein sequence, with the N-terminal region (residues 3-25) showing the largest attenuation correlating with its strongest binding to the lipids (Figure 8A, black traces) [69]. The “invisible” lipid-bound form of α-synuclein can be studied via transferred NOE experiments since the exchange between the lipid-bound ordered state and the unbound disordered state is comparable to or faster than T1. There appear to be multiple binding modes within the previously determined helical regions (residues 1-98) in SDS micelles, providing evidence for a number of distinct lipid-bound species. In addition, α-synuclein variants implicated in Parkinson’s disease show differential phospholipid binding properties [70]. The dynamic equilibrium between these lipid-bound forms may provide a molecular explanation for the biphasic dependence of amyloid formation on lipid concentration [71]. The N-terminal region (1-25) adopts an α-helical conformation in all lipid-bound forms and the 40 most C-terminal residues remain dynamically disordered. However, they also bind to lipids to an extent (Figure 5D), which may be modulated by cis-trans equilibria of the peptide bonds preceding the five prolines in this sequence [69]. The weak binding of the unstructured C-terminal domain to lipid bilayers has been confirmed in more recent combined solution and solid-state NMR studies [72–73]. These latter studies also found that the region comprised of residues 65-97, which has significant overlap with the non-amyloid-β component (NAC) region (residues 61-95), has membrane-binding properties that do not depend on the N-terminal membrane anchor (residues 1-25). These findings support a “double-anchor” mechanism, in which a single α-synuclein can crosslink two vesicles with the N-terminal (membrane anchor) and central (membrane sensor) regions each binding to separate membranes. This model provides a molecular and structural explanation for the suggested loss of function that correlates with an impairment of vesicle clustering by the A30P mutated form of familial Parkinson’s disease. Furthermore, the model also suggests that α-synuclein may act as a molecular chaperone for the formation of SNARE complexes by crosslinking two membranes prior to SNARE complex formation. This proposed function attributed to the amyloidogenic NAC domain would provide a direct mechanistic link between the physiological and pathological roles of α-synuclein [73].
Figure 8.
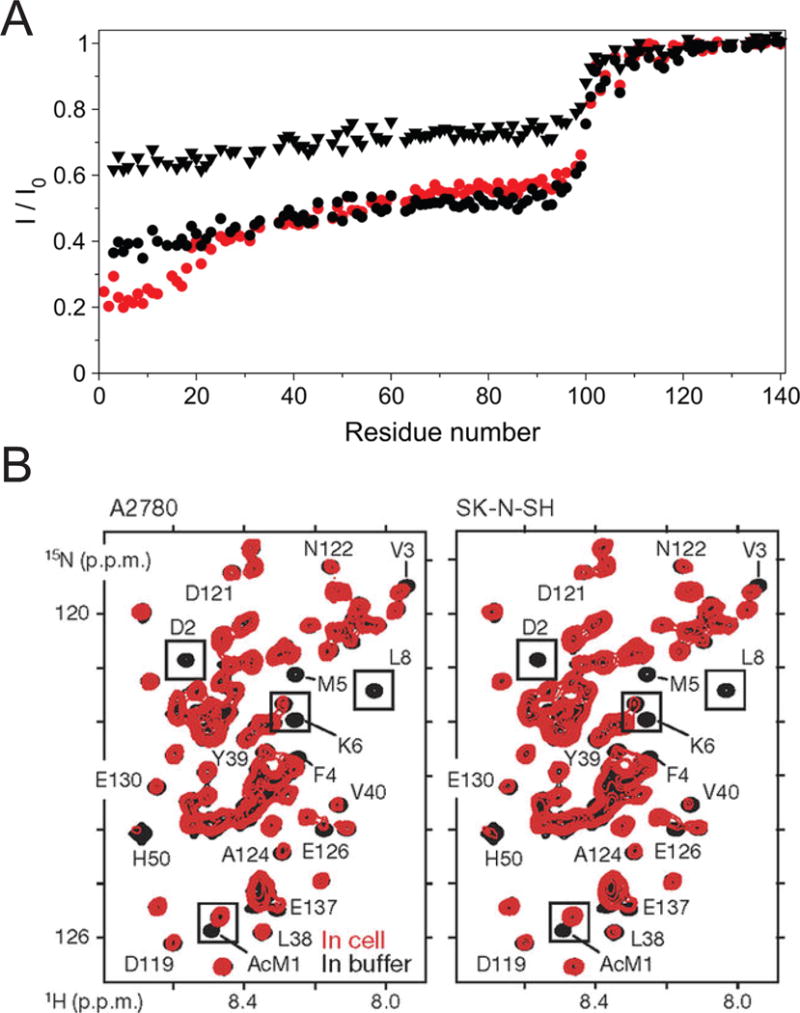
Solution NMR of α-synuclein. (A) Ratios of wild-type α-synuclein TROSY-HSQC peak heights in the presence (I) and absence (Io) of lipid bilayer vesicles. Data from the following three samples are presented: N-terminally acetylated α-synuclein at a lipid:protein (L:P) ratio of 22 (red circles), nonacetylated α-synuclein at an L:P ratio of 22 (black triangles), and nonacetylated α-synuclein at an L:P ratio of 44 (black circles). Reproduced with permission from reference [75]. (B) In-cell HSQC NMR spectra of α-synuclein in A2780 and SK-N-SH cells (red) and of isolated N-terminally acetylated α-synuclein in buffer (black). AcM1, acetylated Met1. Reproduced with permission from reference [81].
α-Synucleins that were isolated from Lewy bodies from patients with dementia were found to be acetylated at the N-terminus [74]. When α-synuclein is N-terminally acetylated, the chemical shifts of the first 12 residues are perturbed in aqueous buffer, but the rest of the sequence is not affected, indicating an absence of specific interactions between the C- and N-terminal regions [75]. The lipid binding of α-synuclein is strongly enhanced by N-terminal acetylation, with the strongest enhancement localized in the first 12 residues (Figure 8A, red circles). This stronger binding of the N-terminus also leads to a 2-fold enhancement of lipid binding of residues 25-90. Similar changes were observed in two different detergents, and in liposomes with a lower content of negatively charged lipids [76]. Apparently, N-terminal acetylation stabilizes the N-terminal helicity, which leads to an enhanced affinity for lipid-water interfaces. It is possible that the acetylated N-terminal region plays an initiation role in an “initiation-elongation” process of α-synuclein binding to membranes.
It has been debated whether native α-synuclein is a monomer or a tetramer, or a dynamic mixture of the two [77–80]. A recent in-cell NMR study showed that exogenously delivered α-synuclein exists as an N-terminally acetylated, disordered and highly dynamic monomer in mammalian cells, without detectable signs of oligomerization, spontaneous aggregation, or targeted degradation (Figure 8B) [81]. In this crowded intracellular environment with an abundance of membranes, α-synuclein adopts compact, but not stably folded or fully membrane-associated structures. It is likely that the compact form of α-synuclein protects the hydrophobic residues of the amyloidogenic NAC domain from interactions leading to oligomerization. Critical residues acting as “interaction hotspots” may be required to form nucleating oligomers of α-synuclein, possibly through transient binding to cellular membranes.
4. Conclusions and outlook
SNARE-mediated membrane fusion is a dynamic process that is orchestrated by both weak and strong interactions of many proteins and lipids. Using a range of solution NMR techniques, we have learned a lot in recent years about the structure and dynamics of a number of SNARE and related proteins in membrane environments. In the four examples summarized in this review, these proteins all show the formation of ordered structures induced by the presence of membrane lipids. In the case of synaptobrevin and syntaxin, ordered regions are present in the SNARE motif before SNARE complex assembly, and the population of ordered conformations may be enhanced by increased membrane curvature. These pre-existing ordered domains may serve as nucleation sites for SNARE assembly and support the N- to C-terminal zippering of SNAREs and the formation of the four-helix bundle of the final folded SNARE complex. In the case of complexin and α-synuclein, N- and C-terminal regions show membrane-induced and curvature-dependent helix formation to various degrees. The presence of these membrane-binding, curvature-sensing domains leads to synaptic vesicle-recognition models, which in turn have been used to explain the clamping or vesicle clustering functions of these two proteins. In addition to protein-membrane interaction, protein-protein interactions can also be modulated by the close proximity of membranes. At least in two cases, these interactions are different than what had been observed with soluble domains alone in aqueous buffers. In DPC micelles, Munc18 interacts specifically with the H3N helix of syntaxin, which may indicate that the role of Munc18 in facilitating SNARE assembly involves its preferential binding to that helix of syntaxin. In lipid vesicles, the cooperative binding of complexin to the 1:1 binary t-SNARE complex and the membrane provides a straightforward explanation of the clamping activity of complexin in regulated exocytosis.
There is no doubt that solution NMR of membrane-associated SNAREs and related proteins has re-shaped our view of how these proteins facilitate membrane fusion. As summarized in this review, the combination of NMR with other techniques, such as EPR and fluorescence spectroscopies, as well as functional in vitro and in vivo studies, provides a very powerful approach to elucidate the elusive molecular mechanism of SNARE-mediated membrane fusion. Since the proteins discussed share many characteristics of intrinsically disordered proteins (IDPs), solution NMR techniques are highly suitable and perhaps the only high-resolution methods available to study the structures and dynamics of this class of proteins [82–83]. We envision that other SNAREs and related proteins, SNARE assembly intermediates, and complexes between SNARE and SNARE-effector proteins undergo conformational changes in membranes, perhaps with different twists and new surprises. Therefore, we expect that solution NMR of this class of proteins in membrane-mimetic environments will continue to play a major role in elucidating the dynamic nature of SNARE-mediated membrane fusion.
Highlights.
SNARE and related proteins display different structures and dynamics in membranes.
Syntaxin and synaptobrevin have ordered and disordered regions before assembly.
Complexin binds cooperatively to t-SNARE complex and membranes.
α-synuclein shows membrane-induced and curvature-dependent helix formation.
Acknowledgments
This work is supported by NIH grants P01 GM72694, R01 GM51329, and R01 AI30557. We thank members of the Tamm lab, past and present, for their valuable contributions, and Dr. Volker Kiessling and Dr. David Cafiso for their helpful discussions.
Glossary
- ATP
adenosine triphosphate
- CD
circular dichroism
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- CHAPSO
3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate
- CMC
critical micelle concentration
- DDM
n-dodecyl-β-maltoside
- DH6PC
1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DH7PC
1,2-diheptanoyl-sn-glycero-3-phosphocholine
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DOPS
1,2-dioleoyl-sn-glycero-3-phospho-L-serine
- DPC
Dodecylphosphocholine
- EM
Electron Microscopy
- EPR
electron paramagnetic resonance
- FLIC
fluorescence interference contrast
- FTIR
Fourier-transform infrared spectroscopy
- HSQC
heteronuclear single quantum correlation
- IDP
intrinsically disordered protein
- LDAO
lauryldimethylamine oxide
- L
P: lipid:protein
- LUV
large unilamellar vesicle
- MSP
membrane scaffold protein
- NAC
non-amyloid-β component
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- NSF
N-ethylmaleimide sensitive factor
- β-OG
octyl-β-glucoside
- OmpX
outer membrane protein X
- PC
phosphatidylcholine
- PG
phosphatidylglycerol
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPG
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol)
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine
- PRE
paramagnetic relaxation enhancement
- PS
phosphatidylserine
- RDC
residual dipolar coupling
- SDS
sodium dodecyl sulfate
- SM
Sec1/Munc18
- SMA
styrene-maleic acid
- SMALP
SMA lipid particle
- SNAP
soluble NSF attachment protein
- SNAP-25
synaptosomal-associated protein 25
- SNARE
soluble N-ethylmaleimide sensitive factor attachment protein receptor
- SUV
small unilamellar vesicles
- TM
transmembrane
- TRACT
TROSY for rotational correlation times
- TROSY
transverse relaxation optimized spectroscopy
- t-SNAREs
target membrane SNAREs
- VAMP
vesicle associated membrane protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 2.Jahn R, Scheller RH. SNAREs–engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 3.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 4.Südhof TC, Rizo J. Synaptic vesicle exocytosis. Cold Spring Harbor perspectives in biology. 2011;3 doi: 10.1101/cshperspect.a005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jahn R, Fasshauer D. Molecular machines governing exocytosis of synaptic vesicles. Nature. 2012;490:201–207. doi: 10.1038/nature11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rizo J, Xu J. The Synaptic Vesicle Release Machinery. Annual Review of Biophysics. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- 7.Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerber SH, Rah JC, Min SW, Liu X, de Wit H, Dulubova I, Meyer AC, Rizo J, Arancillo M, Hammer RE, Verhage M, Rosenmund C, Sudhof TC. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321:1507–1510. doi: 10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou P, Pang ZP, Yang X, Zhang Y, Rosenmund C, Bacaj T, Südhof TC. Syntaxin-1 N-peptide and H-abc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 2013;32:159–171. doi: 10.1038/emboj.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jakhanwal S, Lee CT, Urlaub H, Jahn R. An activated Q-SNARE/SM protein complex as a possible intermediate in SNARE assembly. The EMBO Journal. 2017;36:1788–1802. doi: 10.15252/embj.201696270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker RW, Jeffrey PD, Zick M, Phillips BP, Wickner WT, Hughson FM. A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly. Science. 2015;349:1111–1114. doi: 10.1126/science.aac7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sitarska E, Xu J, Park S, Liu X, Quade B, Stepien K, Sugita K, Brautigam CA, Sugita S, Rizo J. Autoinhibition of Munc18-1 modulates synaptobrevin binding and helps to enable Munc13-dependent regulation of membrane fusion. eLife. 2017;6:e24278. doi: 10.7554/eLife.24278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18:542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai Y, Choi UB, Leitz J, Rhee HJ, Lee C, Altas B, Zhao M, Pfuetzner RA, Wang AL, Brose N, Rhee J, Brunger AT. Molecular Mechanisms of Synaptic Vesicle Priming by Munc13 and Munc18. Neuron. 2017;95:591–607. doi: 10.1016/j.neuron.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Camacho M, Xu Y, Esser V, Liu X, Trimbuch T, Pan YZ, Ma C, Tomchick DR, Rosenmund C, Rizo J. Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. eLife. 2017;6:e22567. doi: 10.7554/eLife.22567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai J, Tucker WC, Chapman ER. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat Struct Mol Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 17.Kuo W, Herrick DZ, Cafiso DS. Phosphatidylinositol 4,5-Bisphosphate Alters Synaptotagmin 1 Membrane Docking and Drives Opposing Bilayers Closer Together. Biochemistry. 2011;50:2633–2641. doi: 10.1021/bi200049c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou Q, Lai Y, Bacaj T, Zhao M, Lyubimov AY, Uervirojnangkoorn M, Zeldin OB, Brewster AS, Sauter NK, Cohen AE, Soltis SM, Alonso-Mori R, Chollet M, Lemke HT, Pfuetzner RA, Choi UB, Weis WI, Diao J, Sudhof TC, Brunger AT. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015;525:62–67. doi: 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park Y, Seo JB, Fraind A, Perez-Lara A, Yavuz H, Han K, Jung SR, Kattan I, Walla PJ, Choi M, Cafiso DS, Koh DS, Jahn R. Synaptotagmin-1 binds to PIP2-containing membrane but not to SNAREs at physiological ionic strength. Nat Struct Mol Biol. 2015;22:815–823. doi: 10.1038/nsmb.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trimbuch T, Rosenmund C. Should I stop or should I go? The role of complexin in neurotransmitter release. Nat Rev Neurosci. 2016;17:118–125. doi: 10.1038/nrn.2015.16. [DOI] [PubMed] [Google Scholar]
- 21.Mohrmann R, Dhara M, Bruns D. Complexins: small but capable. Cellular and Molecular Life Sciences. 2015;72:4221–4235. doi: 10.1007/s00018-015-1998-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 23.Zhao M, Wu S, Zhou Q, Vivona S, Cipriano DJ, Cheng Y, Brunger AT. Mechanistic insights into the recycling machine of the SNARE complex. Nature. 2015;518:61–67. doi: 10.1038/nature14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heider MR, Gu M, Duffy CM, Mirza AM, Marcotte LL, Walls AC, Farrall N, Hakhverdyan Z, Field MC, Rout MP, Frost A, Munson M. Subunit connectivity, assembly determinants and architecture of the yeast exocyst complex. Nat Struct Mol Biol. 2016;23:59–66. doi: 10.1038/nsmb.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang B, Tamm LK. NMR as a tool to investigate the structure, dynamics and function of membrane proteins. Nat Struct Mol Biol. 2016;23:468–474. doi: 10.1038/nsmb.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oliver RC, Lipfert J, Fox DA, Lo RH, Doniach S, Columbus L. Dependence of Micelle Size and Shape on Detergent Alkyl Chain Length and Head Group. PLoS ONE. 2013;8:e62488. doi: 10.1371/journal.pone.0062488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez C, Hilty C, Wider G, Wuthrich K. Lipid-protein interactions in DHPC micelles containing the integral membrane protein OmpX investigated by NMR spectroscopy. Proc Natl Acad Sci USA. 2002;99:13533–13537. doi: 10.1073/pnas.212515099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang B, Arora A, Tamm LK. Fast-time scale dynamics of outer membrane protein A by extended model-free analysis of NMR relaxation data. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2010;1798:68–76. doi: 10.1016/j.bbamem.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gautier A, Mott HR, Bostock MJ, Kirkpatrick JP, Nietlispach D. Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat Struct Mol Biol. 2010;17:768–774. doi: 10.1038/nsmb.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reckel S, Gottstein D, Stehle J, Löhr F, Verhoefen MK, Takeda M, Silvers R, Kainosho M, Glaubitz C, Wachtveitl J, Bernhard F, Schwalbe H, Güntert P, Dötsch V. Solution NMR Structure of Proteorhodopsin. Angewandte Chemie International Edition. 2011;50:11942–11946. doi: 10.1002/anie.201105648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poget SF, Girvin ME. Solution NMR of membrane proteins in bilayer mimics: small is beautiful, but sometimes bigger is better. Biochim Biophys Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett PJ, Song Y, Van Horn WD, Hustedt EJ, Schafer JM, Hadziselimovic A, Beel AJ, Sanders CR. The Amyloid Precursor Protein Has a Flexible Transmembrane Domain and Binds Cholesterol. Science. 2012;336:1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Letters. 2002;2:853–856. [Google Scholar]
- 34.Hagn F, Etzkorn M, Raschle T, Wagner G. Optimized Phospholipid Bilayer Nanodiscs Facilitate High-Resolution Structure Determination of Membrane Proteins. Journal of the American Chemical Society. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nasr ML, Baptista D, Strauss M, Sun ZYJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, Wagner G. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Meth. 2017;14:49–52. doi: 10.1038/nmeth.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Planchard N, Point É, Dahmane T, Giusti F, Renault M, Le Bon C, Durand G, Milon A, Guittet É, Zoonens M, Popot JL, Catoire LJ. The Use of Amphipols for Solution NMR Studies of Membrane Proteins: Advantages and Constraints as Compared to Other Solubilizing Media. The Journal of Membrane Biology. 2014;247:827–842. doi: 10.1007/s00232-014-9654-z. [DOI] [PubMed] [Google Scholar]
- 37.Etzkorn M, Raschle T, Hagn F, Gelev V, Rice Amanda J, Walz T, Wagner G. Cell-free Expressed Bacteriorhodopsin in Different Soluble Membrane Mimetics: Biophysical Properties and NMR Accessibility. Structure. 2013;21:394–401. doi: 10.1016/j.str.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowles TJ, Finka R, Smith C, Lin YP, Dafforn T, Overduin M. Membrane Proteins Solubilized Intact in Lipid Containing Nanoparticles Bounded by Styrene Maleic Acid Copolymer. Journal of the American Chemical Society. 2009;131:7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 39.Orwick MC, Judge PJ, Procek J, Lindholm L, Graziadei A, Engel A, Gröbner G, Watts A. Detergent-Free Formation and Physicochemical Characterization of Nanosized Lipid–Polymer Complexes: Lipodisq. Angewandte Chemie International Edition. 2012;51:4653–4657. doi: 10.1002/anie.201201355. [DOI] [PubMed] [Google Scholar]
- 40.Dörr JM, Scheidelaar S, Koorengevel MC, Dominguez JJ, Schäfer M, van Walree CA, Killian JA. The styrene–maleic acid copolymer: a versatile tool in membrane research. European Biophysics Journal. 2016;45:3–21. doi: 10.1007/s00249-015-1093-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hazzard J, Sudhof TC, Rizo J. NMR analysis of the structure of synaptobrevin and of its interaction with syntaxin. J Biomol NMR. 1999;14:203–207. doi: 10.1023/a:1008382027065. [DOI] [PubMed] [Google Scholar]
- 43.Bowen M, Brunger AT. Conformation of the synaptobrevin transmembrane domain. Proc Natl Acad Sci USA. 2006;103:8378–8383. doi: 10.1073/pnas.0602644103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ellena JF, Liang B, Wiktor M, Stein A, Cafiso DS, Jahn R, Tamm LK. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci USA. 2009;106:20306–20311. doi: 10.1073/pnas.0908317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brewer KD, Li W, Horne BE, Rizo J. Reluctance to membrane binding enables accessibility of the synaptobrevin SNARE motif for SNARE complex formation. Proceedings of the National Academy of Sciences. 2011;108:12723–12728. doi: 10.1073/pnas.1105128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang B, Dawidowski D, Ellena JF, Tamm LK, Cafiso DS. The SNARE motif of synaptobrevin exhibits an aqueous-interfacial partitioning that is modulated by membrane curvature. Biochemistry. 2014;53:1485–1494. doi: 10.1021/bi401638u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Misura KMS, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404:355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 48.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the Vital Functions of Munc18 and Munc13 in Neurotransmitter Release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dawidowski D, Cafiso DS. Allosteric Control of Syntaxin 1a by Munc18-1: Characterization of the Open and Closed Conformations of Syntaxin. Biophys J. 2013;104:1585–1594. doi: 10.1016/j.bpj.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Bogaart G, Lang T, Jahn R. Microdomains of SNARE Proteins in the Plasma Membrane. Current Topics in Membranes. 2013;72:193–230. doi: 10.1016/B978-0-12-417027-8.00006-4. [DOI] [PubMed] [Google Scholar]
- 51.Fernandez I, Ubach J, Dulubova I, Zhang XY, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- 52.Lerman JC, Robblee J, Fairman R, Hughson FM. Structural Analysis of the Neuronal SNARE Protein Syntaxin-1A. Biochemistry. 2000;39:8470–8479. doi: 10.1021/bi0003994. [DOI] [PubMed] [Google Scholar]
- 53.Chen X, Lu J, Dulubova I, Rizo J. NMR analysis of the closed conformation of syntaxin-1. J Biomol NMR. 2008;41:43–54. doi: 10.1007/s10858-008-9239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liang B, Kiessling V, Tamm LK. Prefusion structure of syntaxin-1A suggests pathway for folding into neuronal trans-SNARE complex fusion intermediate. Proceedings of the National Academy of Sciences. 2013;110:19384–19389. doi: 10.1073/pnas.1314699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Starai VJ, Hickey CM, Wickner W. HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol Biol Cell. 2008;19:2500–2508. doi: 10.1091/mbc.E08-01-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McMahon HT, Missler M, Li C, Südhof TC. Complexins: Cytosolic proteins that regulate SNAP receptor function. Cell. 1995;83:111–119. doi: 10.1016/0092-8674(95)90239-2. [DOI] [PubMed] [Google Scholar]
- 57.Wragg Rachel T, Snead D, Dong Y, Ramlall Trudy F, Menon I, Bai J, Eliezer D, Dittman Jeremy S. Synaptic Vesicles Position Complexin to Block Spontaneous Fusion. Neuron. 2013;77:323–334. doi: 10.1016/j.neuron.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snead D, Wragg RT, Dittman JS, Eliezer D. Membrane curvature sensing by the C-terminal domain of complexin. Nature Communications. 2014;5:4955. doi: 10.1038/ncomms5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zdanowicz R, Kreutzberger A, Liang B, Kiessling V, Tamm LK, Cafiso DS. Complexin Binding to Membranes and Acceptor t-SNAREs Explains Its Clamping Effect on Fusion. Biophysical Journal. 2017;113:1235–1250. doi: 10.1016/j.bpj.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Snead D, Lai AL, Wragg RT, Parisotto DA, Ramlall TF, Dittman JS, Freed JH, Eliezer D. Unique Structural Features of Membrane-Bound C-Terminal Domain Motifs Modulate Complexin Inhibitory Function. Frontiers in Molecular Neuroscience. 2017;10 doi: 10.3389/fnmol.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pabst S, Hazzard JW, Antonin W, Südhof TC, Jahn R, Rizo J, Fasshauer D. Selective Interaction of Complexin with the Neuronal SNARE Complex: Determination of the Binding Regions. Journal of Biological Chemistry. 2000;275:19808–19818. doi: 10.1074/jbc.M002571200. [DOI] [PubMed] [Google Scholar]
- 62.Chen X, Tomchick DR, Kovrigin E, Arac D, Machius M, Sudhof TC, Rizo J. Three-dimensional structure of the complexin/SNARE complex. Neuron. 2002;33:397–409. doi: 10.1016/s0896-6273(02)00583-4. [DOI] [PubMed] [Google Scholar]
- 63.Kreutzberger AJB, Liang B, Kiessling V, Tamm LK. Assembly and Comparison of Plasma Membrane SNARE Acceptor Complexes. Biophysical Journal. 2016;110:2147–2150. doi: 10.1016/j.bpj.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burré J, Sharma M, Tsetsenis T, Buchman V, Etherton MR, Südhof TC. α-Synuclein Promotes SNARE-Complex Assembly in Vivo and in Vitro. Science. 2010;329:1663–1667. doi: 10.1126/science.1195227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Darios F, Ruipérez V, López I, Villanueva J, Gutierrez LM, Davletov B. α-Synuclein sequesters arachidonic acid to modulate SNARE-mediated exocytosis. EMBO reports. 2010;11:528–533. doi: 10.1038/embor.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chandra S, Chen X, Rizo J, Jahn R, Südhof TC. A Broken α-Helix in Folded α-Synuclein. Journal of Biological Chemistry. 2003;278:15313–15318. doi: 10.1074/jbc.M213128200. [DOI] [PubMed] [Google Scholar]
- 67.Bussell R, Eliezer D. A Structural and Functional Role for 11-mer Repeats in α-Synuclein and Other Exchangeable Lipid Binding Proteins. Journal of Molecular Biology. 2003;329:763–778. doi: 10.1016/s0022-2836(03)00520-5. [DOI] [PubMed] [Google Scholar]
- 68.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and Dynamics of Micelle-bound Human α-Synuclein. Journal of Biological Chemistry. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 69.Bodner CR, Dobson CM, Bax A. Multiple Tight Phospholipid-Binding Modes of α-Synuclein Revealed by Solution NMR Spectroscopy. Journal of Molecular Biology. 2009;390:775–790. doi: 10.1016/j.jmb.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bodner CR, Maltsev AS, Dobson CM, Bax A. Differential Phospholipid Binding of α-Synuclein Variants Implicated in Parkinson’s Disease Revealed by Solution NMR Spectroscopy. Biochemistry. 2010;49:862–871. doi: 10.1021/bi901723p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galvagnion C, Buell AK, Meisl G, Michaels TCT, Vendruscolo M, Knowles TPJ, Dobson CM. Lipid vesicles trigger α-synuclein aggregation by stimulating primary nucleation. Nat Chem Biol. 2015;11:229–234. doi: 10.1038/nchembio.1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fusco G, De Simone A, Gopinath T, Vostrikov V, Vendruscolo M, Dobson CM, Veglia G. Direct observation of the three regions in α-synuclein that determine its membrane-bound behaviour. Nature Communications. 2014;5:3827. doi: 10.1038/ncomms4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fusco G, Pape T, Stephens AD, Mahou P, Costa AR, Kaminski CF, Kaminski Schierle GS, Vendruscolo M, Veglia G, Dobson CM, De Simone A. Structural basis of synaptic vesicle assembly promoted by α-synuclein. Nature Communications. 2016;7:12563. doi: 10.1038/ncomms12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, Barbour R, Huang J, Kling K, Lee M, Diep L, Keim PS, Shen X, Chataway T, Schlossmacher MG, Seubert P, Schenk D, Sinha S, Gai WP, Chilcote TJ. Phosphorylation of Ser-129 Is the Dominant Pathological Modification of α-Synuclein in Familial and Sporadic Lewy Body Disease. Journal of Biological Chemistry. 2006;281:29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 75.Maltsev AS, Ying J, Bax A. Impact of N-Terminal Acetylation of α-Synuclein on Its Random Coil and Lipid Binding Properties. Biochemistry. 2012;51:5004–5013. doi: 10.1021/bi300642h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dikiy I, Eliezer D. N-terminal Acetylation Stabilizes N-terminal Helicity in Lipid- and Micelle-bound α-Synuclein and Increases Its Affinity for Physiological Membranes. Journal of Biological Chemistry. 2014;289:3652–3665. doi: 10.1074/jbc.M113.512459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartels T, Choi JG, Selkoe DJ. α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation. Nature. 2011;477:107–110. doi: 10.1038/nature10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Burre J, Vivona S, Diao J, Sharma M, Brunger AT, Sudhof TC. Properties of native brain α-synuclein. Nature. 2013;498:E4–E6. doi: 10.1038/nature12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fauvet B, Mbefo MK, Fares MB, Desobry C, Michael S, Ardah MT, Tsika E, Coune P, Prudent M, Lion N, Eliezer D, Moore DJ, Schneider B, Aebischer P, El-Agnaf OM, Masliah E, Lashuel HA. α-Synuclein in Central Nervous System and from Erythrocytes, Mammalian Cells, and Escherichia coli Exists Predominantly as Disordered Monomer. Journal of Biological Chemistry. 2012;287:15345–15364. doi: 10.1074/jbc.M111.318949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dettmer U, Newman AJ, Soldner F, Luth ES, Kim NC, von Saucken VE, Sanderson JB, Jaenisch R, Bartels T, Selkoe D. Parkinson-causing α-synuclein missense mutations shift native tetramers to monomers as a mechanism for disease initiation. Nature Communications. 2015;6:7314. doi: 10.1038/ncomms8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Theillet FX, Binolfi A, Bekei B, Martorana A, Rose HM, Stuiver M, Verzini S, Lorenz D, van Rossum M, Goldfarb D, Selenko P. Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature. 2016;530:45–50. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- 82.Csizmok V, Follis AV, Kriwacki RW, Forman-Kay JD. Dynamic Protein Interaction Networks and New Structural Paradigms in Signaling. Chemical Reviews. 2016;116:6424–6462. doi: 10.1021/acs.chemrev.5b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berlow RB, Dyson HJ, Wright PE. Hypersensitive termination of the hypoxic response by a disordered protein switch. Nature. 2017;543:447–451. doi: 10.1038/nature21705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kreutzberger AJB, Kiessling V, Liang B, Seelheim P, Jakhanwal S, Jahn R, Castle JD, Tamm LK. Reconstitution of calcium-mediated exocytosis of dense-core vesicles. Science Advances. 2017;3:e1603208. doi: 10.1126/sciadv.1603208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cierpicki T, Liang B, Tamm LK, Bushweller JH. Increasing the accuracy of solution NMR structures of membrane proteins by application of residual dipolar couplings. High-resolution structure of outer membrane protein A. J Am Chem Soc. 2006;128:6947–6951. doi: 10.1021/ja0608343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mesleh MF, Veglia G, DeSilva TM, Marassi FM, Opella SJ. Dipolar Waves as NMR Maps of Protein Structure. Journal of the American Chemical Society. 2002;124:4206–4207. doi: 10.1021/ja0178665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Evenas J, Tugarinov V, Skrynnikov NR, Goto NK, Muhandiram R, Kay LE. Ligand-induced structural changes to maltodextrin-binding protein as studied by solution NMR spectroscopy. J Mol Biol. 2001;309:961–974. doi: 10.1006/jmbi.2001.4695. [DOI] [PubMed] [Google Scholar]


