Abstract
The detailed, atomistic-level understanding of molecular signaling along the tumor-suppressive Hippo signaling pathway that controls tissue homeostasis by balancing cell proliferation and death through apoptosis is a promising avenue for the discovery of novel anticancer drug targets. The activation of kinases such as Mammalian STE20-Like Protein Kinases 1 and 2 (MST1 and MST2)—modulated through both homo- and heterodimerization (e.g. interactions with Ras association domain family, RASSF, enzymes)—is a key upstream event in this pathway and remains poorly understood. On the other hand, RASSFs (such as RASSF1A or RASSF5) act as important apoptosis activators and tumor suppressors, although their exact regulatory roles are also unclear. We present recent molecular studies of signaling along the Ras-RASSF-MST pathway, which controls growth and apoptosis in eukaryotic cells, including a variety of modern molecular modeling and simulation techniques. Using recently available structural information, we discuss the complex regulatory scenario according to which RASSFs perform dual signaling functions, either preventing or promoting MST2 activation, and thus control cell apoptosis. Here, we focus on recent studies highlighting the special role being played by the specific interactions between the helical Salvador/RASSF/Hippo (SARAH) domains of MST2 and RASSF1a or RASSF5 enzymes. These studies are crucial for integrating atomistic-level mechanistic information about the structures and conformational dynamics of interacting proteins, with information available on their system-level functions in cellular signaling.
Keywords: competing protein interactions, SARAH domains, signaling switches, apoptosis, cell fate decision, molecular dynamics
Molecular modeling in cancer research
The complexity of cancer and the vast amount of experimental data available have made the use of modern computer-aided approaches crucial for investigating specific molecular interactions and their corresponding molecular mechanisms. For this reason, there is an increasing interest in these computational methods. The large variety of available computational tools ranges from sequence-based, bioinformatics-based approaches to advanced ab initio calculations. Some of the most used methods include docking proteins with associated drugs to estimate the interaction, coarse-grained models, elastic network models and atomistic molecular dynamics among others [1, 2]. To combat cancer and other diseases, several system-related approaches have become the focus of attention, a key element corresponding to the regulation of the Hippo pathway that controls cell death, in an attempt to induce apoptosis in cancer cells.
The Hippo pathway
The Hippo pathway was discovered in genetic screens in the fruit fly Drosophila melanogaster, where knocking out its name-giving component, the kinase Hippo, causes tissue overgrowth [3, 4]. This phenotype is owing to a reduction of cell death and stimulation of proliferation combined with a loss of proper organ size control. In Drosophila the core pathway consists of Hippo, which phosphorylates and activates Warts, another kinase, which phosphorylates the Yorkie protein. This phosphorylation sequesters Yorkie in the cytosol preventing it from transcribing antiapoptotic and growth stimulatory genes in the nucleus, such as cyclin E, Diap1 and other targets [5]. The peripheral stimuli that regulate the Hippo pathway in D. melanogaster include cytoskeletal cues and cell–cell contacts, but although several components are known, the biochemical mechanisms of this regulation remains to be discovered [6].
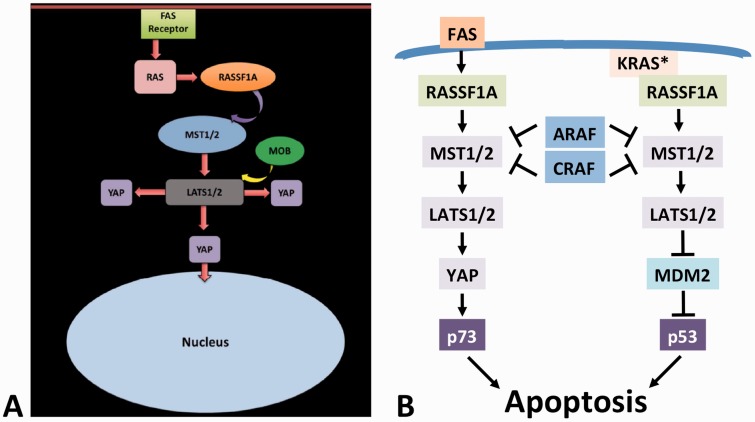
In mammalian cells, the core components are conserved: the Mammalian STE20-Like Protein Kinases 1 and 2 (MST1 and MST2) correspond to Hippo, the Large Tumor Suppressor Kinases 1 and 2 (LATS1 and LATS2) correspond to Warts, and the Yes-associated proteins 1 and 2 (YAP1 and YAP2) are homologues of Yorkie [4]. However, the functional wiring of the pathway is different in mammalian cells [7], where MST1/2 can directly regulate YAP [8] (Figure 1A), and where the regulation of MST1/2 includes inhibition by ARAF [9] and CRAF [10] kinases, which have no direct homologues in D. melanogaster (Figure 1B). Another example is RASSF proteins, which inhibit Hippo in D. melanogaster, but activate MST1/2 kinases in mammalian cells [11, 12]. The reasons for these discrepancies are unclear, but may be related to the fact that the RASSF–MST1/2 signaling pathway in mammals has evolved to become a major tumor suppressor pathway [13, 14]. By contrast, D. melanogaster has no naturally occurring tumors, and hence, the Hippo pathway plausibly may serve different functions in the fruit fly.
Figure 1.
(A) Schematic representation of apoptosis regulation by the mammalian Hippo signaling pathway, along which (B) the regulation of MST1/2 involves the inhibition by ARAF and CRAF kinases, which do not contain a SARAH domain, leading to p73- and p53-mediated apoptosis (see text for details).
The mammalian Hippo pathway contains several tumor suppressors, such as RASSF1A, LATS1, p73 and p53 (Figure 1B). Thus, it is no surprise that in addition to the regulation of cell proliferation and organ size, the mammalian Hippo pathway is also an important mediator of apoptosis (Figure 1B). It contributes to apoptosis initiated by death receptors, such as Fas, by inducing the formation of a YAP/p73 transcription factor complex that activates the expression of the pro-apoptotic gene PUMA [12]. It also participates in apoptosis induced by oncogenic KRAS, which is the only RAS gene family member that when mutated can not only cause cell transformation and cancer, but also apoptosis [15]. In this scenario, mutated KRAS binds RASSF1A, leading to the activation of MST2 and LATS1, which inactivate the murine double minute protein MDM2 (an inhibitor of p53), thus promoting p53 accumulation and apoptosis [16]. These functions, where the same core pathway processes different upstream inputs into different effector mechanisms of apoptosis illustrate the versatility of RASSF and MST1/2 signaling and its potentially widespread roles in apoptosis regulation in mammalian cancer cells [17].
Thus, great efforts are being made to understand the Hippo pathway not only in terms of genetic interactions but also in terms of its biochemical and mechanistic regulation at the level of its protein components, in particular the core kinases MST1/2 and LATS1/2 [4, 18]. The regulation of protein–protein interactions seems to play an important role in the control and function of this pathway. Recent data show that RASSF1A can release MST2 from its inhibitory complex with CRAF. The competition between RASSF1A and CRAF for MST2 binding, combined with phosphorylation of key residues that modulate the binding affinities, causes switch-like transitions between MST2-RASSF1A and MST2-CRAF protein complexes, which coordinate the mutually exclusive decision between apoptosis and proliferation [19]. These interesting biochemical properties and their biological consequences are closely linked to structural features of the protein interactions that allow dynamic regulation and the adjustment of signal flux through the pathway appropriate for triggering highly specific biological responses. Here, we will focus our attention on the MST1/2 kinases, which once phosphorylated, transduce the cell signal toward the LATS1/2 kinases [20], and their interactions with RASSF scaffolds, which regulates their activity.
The activation of kinases such as MST1 and MST2 is modulated through both homo- and heterodimerization (Figure 2A and B respectively; e.g. interactions with RASSF scaffold proteins). While this is a key upstream event in this pathway, it remains poorly understood. On the other hand, RASSFs (such as RASSF1A or RASSF5) act as important apoptosis activators and tumor suppressors, though their exact regulatory roles are also unclear.
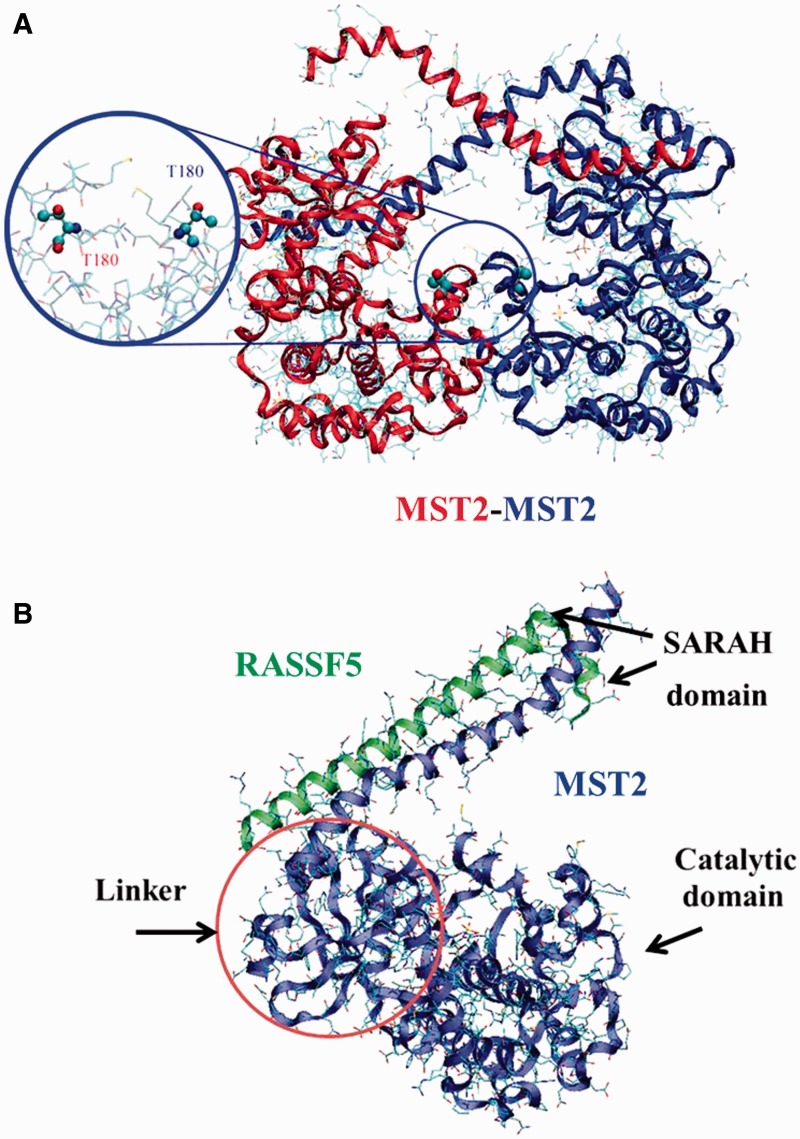
Figure 2.
(A) MST2-MST2 dimer structure (PDB ID: 4LG4). Activation loop and T180 have been highlighted. (B) MTS2-RASSF5 complex from crystal structure (PDB ID: 4LGD) showing the direct interaction between the RASSF5 (green) and the MST2 (blue) SARAH domains. The MST2 kinase domain (blue) is also resolved in the 4LGD crystal structure. Representation of the possible linker between the MST2 kinase and SARAH domains is also shown though, due to its intrinsically disordered nature, it cannot be resolved experimentally. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
SARAH domains
An important characteristic of MST proteins is a particular helical segment, known as the SARAH (Salvador/RASSF/Hippo) domain motif, which has been found to be essential in the activation process of those proteins, and therefore in the initiation of signal transduction [5].
The SARAH domain is located in the C terminus region in three types of eukaryotic proteins denoted in its name (i.e. Salvador, RASSF and Hippo), which are known to be tumor suppressors [5]. The SARAH domain mediates signal transduction from Hippo via the Sav (Salvador) scaffolding protein toward the protein Wts (Warts), downstream along the signaling pathway, by acting as a scaffold that facilitates the Wts phosphorylation by Hippo [3–5, 21]. The SARAH domain is also involved in the dimerization of the mammalian MST1/2 kinases, which form homodimers via their C-terminal SARAH domain. SARAH domains are also known to be associated with other protein domains [5, 21], such as in kinase domains, the WW/rsp5/WWP domain [22–24], the C1 domain [25], the LIM domain [26] or the Ras-associating (RA) domain [27, 28].
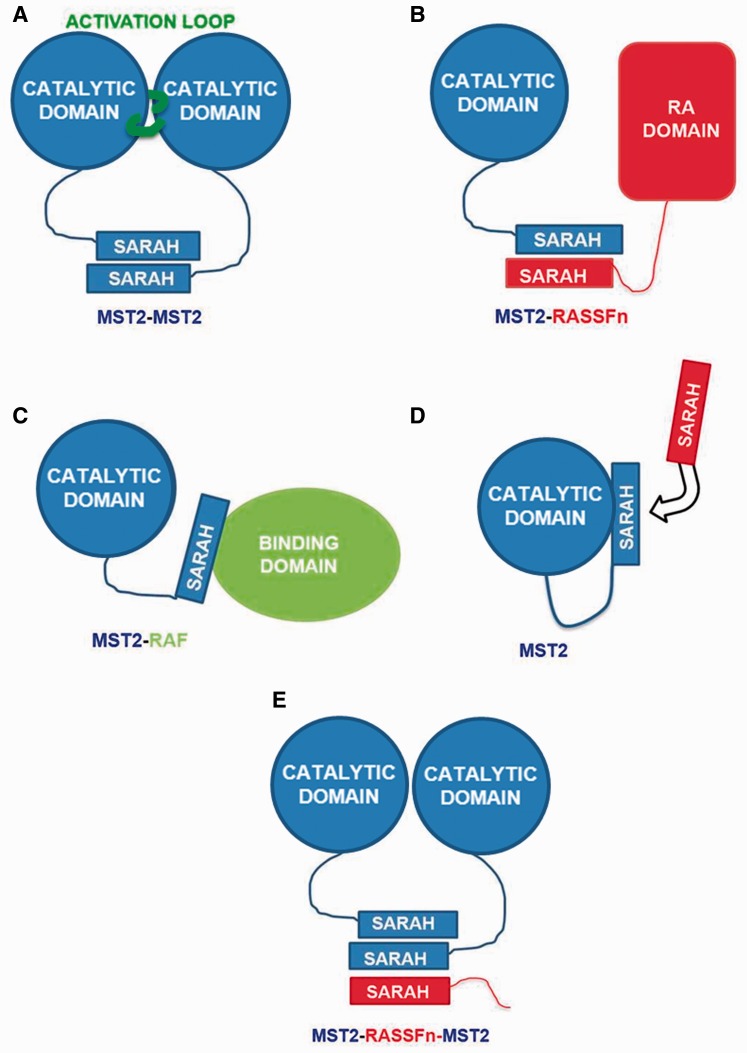
The process of signal transduction involves the formation of MST2 dimers, and the dimerization process is governed by the interaction through SARAH domains [12, 19, 29–32] (Figure 3A). In addition, RASSF scaffolds also interact with and regulate the activity of MST proteins through protein interactions mediated by their SARAH domain (Figure 3B). However, despite the fact that the SARAH domain is essential in the dimerization process, MST1/2 proteins may interact using their SARAH domain with another type of suppressors that do not contain a SARAH domain, such as ARAF [9] and CRAF (Figures 1B and 3C). One example is CRAF, which controls cell proliferation, oncogenic transformation, differentiation and apoptosis. CRAF binds to a small segment of the MST2 SARAH domain competing with RASSF1A [12, 19, 29] and RASSF5 scaffolds [33]. Other possibilities for the SARAH domain function have been considered and are still in progress of investigation by our group, for example, covering the activation loop in the MST itself (Figure 3D), and even interactions with three SARAH domain members. Trimers, have been postulated for Salvador [5] (Figure 3E) and between MST2, WW6 and RASSF1A (or RASSF6) [34, 35]. However, some studies suggest that Sav-RASSF5-MST1 trimers may not be likely to form stable complexes under experimental conditions [36].
Figure 3.
Schematic representation of possible SARAH-mediated intermolecular interactions. (A) MST2 (blue) homodimers can be subject to autophosphorylation mediated by an active loop (green). (B) RASSFn scaffolding proteins (red) can form competitively heterodimers with MST2 SARAH domains. (C) MST2 SARAH domains can interact with other binding domains from partners such as RAF. (D) Owing to its large and flexible linker, MST2 may present self-interactions of its SARAH and catalytic domains. (E) MST2 SARAH homodimers (blue) may be affected by tertiary (e.g. RASSFn) SARAH domains that could modulate the MST2 homo-interactions. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
We focus our attention on those SARAH domains that belong to proteins in the Hippo pathway, in particular to those from MST1/2 kinases and RASSF scaffolds. Despite intensive research in the past decade, the role of SARAH domains is still poorly understood. It is known that these long helixes play a crucial role in the MST dimerization, and through them, RASSF scaffolds control the activity of such kinases. In particular, members of the RASSF family (RASSF1-10) have been denoted as tumor suppressors scaffolds, which are frequently down-regulated by promoter hypermethylation in several types of cancer. These proteins present common types of RA and SARAH domains, which may potentially bind Ras oncoproteins and play an important role in protein-protein interactions with other proteins through their SARAH domains (e.g., with MST). However, among the entire RASSF family scaffold, only RASSF1-6 possess SARAH domains, while the SARAH domain is absent in the remainder (RASSF7-10) [37]. A multiple sequence alignment of the SARAH domains using Clustal Omega [38–40] is shown in Figure 4A. By comparing the primary structure of the SARAH domains from the RASSFn family, we observe a moderate to high level of sequence identity and sequence similarity among all of the SARAH domains belonging to the RASSF scaffolds family. In particular, RASSF5 (also known as Nore1 or RAPL) and RASSF1A are highly conserved and present a high percentage of sequence identity (54.1%) and similarity (89.4%) (Figure 4B) (G. Sánchez-Sanz et al., unpublished data). In addition, MST and RASSF SARAH domains are also highly homologous, as both helical structures have fair sequence identity and similarity [41]. For example, MST2 and RASSF1 (Figure 4C) have 31.4% identity and 64.6% sequence similarity, which is reflected in their structural similarities (G. Sánchez-Sanz et al., unpublished data). This is important because RASSF1A has been identified to mediate proapoptotic signals through binding of the mammalian sterile 20-like kinases 1 and 2 (MST1 and MST2) [42, 43].
Figure 4.
Multiple and pairwise alignments of SARAH domains using Clustal Omega software of (A) RASSFn, with n = 1–6, (B) RASSF1 and RASSF5 alone, and (C) MST2–RASSF1A. Color coding: red = small and amino acids (including aromatic - Y); blue = acidic; green = hydroxyl + sulfhydryl + amine + G; magenta = basic (without H). In the last row, the alignment results are represented as follows: an asterisk (*) = indicates positions that have a single, fully conserved residue, a colon (:) = indicates conservation between groups with strongly similar properties, a period (.) = indicates conservation between groups with weakly similar properties. In (C), the two domains have 31.4% sequence identity and 64.6% sequence similarity. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
It is also worth mentioning the significant pioneering efforts that have been made to crystallize SARAH domain structures. A thorough search on the UniProt [44] and RCSB PDB data banks reveals several crystal structures of SARAH domain monomers and homo- and heterodimers. The crystal structure of MST SARAH domains have been resolved by X-ray diffraction, for MST1, PDB ID: 2JO8 [36], 4OH8 [45], 4NR2 (A. Chaikuad et al., unpublished data), 2YMY [41] and for MST2 PDBID: 4LGD [32]. In the case of RASSF family, there is a lack of crystal structures for any of the RASSF scaffolds, with the exception of RASSF5 (PDB ID: 4LGD [32] and 2YMY [41]). However, more important than studying the protomers alone is to look at dimeric structures, as the dimerization process between two SARAH domains is crucial in understanding the activation of MST kinases. As suggested by recent experiments, here we focus on understanding the specific dimeric interactions between the helical SARAH domains of MST2 and RASSF1A or RASSF5 scaffolds. Our discussion is aimed at the specific part of those proteins, SARAH domains and, in particular, at the structure of dimers formed between MSTn and RASSFn.
SARAH–SARAH domain interactions
As stated above, SARAH domains have been shown to form antiparallel, coiled coil dimers. In general, long helix–helix interactions, including dimer, tetramers and barrels of helices have been intensely studied [46] and the importance and nature of the intermolecular interactions between each have been highlighted. For example, the MST1-SARAH domain monomeric conformation has been shown to be thermodynamically unstable, and may unfold and later on dissociate without presenting any stable intermediate state [41]. More interactions of MST1 SARAH dimers and the influence of the intrinsically disordered inhibitory domain in the dimerization process have been previously studied [47].
The number of studies devoted to the SARAH–SARAH domains interactions is significant because they may govern the activity of MST kinases and therefore control cell signaling. Thus, SARAH domain interactions seem to act as a main driver for the activation of apoptosis. Here, we highlight some of those studies, which may contain not only experimental but also computational procedures related to crystal structures. Homodimers of MST1–MST1 have been studied and their crystal structures resolved (PDBID: 2JO8 [36], 4OH8 [46], 4NR2 (A. Chaikuad et al., unpublished data)). Much attention has been attracted by MST2–MST2 homodimers, in which several crystal structures have been published (PDBID: 4OH9 [45], 4HKD (G.G. Liu et al., unpublished data), 4L0N (A. Chaikuad et al., unpublished data)). RASSF5-RASSF5 homodimers [41] have also been the subject of many studies. The structure has been successfully crystallized, and its structure in solution investigated including circular dichroism. Additionally, the dissociation energetics of RASSF5–RASFF5 dimers was also probed by isothermal titration calorimetry.
In the case of heterodimers, the majority of the experimental studies have been devoted to the MST–RASSF1 or MST–RASSF5 interactions [12, 19, 29, 30, 48]. For example, interactions between RASSF1A (and RASSF1C) and MST1 and MST2 have been identified and analyzed showing that this interaction depends on the C-terminal SARAH domain, i.e. dimerization of both SARAH domains of each protein [31]. Nevertheless, few crystal structures are available for heterodimers, i.e. MST1–RASSF5 dimers have been resolved (PDBID: 4OH8 [45], 2YMY [41]) and more recently MST2–RASSF5 SARAH dimers crystal structures have been published, 4LGD [32].
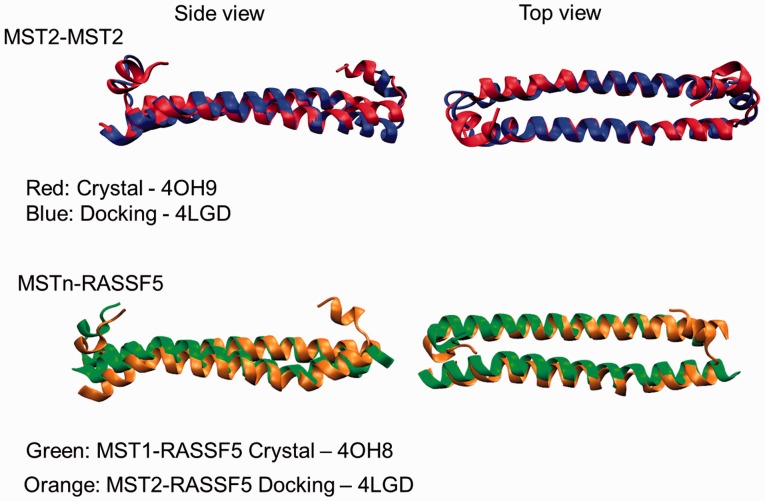
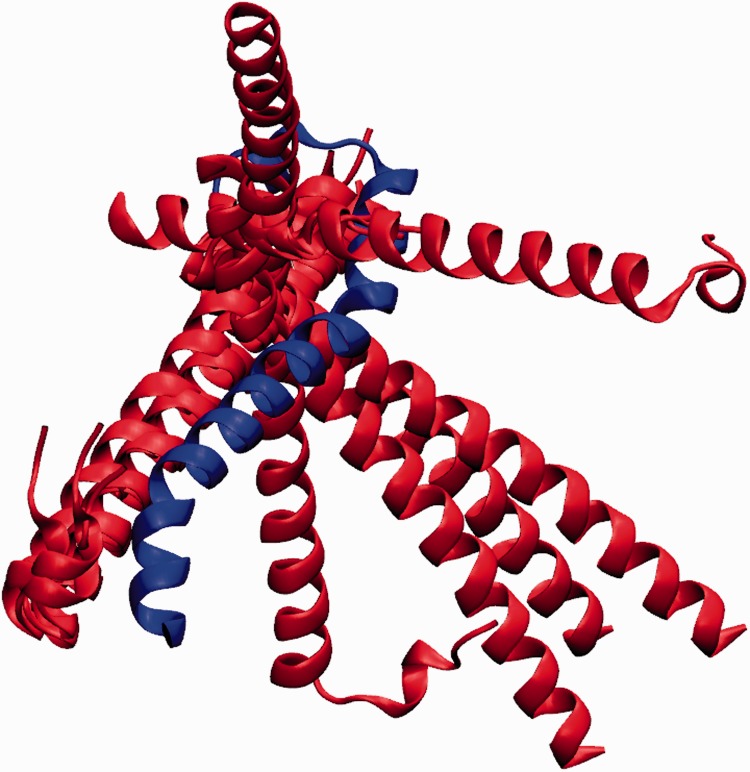
We have been actively researching the homo- and heterodimerization of MST2 and RASSF1 and 5 SARAH domains, paying special attention to the driving forces that govern the stability of such dimers (G. Sánchez Sanz et al., unpublished data). We would like to highlight the importance of computational modeling for such tasks, in particular, those cases in which the crystal structure of a specific system is not available. Figure 5 shows a comparison between MST2-MST2 crystal structure (4OH9) and the one resulting from molecular docking simulation (Zdock [49–51]) using the MST2 monomer from 4LGD. As it is shown, the alignment of both structures match, revealing how the computational approach may reproduce the experimental structures. Furthermore, in Figure 5, two different heterodimers have been also compared; the MST1-RASSF5 SARAH crystal structure from 4OH8 and the MST2-RASSF5 modeled using 4LGD as template. This again shows that the computational approach predicts reliable structures directly comparable with experimental structures.
Figure 5.
Structural alignment of crystal and docked structures of SARAH dimers. The top row shows MST2-MST2 SARAH homodimer structures in red: crystal structure from 4OH9, and in blue: based on the MST2 protomer structures from 4LGD. The bottom row illustrates in green: crystal structures of MST1-RASSF5 SARAH dimer (4OH9), and in orange: MST2-RASSF5 docked structures (based on the MST2 structures from 4LGD). A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
Recently, we have carried out an in-depth analysis not only of the possible poses arising from docking (Figure 6), but also using full atomistic molecular dynamic simulations to describe the behavior of those dimers in solvation and to study their intermolecular interactions (G. Sánchez Sanz et al., unpublished data).
Figure 6.
Top 10 highest scoring SARAH heterodimer structures from a docking study (using Zdock) of RASSF5 SARAH domains (red) docked on a target MST2 SARAH domain (blue, from the 4LGD structure). A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
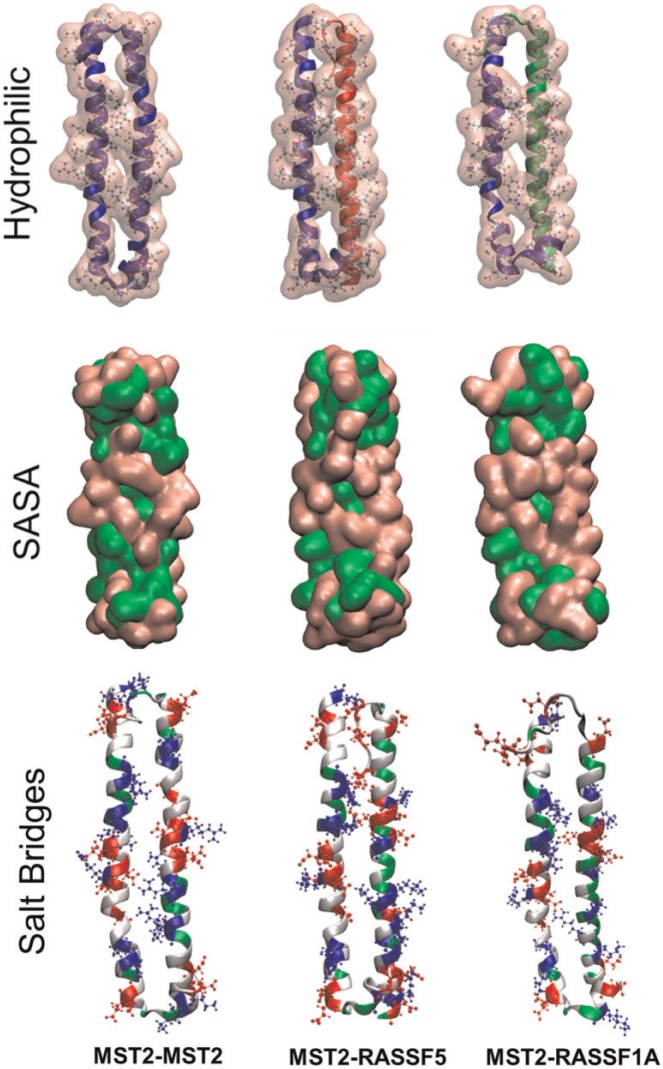
Figure 7 shows the different homo- and heterodimers investigated, MST2-MST2 (blue-blue), MST2-RASSF5 (blue-red) and MST2-RASSF1A (blue-green). It is worth noting that the MST2-MST2 and MST2-RASSF5 dimers were obtained from docking studies using 4LGD monomers as receptors and ligands. However, in the case of MST2-RASSF1A, there is no crystal structure available, so homology modeling was used using RASSF5 as a template, with RASSF1A sequence (Uniprot Q9NS23) using the SwissProt program [52]. Once again, computational modeling is shown to be a useful tool that gives insight into the biophysical properties, functions and biological mechanisms of proteins and their interactions within the cell (G. Sánchez Sanz et al., unpublished data) [2, 53–55].
Figure 7.
Molecular representations of MST2 homo- (left, blue backbone) and heterodimers with RASSF5 (center, red backbone) and RASSF1A (right, green backbone) SARAH domains. The first row illustrates the backbone and hydrophilic interactions. The middle row shows the solvent accessible surface area (SASA) for each dimer with the corresponding hydrophilic (pink) and hydrophobic (green) SASA fractions. The bottom row illustrates the same SARAH dimers with their characteristic electrostatic interactions (red: acidic, blue: basic, gray: hydrophobic, and green: hydrophilic amino acids). A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
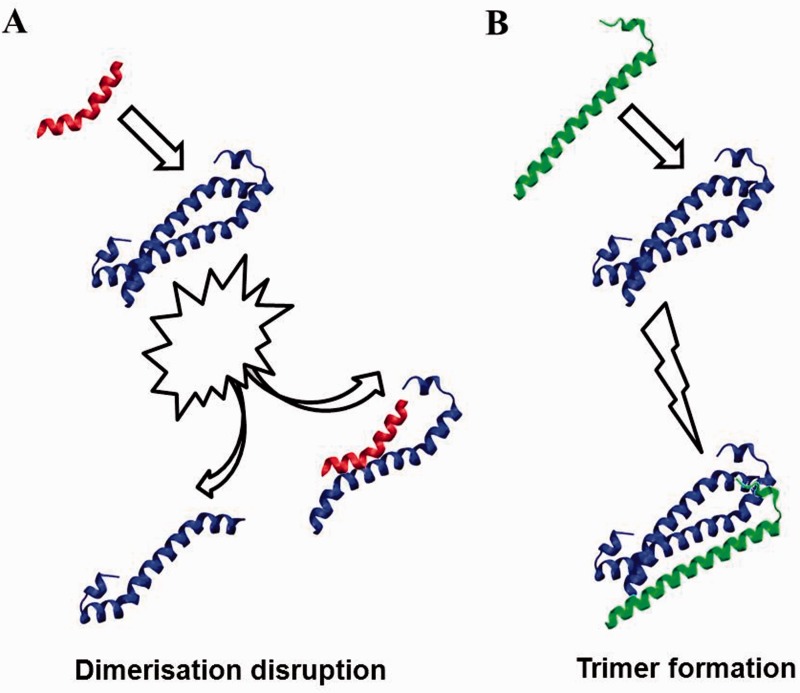
Interestingly, we also note that the SARAH–SARAH domain interactions are not restricted to dimers. In fact, the existence of SARAH domain trimeric complexes has been suggested by Scheel [5], though it has been subsequently questioned by Makbul et al. [41]. However, recent studies have successfully designed novel ‘disruptor’ peptides that can interfere efficiently with dimer formation, MST-MST, MST-RAF and MST2-RASSF1A [22] and have also probed the possibility of trimer formation, as illustrated in Figure 8.
Figure 8.
Schematic representation of possible molecular mechanisms involving MST2 homodimers (blue) and tertiary binding partners. (A) Small peptides can be designed to bind competitively and disrupt SARAH homodimers. (B) Sarah domains of other kinases (e.g. RASSFn, green) may bind noncompetitively to MST monomers or homomodimers and could facilitate and/or stabilize homodimeric interactions. A colour version of this figure is available at BIB online: http://bib.oxfordjournals.org.
Concluding remarks and outlook
The investigation of signaling pathways involved in cancer is one of the most important modern biomedical research avenues. Among all the possible complex signaling pathways that are under consideration in current cancer research, the Hippo pathway, which controls cell death, has been the objective of numerous investigations in the recent years. The MST protein family is particularly important for the activation of apoptosis and, therefore, for the possibility of controlling cancer cell death.
One specific domain of MST kinases has been highlighted to play a key role in the activation of the MST proteins and the subsequent signal transduction—the long terminal helix known as a SARAH domain. It has been also shown that the regulation of the MST kinases by its scaffolds, the RASSF protein family, is mediated by the interaction between MST and RASSF through their corresponding SARAH domains. Several crystal structures and publications have identified monomeric and dimeric (both homo- and heterodimers) structures involving MST and RASSF SARAH domains. Despite increasing efforts to characterize the structural interactions between SARAH domains, their interface and the implications of the dimerization on the activation process, the particular role of the SARAH domains remains poorly understood, both from an experimental and from a computational point of view.
Here, we have presented recent studies of SARAH-mediated interactions, describing the current state of the field in relation to the available crystal structures and modern computational molecular modeling approaches available. While there are several crystal structures for homo- and heterodimers, computational and mechanistic modeling is only recently catching up. We have focused our attention on describing structural properties of SARAH domain dimeric interactions both from the computational and experimental point of view. Recently, the molecular interactions between RAF1 and MST2 proteins has been studied both experimentally and computationally, showing that MST2 modulates the crosstalk between the mitogenic Raf and the pro-apoptotic MST2 pathway [19]. This study showed that a designed 17-mer peptide can disrupt effectively RAF1-MST2 dimerization [19]. Understanding at a molecular level how such ‘disruptor’ peptides affect the dimerization process can be crucial for future development of novel anti-cancer drugs that can activate MST2 by changing its inhibitory interaction with RAF1.
While experimental studies have provided a strong foundation to lead the cancer research investigation in this area, computational approaches are becoming both increasingly available and able to provide unique atomistic-level insights on the underlying molecular mechanisms, with reliable and experimentally testable results.
Key Points
Hippo signaling pathway controls tissue homeostasis by balancing cell proliferation and death through apoptosis.
MST1/2 kinase are key components of the mammalian Hippo signaling pathway.
MST kinases are regulated by RASSF scaffolds by interacting through SARAH domains.
Few crystal structures are available for MST-RASSF SARAH or MST-MST dimers, and exclusively for MST2, MST1 and RASSF5.
Molecular modeling (e.g., homology-based methods, docking and molecular dynamics) provides essential tools to probe and understand protein structure and activity.
Funding
The authors are grateful for financial support from the Human Science Frontier Program and from the Irish Research Council, and wish to thank the DJEI/DES/SFI/HEA Irish Centre for High-End Computing (ICHEC), and the Biowulf Linux cluster at the National Institutes of Health, USA (http://biowulf.nih.gov) for the provision of computational support and facilities. The authors also acknowledge funding from the Science Foundation Ireland under grant No. 06/CE/B1129 (LKN, DM, BNK,WK), the European Union Seventh Framework Program (FP7/2007- 2013) ASSET project under grant agreement number FP7-HEALTH-2010-259348 (BNK, WK), and the PRIMES project under grant agreement No. FP7-HEALTH-2011-278568 (LNK, BNK, WK).
References
- 1. Friedman R, Boye K, Flatmark K. Molecular modelling and simulations in cancer research. BBA-Rev Cancer 2013;1836:1–14. [DOI] [PubMed] [Google Scholar]
- 2. Jambrina PG, Bohuszewicz O, Buchete N-V, et al. Molecular mechanisms of asymmetric RAF dimer activation. Biochem Soc Trans 2014;42:784–90. [DOI] [PubMed] [Google Scholar]
- 3. Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010;19:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Avruch J, Zhou D, Fitamant J, et al. Protein kinases of the hippo pathway: regulation and substrates. Semin Cell Dev Biol 2012;23:770–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scheel H, Hofmann K. A novel inter action motif, SARAH, connects three classes of tumor suppressor. Curr Biol 2003;13:R899–900. [DOI] [PubMed] [Google Scholar]
- 6. Gaspar P, Tapon N. Sensing the local environment: actin architecture and Hippo signalling. Curr Opin Cell Biol 2014;31:74–83. [DOI] [PubMed] [Google Scholar]
- 7. Matallanas D, Romano D, Hamilton G, et al. A hippo in the ointment: MST signalling beyond the fly. Cell Cycle 2008;7:879–84. [DOI] [PubMed] [Google Scholar]
- 8. Zhou D, Conrad C, Xia F, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 2009;16:425–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rauch J, O'Neill E, Mack B, et al. Heterogeneous nuclear ribonucleoprotein H Blocks MST2-dependent apoptosis in cancer cells via regulation of A-Raf transcription. Cancer Res 2010;70:1679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O'Neill E, Rushworth L, Baccarini M, et al. Role of the kinase MST2 in suppression of apoptosis by the proto-oncogene product Raf-1. Science 2004;306:2267–70. [DOI] [PubMed] [Google Scholar]
- 11. Khokhlatchev A, Rabizadeh S, Xavier R, et al. Identification of a novel ras-regulated proapoptotic pathway. Curr Biol 2002;12:253–65. [DOI] [PubMed] [Google Scholar]
- 12. Matallanas D, Romano D, Yee K, et al. RASSF1A elicits apoptosis through an MST2 pathway directing proapoptotic transcription by the p73 tumor suppressor protein. Mol Cell 2007;27:962–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Avruch J, Xavier R, Bardeesy N, et al. Rassf family of tumor suppressor polypeptides. J Biol Chem 2009;284:11001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fausti F, Di Agostino S, Sacconi A, et al. Hippo and rassf1a pathways: a growing affair. Mol Biol Int 2012;2012:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cox AD, Der CJ. The dark side of Ras: regulation of apoptosis. Oncogene 2003;22:8999–9006. [DOI] [PubMed] [Google Scholar]
- 16. Matallanas D, Romano D, Al-Mulla F, et al. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell 2011;44:893–906. [DOI] [PubMed] [Google Scholar]
- 17. Gomez M, Gomez V, Hergovich A. The Hippo pathway in disease and therapy: cancer and beyond. Clin Transl Med 2014;3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rawat SJ, Chernoff J. Regulation of mammalian Ste20 (Mst) kinases. Trends Biochem Sci 2015;40:149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Romano D, Nguyen LK, Matallanas D, et al. Protein interaction switches coordinate Raf-1 and MST2/Hippo signalling. Nat Cell Biol 2014;16:673–84. [DOI] [PubMed] [Google Scholar]
- 20. Hao Y, Chun A, Cheung K, et al. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 2008;283:5496–509. [DOI] [PubMed] [Google Scholar]
- 21. Schagdarsurengin U, Richter AM, Hornung J, et al. Frequent epigenetic inactivation of RASSF2 in thyroid cancer and functional consequences. Mol Cancer 2010;9:264–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bork P, Sudol M. The WW domain: a signalling site in dystrophin?. Trends Biochem Sci 1994;19:531–3. [DOI] [PubMed] [Google Scholar]
- 23. Hofmann K, Bucher P. The rsp5-domain is shared by proteins of diverse functions. FEBS Lett 1995;358:153–7. [DOI] [PubMed] [Google Scholar]
- 24. Sudol M, Chen HI, Bougeret C, et al. Characterization of a novel protein-binding module — the WW domain. FEBS Lett 1995;369:67–71. [DOI] [PubMed] [Google Scholar]
- 25. Azzi A, Boscoboinik D, Hensey C. The protein kinase C family. EurJ Biochem 1992;208:547–57. [DOI] [PubMed] [Google Scholar]
- 26. Klug A. Zinc finger peptides for the regulation of gene expression. J Mol Biol 1999;293:215–18. [DOI] [PubMed] [Google Scholar]
- 27. Hofer F, Fields S, Schneider C, et al. Activated Ras interacts with the Ral guanine nucleotide dissociation stimulator. Proc Natl Acad Sci 1994;91:11089–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ponting CP, Benjamin DR. A novel family ofras-binding domains. Trends Biochem Sci 1996;21:422–5. [DOI] [PubMed] [Google Scholar]
- 29. Nguyen LK, Matallanas DG, Romano D, et al. Competing to coordinate cell fate decisions: the MST2-Raf-1 signaling device. Cell Cycle 2015;14:189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Romano D, Matallanas D, Frederick DT, et al. One Hippo and many masters: differential regulation of the Hippo pathway in cancer. Biochem Soc Trans 2014;43:816–12. [DOI] [PubMed] [Google Scholar]
- 31. Dittfeld C, Richter AM, Steinmann K, et al. The SARAH domain of RASSF1A and Its tumor suppressor function. Mol Biol Int 2012;2012:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ni L, Li S, Yu J, et al. Structural basis for autoactivation of human Mst2 kinase and its regulation by RASSF5. Structure 2013;21:1757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stieglitz B, Bee C, Schwarz D, et al. Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J 2008;27:1995–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo C, Tommasi S, Liu L, et al. RASSF1A is part of a complex similar to the Drosophila Hippo/Salvador/Lats tumor-suppressor network. Curr Biol 2007;17:700–5. [DOI] [PubMed] [Google Scholar]
- 35. Ikeda M, Kawata A, Nishikawa M, et al. Hippo pathway–dependent and –independent roles of RASSF6. Sci Signal 2009;2:ra59. [DOI] [PubMed] [Google Scholar]
- 36. Hwang E, Ryu K-S, Pääkkönen K, et al. Structural insight into dimeric interaction of the SARAH domains from Mst1 and RASSF family proteins in the apoptosis pathway. Proc Natl Acad Sci 2007;104:9236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan JJ, Flatters D, Rodrigues-Lima F, et al. Comparative analysis of interactions of RASSF1-10. Adv Biol Regul 2012;53:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Goujon M, McWilliam H, Li W, et al. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 2010;38:W695–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McWilliam H, Li W, Uludag M, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 2013;41:W597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Makbul C, Constantinescu Aruxandei D, Hofmann E, et al. Structural and thermodynamic characterization of Nore1-SARAH: a small, helical module important in signal transduction networks. Biochemistry 2013;52:1045–54. [DOI] [PubMed] [Google Scholar]
- 42. Praskova M, Khoklatchev A, Ortiz-Vega S, et al. Regulation of the MST1 kinase by autophosphorylation, by the growth inhibitory proteins, RASSF1 and NORE1, and by Ras. Biochem J 2004;381:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oh HJ, Lee K-K, Song SJ, et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res 2006;66:2562–9. [DOI] [PubMed] [Google Scholar]
- 44. Apweiler R, Bairoch A, Wu CH, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res 2004;32:D115–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hwang E, Cheong H-K, Mushtaq AU, et al. Structural basis of the heterodimerization of the MST and RASSF SARAH domains in the Hippo signalling pathway. Acta Crystallogr Sect D 2014;70:1944–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomson AR, Wood CW, Burton AJ, et al. Computational design of water-soluble α-helical barrels. Science 2014;346:485–8. [DOI] [PubMed] [Google Scholar]
- 47. Constantinescu Aruxandei D, Makbul C, Koturenkiene A, et al. Dimerization-induced folding of MST1 SARAH and the influence of the intrinsically unstructured inhibitory domain: low thermodynamic stability of monomer. Biochemistry 2011;50:10990–11000. [DOI] [PubMed] [Google Scholar]
- 48. Romano D, Matallanas D, Weitsman G, et al. Proapoptotic kinase MST2 coordinates signaling crosstalk between RASSF1A, Raf-1, and Akt. Cancer Res 2010;70:1195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mintseris J, Pierce B, Wiehe K, et al. Integrating statistical pair potentials into protein complex prediction. Proteins 2007;69:511–20. [DOI] [PubMed] [Google Scholar]
- 50. Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS ONE 2011;6:e24657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pierce BG, Wiehe K, Hwang H, et al. ZDOCK server: interactive docking prediction of protein–protein complexes and symmetric multimers. Bioinformatics 2014;30:1771–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boeckmann B, Bairoch A, Apweiler R, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 2003;31:365–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kelly CM, Muzard J, Brooks BR, et al. Structure and dynamics of the fibronectin-III domains of aplysia californica cell adhesion molecules. Phys Chem Chem Phys 2015;17:9634–43. [DOI] [PubMed] [Google Scholar]
- 54. Milac AL, Buchete NV, Fritz TA, et al. Substrate-induced conformational changes and dynamics of UDP-N-acetylgalactosamine: polypeptide N-acetylgalactosaminyltransferase-2. J Mol Biol 2007;373:439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tofoleanu F, Brooks BR, Buchete N-V. Modulation of Alzheimer’s Aβ Protofilament-membrane interactions by lipid headgroups. ACS Chem Neurosci 2015;6:446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]