Abstract
Background
It is unclear whether germline breast cancer susceptibility gene mutations affect breast cancer related outcomes. We wanted to evaluate mutation patterns in 20 breast cancer susceptibility genes and correlate the mutations with clinical characteristics to determine the effects of these germline mutations on breast cancer prognosis.
Methods
The study cohort included 480 ethnic Chinese individuals in Taiwan with at least one of the six clinical risk factors for hereditary breast cancer: family history of breast or ovarian cancer, young age of onset for breast cancer, bilateral breast cancer, triple negative breast cancer, both breast and ovarian cancer, and male breast cancer. PCR-enriched amplicon-sequencing on a next generation sequencing platform was used to determine the germline DNA sequences of all exons and exon-flanking regions of the 20 genes. Protein-truncating variants were identified as pathogenic.
Results
We detected a 13.5% carrier rate of pathogenic germline mutations, with BRCA2 being the most prevalent and the non-BRCA genes accounting for 38.5% of the mutation carriers. BRCA mutation carriers were more likely to be diagnosed of breast cancer with lymph node involvement (66.7% vs 42.6%; P = 0.011), and had significantly worse breast cancer specific outcomes. The 5-year disease-free survival was 73.3% for BRCA mutation carriers and 91.1% for non-carriers (hazard ratio for recurrence or death 2.42, 95% CI 1.29–4.53; P = 0.013). After adjusting for clinical prognostic factors, BRCA mutation remained an independent poor prognostic factor for cancer recurrence or death (adjusted hazard ratio 3.04, 95% CI 1.40–6.58; P = 0.005). Non-BRCA gene mutation carriers did not exhibit any significant difference in cancer characteristics or outcomes compared to those without detected mutations. Among the risk factors for hereditary breast cancer, the odds of detecting a germline mutation increased significantly with having bilateral breast cancer (adjusted odds ratio 3.27, 95% CI 1.64–6.51; P = 0.0008) or having more than one risk factor (odds ratio 2.07, 95% CI 1.22–3.51; P = 0.007).
Conclusions
Without prior knowledge of the mutation status, BRCA mutation carriers had more advanced breast cancer on initial diagnosis and worse cancer-related outcomes. Optimal approach to breast cancer treatment for BRCA mutation carriers warrants further investigation.
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4229-5) contains supplementary material, which is available to authorized users.
Keywords: Cancer susceptibility gene, Breast cancer, BRCA1 & BRCA2, Breast cancer prognosis, Next generation sequencing
Background
Multigene panel testing is increasingly adopted for managing breast cancer susceptibility in high risk individuals suspected of having hereditary breast cancer, but the evidence-based practice guidelines remain far from being comprehensive. The advent of next generation sequencing (NGS) technologies is making multigene panel testing easier and affordable [1–4]. In addition, multigene panel testing could identify up to 50% more individuals with cancer susceptibility gene mutations in comparison with testing only for BRCA1 and BRCA2 (BRCA) [5]. Most of these additional mutations are from moderate risk genes, many of which could result in alterations of cancer risk estimation and clinical action [5, 6]. However, arriving at consistent and optimal clinical recommendations on the basis of the interpretations of the multigene panel testing and associated variants of uncertain significance (VUS) could be challenging due to lack of comprehensive understanding on the consequences of the genetic alterations [7]. As the multigene panel testing is becoming widely adopted, studies are needed to develop evidence-based practice guidelines.
In addition to risk assessment of breast cancer susceptibility in germline mutation carriers, understanding prognosis after breast cancer diagnosis will also impact practice guidelines for breast cancer. With the efficacy of poly (ADP-ribose) polymerase (PARP) inhibitors in controlling BRCA mutation positive tumors, many clinical trials are now underway evaluating their use in breast cancer [8]; their incorporation into systemic therapy in clinical practice is highly anticipated. It has not been established whether BRCA or any cancer susceptibility gene mutation is an independent prognostic factor after breast cancer diagnosis. Despite the suspicion for a more aggressive tumor phenotype, most studies have fallen short of showing differences in clinical outcomes in BRCA mutation carriers [9–12]. Systematic reviews with larger pooled sample size have yielded conflicting conclusions, possibly due to variability of included studies [13–15]. Consequently, conventional decisions regarding systemic therapy for BRCA mutation-associated breast cancer have been based on disease characteristics rather than BRCA mutation status. As such, it would be informative to discover causal or statistical correlations of germline breast cancer susceptibility gene mutations to breast cancer prognosis.
A panel of 20 known and candidate breast cancer susceptibility genes were selected herein for the multigene panel testing study. Among the 20 genes, BRCA1, BRCA2, PALB2, TP53, CDH1, PTEN, ATM, CHEK2, BARD1, STK11, NBN have been well established as breast cancer susceptibility genes [7, 16, 17]. Some are part of rare high-penetrance cancer predisposing syndromes (e.g. BRCA1, BRCA2, TP53, CDH1, PTEN, STK11, PALB2) while others are moderate-penetrance genes (e.g. ATM, NBN, CHEK2, BARD1). The impacts of the mutations in RAD50, RAD51C, and RAD51D on breast cancer susceptibility and survival are controversial: Mutations in RAD50 have been found not associated with breast cancer risk [17]. Also mutations in RAD51C have not been found to increase the risk of breast cancer [18–20] and mutations in RAD51D have been associated with high risk of ovarian cancer but not with breast cancer [21]. Nevertheless, other studies have indicated that mutations in RAD51C [22] and RAD51D [17, 23, 24] contribute to the risk of both breast and ovarian cancer, and that RAD50 is an intermediate-risk breast cancer susceptibility gene [25]. Although germline mutations in the DNA mismatch repair genes (MLH1, MSH2, MSH6, PMS2) have been mostly associated with Lynch syndrome, evidence has been established to support the connections between the mutations in the DNA mismatch repair genes and the risk or survival of breast cancer [26–29]. Similarly, whether BRIP1 is a breast cancer susceptibility gene remains controversial [30], and perhaps is dependent on the ethnicity of the cohort studied [22]. NF1 mutations have been known to associate with increased risk of breast cancer in younger population [31] and poor breast cancer survival [32]. To clarify the controversies, we included in the panel the potentially relevant genes above to explore the germline mutation-dependence of breast cancer predisposition and outcomes in our local high risk population.
Different ethnic populations need respective studies on cancer risks pertinent to germline mutations. In the western populations, about 5% of the breast cancer patients may carry heritable cancer susceptibility gene mutations [33, 34]. BRCA1 and BRCA2 account for the majority of these gene mutations, with BRCA1 being the most common [34, 35]. However, studies in Asian populations have indicated somewhat different conclusions: available results show that BRCA mutation rates in Asians are lower than those in Whites, and that the distributions of the gene mutations are also different [36–43]. It is imperative to enrich mutation databases on different ethnic populations, so as to better interpret ethnically specific germline mutations and better manage cancer risks among corresponding ethnic populations.
In this study, we analyzed germline mutations in the 20 breast cancer susceptibility genes using NGS-based technique in a cohort of high risk ethnic Chinese population. We evaluated the correlation of mutations with clinical characteristics and cancer outcomes. We aimed to clarify the prognostic value of BRCA and other breast cancer susceptibility gene mutations on breast cancer specific outcomes after conventional cancer treatment.
Methods
Study participants and data collection
Koo Foundation Sun Yat-Sen Cancer Center (KF-SYSCC) treats over 1000 newly diagnosed breast cancer patients annually. Between July 30, 2015 and March 31, 2016, we enrolled 480 individuals fulfilling at least one of the six eligibility criteria: family history of breast or ovarian cancer at any age (2 or more individuals on the same lineage of the family), personal history of breast cancer with age of diagnosis less than or equal to 40, bilateral breast cancer diagnosed at the same time or sequentially, triple negative (ER/PR/HER2 negative) breast cancer, breast and ovarian cancer in the same individual, and male breast cancer. None of the participants had known mutation status in any cancer susceptibility genes prior to enrollment. Clinical information was collected through participant surveys, electronic medical records, and the institutional breast cancer database.
Sequencing and variant analyses of cancer susceptibility genes in genomic DNA
Germline DNA sequencing of all exonal regions was done in twenty breast cancer susceptibility genes: BRCA1, BRCA2, PTEN, TP53, CDH1, STK11, NF1, NBN, MLH1, MSH2, MSH6, PMS2, ATM, BRIP1, CHEK2, PALB2, RAD50, BARD1, RAD51C, and RAD51D. Polymerase chain reaction (PCR)-enriched amplicon-sequencing on an NGS platform was used to sequence genomic DNA extracted from whole blood or frozen buffy coat samples using the Gentra Puregene Blood kit (Qiagen, Minneapolis, MN, USA). The DNA samples were first PCR amplified using the Qiagen GeneRead DNAseq custom panel primer sets for the 20 genes, covering all exons as well as at least 10-base exon-flanking regions. The Qiagen primer set included 1184 amplicons and provided at least 90% coverage for most genes except for STK11 (59%), PMS2 (74%) and MSH2 (89%). PCR enriched amplicons were end-repaired, adenylated, and ligated to NEXTflex-96 DNA barcodes (Bioo Scientific, Austin, Texas, USA) using the Qiagen GeneRead DNA Library I Core Kit. Barcoded libraries were amplified using the Qiagen GeneRead DNA I Amp Kit and NEXTflex primers (Bioo Scientific). Quality control and quantification of libraries were performed using the Qubit dsDNA HS Assay kit and the Agilent DNA 1000 kit. The barcoded DNA libraries were pooled in equal amounts and underwent 2x150bp paired-end sequencing on an Illumina MiSeq platform. The average base call error rates were less than 1.0%.
We constructed a pipeline based on public domain software and databases for alignment, variant calling, and annotation, using GRCh37 as the reference genome. BWA (http://bio-bwa.sourceforge.net/) was used to map reads to the reference genome. Bam-readcount (https://github.com/genome/bam-readcount) was used to count variants for each aligned position. Variant calling protocols were carried out either based on GATK Best Practices (https://software.broadinstitute.org/gatk/best-practices/) or using a non-GATK based algorithm, where lower limits of 50 for read depth and 10% for proportion of raw reads with a variant were used for variant calling. Variants that were intergenic, intronic (except for the 10 bp exon-flanking regions), or synonymous (sense) were excluded. All other variants identified with the two algorithms were compared, and discrepant variants were manually inspected by viewing the BAM reads using the Integrative Genomics Viewer (IGV, Broad Institute, Inc.) to decide on the validity of the variant. The variants were searched in the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) and the ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/). Variants were annotated as pathogenic, uncertain significance, or benign, using variant-dependent methods and disease-dependent methods. Nonsense, frameshift, and splice-site mutations that result in a truncated protein product were classified as pathogenic. The clinical significance interpretation on ClinVar, if available, was referenced for categorization. Novel missense mutations not found in the public databases were classified as variants of uncertain significance. We used various in silico models (Align-GVGD [44], PolyPhen-2 [45], SIFT [46], PROVEAN [47], CADD [48]) to evaluate the deleteriousness of the variants, especially missense variants. However, we did not change classification based on the in silico models. As the interpretation of missense variants is often controversial [7], we took a more conservative approach of only including the protein-truncating variants in the clinical correlation of this study. All variants classified as pathogenic were further verified using the Sanger sequencing method, confirming they were germline mutations.
Detection of large genomic rearrangement using copy number variation (CNV) analyses
Coverage or read depth has been used to detect CNVs in genome-scale (whole genome sequencing) datasets. Multiplex PCR-based enrichment focuses sequencing efforts on a very small fraction of the genome, and the observed read depth for each of the regions can differ due to varying number of PCR amplicons, sequence variation, or PCR enrichment efficiency. For CNV detection in our PCR-enriched amplicon sequencing data of the 20 genes, we used two algorithms, Quandico [49] and ONCOCNV [50], specifically developed for CNV analysis of amplicon sequencing data. The CNVs detected with these algorithms were then verified experimentally using the multiplex ligation-dependent probe amplification (MLPA) technique. This analysis resulted in the discovery of two carriers of a BRCA1 large genomic rearrangement (LGR) in the study cohort.
Clinical correlation and statistical analyses
For study participants who have had breast cancer, tumor characteristics and clinical outcomes were extracted from the institutional breast cancer database and participant survey. Correlation statistics between clinical characteristics and BRCA mutation or non-BRCA mutation status were performed using the Chi-square test or t test.
For the correlation analyses of clinical outcomes and germline mutations, we performed survival analyses using Kaplan-Meier curves and Cox proportional hazards regression analysis. The primary end point was disease-free survival, defined as the time from breast cancer surgery to the first appearance of one of the following: invasive recurrence of breast cancer (local, regional, or distant) or death without breast-cancer recurrence. Secondary end points included the following: time interval without breast cancer recurrence, defined as the time from breast cancer surgery to the recurrence of invasive breast cancer (local, regional, or distant); time interval before a recurrence of breast cancer at a distant site, defined as the time from breast cancer surgery to the recurrence of breast cancer at a distant site; and overall survival, defined as the time from breast cancer surgery to death from any cause. For patients who did not have an end-point event, the times were censored at the date of the last follow-up visit (or for the analysis of overall survival, the date at which the patient was last known to be alive).
The above primary and secondary end points of the groups with different mutation status were compared using Kaplan-Meier curves, and the statistical significance was evaluated using the log-rank test. Cox proportional hazards regression analysis was used to evaluate univariate and multivariate hazard ratios (HR) for BRCA germline mutation for the end points. The covariates for the multivariate analyses included: tumor size > 2 cm, lymph node positivity, triple negative tumor type, young age of onset (≤ 40), mastectomy (vs breast-conserving surgery), adjuvant chemotherapy, adjuvant radiotherapy, and hormonal therapy.
The six eligibility criteria for study enrollment were considered clinical risk factors for having a germline mutation in the cancer susceptibility genes sequenced. The odds ratios (OR) of having a pathogenic germline mutation were calculated using multivariable logistic regression with the six dichotomous risk factors as independent variables, and having a pathogenic germline mutation as the dependent variable. The odds ratios for the number of risk factors were obtained by logistic regression. Statistical significance was represented as 95% confidence intervals and P-values. An alpha level of 0.05 was defined as statistically significant for rejecting the null hypothesis. All analyses were performed using SAS 9.4® (SAS Institute, Cory, NC, USA).
Results
Study population
All 480 individuals in the study cohort were ethnically Chinese. In the cohort, 95.4% (458 individuals) had a personal history of breast cancer. The mean age of onset for breast cancer was 41.8 (range 17–82). The proportions (numbers) of individuals with each risk factor were: family history 47.5% (228), age of onset ≤40 54.2% (260), bilateral breast cancer 11.3% (54), triple-negative breast cancer 26.5% (127), breast and ovarian cancer in the same woman 0.8% (4), and male breast cancer 1.3% (6).
Characteristics of the germline mutations
To survey the breast cancer susceptibility gene mutations, we sequenced the panel of 20 genes from all individuals in the study cohort. We identified 47 pathogenic mutations carried in 65 individuals - a detection rate of 13.5% in 11 genes and 8.3% (40) in BRCA genes in this high risk population. Pathogenic mutations in BRCA2 were the most prevalent, comprising 52.3% (34) of the 65 pathogenic mutation carriers (Fig. 1); individuals with BRCA1 mutations were only 9.2% (6). PALB2 was the second most common gene to have pathogenic mutations with 13.8% (9) carrier rate. Non-BRCA gene mutations contributed to 38.5% (25) - a significant portion of the pathogenic mutation carriers.
Fig. 1.
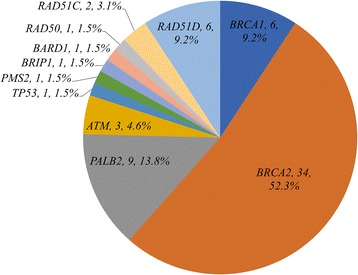
Distribution of the 20 breast cancer susceptibility genes. Genes not shown (PTEN, CDH1, STK11, NF1, NBN, MLH1, MSH2, MSH6, CHEK2) are those without identified pathogenic mutations in the study cohort
A substantial portion of the pathogenic mutations are novel. Table 1 summarizes the 47 identified pathogenic mutations, which contain 15 nonsense, 28 frameshift, 3 splice-site variants, and 1 large genomic rearrangement (multiple exon deletion). Seventeen (36.2%) of the pathogenic mutations have not been recorded in the dbSNP database, designated as novel variants. We also identified 173 variants of uncertain significance (VUS), of which 45 (26.0%) were novel; 183 (38.1%) in the study cohort carried at least one VUS.
Table 1.
Pathogenic germline mutations detected in the 20 breast cancer susceptibility genes in a high risk population for hereditary breast cancer in Taiwan (N = 480)
| Gene | HGVS notation | Type of variant | No. of carriers | SNP ID |
|---|---|---|---|---|
| BRCA1 | NM_007294.3:c.5075-1G > A | Splice site | 1 | rs1800747 |
| BRCA1 | NM_007294.3:c.4678_4679delGG (p.Gly1560Asnfs) | Frameshift | 2 | Novel |
| BRCA1 | NM_007294.3:c.3644_3648delACTTA (p.Asn1215Ilefs) | Frameshift | 1 | Novel |
| BRCA1 | deletion of exon 1 to 16 | LGR | 2 | Novel |
| BRCA2 | NM_000059.3:c.-7_9del16 | Frameshift | 1 | Novel |
| BRCA2 | NM_000059.3:c.469_470delAA (p.Lys157Valfs) | Frameshift | 2 | rs397507739 |
| BRCA2 | NM_000059.3:c.755_758delACAG (p.Asp252Valfs) | Frameshift | 1 | rs80359659 |
| BRCA2 | NM_000059.3:c.799dupG (p.Thr269Asnfs) | Frameshift | 1 | Novel |
| BRCA2 | NM_000059.3:c.857C > G (p.Ser286Ter) | Nonsense | 1 | Novel |
| BRCA2 | NM_000059.3:c.2095C > T (p.Gln699Ter) | Nonsense | 1 | rs878853559 |
| BRCA2 | NM_000059.3:c.2442delC (p.Met815Trpfs) | Frameshift | 1 | rs397507627 |
| BRCA2 | NM_000059.3:c.2754delC (p.Asn918Lysfs) | Frameshift | 1 | Novel |
| BRCA2 | NM_000059.3:c.2808_2811delACAA (p.Ala938Profs) | Frameshift | 1 | rs80359351 |
| BRCA2 | NM_000059.3:c.2990 T > G (p.Leu997Ter) | Nonsense | 1 | Novel |
| BRCA2 | NM_000059.3:c.3109C > T (p.Gln1037Ter) | Nonsense | 3 | rs80358557 |
| BRCA2 | NM_000059.3:c.3322A > T (p.Lys1108Ter) | Nonsense | 1 | Novel |
| BRCA2 | NM_000059.3:c.3883C > T (p.Gln1295Ter) | Nonsense | 1 | rs879255309 |
| BRCA2 | NM_000059.3:c.4914dupA (p.Val1639Serfs) | Frameshift | 1 | rs786203494 |
| BRCA2 | NM_000059.3:c.5141_5144delATTT (p.Tyr1714Cysfs) | Frameshift | 1 | rs80359487 |
| BRCA2 | NM_000059.3:c.5164_5165delAG (p.Ser1722Tyrfs) | Frameshift | 6 | rs80359490 |
| BRCA2 | NM_000059.3:c.5621_5624delTTAA (p.Ile1874Argfs) | Frameshift | 1 | rs80359526 |
| BRCA2 | NM_000059.3:c.6275_6276delTT (p.Leu2092Profs) | Frameshift | 1 | rs11571658 |
| BRCA2 | NM_000059.3:c.6490C > T (p.Gln2164Ter) | Nonsense | 1 | rs397507860 |
| BRCA2 | NM_000059.3:c.6800C > A (p.Ser2267Ter) | Nonsense | 1 | rs377698594 |
| BRCA2 | NM_000059.3:c.8203delC (p.Leu2736Serfs) | Frameshift | 1 | Novel |
| BRCA2 | NM_000059.3:c.8234dupT (p.Thr2746Aspfs) | Frameshift | 1 | rs276174903 |
| BRCA2 | NM_000059.3:c.8400_8402delTTTinsAAAA (p.Phe2801Lysfs) | Frameshift | 1 | rs483353077 |
| BRCA2 | NM_000059.3:c.8485C > T (p.Gln2829Ter) | Nonsense | 1 | rs80359099 |
| BRCA2 | NM_000059.3:c.8961_8964delGAGT (p.Ser2988Phefs) | Frameshift | 1 | rs80359734 |
| BRCA2 | NM_000059.3:c.9227delG (p.Gly3076Aspfs) | Frameshift | 1 | rs397508040 |
| PALB2 | NM_024675.3:c.3143delA (p.Lys1048Argfs) | Frameshift | 1 | Novel |
| PALB2 | NM_024675.3:c.2968G > T (p.Glu990Ter) | Nonsense | 1 | rs876659036 |
| PALB2 | NM_024675.3:c.2480_2481delCA (p.Thr827Metfs) | Frameshift | 1 | Novel |
| PALB2 | NM_024675.3:c.2257C > T (p.Arg753Ter) | Nonsense | 1 | rs180177110 |
| PALB2 | NM_024675.3:c.1059delA (p.Lys353Asnfs) | Frameshift | 1 | rs730881872 |
| PALB2 | NM_024675.3:c.1050_1051delAAinsTCT (p.Gln350Hisfs) | Frameshift | 2 | rs180177098 |
| PALB2 | NM_024675.3:c.643G > T (p.Glu215Ter) | Nonsense | 2 | Novel |
| ATM | NM_000051.3:c.2284_2285delCT (p.Leu762Valfs) | Frameshift | 2 | rs587781658 |
| ATM | NM_000051.3:c.6312G > A (p.Trp2104Ter) | Nonsense | 1 | Novel |
| TP53 | NM_000546.5:c.416_420dupAGACC (p.Cys141Argfs) | Frameshift | 1 | Novel |
| PMS2 | NM_000535.5:c.1144 + 1G > A | Splice site | 1 | rs373885654 |
| BRIP1 | NM_032043.2:c.2244C > G (p.Tyr748Ter) | Nonsense | 1 | Novel |
| BARD1 | NM_000465.3:c.654G > A (p.Trp218Ter) | Nonsense | 1 | Novel |
| RAD50 | NM_005732.3:c.2157dupA (p.Glu723Glyfs) | Frameshift | 1 | rs397507178 |
| RAD51C | NM_058216.2:c.394dupA (p.Thr132Asnfs) | Frameshift | 1 | rs730881940 |
| RAD51C | NM_058216.2:c.905-2A > C | Splice site | 1 | rs779582317 |
| RAD51D | NM_002878.3:c.270_271dupTA (p.Lys91Ilefs) | Frameshift | 6 | rs753862052 |
LGR: large genomic rearrangement
Correlation of mutations with tumor characteristics
To characterize the clinical feature upon cancer diagnosis in the germline mutation carriers, we compared clinical characteristics among 3 subgroups of the cohort with different mutation status. Among the 457 patients with breast cancer characteristics and clinical outcomes available, the tumor characteristics of those with BRCA mutation or with non-BRCA mutation are compared with those without any detected pathogenic germline mutation in Table 2A. None of the patients had prior knowledge of cancer susceptibility gene mutation status at the time of their breast cancer diagnosis or treatment. Those carrying BRCA mutations were more likely to have lymph node involvement upon initial breast cancer diagnosis when compared with those without mutations (66.7% vs 42.6%; P = 0.011). Probably as a result, more BRCA mutation carriers received chemotherapy (100% vs 82%; P = 0.005) and had mastectomy as opposed to breast conserving surgery (80.6% vs 62.1%; P = 0.028). Other cancer characteristics, including age of onset, tumor size, overall stage, estrogen receptor, HER2 overexpression, nuclear grade, lymphovascular invasion, receipt of radiotherapy or hormonal therapy, did not show statistically significant difference in the BRCA mutation carriers, as compared with those without mutations. In the non-BRCA mutation carrying breast cancer patients, all tumor characteristics were statistically indistinguishable from those of the breast cancer patients without germline mutations.
Table 2.
Clinical characteristics (A) and outcomes (B) in the cohort of breast cancer patients (N = 457) and their correlation with BRCA mutation and non-BRCA mutation carrying status
| No mutation | BRCA mutation | Non-BRCA mutation | ||||
|---|---|---|---|---|---|---|
| N = 397 | N = 36 | P-value* | N = 24 | P-value* | ||
| A. Clinical characteristics | ||||||
| Age of onset, mean (SD) | 41.7 (9.9) | 42.1 (10.1) | 0.846 | 42.3 (11.4) | 0.771 | |
| no. (%) | no. (%) | no. (%) | ||||
| Lymph node | positive | 160 (42.6) | 20 (66.7) | 0.011 | 8 (36.4) | 0.568 |
| negative | 216 (57.5) | 10 (33.3) | 14 (63.6) | |||
| Tumor size | ≤ 2 cm | 194 (52.3) | 16(55.2) | 0.765 | 13 (59.1) | 0.535 |
| > 2 cm | 177 (47.7) | 13 (44.8) | 9 (40.9) | |||
| Surgery type | MRM | 246 (62.1) | 29 (80.6) | 0.028 | 14 (58.3) | 0.711 |
| BCT | 150 (37.9) | 7 (19.4) | 10 (41.7) | |||
| Chemotherapy | yes | 323 (82.0) | 36 (100) | 0.005 | 17 (70.8) | 0.174 |
| no | 71 (18.0) | 0 (0) | 7 (29.2) | |||
| Radiotherapy | yes | 275 (69.8) | 29 (82.9) | 0.103 | 16 (66.7) | 0.746 |
| no | 119 (30.2) | 6 (17.1) | 8 (33.3) | |||
| Hormonal therapy | yes | 217 (57.7) | 21 (70) | 0.189 | 12 (54.5) | 0.770 |
| no | 159 (42.3) | 9 (30) | 10 (45.5) | |||
| Stage | 0 (DCIS) | 11 (2.8) | 0 (0) | 0.307 | 1 (4.2) | 0.704 |
| 1 | 138 (35.0) | 8 (23.5) | 11 (45.8) | |||
| 2 | 159 (40.4) | 15 (44.1) | 10 (41.7) | |||
| 3 | 48 (12.2) | 8 (23.5) | 1 (4.2) | |||
| 4 | 7 (1.8) | 0 (0) | 0 (0) | |||
| LABC | 31 (7.9) | 3 (8.8) | 1 (4.2) | |||
| Triple negative | yes | 108 (27.2) | 9 (25.0) | 0.776 | 9 (37.5) | 0.274 |
| no | 289 (72.8) | 27 (75.0) | 15 (62.5) | |||
| ER | positive | 234 (59.5) | 24 (66.7) | 0.403 | 12 (52.2) | 0.485 |
| negative | 159 (40.5) | 12 (33.3) | 11 (47.8) | |||
| HER2 overexpression | yes | 72 (19.8) | 3 (8.8) | 0.118 | 3 (14.3) | 0.537 |
| no | 292 (80.2) | 31 (91.2) | 18 (85.7) | |||
| Nuclear grade | 1 | 51 (13.9) | 1 (3.5) | 0.276 | 3 (14.3) | 0.896 |
| 2 | 123 (33.5) | 11 (37.9) | 8 (38.1) | |||
| 3 | 193 (52.6) | 17 (58.6) | 10 (47.6) | |||
| Lymphovascular invasion | prominent | 82 (22.1) | 11 (37.9) | 0.122 | 2 (9.5) | 0.259 |
| focal | 104 (28.0) | 8 (27.6) | 5 (23.8) | |||
| absent | 185 (49.9) | 10 (34.5) | 14 (66.7) | |||
| B. Outcomes | ||||||
| Recurrence† | 53 (13.4) | 11 (30.6) | 0.005 | 1 (4.2) | 0.191 | |
| Distant metastasis | 38 (9.6) | 9 (25.0) | 0.004 | 0 (0) | 0.112 | |
| Locoregional recurrence only | 15 (3.8) | 2 (5.6) | 0.599 | 1 (4.2) | 0.923 | |
| Death | 12 (3.0) | 3 (8.3) | 0.095 | 0 (0) | 0.388 | |
SD: standard deviation; MRM: modified radical mastectomy; BCT: breast conserving therapy; DCIS: ductal carcinoma in situ; LABC: locally advanced breast cancer; ER: estrogen receptor; HER2: human epidermal growth factor receptor 2. * P-values were calculated using Chi-square test for categorical variables and t-test for continuous variables (age). The statistically significant values (< 0.05) are shown in bold. † Recurrence includes distant metastasis, local/ipsilateral breast and regional recurrence, and does not include contralateral breast cancer or second primary cancer
Clinical outcomes
To evaluate the prognostic value of BRCA germline mutation in breast cancer patients, we performed survival analyses comparing carriers and non-carriers of BRCA germline mutation for various clinical end points (Fig. 2). After a median follow-up of 66.9 months, 66 patients (14.4%) had breast cancer recurrence or died. The rate of disease-free survival at 5 years was 73.3% among BRCA mutation carriers, as compared with 91.1% among non-carriers (hazard ratio for recurrence or death 2.42, 95% CI 1.29–4.53; P = 0.013) (Fig. 2a). At 5 years, 73.3% of the BRCA mutation carriers were free from any breast cancer recurrence, as compared with 91.0% of the non-carriers (hazard ratio for recurrence 2.22, 95% CI 1.20–4.47; P = 0.016) (Fig. 2b). Distant recurrence of breast cancer was reported in 47 patients (10.3%), and at 5 years, 79.7% of the BRCA mutation carriers were free from distant metastasis, as compared with 94.2% of the non-carriers (hazard ratio for distant recurrence 2.58, 95% CI 1.24–5.94; P = 0.011) (Fig. 2c). Death was reported in 15 patients (3.3%); 1 patient died without breast cancer recurrence. Overall survival at 5 years was 96.4% among BRCA mutation carriers, as compared with 100% among non-carriers (hazard ratio for death 1.84, 95% CI 0.52–6.54; P = 0.35) (Fig. 2d). Survival analyses comparing BRCA1 and BRCA2 separately showed that BRCA1 mutation carriers appeared to have the worst outcomes in the cohort, followed by BRCA2 mutation carriers (Additional file 1: Fig. S1). The BRCA mutation carriers had significantly poorer breast cancer specific outcomes, most significantly in distant recurrence.
Fig. 2.
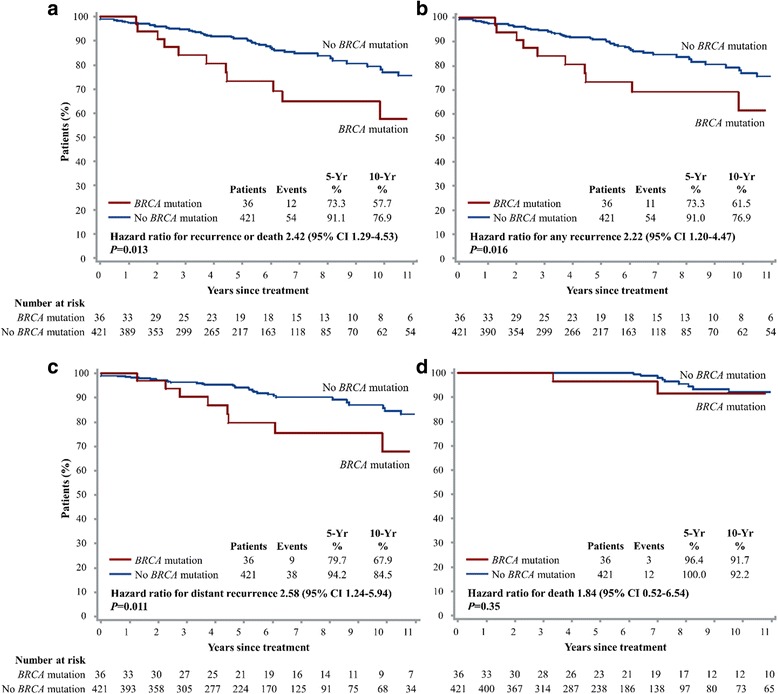
Kaplan-Meier estimates of disease-free survival (a), freedom from breast cancer recurrence (b), freedom from distant recurrence of breast cancer (c), and overall survival (d), according to BRCA mutation carrier status. The 5-year and 10-year values are based on Kaplan–Meier estimates of the time to an event. The hazard ratios are for breast cancer recurrence or death (a), any recurrence (b), distant recurrence (c), and death from any cause (d), respectively, based on Cox proportional hazards univariate analysis
In the multivariate Cox proportional hazards model, BRCA mutation remained an independent poor prognostic factor for all cancer outcomes after adjustment for covariates including tumor size, lymph node status, triple negative tumor type, age, and treatment modalities (Table 3). The adjusted hazard ratios for BRCA mutation carriers were 3.04 (95% CI 1.40–6.58; P = 0.005) for recurrence or death, 2.70 (95% CI 1.20–6.06; P = 0.016) for any recurrence, 2.86 (95% CI 1.11–7.35; P = 0.029) for distant recurrence, and 8.01 (95% CI 1.44–44.7; P = 0.018) for any death. Among the clinical covariates, multivariate analysis showed poorer prognosis in large tumors, and favorable prognosis for those having had chemotherapy or with triple negative tumor types (Additional file 2: Table S2).
Table 3.
Effects of BRCA germline mutation on breast cancer end points by multivariate Cox proportional hazards analysis
| End Point | Adjusted Hazard Ratio* (95% CI) | P-value |
|---|---|---|
| Breast cancer recurrence or death | 3.04 (1.40–6.58) | 0.005 |
| Locoregional or distant recurrence | 2.70 (1.20–6.06) | 0.016 |
| Distant recurrence | 2.86 (1.11–7.35) | 0.029 |
| Death | 8.01 (1.44–44.7) | 0.018 |
*The hazard ratios were calculated using multivariate Cox proportional hazards model adjusted for tumor size, lymph node status, triple negative status, age of breast cancer onset, surgery type, chemotherapy, radiotherapy, and hormonal therapy
To evaluate the prognostic consequences of the non-BRCA gene mutations, we compared the clinical outcomes between breast cancer patients with BRCA mutation, with non-BRCA mutation, and without mutation (Table 2B). The BRCA mutation carriers had a higher rate of any recurrence (30.6% vs 13.4%; P = 0.005) or distant recurrence (25.0% vs 9.6%; P = 0.004) than those with no mutation. In contrast, the non-BRCA mutation group had no distant metastasis or death and only one locoregional recurrence. No statistically significant difference in outcome was detected in this group as compared with those with no mutation. PALB2 mutation was the most prevalent in the non-BRCA mutation group, and survival analyses comparing PALB2 to BRCA1, BRCA2 mutation carriers and to the remaining patients showed that PALB2 mutation carriers had favorable prognosis (Additional file 1: Figure S1).
Significance of risk factors for breast cancer susceptibility gene mutation
Correlations between the hereditary breast cancer risk factors and having a detectable germline mutation in any of the 20 genes were shown in Table 4. Multivariate analysis showed that the only significant risk factor in this high risk cohort was having bilateral breast cancer (synchronous or metachronous), with an adjusted odds ratio of 3.27 (95% CI 1.64–6.51; P = 0.0008). Family history, age of onset for breast cancer, and triple-negative breast cancer did not show significant correlation with the presence of mutation. The number of individuals with both breast and ovarian cancer or with male breast cancer was too small to detect any statistical significance.
Table 4.
Correlation between clinical risk factors of hereditary breast cancer and having pathogenic germline mutations in the 20 breast cancer susceptibility genes
| Risk factors | Mutation rate | OR (95% CI)* | P-value | |
|---|---|---|---|---|
| Family history† | Yes | 16.2% (37/228) | 1.60 (0.90–2.88) | 0.113 |
| No | 11.1% (28/252) | |||
| Age of onset | ≤ 40 | 12.7% (33/260) | 1.26 (0.69–2.31) | 0.452 |
| > 40 | 13.6% (27/198) | |||
| Triple negative breast cancer | Yes | 14.2% (18/127) | 1.27 (0.67–2.42) | 0.469 |
| No | 13.3% (47/353) | |||
| Bilateral breast cancer | Yes | 29.6% (16/54) | 3.27 (1.64–6.51) | 0.0008 |
| No | 11.5% (49/426) | |||
| Breast and ovarian cancer | Yes | 50% (2/4) | 5.90 (0.70–49.6) | 0.103 |
| No | 13.0% (63/476) | |||
| Male breast cancer | Yes | 16.7% (1/6) | 2.35 (0.25–21.9) | 0.454 |
| No | 13.1% (64/474) | |||
| No. of risk factors | 1 | 10.4% (32/309) | 1.82 (1.25–2.64) | 0.002 |
| 2 | 18.4% (27/147) | |||
| 3 | 19.1% (4/21) | |||
| 4 | 50% (1/2) | |||
| 5 | 100% (1/1) | |||
| All subjects | 13.5% (65/480) | |||
* The adjusted odds ratios (OR) of having a pathogenic mutation in the 20 genes were calculated using multivariable logistic regression with the six dichotomous risk factors; the odds ratio for the no. of risk factors was obtained using univariate logistic regression. The statistically significant P-values (< 0.05) are shown in bold. † The presence of family history was defined as two or more persons on the same lineage of the family having breast or ovarian cancer. If the study participant had breast cancer, only one family member with breast/ovarian cancer was needed to qualify as having family history
Having higher number of risk factors in an individual was also significantly correlated with having a germline mutation. For each additional risk factor, the odds ratio of having a mutation was 1.82 (95% CI 1.25–2.64; P = 0.002). Having more than one risk factor was associated with an odds ratio of 2.07 (95% CI 1.22–3.51; P = 0.007) of detecting a mutation.
To focus on the impact of high penetrance breast cancer susceptibility genes, we repeated the correlation analysis for four high penetrance genes (BRCA1, BRCA2, PALB2, TP53), and for the two most prominent breast cancer susceptibility genes (BRCA1, BRCA2) (Additional file 3: Table S3). The results shown in Additional file 3: Table S3 did not lead to conclusions disagreeing with those based on the 20-gene analysis shown in Table 4, maintaining similar statistical correlations of having germline mutations with bilateral breast cancer and with more clinical risk factors.
Discussion
We showed that BRCA germline mutation carriers in this large ethnic Chinese cohort were more likely to be diagnosed with breast cancer already spread to regional lymph nodes, and their breast cancer related outcomes were significantly worse. The 5-year disease-free survival rate was only 73.3% for BRCA mutation carriers, in contrast to 91.1% for non-mutation carriers. The BRCA mutation status was an independent prognostic factor with an adjusted hazard ratio of 3.04 (95% CI 1.40–6.58) for cancer recurrence or death. The poor clinical outcome in BRCA mutation carriers mainly resulted from recurrence as distant metastasis, therefore excluding the contribution by new primary cancer in the ipsilateral or contralateral breast, of which the risk had been known to be elevated in BRCA mutation carriers. Our result implied the more aggressive nature of breast tumors in BRCA germline mutation carriers. Most previous studies on clinical outcomes of BRCA mutation carriers have failed to show a significant prognostic effect by BRCA mutation [9–11, 13, 14]. However, a recent systematic review showed that both BRCA1 and BRCA2 mutation carriers had significantly worse breast cancer specific survival [15]. The discrepancy in these studies may in part result from limitations due to small sample sizes, lack of adjusting for disease characteristics, variations in mutation assay techniques, mutation types, cancer treatment modalities, or lengths of follow-up. Our study was conducted in an all-Chinese cohort where all study participants underwent the same NGS-based complete sequencing of the coding regions of BRCA genes among other genes. The majority of the mutations were in the BRCA2 gene. The follow-ups were extensive in terms of length, with median duration over 5 years, and completeness. The tumor characteristics and outcome data had been collected in a prospective manner in a breast cancer registry as well as after the participants were enrolled. The majority of the breast cancer patients in the cohort (94%) had undergone treatment in a single cancer center where cancer characteristic-based treatment guideline was consistently adhered to. The homogeneity on data collection may have strengthened the validity of the prognostic analysis.
Studies have shown that tumor cells with BRCA mutations may show different response to different chemotherapy agents; they may have enhanced sensitivity to platinum while more resistant to taxanes. However, clinical studies on comparison of chemotherapy regimens in the BRCA mutation populations are limited [12]. For the breast cancer patients in our cohort, less than 10% received cisplatin in the neoadjuvant setting and none in the adjuvant setting, while about a third received a taxane (docetaxel) in the adjuvant setting. There were no significant differences in chemotherapy choice between the groups with and without BRCA mutation. Despite a higher rate of BRCA mutation carriers receiving chemotherapy, they had poorer cancer outcomes. Further prospective studies are needed to determine the optimal chemotherapy for BRCA mutation carriers. With the anticipated efficacy of incorporation of PARP inhibitors in the treatment of BRCA mutation associated breast tumors, knowledge of BRCA mutation status prior to initial cancer treatment becomes even more crucial.
In this Chinese cohort of high risk individuals for hereditary breast cancer, we found an overall prevalence of 13.5% for carriers of germline mutations in 11 of the 20 breast cancer susceptibility genes. In contrast to western populations, BRCA2 mutations (52.3%) were much more common than BRCA1 mutations (9.2%) in our cohort, similar to findings in other studies in the Asian population [34]. Non-BRCA genes contributed to 38.5% of the mutation carriers, with PALB2 (13.8%), RAD51D (9.2%), and ATM (4.6%) being the majority. PALB2 is particularly important since lifetime risk for breast cancer can reach 58% in those with family history [51], and NCCN guideline recommends consideration of risk-reducing mastectomy [52]. Among the 8 cases with RAD51C and RAD51D mutations, 6 (75%) were triple negative breast cancer, in agreement with the recent studies suggesting that mutations in these two genes may confer higher risks of triple-negative or basal subtypes of breast cancer [23, 24]. We also found 2 individuals with protein-truncating mutations in TP53 and PMS2 genes, which are high-penetrance cancer predisposing genes and would result in significantly high risk for other cancers. These results showed that testing more than BRCA1 and BRCA2 increased the detection rate of clinically actionable high and moderate risk gene mutations, therefore may be an important strategy in the Chinese population. In a study by Thompson et al., significant excess of mutations was only observed for PALB2 and TP53 in familial breast cancer cases compared to cancer-free controls [6]. We similarly only found a small number of genes contributing to the majority of mutation carriers. To overcome the challenge of high rates of VUS and questionable clinical actionability, we recommend limiting cancer susceptibility multigene panel in clinical testing to include only a handful of genes with high clinical impact.
Among the six risk factors for hereditary breast cancer in this cohort of all high risk individuals, only bilateral breast cancer showed a statistically significant odds ratio of 3.27 for having a germline mutation in multivariate analysis. In addition, having more risk factors was also associated with a high detection rate of mutations (OR 2.07 for having more than one risk factor). These results suggest that these known risk factors were helpful in identifying individuals for genetic testing and we may need to pay particular attention to those with bilateral breast cancer, even in the absence of family history or young age of onset. Larger cohorts are needed to clarify the significance of ovarian cancer and male breast cancer on breast cancer susceptibility gene mutations in the Asian population. Results from the correlation analysis done with the few high penetrance genes were similar to those done with all the studied genes, suggesting that the correlations were driven by these high penetrance genes including BRCA1, BRCA2, PALB2, and TP53, which was expected since they represented the majority of the pathogenic variants.
There were some limitations in our study. First, we did not conduct experiments to detect large genome rearrangement (LGR) in all study participants, but used bioinformatics analytical tools to detect copy number variations on the NGS data. This could underestimate the prevalence of LGR in this cohort. However, LGRs have not been shown to contribute significantly to germline mutations in BRCA genes in East Asian populations [53, 54]. Second, we were conservative in classifying variants as pathogenic and limited those to protein-truncating variants, which were without ambiguity in assignment of pathogenicity. There were two missense variants classified as likely pathogenic in the ClinVar database, and many missense variants deemed damaging by multiple in silico models. However, we did not assign those as pathogenic mutations in this study. We could therefore have underestimated the prevalence of pathogenic mutations. Further studies are underway to evaluate variant segregation with cancer in families, and the accuracy of in silico models.
Conclusions
In this high risk ethnic Chinese cohort, 13.5% had a germline pathogenic mutation in one of twenty breast cancer susceptibility genes, and 8.3% had a BRCA1or BRCA2 mutation. BRCA mutation carriers, when diagnosed with breast cancer, were more likely to have lymph node involvement. Their breast cancer specific outcomes were significantly worse even after adjusting for clinical prognostic factors, suggesting BRCA mutation to be an independent factor for poor prognosis. Our results highlighted the importance of early testing for breast cancer susceptibility genes, not only for prevention and earlier diagnosis of breast cancer, but also for optimal treatment and surveillance strategies after breast cancer is diagnosed. Further studies are needed to evaluate different treatment approaches for breast cancer in BRCA mutation carriers to improve outcome.
Additional files
Figure S1. Kaplan-Meier estimates of disease-free survival (A), freedom from breast cancer recurrence (B), freedom from distant recurrence of breast cancer (C), and overall survival (D), according to BRCA1, BRCA2, PALB2 mutation carrier status and all others in the breast cancer cohort. The 5-year and 10-year values are based on Kaplan–Meier estimates of the time to an event. P-values are calculated using the log-rank test. (TIFF 520 kb)
Table S2. Effects of BRCA germline mutation and other prognostic factors (all covariates) on clinical end points by Cox proportional hazards analysis. The four clinical end points are: breast cancer (BC) recurrence or death, BC recurrence (locoregional or distant), distant recurrence, and death from any cause. Both univariate and multivariate analyses are shown. Statistically significant P-values (< 0.05) in the multivariate analyses are shown in bold. (XLSX 14 kb)
Table S3. Correlation between clinical risk factors of hereditary breast cancer and having pathogenic germline mutations in different gene combinations: A. Correlation with all 20 genes; B. Correlation with BRCA1, BRCA2, PALB2, TP53 only; C. Correlation with BRCA1, BRCA2 only. (XLSX 14 kb)
Table S1. Carriers of pathogenic mutations with their respective clinical risk factors for hereditary breast cancer, and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) accession numbers for novel pathogenic variants. (XLSX 15 kb)
Table S4. Clinical Dataset. Spreadsheet with code-book, containing pathogenic germline gene mutation, hereditary breast cancer risk factors, clinical characteristics, and outcomes in the cohort of breast cancer patients (N = 457). (XLSX 65 kb)
Acknowledgements
The authors wish to thank the National Center for Genome Medicine at Academia Sinica (Taipei, Taiwan) for their technical and bioinformatics support in the next generation sequencing experiments and data analyses. We would also like to thank Ms. Ling-ling Hsieh for coordinating the study and data collection, Dr. Fei-Hung Hung for helpful review of the literature, and Ms. Pei-Hsuan Chen for assistance in processing the sequencing data.
Funding
This work was supported by Ministry of Science and Technology, Taiwan [MOST104–3114-Y-368-002 to SHCC, MOST106–0210–01-15-02 to ASY, MOST106–0210–01-15-04 to ASY]; and the health and welfare surcharge of tobacco products in Taiwan (Ministry of Health and Welfare) [MOHW106-TDU-B-212-134004 to SHCC]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
The genomic variants with clinical assertions identified in the current study are available in the ClinVar repository (https://www.ncbi.nlm.nih.gov/clinvar/) and can be searched using the HGVS notation or the accession number for each submitted variant. The accession numbers are listed in Additional file 4: Table S1. The clinical dataset acquired during the current study are provided in Additional file 5: Table S4.
Authors’ contributions
YAW conceptualized and designed the study, and was a major contributor in writing the manuscript. JWJ completed a significant amount of data analyses and manuscript writing. CFH acquired and did statistical analyses of the clinical data. HPP, JWJ and ASY set up the bioinformatics pipeline to identify and annotate the genomic variants. CFY coordinated and performed the sequencing experiments. SHCC and ASY acquired funding and made substantial contributions in writing the manuscript. All authors read, contributed to, and approved the final manuscript.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and the study protocol was approved by the Institutional Review Board Committee at Koo Foundation Sun Yat-Sen Cancer Center (case No. 20141222A). Written informed consent was obtained from each study participant.
Consent for publication
Not applicable.
Competing interests
None of the authors have any competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12885-018-4229-5) contains supplementary material, which is available to authorized users.
Contributor Information
Yong Alison Wang, Phone: +886-2-2897-0011 ext 1746, Email: wang@kfsyscc.org.
Jhih-Wei Jian, Email: bioinfowei@gmail.com.
Chen-Fang Hung, Email: cfhorng@kfsyscc.org.
Hung-Pin Peng, Email: peng1234@gate.sinica.edu.tw.
Chi-Fan Yang, Email: cfyang@ibms.sinica.edu.tw.
Hung-Chun Skye Cheng, Email: skye@kfsyscc.org.
An-Suei Yang, Phone: +886-2-2787-1232, Email: yangas@gate.sinica.edu.tw.
References
- 1.Stanislaw C, Xue Y, Wilcox WR. Genetic evaluation and testing for hereditary forms of cancer in the era of next-generation sequencing. Cancer biology & medicine. 2016;13:55–67. doi: 10.20892/j.issn.2095-3941.2016.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cobain EF, Milliron KJ, Merajver SD. Updates on breast cancer genetics: clinical implications of detecting syndromes of inherited increased susceptibility to breast cancer. Semin Oncol. 2016;43:528–535. doi: 10.1053/j.seminoncol.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, et al. American Society of Clinical Oncology policy statement update: genetic and genomic testing for Cancer susceptibility. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2015;33:3660–3667. doi: 10.1200/JCO.2015.63.0996. [DOI] [PubMed] [Google Scholar]
- 4.Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32:2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desmond A, Kurian AW, Gabree M, Mills MA, Anderson MJ, Kobayashi Y, et al. Clinical Actionability of multigene panel testing for hereditary breast and ovarian Cancer risk assessment. JAMA oncology. 2015;1:943–951. doi: 10.1001/jamaoncol.2015.2690. [DOI] [PubMed] [Google Scholar]
- 6.Thompson ER, Rowley SM, Li N, McInerny S, Devereux L, Wong-Brown MW, et al. Panel testing for familial breast Cancer: calibrating the tension between research and clinical care. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34:1455–1459. doi: 10.1200/JCO.2015.63.7454. [DOI] [PubMed] [Google Scholar]
- 7.Easton DF, Pharoah PDP, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-Cancer risk. N Engl J Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roviello G, Milani M, Gobbi A, Dester M, Cappelletti MR, Allevi G, et al. A phase II study of olaparib in breast cancer patients: biological evaluation from a ‘window of opportunity’ trial. Future oncology (London, England) 2016;12:2189–2193. doi: 10.2217/fon-2016-0116. [DOI] [PubMed] [Google Scholar]
- 9.Rennert G, Bisland-Naggan S, Barnett-Griness O, Bar-Joseph N, Zhang S, Rennert HS, et al. Clinical outcomes of breast cancer in carriers of BRCA1 and BRCA2 mutations. N Engl J Med. 2007;357:115–123. doi: 10.1056/NEJMoa070608. [DOI] [PubMed] [Google Scholar]
- 10.Brekelmans CT, Tilanus-Linthorst MM, Seynaeve C, vd Ouweland A, Menke-Pluymers MB, Bartels CC, et al. Tumour characteristics, survival and prognostic factors of hereditary breast cancer from BRCA2-, BRCA1- and non-BRCA1/2 families as compared to sporadic breast cancer cases. Eur J Cancer. 2007;43:867–876. doi: 10.1016/j.ejca.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 11.Robson ME, Chappuis PO, Satagopan J, Wong N, Boyd J, Goffin JR, et al. A combined analysis of outcome following breast cancer: differences in survival based on BRCA1/BRCA2 mutation status and administration of adjuvant treatment. Breast cancer research: BCR. 2004;6:R8–r17. doi: 10.1186/bcr658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith KL, Isaacs C. BRCA mutation testing in determining breast cancer therapy. Cancer journal (Sudbury, Mass) 2011;17:492–499. doi: 10.1097/PPO.0b013e318238f579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebens FP, Carly B, Pastijn A, Rozenberg S. Management of BRCA1/2 associated breast cancer: a systematic qualitative review of the state of knowledge in 2006. Eur J Cancer. 2007;43:238–257. doi: 10.1016/j.ejca.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 14.Bordeleau L, Panchal S, Goodwin P. Prognosis of BRCA-associated breast cancer: a summary of evidence. Breast Cancer Res Treat. 2009;119:13. doi: 10.1007/s10549-009-0566-z. [DOI] [PubMed] [Google Scholar]
- 15.Baretta Z, Mocellin S, Goldin E, Olopade OI, Huo D. Effect of BRCA germline mutations on breast cancer prognosis: a systematic review and meta-analysis. Medicine. 2016;95:e4975. doi: 10.1097/MD.0000000000004975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graffeo R, Livraghi L, Pagani O, Goldhirsch A, Partridge AH, Garber JE. Time to incorporate germline multigene panel testing into breast and ovarian cancer patient care. Breast Cancer Res Treat. 2016;160:393–410. doi: 10.1007/s10549-016-4003-9. [DOI] [PubMed] [Google Scholar]
- 17.Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, et al. Associations between Cancer predisposition testing panel genes and breast Cancer. JAMA oncology. 2017;3:1190–1196. doi: 10.1001/jamaoncol.2017.0424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Romero A, Perez-Segura P, Tosar A, Garcia-Saenz JA, Diaz-Rubio E, Caldes T, et al. A HRM-based screening method detects RAD51C germ-line deleterious mutations in Spanish breast and ovarian cancer families. Breast Cancer Res Treat. 2011;129:939–946. doi: 10.1007/s10549-011-1543-x. [DOI] [PubMed] [Google Scholar]
- 19.Vuorela M, Pylkas K, Hartikainen JM, Sundfeldt K, Lindblom A, von Wachenfeldt Wappling A, et al. Further evidence for the contribution of the RAD51C gene in hereditary breast and ovarian cancer susceptibility. Breast Cancer Res Treat. 2011;130:1003–1010. doi: 10.1007/s10549-011-1677-x. [DOI] [PubMed] [Google Scholar]
- 20.Thompson ER, Boyle SE, Johnson J, Ryland GL, Sawyer S, Choong DY, et al. Analysis of RAD51C germline mutations in high-risk breast and ovarian cancer families and ovarian cancer patients. Hum Mutat. 2012;33:95–99. doi: 10.1002/humu.21625. [DOI] [PubMed] [Google Scholar]
- 21.Pelttari LM, Kiiski J, Nurminen R, Kallioniemi A, Schleutker J, Gylfe A, et al. A Finnish founder mutation in RAD51D: analysis in breast, ovarian, prostate, and colorectal cancer. J Med Genet. 2012;49:429–432. doi: 10.1136/jmedgenet-2012-100852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato K, Koyasu M, Nomura S, Sato Y, Kita M, Ashihara Y, et al. Mutation status of RAD51C, PALB2 and BRIP1 in 100 Japanese familial breast cancer cases without BRCA1 and BRCA2 mutations. Cancer Sci. 2017;108:2287–2294. doi: 10.1111/cas.13350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Couch FJ, Hart SN, Sharma P, Toland AE, Wang X, Miron P, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J Clin Oncol. 2015;33:304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ollier M, Radosevic-Robin N, Kwiatkowski F, Ponelle F, Viala S, Privat M, et al. DNA repair genes implicated in triple negative familial non-BRCA1/2 breast cancer predisposition. Am J Cancer Res. 2015;5:2113–2126. [PMC free article] [PubMed] [Google Scholar]
- 25.Damiola F, Pertesi M, Oliver J, Le Calvez-Kelm F, Voegele C, Young EL, et al. Rare key functional domain missense substitutions in MRE11A, RAD50, and NBN contribute to breast cancer susceptibility: results from a breast Cancer family registry case-control mutation-screening study. Breast Cancer Res. 2014;16:R58. doi: 10.1186/bcr3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harkness EF, Barrow E, Newton K, Green K, Clancy T, Lalloo F, et al. Lynch syndrome caused by MLH1 mutations is associated with an increased risk of breast cancer: a cohort study. J Med Genet. 2015;52:553–556. doi: 10.1136/jmedgenet-2015-103216. [DOI] [PubMed] [Google Scholar]
- 27.Maresca L, Spugnesi L, Lodovichi S, Cozzani C, Naccarato AG, Tancredi M, et al. MSH2 role in BRCA1-driven tumorigenesis: a preliminary study in yeast and in human tumors from BRCA1-VUS carriers. Eur J Med Genet. 2015;58:531–539. doi: 10.1016/j.ejmg.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Goldberg M, Bell K, Aronson M, Semotiuk K, Pond G, Gallinger S, et al. Association between the lynch syndrome gene MSH2 and breast cancer susceptibility in a Canadian familial cancer registry. J Med Genet. 2017;54:742–746. doi: 10.1136/jmedgenet-2017-104542. [DOI] [PubMed] [Google Scholar]
- 29.Haricharan S, Bainbridge MN, Scheet P, Brown PH. Somatic mutation load of estrogen receptor-positive breast tumors predicts overall survival: an analysis of genome sequence data. Breast Cancer Res Treat. 2014;146:211–220. doi: 10.1007/s10549-014-2991-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Easton DF, Lesueur F, Decker B, Michailidou K, Li J, Allen J, et al. No evidence that protein truncating variants in BRIP1 are associated with breast cancer risk: implications for gene panel testing. J Med Genet. 2016;53:298–309. doi: 10.1136/jmedgenet-2015-103529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharif S, Moran A, Huson SM, Iddenden R, Shenton A, Howard E, et al. Women with neurofibromatosis 1 are at a moderately increased risk of developing breast cancer and should be considered for early screening. J Med Genet. 2007;44:481–484. doi: 10.1136/jmg.2007.049346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uusitalo E, Kallionpaa RA, Kurki S, Rantanen M, Pitkaniemi J, Kronqvist P, et al. Breast cancer in neurofibromatosis type 1: overrepresentation of unfavourable prognostic factors. Br J Cancer. 2017;116:211–217. doi: 10.1038/bjc.2016.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ripperger T, Gadzicki D, Meindl A, Schlegelberger B. Breast cancer susceptibility: current knowledge and implications for genetic counselling. European journal of human genetics : EJHG. 2009;17:722–731. doi: 10.1038/ejhg.2008.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, et al. Frequency of mutations in individuals with breast cancer referred for BRCA1 and BRCA2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015;121:25–33. doi: 10.1002/cncr.29010. [DOI] [PubMed] [Google Scholar]
- 35.Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The breast Cancer linkage consortium. Am J Hum Genet. 1998;62:676–689. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J, Pei R, Pang Z, Ouyang T, Li J, Wang T, et al. Prevalence and characterization of BRCA1 and BRCA2 germline mutations in Chinese women with familial breast cancer. Breast Cancer Res Treat. 2012;132:421–428. doi: 10.1007/s10549-011-1596-x. [DOI] [PubMed] [Google Scholar]
- 37.Cao W, Wang X, Li JC. Hereditary breast cancer in the Han Chinese population. Journal of epidemiology / Japan Epidemiological Association. 2013;23:75–84. doi: 10.2188/jea.JE20120043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao NY, Hu Z, Yu JM, Li WF, Zhang B, Su FX, et al. Evaluating the performance of models for predicting the BRCA germline mutations in Han Chinese familial breast cancer patients. Breast Cancer Res Treat. 2009;116:563–570. doi: 10.1007/s10549-008-0181-4. [DOI] [PubMed] [Google Scholar]
- 39.Kuo WH, Lin PH, Huang AC, Chien YH, Liu TP, Lu YS, et al. Multimodel assessment of BRCA1 mutations in Taiwanese (ethnic Chinese) women with early-onset, bilateral or familial breast cancer. J Hum Genet. 2012;57:130–138. doi: 10.1038/jhg.2011.142. [DOI] [PubMed] [Google Scholar]
- 40.Kim H, Choi DH. Distribution of BRCA1 and BRCA2 mutations in Asian patients with breast Cancer. J Breast Cancer. 2013;16:357–365. doi: 10.4048/jbc.2013.16.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin PH, Kuo WH, Huang AC, Lu YS, Lin CH, Kuo SH, et al. Multiple gene sequencing for risk assessment in patients with early-onset or familial breast cancer. Oncotarget. 2016;7:8310–8320. doi: 10.18632/oncotarget.7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong ESY, Shekar S, Met-Domestici M, Chan C, Sze M, Yap YS, et al. Inherited breast cancer predisposition in Asians: multigene panel testing outcomes from Singapore. Npj. Genomic Medicine. 2016;1:15003. doi: 10.1038/npjgenmed.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Bruin MA, Kwong A, Goldstein BA, Lipson JA, Ikeda DM, McPherson L, et al. Breast cancer risk factors differ between Asian and white women with BRCA1/2 mutations. Familial Cancer. 2012;11:429–439. doi: 10.1007/s10689-012-9531-9. [DOI] [PubMed] [Google Scholar]
- 44.Mathe E, Olivier M, Kato S, Ishioka C, Hainaut P, Tavtigian SV. Computational approaches for predicting the biological effect of p53 missense mutations: a comparison of three sequence analysis based methods. Nucleic Acids Res. 2006;34:1317–1325. doi: 10.1093/nar/gkj518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 47.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the functional effect of amino acid substitutions and Indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reinecke F, Satya RV, DiCarlo J. Quantitative analysis of differences in copy numbers using read depth obtained from PCR-enriched samples and controls. BMC bioinformatics. 2015;16:17. doi: 10.1186/s12859-014-0428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boeva V, Popova T, Lienard M, Toffoli S, Kamal M, Le Tourneau C, et al. Multi-factor data normalization enables the detection of copy number aberrations in amplicon sequencing data. Bioinformatics. 2014;30:3443–3450. doi: 10.1093/bioinformatics/btu436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.NCCN Clinical Practice Guidelines in Oncology - Genetic/Familial High-Risk Assessment: Breast and Ovarian Version 2.2017. Available from: https://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf.
- 53.Kwong A, Ng EKO, Law FBF, Wong HN, Wa A, Wong CLP, et al. Novel BRCA1 and BRCA2 genomic rearrangements in southern Chinese breast/ovarian cancer patients. Breast Cancer Res Treat. 2012;136:931–933. doi: 10.1007/s10549-012-2292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seong MW, Cho SI, Noh DY, Han W, Kim SW, Park CM, et al. Low contribution of BRCA1/2 genomic rearrangement to high-risk breast cancer in the Korean population. Familial Cancer. 2009;8:505–508. doi: 10.1007/s10689-009-9279-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Kaplan-Meier estimates of disease-free survival (A), freedom from breast cancer recurrence (B), freedom from distant recurrence of breast cancer (C), and overall survival (D), according to BRCA1, BRCA2, PALB2 mutation carrier status and all others in the breast cancer cohort. The 5-year and 10-year values are based on Kaplan–Meier estimates of the time to an event. P-values are calculated using the log-rank test. (TIFF 520 kb)
Table S2. Effects of BRCA germline mutation and other prognostic factors (all covariates) on clinical end points by Cox proportional hazards analysis. The four clinical end points are: breast cancer (BC) recurrence or death, BC recurrence (locoregional or distant), distant recurrence, and death from any cause. Both univariate and multivariate analyses are shown. Statistically significant P-values (< 0.05) in the multivariate analyses are shown in bold. (XLSX 14 kb)
Table S3. Correlation between clinical risk factors of hereditary breast cancer and having pathogenic germline mutations in different gene combinations: A. Correlation with all 20 genes; B. Correlation with BRCA1, BRCA2, PALB2, TP53 only; C. Correlation with BRCA1, BRCA2 only. (XLSX 14 kb)
Table S1. Carriers of pathogenic mutations with their respective clinical risk factors for hereditary breast cancer, and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) accession numbers for novel pathogenic variants. (XLSX 15 kb)
Table S4. Clinical Dataset. Spreadsheet with code-book, containing pathogenic germline gene mutation, hereditary breast cancer risk factors, clinical characteristics, and outcomes in the cohort of breast cancer patients (N = 457). (XLSX 65 kb)
Data Availability Statement
The genomic variants with clinical assertions identified in the current study are available in the ClinVar repository (https://www.ncbi.nlm.nih.gov/clinvar/) and can be searched using the HGVS notation or the accession number for each submitted variant. The accession numbers are listed in Additional file 4: Table S1. The clinical dataset acquired during the current study are provided in Additional file 5: Table S4.


