Abstract
Z-DNA binding protein 1 (ZBP1), initially reported as an IFN-inducible, tumor-associated protein, harbors nucleic acid binding domains for left handed helix (Z-form) and RHIM domains for protein homotypic interactions. Recent studies have identified ZBP1 as an innate sensor of viral infections and a target of viral evasion strategies, regulating cell death, inflammasome activation and proinflammatory responses. ZBP1 also functions during development and can trigger perinatal lethality when its RHIM-dependent interactions are not restricted. Here, we review the history and emergence of ZBP1 as a pathogen sensor and a central regulator of cell death and inflammatory responses. We also discuss the gaps in our knowledge regarding the regulation and functions of ZBP1 and highlight potential avenues for future research.
Keywords: ZBP1, DAI, cell death, inflammation, RIPK1, RIPK3, Caspase-8, innate immunity, inflammasome
ZBP1: A rising star in innate immunity
Innate immune sensing of pathogen-associated or danger-associated molecular patterns (PAMPs and DAMPs) activates various intracellular signaling cascades, which transduces signals that elicit a proinflammatory immune response. In addition, many of the innate sensors also trigger programmed cell death in response to pathogens or other immunological stimuli, either by directly interacting with other mediators of cell death or indirectly via the downstream effects of secreted cytokines. Inflammation and cell death are two distinct, but highly interconnected and mutually regulated phenomena that are important in immune defense, pathogenesis and in maintaining organismal homeostasis [1]. Both inflammation and cell death are also recognized as host-mediated mechanisms that facilitate elimination of invading pathogens [2]. Some of the best-known examples of innate sensors initiating cell death and proinflammatory response upon ligand recognition are the inflammasome-forming NLR and AIM2 receptors [3]. Z-DNA binding protein 1 (ZBP1) is another host protein recently recognized as an innate immune sensor regulating activation of both programmed cell death and inflammation in diverse conditions including infection and embryonic development. Emerging studies also suggest a unique role for ZBP1 in sensing microbial signatures that are not previously identified as PAMPs. In this review, we discuss the identification, characterization and emergence of ZBP1 as a central regulator of innate immune responses and programmed cell death.
From DLM1 to ZBP1: Identification and characterization
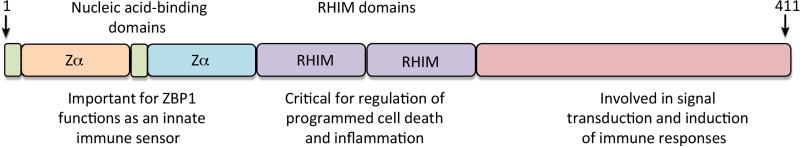
The ZBP1 story began in 1999 when a novel gene was identified during studies investigating the interactions between tumors and their associated cells and tissues. This newly described interferon (IFN) γ-inducible gene was upregulated in tumor stroma and activated macrophages and was initially named DLM-1 [4]. ZBP1 was grouped among a conserved family of Z-DNA binding proteins after characterization of the N terminal Z-DNA binding domain (ZBD) which binds to Z-DNA, a left-handed (instead of the most common right-handed) double-strand DNA helix [5], in a structure-specific manner [6] (Figure 1). The protein shows high degree of conservation between the human and murine homologs with 47% protein sequence identity. The Zα domain of ZBP1 also harbors residues conserved between other Zα family members including human and mouse RNA editing enzyme ADAR1 and the E3L proteins from vaccinia virus, Yaba-like disease virus and orf virus [6]. Mouse ZBP1 is detected in high levels in normal lung, spleen and liver tissues whereas the human protein is strongly expressed in small intestine and lymphatic tissues [4, 7]. Biochemical characterization and crystal structure of Zα domain bound to Z-DNA led to the renaming of the protein as Z-DNA binding protein 1 (ZBP1) [6, 8, 9]. ZBP1 is the current recommended name for the tumor stroma and activated macrophage protein DLM1 as per the Universal Protein Resource (UniProt).
Figure 1. Domain structure of ZBP1.
ZBP1 encodes two N-terminal Z-DNA binding domains, which is reported to bind with Z-DNA, B-DNA and RNA. ZBP1 also has two RHIM domains at the center part that facilitate interactions with other RHIM domain-containing proteins. These RHIM domains are important in mediating ZBP1—dependent cell death and inflammatory responses. The conserved C-terminal domains of ZBP1 were reported to interact with TBK1 and IRF3 to induce type I IFN responses to immunostimulatory DNA.
ZBP1 can form nuclear foci in activated cells; however it is primarily localized in the cytoplasm and is associated with cytoplasmic stress granules and processing bodies that are important in regulation of mRNA turnover [10, 11]. Although the earlier studies did not delve into the functional aspects of the protein, they predicted a possible role for ZBP1 in host defense against tumors and viral infections based on the IFN-dependent induction, expression in lymphatic tissues and the involvement of other Zα family proteins in viral pathogenesis [6, 7, 10, 12]. It is interesting to note that these pioneering studies also predicted cellular RNA in Z-confirmation, viral Z-DNA or Z-RNA as the targets of ZBP1 [10].
The DAI proposal: ZBP1 as a DNA sensor
Despite detailed structural and biochemical characterization and general interest in the protein as a potential key to unravel the mysteries of Z-DNA, the biological functions of ZBP1 have remained elusive for many years. During their studies investigating DNA-sensing mechanisms, Taniguchi and colleagues found a role for ZBP1 in DNA-mediated innate immune responses and characterized the protein as a cytosolic DNA sensor [13]. At the molecular level, ZBP1 after binding to dsDNA, associated with TBK1 and IRF3 and regulated activation of IRF3 and induction of type I IFNs. A role for ZBP1 in antiviral responses during DNA virus, but not RNA virus infection was also demonstrated. An alternative name, DNA-dependent activator of IFN-regulatory factors (DAI) was proposed for ZBP1 based on these observations [13]. A subsequent study found the requirement for N-terminal DNA-interacting regions and dimerization/oligomerization and phosphorylation of ZBP1 to evoke DNA-mediated innate responses [14]. ZBP1 was also shown to promote NFκB activation and proinflammatory cytokine production in response to immunostimulatory DNA [13, 15]. Although these studies collectively suggest DNA-sensing functions for ZBP1, this was disputed after generation of Zbp1−/− mice [16]. ZBP1-deficient cells or mice responded normally to DNA and DNA virus infection and induced normal levels of IFNs and other cytokines. ZBP1 was also dispensable for proper induction of adaptive immune responses to DNA vaccine [16]. These findings questioned the role of ZBP1 as a DNA sensor and suggested no role or redundant roles for ZBP1 in DNA-mediated responses. Moreover, most of the recent studies with cell lines or Zbp1−/− mice or cells (discussed below) argue against the role of ZBP1 as a DNA sensor and calls for additional validation of the standing of ZBP1 as ‘DAI’.
Induction into a unique family of four: Identification of RHIM
Another twist in the story came with the identification of ZBP1 as the fourth mammalian protein harboring receptor-interacting protein homotypic interaction motif (RHIM) domains. Two independent studies investigating the mechanisms regulating ZBP1-dependent NFκB activation reported conserved regions in the central portion of the protein with sequence similarity to the RHIM domains of RIPK1, RIPK3 and TRIF [15, 17] (Figure 1). Studies from the Mocarski group using expression plasmids or RNA interference demonstrated RHIM-dependent interactions between ZBP1 and RIPK1 that trigger NFκB activation [15]. ZBP1-RIPK1 interaction was also shown to be necessary for Ifnb expression in response to immunostimulatory DNA. Tschopp and colleagues also reported RHIM-dependent interactions between ZBP1, RIPK1 and RIPK3 leading to NFκB activation in HEK293 T cells transfected with expression plasmids [17]. Both these studies found a role for RIPK3 in ZBP1 and RIPK1-dependent signal transduction, which is in contrast to RIPK3 inhibition of RIPK1-mediated signaling downstream of TNF receptor or of the TLR adaptor TRIF [18, 19].
RHIM-dependent interactions in virus-induced necrosis
The identification of the RHIM domain provided the first indication for involvement of ZBP1 in cell death and antiviral responses. Tschopp and colleagues found RHIM-dependent interactions between ZBP1 and the M45 protein of the DNA virus murine cytomegalovirus (MCMV) [17]. They further demonstrated inhibition of ZBP1-RIPK1/RIPK3 interaction and downstream signaling by M45 in a RHIM-dependent manner. Based on previous studies demonstrating M45-mediated suppression of both cell death and RIPK1 signaling during MCMV infection [20, 21], the authors proposed a role for M45 targeting of ZBP1-RIPK1/RIPK3 complex in MCMV replication and pathogenesis [17]. Indeed, this hypothesis was proven right by a subsequent study from Mocarski and colleagues. The role of ZBP1-RIPK3 interaction in virus-induced necrosis independently of IFNβ and NFκB signaling was demonstrated in this study [22]. Whereas ZBP1-RIPK3 complex was undetectable in WT MCMV-infected cells, the complex formation occurs during infection with M45mutRHIM MCMV confirming ZBP1-RIPK3 complex as the target of M45 protein. Notably, M45mutRHIM MCMV, which is unable to productively infect and replicate in WT mice, restored footpad swelling and virus replication in Zbp1−/− mice demonstrating the relevance for RHIM-dependent interaction in virus replication and pathogenesis of MCMV infection [22]. These findings identified ZBP1 as a molecular component regulating virus-induced necrosis, however, the upstream events leading to the formation of ZBP1-RIPK3 complex were not explored. Nevertheless, these studies support a role for ZBP1 in antiviral host defense and identified ZBP1 as a target of viral evasion strategies.
ZBP1 in the limelight as a sensor of influenza virus
Even though ZBP1 was studied over the years and was biochemically and functionally characterized to a great extent, it has remained unresolved whether ZBP1 functions as a pathogen sensor during an infection. The much-awaited answer came from an unexpected model when ZBP1 was identified as an innate sensor of influenza virus (IAV), a single-stranded negative sense RNA virus [23]. Studies aiming to characterize the upstream receptors inducing programed cell death during IAV infection found ZBP1 as a unique sensor of IAV regulating cell death and inflammatory responses [23] (Figure 2). Unlike other well-known viral sensors that detect patterns associated with infecting virions, ZBP1-mediated sensing occurs later during infection, after virus replication, and is dependent on type I IFN signaling. Translocation of ZBP1 to nuclear compartment where IAV replication occurs indicates that ZBP1 sensing of IAV can occur either in the nuclear or cytoplasmic compartments [23]. IAV infection also induces ubiquitylation of ZBP1, however, the functional relevance of this modification is currently unknown [24]. Whereas the initial study report ZBP1 interaction with IAV nucleoprotein (NP) and polymerase subunit PB1, two subsequent studies report genomic viral RNA or the viral ribonuleoprotein (vRNP) complexes as the ligands for ZBP1 [24, 25]. Although these studies show ZBP1 interaction with different components of IAV and agree on the role of ZBP1 as an IAV sensor, none of them provide conclusive evidence to confirm the actual ligand inducing ZBP1 activation. This will be technically challenging because of the requirement for actively replicating virus to initiate ZBP1-dependent responses.
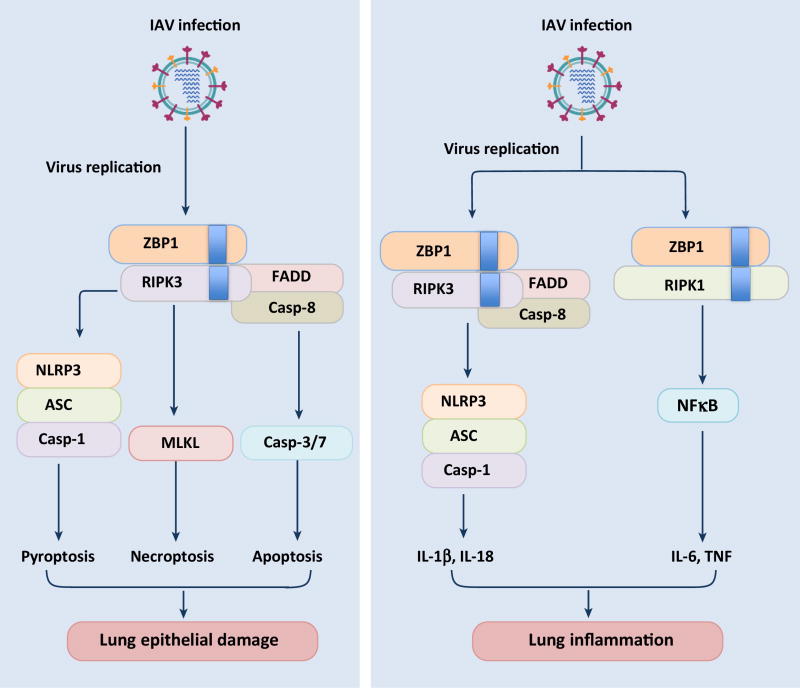
Figure 2. ZBP1 regulation of cell death and inflammatory responses during influenza virus infection.
ZBP1 after sensing IAV infection associates with RIPK1 and RIPK3 kinases via RHIM domain interactions. ZBP1-RIPK3 complex activates parallel pathways of caspase-8-dependent apoptosis and MLKL-mediated necroptosis. In addition, ZBP1 regulates the NLRP3 inflammasome activation, pyroptosis and secretion of IL-1β and IL-18 via the RIPK3-caspase-8 axis. ZBP1-RIPK1 axis transduces downstream signaling and mediates secretion of various proinflammatory cytokines during IAV infection. ZBP1 regulation of cell death and inflammatory responses is also evident during in vivo infections and ZBP1 controls lung epithelial damage and inflammation during IAV infection.
ZBP1 regulation of influenza virus-induced cell death
IAV infection is known to induce activation of multiple programmed cell death pathways including apoptosis (a non-inflammatory form of cell death mediated by executioner caspases [2, 26]), necroptosis (programmed form of necrosis mediated by RIPK3 and downstream effector MLKL [27]) and pyroptosis (inflammasome-dependent cell death executed by inflammatory caspases and gasdermin D [28–30]) [31–33]. ZBP1 functions as a central regulator of all these programmed cell death pathways and cell death is abrogated in ZBP1-deficient cells infected with IAV [23, 25] (Figure 2). Induction of ZBP1 and IAV-induced cell death is also dependent on the virus sensing TLR and RIG-I receptors as well as components of IFN signaling pathway in a cell type specific manner [23, 24]. After sensing IAV ligands, ZBP1 interacts with RIPK3 via RHIM-domains and induce activation of complementary pathways of apoptosis, necroptosis and pyroptosis. Consistent with this parallel activation, IAV-induced cell death can be prevented only by simultaneous inhibition of all these cell death pathways [23, 25]. Although ZBP1 regulates cell death regardless of IAV strain, it does not seem to be involved in regulation of cell death during infection with other RNA viruses [23, 25]. Influenza virus is unique among RNA viruses because virus replication occurs within the nucleus. Inhibition of nuclear export blocks cell death in IAV-infected cells either due to impairment in virus replication or due to accumulation of ZBP1 complex within the nuclear compartment [24]. Of note, the association of ZBP1 with vRNP complexes was directly observed in the cytoplasmic compartment [24]. A more detailed understanding regarding virus sensing and execution of cell death, including the role of viral effectors of cell death like PB1-F2 is necessary to unravel the mysteries behind ZBP1 regulation of IAV-induced cell death.
Not just cell death: Regulator of NLRP3 inflammasome and proinflammatory responses
ZBP1 has also been identified as a critical upstream regulator of nucleotide and oligomerization domain, leucine-rich repeat-containing protein family, pyrin domain containing 3 (NLRP3) inflammasome, a multiprotein complex important in mediating immune and healing responses during IAV infection [23, 34–36]. ZBP1 regulation of NLRP3 inflammasome is IAV-specific and ZBP1 is not involved in the regulation of inflammasome activation in response to other stimuli including MCMV infection. Whereas ZBP1-RIPK3-caspase-8 axis mediates inflammasome-dependent responses, ZBP1–RIPK1 signaling mediates secretion of various proinflammatory cytokines in IAV-infected cells [23] (Figure 2). ZBP1–RIPK1 regulation of cytokine production is dependent on the scaffolding function of RIPK1, but occurs independently of its kinase activity [23]. The involvement of RIPK1 in proinflammatory responses is consistent with its role in transducing NFκB activation signals in response to other stimuli, however the signaling pathways regulated by ZBP1-RIPK1 axis in IAV-infected cells is not studied so far.
ZBP1 in pathogenesis of influenza virus infection
The physiological relevance of ZBP1 in modulating susceptibility to IAV infection is evident during in vivo infection [23, 25]. Two independent studies found decrease in epithelial cell death and increase in virus replication in Zbp1−/− mice, however, they report contrasting results with regard to susceptibility to infection. Consistent with in vitro studies, Kuriakose et al reported reduction in respiratory epithelial damage and lung inflammation in Zbp1−/− mice, which seems to be advantageous to the host. However, the virus persists and replicates within the infected cells leading to a delay in recovery from infection but the animals were protected from mortality [23]. This observation aligns with several studies from humans and animal models demonstrating exaggerated inflammatory response and lung epithelial damage as the major factors augmenting mortality during IAV infection [37–41]. In contrast to this, Thapa et al found increased mortality in Zbp1−/− mice, which is attributed to increased virus burden [25]. This study did not assess the inflammatory response in the lungs and therefore it is difficult to compare ZBP1 regulation of lung inflammation between these two studies. The contrasting results observed in these studies could also be due to differences in dose of infection, humane end points used or other host susceptibility factors. Overall, the observations from these studies implicate ZBP1 as a key modulator of pathogenesis and disease outcome during IAV infection, although this still needs to be investigated and substantiated in the context of IAV infection in humans.
ZBP1 in development, perinatal lethality and skin inflammation
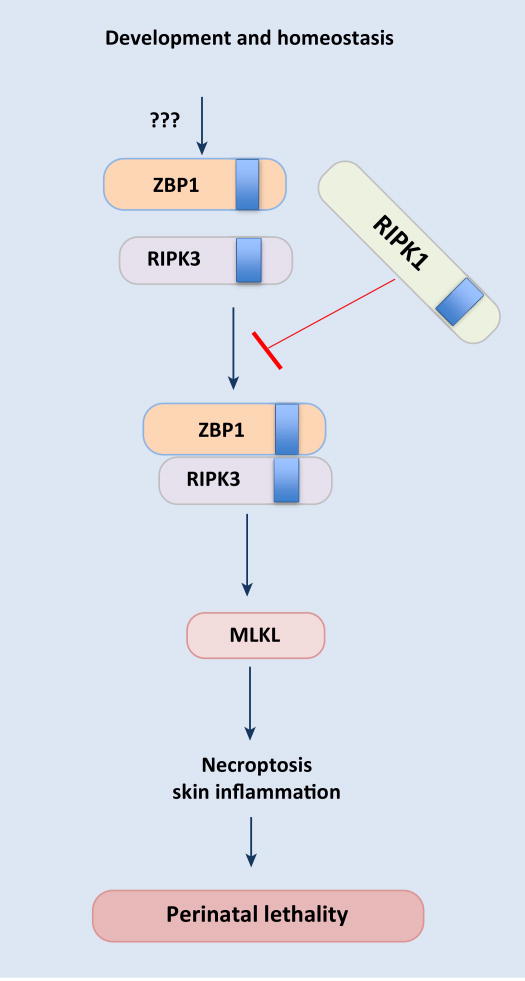
The importance of ZBP1 function in development and homeostasis is also becoming evident with emerging studies. The Dixit and Pasparakis groups independently showed aberrant activation of ZBP1 in a newly generated mouse strain expressing RIPK1 with mutation in the conserved RHIM domain (the core residues of RHIM domain IQIG substituted with alanines; Ripk1RHIM/RHIM) [42, 43]. ZBP1 activity is kept under check in normal conditions by RIPK1 via RHIM-dependent interactions and lack of inhibition triggers interaction between ZBP1 and RIPK3 leading to induction of necroptosis and perinatal lethality (Figure 3). These observations also provide molecular insights regarding the kinase-independent pro-survival function of RIPK1 in late embryonic development [44]. Both the RHIM and kinase activity of RIPK3 is necessary for induction of necroptosis in Ripk1RHIM/RHIM mice. Consistent with ZBP1-mediated triggering of necroptosis, development of lesions and perinatal lethality in Ripk1RHIM/RHIM mice can be prevented by deletion of ZBP1, RIPK3 or MLKL [42, 43]. Immunoprecipitation experiments revealed ZBP1–RIPK3 interaction in Ripk1RHIM/RHIM cells; however, these experiments failed to demonstrate constitutive association between ZBP1 and RIPK1 in wild type cells. The mechanism by which RIPK1 RHIM counteracts ZBP1–RIPK3 complex formation is not understood yet. Moreover, the trigger that initiates ZBP1 activation and its interaction with RIPK3 is also not known.
Figure 3. ZBP1-RIPK1 axis in embryonic development and homeostasis.
Aberrant activation of ZBP1 during embryonic development leads to necroptosis and perinatal lethality. When ZBP1 activity is not under check, it interacts with RIPK3 and activates MLKL-mediated necroptosis. The RHIM domain of RIPK1 inhibits activation of ZBP1 and its interaction with RIPK3 during development via yet unidentified mechanisms. The endogenous stimulus activating ZBP1 during development is not known.
ZBP1 was also identified as a potent inducer of keratinocyte necroptosis and skin inflammation in mice with epidermis-specific RIPK1 deficiency (RIPK1mRHIM/E-KO) [43]. Expression of Ifnb and Zbp1 is increased in the skin of RIPK1mRHIM/E-KO mice and deficiency of ZBP1 prevented skin inflammation in these animals. This shows that inhibition of ZBP1 function by RIPK1 RHIM is critical for maintaining skin homeostasis [43]. This contrasts with IAV infection where the scaffolding function of RIPK1 is required to promote proinflammatory cytokine production, which is also dependent on ZBP1. The involvement of ZBP1 in intestinal homeostasis is not investigated yet, and it is not known whether ZBP1 is also involved in the induction of inflammation and cell death in intestinal epithelial cells observed in the absence of RIPK1 [45]. The observations from Ripk1RHIM/RHIM mice unveiled the importance of ZBP1 during development and further identified RIPK1 as a brake regulating ZBP1 activity.
ZBP1 as a sensor of viral and endogenous RNA transcripts
Two recent studies further explored ZBP1 regulation of MCMV-induced cell death and reported newly synthesized RNA transcripts as the ligands activating ZBP1-dependent cell death during infection with M45mutRHIM MCMV [46, 47] (Figure 4). Induction of cell death in M45mutRHIM MCMV-infected cells requires intact ZBDs of ZBP1. Using recombinant viruses that lack a functional immediate-early viral transactivator IE3, Upton and colleagues demonstrated the requirement for either IE3 protein or its transcriptional activity in promoting cell death during M45mutRHIM MCMV infection [46]. Rehwinkel and colleagues also demonstrated a requirement for viral RNA synthesis and an intact ZBD of ZBP1 for MCMV-induced cell death [47]. ZBP1 binding to both newly synthesized RNA in virus-infected cells and endogenous RNA was demonstrated in this study [47]. These studies hypothesize that RNA in Z-confirmation is the molecular pattern recognized by ZBP1 in virus-infected cells. The binding of ZBP1 to endogenous RNA also provides hints regarding the activation of ZBP1 in Ripk1RHIM/RHIM mice. Although these studies indicate an interesting possibility of ZBP1 binding to Z-RNA, further experimental evidence is needed to confirm and validate this hypothesis.
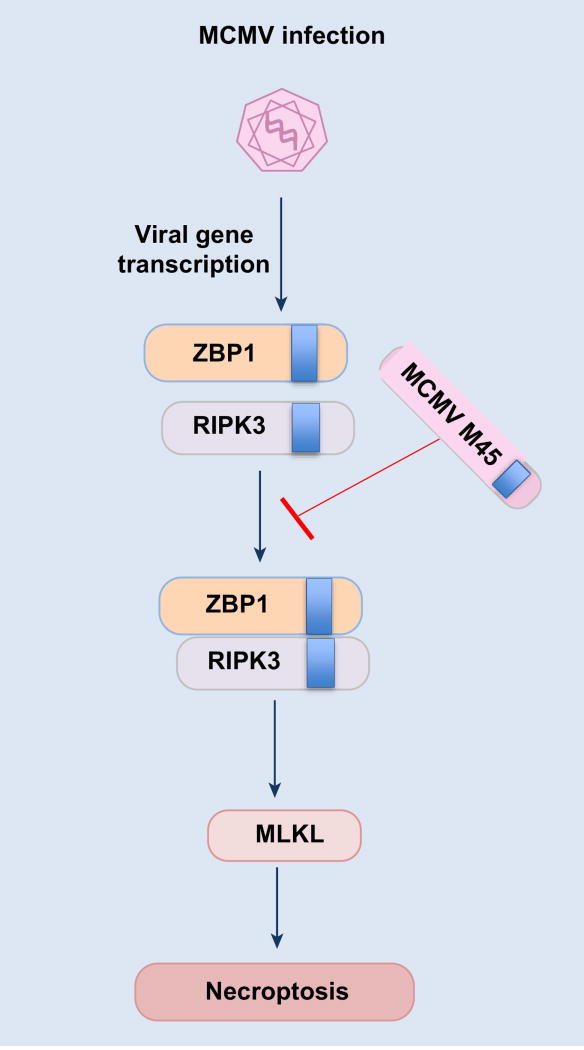
Figure 4. ZBP1 sensing and regulation of necroptosis during MCMV infection.
In addition to IAV, ZBP1 also functions as a sensor of MCMV infection and it interacts with newly transcribed viral gene products. Sensing and activation of ZBP1 leads to its interaction with RIPK3 and subsequent induction of necroptosis. The RHIM domain-containing virus protein M45 inhibits the ZBP1-RIPK3 complex formation and cell death during MCMV infection.
Vaccinia virus (VV) encodes the Zα domain containing protein E3L and various other inhibitors of cell death [6, 48, 49] and is known to induce programmed necrosis and inflammation via the RIPK1-RIPK3 complex [50]. A recent study demonstrated the role of Zα domain of E3 in inhibiting IFN and RIPK3-dependent necroptosis during VV infection, which is also dependent on ZBP1 [51]. How E3 Zα domain inhibits ZBP1/RIPK3-mediated necroptosis is unclear and the authors hypothesize competitive binding and sequestration of the viral PAMP as the potential mechanism by which E3 inhibits ZBP1 activation. This study also extends the current knowledge on ZBP1 as a target of viral immune evasion strategies and further helps to explain the enhanced oncolytic activity of VV expressing ZBP1 [52].
Human ZBP1: Still in the shadows
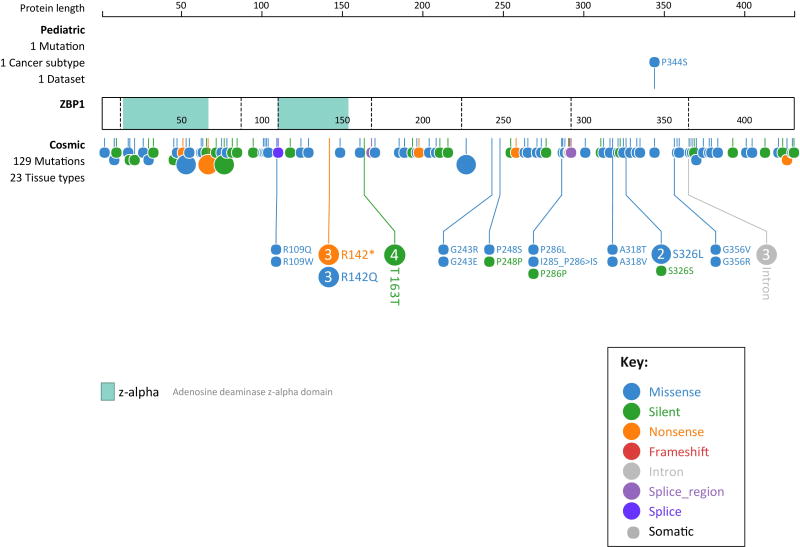
Earlier studies reported a high degree of conservation between amino acid sequences of human and murine ZBP1, however the functions of human ZBP1 have not been explored in the light of recent studies. Human ZBP1 shows highly heterogeneous transcripts suggesting complex regulation of this gene resulting in multiple protein isoforms [7]. As per the protein coding genetic variation data provided by the Exome Aggregation Consortium (ExAC), 71 synonymous, 173 missense and 15 loss of function variants were observed for ZBP1 in healthy subjects [53]. The Catalogue of Somatic Mutations in Cancer (COSMIC) lists 129 cancer-associated mutations in ZBP1 from 23 tissue types and one of these mutations is specifically linked to pediatric cancer [54] (Figure 5). These mutations in ZBP1 associated with human cancer suggest a possible role for genetic lesions in ZBP1 in the onset and progression of different cancers. Proteomics analysis and the mapping of the human innate immunity interactome for type I IFN identified ZBP1 as a member of this interactome endowed with antiviral functions [55]. Consistent with this, ZBP1 overexpression inhibits HCMV replication in human foreskin fibroblasts and hepatitis B virus in human hepatoma cell line Huh7 [56, 57]. In contrast, replication of HIV-1 is increased in 293T cells expressing ZBP1 [58]. Experiments using lentiviral vectors expressing shRNAs for ZBP1 demonstrate ZBP1 inhibition of herpes simplex 1(HSV-1) replication in HepG2 cells independently of its DNA-sensing functions [59]. Similar to the recent observations from MCMV-infected cells, ZBP1 activity during HSV-1 infection was linked to expression of viral immediate early genes and ZBP1 co-localized with the viral protein ICP0 in both nuclear and cytoplasmic foci [59]. Whether human ZBP1 mediates other antiviral responses including cell death and inflammation is currently unknown. Another recent study reported cleavage and inactivation of synthetic genes encoding all mammalian RHIM-containing proteins including ZBP1 by purified recombinant effector protein EspL of enteropathogenic E. coli [60]. This suggests a possible role for ZBP1 in pathogenesis of bacterial infections by mediating necroptosis and inflammation and warrants future investigation.
Figure 5. Schematic showing cancer-associated mutations in human ZBP1.
Cancer-associated mutations in ZBP1 as per the Catalogue of Somatic Mutations in Cancer (COSMIC) database and Pediatric Cancer genome project depicted using ProteinPaint, (a web application for visualizing genomic data). The interactive tool is available at https://pecan.stjude.org/proteinpaint/ZBP1. Each color-coded circle depicts a mutation and the larger circles with numbers represent mutations found in more than one sample. This interactive tool allows visualization of major attributes of the mutation including the details of the sample from which the mutation was identified.
Concluding Remarks and future perspectives
The identification of ZBP1 as a pathogen sensor and its emergence as a key regulator of cell death and inflammation followed a rather tortuous route. ZBP1 still stands as an enigmatic molecule although recent studies have unveiled multiple aspects of its activation and functions. The diverse conditions and nature of ligands that are reported to interact with ZBP1 makes the mechanism leading to ZBP1 activation quite puzzling. ZBP1 in fact provides a very good example of a receptor that senses ‘patterns of pathogenesis’ rather than true molecular patterns presented by invading pathogens [61]. It is also likely that a common upstream event or other contextual cues leads to ZBP1 activation and downstream signal transduction, or perhaps ZBP1 might be functioning both in a ligand-dependent and ligand-independent manner. The physiological relevance of the nuclear localization of ZBP1 and whether this determines the specificity of ZBP1 function is also unclear. The significance of ubiquitylation and other post translational modifications of ZBP1 reported in various studies is not well studied and might be relevant in determining distinct, stimulus-specific responses [14, 24]. The knowledge gained from recent reports also necessitates studies on ZBP1 in the context of other infectious and inflammatory conditions as well in tumorigenesis and cancer metastasis. Ultimately, studies on regulation and functions of ZBP1 and ZBP1-dependent responses may provide greater insights into the molecular mechanisms governing cell death and inflammation in both physiological and pathological conditions and may aid in the development of therapeutic strategies for the treatment of infectious and inflammatory diseases (see Outstanding questions).
Outstanding questions.
What is the actual ligand or molecular pattern triggering ZBP1 activation?
Is there any difference in ZBP1 activation between infectious and non-infectious conditions?
Is nucleic acid sensing necessary for ZBP1 activation?
Which cellular compartment does ZBP1-mediated sensing occurs?
How does RIPK1 keep ZBP1 activity in check under physiological conditions?
What determines the specificity of ZBP1 function?
How does ZBP1 distinguish between different pathogens? Does it have a broader role in regulation of host immune responses during bacterial, viral, fungal and parasitic infections?
Is ZBP1 involved in tumorigenesis and cancer metastasis?
Does ZBP1 have a role in progression of inflammatory diseases?
Does human ZBP1 perform similar functions as that of the murine homolog?
Trends.
Z-DNA binding protein 1 (ZBP1) functions as a central regulator of programmed cell death and inflammatory responses in diverse conditions through RHIM domain-dependent interactions with the kinases RIPK1 and RIPK3.
ZBP1 was recently identified as an innate immune sensor of both RNA (influenza virus) and DNA (MCMV) viruses and a target of viral evasion strategies. ZBP1 interacts with different viral components; however the viral PAMP triggering ZBP1 activation is not confirmed yet.
Activation of ZBP1 during embryonic development leads to necroptosis and perinatal lethality and aberrant activation of ZBP1 is kept in check by the RHIM-dependent scaffolding function of RIPK1.
ZBP1 interaction with DNA as well as viral and endogenous RNA via its N-terminal Z-DNA binding domains have been reported; however the nucleic acid sensing functions of ZBP1 under physiological and pathological conditions remains controversial.
Acknowledgments
Research studies from our laboratory are supported by the US National Institutes of Health (AI101935, AI124346, AR056296 and CA163507 to T.D.K.), the American Lebanese Syrian Associated Charities (to T.D.K.). We also thank Dr. Parimal Samir for helping with ProteinPaint web application.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wallach D, et al. Concepts of tissue injury and cell death in inflammation: a historical perspective. Nat Rev Immunol. 2014;14(1):51–9. doi: 10.1038/nri3561. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen I, et al. Programmed cell death as a defence against infection. Nat Rev Immunol. 2017;17(3):151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma D, Kanneganti TD. The cell biology of inflammasomes: Mechanisms of inflammasome activation and regulation. J Cell Biol. 2016;213(6):617–29. doi: 10.1083/jcb.201602089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y, et al. Cloning of DLM-1, a novel gene that is up-regulated in activated macrophages, using RNA differential display. Gene. 1999;240(1):157–63. doi: 10.1016/s0378-1119(99)00419-9. [DOI] [PubMed] [Google Scholar]
- 5.Herbert A, Rich A. The biology of left-handed Z-DNA. J Biol Chem. 1996;271(20):11595–8. doi: 10.1074/jbc.271.20.11595. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz T, et al. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat Struct Biol. 2001;8(9):761–5. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 7.Rothenburg S, et al. Complex regulation of the human gene for the Z-DNA binding protein DLM-1. Nucleic Acids Res. 2002;30(4):993–1000. doi: 10.1093/nar/30.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha SC, et al. Biochemical characterization and preliminary X-ray crystallographic study of the domains of human ZBP1 bound to left-handed Z-DNA. Biochim Biophys Acta. 2006;1764(2):320–3. doi: 10.1016/j.bbapap.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Ha SC, et al. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc Natl Acad Sci U S A. 2008;105(52):20671–6. doi: 10.1073/pnas.0810463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deigendesch N, et al. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 2006;34(18):5007–20. doi: 10.1093/nar/gkl575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pham HT, et al. Intracellular localization of human ZBP1: Differential regulation by the Z-DNA binding domain, Zalpha, in splice variants. Biochem Biophys Res Commun. 2006;348(1):145–52. doi: 10.1016/j.bbrc.2006.07.061. [DOI] [PubMed] [Google Scholar]
- 12.Kim YG, et al. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci U S A. 2003;100(12):6974–9. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448(7152):501–5. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci U S A. 2008;105(14):5477–82. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaiser WJ, et al. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J Immunol. 2008;181(9):6427–34. doi: 10.4049/jimmunol.181.9.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451(7179):725–9. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 17.Rebsamen M, et al. DAI/ZBP1 recruits RIP1 and RIP3 through RIP homotypic interaction motifs to activate NF-kappaB. EMBO Rep. 2009;10(8):916–22. doi: 10.1038/embor.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meylan E, et al. RIP1 is an essential mediator of Toll-like receptor 3-induced NF-kappa B activation. Nat Immunol. 2004;5(5):503–7. doi: 10.1038/ni1061. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, et al. Identification of a novel homotypic interaction motif required for the phosphorylation of receptor-interacting protein (RIP) by RIP3. J Biol Chem. 2002;277(11):9505–11. doi: 10.1074/jbc.M109488200. [DOI] [PubMed] [Google Scholar]
- 20.Upton JW, et al. Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J Biol Chem. 2008;283(25):16966–70. doi: 10.1074/jbc.C800051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mack C, et al. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc Natl Acad Sci U S A. 2008;105(8):3094–9. doi: 10.1073/pnas.0800168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Upton JW, et al. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe. 2012;11(3):290–7. doi: 10.1016/j.chom.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuriakose T, et al. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol. 2016;1(2) doi: 10.1126/sciimmunol.aag2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kesavardhana S, et al. ZBP1/DAI ubiquitination and sensing of influenza vRNPs activate programmed cell death. J Exp Med. 2017;214(8):2217–2229. doi: 10.1084/jem.20170550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thapa RJ, et al. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe. 2016;20(5):674–681. doi: 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Man SM, Kanneganti TD. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat Rev Immunol. 2016;16(1):7–21. doi: 10.1038/nri.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517(7534):311–20. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 28.Vande Walle L, Lamkanfi M. Pyroptosis. Curr Biol. 2016;26(13):R568–72. doi: 10.1016/j.cub.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 29.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–22. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Shi J, et al. Pyroptosis: Gasdermin-Mediated Programmed Necrotic Cell Death. Trends Biochem Sci. 2017;42(4):245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Herold S, et al. Apoptosis signaling in influenza virus propagation, innate host defense, and lung injury. J Leukoc Biol. 2012;92(1):75–82. doi: 10.1189/jlb.1011530. [DOI] [PubMed] [Google Scholar]
- 32.Rodrigue-Gervais IG, et al. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 2014;15(1):23–35. doi: 10.1016/j.chom.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Kanneganti TD, et al. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281(48):36560–8. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- 34.Thomas PG, et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 2009;30(4):566–75. doi: 10.1016/j.immuni.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allen IC, et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity. 2009;30(4):556–65. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuriakose T, Kanneganti TD. Regulation and functions of NLRP3 inflammasome during influenza virus infection. Mol Immunol. 2017 doi: 10.1016/j.molimm.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Jong MD, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–7. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandes M, et al. A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell. 2013;154(1):197–212. doi: 10.1016/j.cell.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuyama S, Kawaoka Y. The pathogenesis of influenza virus infections: the contributions of virus and host factors. Curr Opin Immunol. 2011;23(4):481–6. doi: 10.1016/j.coi.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kash JC, et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature. 2006;443(7111):578–81. doi: 10.1038/nature05181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanders CJ, et al. Compromised respiratory function in lethal influenza infection is characterized by the depletion of type I alveolar epithelial cells beyond threshold levels. Am J Physiol Lung Cell Mol Physiol. 2013;304(7):L481–8. doi: 10.1152/ajplung.00343.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newton K, et al. RIPK1 inhibits ZBP1-driven necroptosis during development. Nature. 2016;540(7631):129–133. doi: 10.1038/nature20559. [DOI] [PubMed] [Google Scholar]
- 43.Lin J, et al. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 2016;540(7631):124–128. doi: 10.1038/nature20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dillon CP, et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157(5):1189–202. doi: 10.1016/j.cell.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dannappel M, et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014;513(7516):90–4. doi: 10.1038/nature13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sridharan H, et al. Murine cytomegalovirus IE3-dependent transcription is required for DAI/ZBP1-mediated necroptosis. EMBO Rep. 2017 doi: 10.15252/embr.201743947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maelfait J, et al. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. 2017 doi: 10.15252/embj.201796476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benedict CA, et al. To kill or be killed: viral evasion of apoptosis. Nat Immunol. 2002;3(11):1013–8. doi: 10.1038/ni1102-1013. [DOI] [PubMed] [Google Scholar]
- 49.Chan FK, et al. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem. 2003;278(51):51613–21. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 50.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137(6):1112–23. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koehler H, et al. Inhibition of DAI-dependent necroptosis by the Z-DNA binding domain of the vaccinia virus innate immune evasion protein, E3. Proc Natl Acad Sci U S A. 2017;114(43):11506–11511. doi: 10.1073/pnas.1700999114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hirvinen M, et al. Expression of DAI by an oncolytic vaccinia virus boosts the immunogenicity of the virus and enhances antitumor immunity. Mol Ther Oncolytics. 2016;3:16002. doi: 10.1038/mto.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou X, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat Genet. 2016;48(1):4–6. doi: 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, et al. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity. 2011;35(3):426–40. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeFilippis VR, et al. Activation of the interferon response by human cytomegalovirus occurs via cytoplasmic double-stranded DNA but not glycoprotein B. J Virol. 2010;84(17):8913–25. doi: 10.1128/JVI.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen QY, et al. DNA-dependent activator of interferon-regulatory factors inhibits hepatitis B virus replication. World J Gastroenterol. 2012;18(22):2850–8. doi: 10.3748/wjg.v18.i22.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi T, et al. DNA-dependent activator of IFN-regulatory factors enhances the transcription of HIV-1 through NF-kappaB. Microbes Infect. 2010;12(12–13):937–47. doi: 10.1016/j.micinf.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 59.Pham TH, et al. DNA sensing-independent inhibition of herpes simplex virus 1 replication by DAI/ZBP1. J Virol. 2013;87(6):3076–86. doi: 10.1128/JVI.02860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pearson JS, et al. EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat Microbiol. 2017;2:16258. doi: 10.1038/nmicrobiol.2016.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vance RE, et al. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6(1):10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]