Abstract
In this study Illumina MiSeq was performed to investigate microbial diversity in soil, leaves, grape, grape juice and wine. A total of 1,043,102 fungal Internal Transcribed Spacer (ITS) reads and 2,422,188 high quality bacterial 16S rDNA sequences were used for taxonomic classification, revealed five fungal and eight bacterial phyla. At the genus level, the dominant fungi were Ascomycota, Sordariales, Tetracladium and Geomyces in soil, Aureobasidium and Pleosporaceae in grapes leaves, Aureobasidium in grape and grape juice. The dominant bacteria were Kaistobacter, Arthrobacter, Skermanella and Sphingomonas in soil, Pseudomonas, Acinetobacter and Kaistobacter in grape and grapes leaves, and Oenococcus in grape juice and wine. Principal coordinate analysis showed structural separation between the composition of fungi and bacteria in all samples. This is the first study to understand microbiome population in soil, grape, grapes leaves, grape juice and wine in Xinjiang through High-throughput Sequencing and identify microorganisms like Saccharomyces cerevisiae and Oenococcus spp. that may contribute to the quality and flavor of wine.
Introduction
Microbial biodiversity is highly essential for harboring a healthy environment with sustainable economy, especially at agricultural field. In recent years, several studies on biodiversity tried to characterize the microbiome in different agricultural ecosystems, as to understand the dynamics of plant and microbe interaction. It is discovered that plant-associated bacteria and fungi colonize on both exterior (epiphytes) and interior surfaces (endophytes) of plants, while the surrounding soil around the plant acts as the major resource for these microbes [1]. As plant species and soil type play a predominant role in soil microbial community, the interaction between plant and soil microbes are highly complex. Vitis vinifera (wine grape) is an economically important agricultural crop. V. vinifera phyllosphere is easy to be colonized by both bacteria and fungi, which in-turn modulates grapevine health, development, and grape qualities [2]. In this case, microbial activity plays a critical role on grape production and quality [2–3].
Due to its nutritive contents like organic acids, amino acids, and trace elements, wine does have beneficial effects on human health. Studies have shown that long term in-take of wine in diet could prevent chronic diseases and cardiovascular diseases [4–7]. The winemaking is a composite process where numerous microorganisms were involved, especially yeast and bacteria [8–11]. Understanding the composition and population dynamics of the microbial population throughout brewing is highly essential for controlling the process, thereby improving the quality and safety of the wine [12–13]. Microbes in wine brewing is mainly from wine grapes, vineyards, brewing equipment, and surrounding environment, impacts the quality of the wine at a larger level [14–15]. In recent years, studies have shown that the unique flavor in wine is mainly generated from the microbial metabolic process during wine making process [16–17]. Therefore, selecting specific microbes for wine fermentation can help to crease the flavor, thereby enhancing the taste and quality of wine.
Current methods for microbial diversity analysis mainly include traditional culture method and non-culture methods. Traditional culture methods such as restriction fragment length polymorphism (RFLP) and random amplified polymorphic analysis (RAPD) are based on polymerase chain reaction (PCR) techniques. Compare to non-culture methods, they are laborious and time consuming, also poor in reliability. Non-culture methods mainly involve denaturing gradient gel electrophoresis (DGGE), real-time fluorescence PCR, and fluorescence in situ hybridization (FISH). They have the capability to detect the dynamic changes of microorganisms in the fermentation process, but they can only detect a certain group of microorganisms, thus in-turn restricts the complete understanding of the microbial community [18–20]. Compare to these methods, metagenomics approach has the ability to reveal the previously hidden diversity of microorganisms. After the extraction of total genetic material from a selected habitat, subsequent sequencing and bioinformatics analysis, metagenomics approach offers a powerful lens to investigate microbial communities in the specific habitat [12]. High-throughput sequencing technology has completely changed the past research model by providing large amount of data with high accuracy and low cost, and making it feasible to understand the microbial diversity at a much greater scale.
Due to the unique climate conditions and high microbial richness in natural environment, Xinjiang is widely known for wine grapes cultivation. Grapes in Xinjiang were mainly cultivated in Fukang, Manasi and Changji. These areas have gravel sandy loam, long sunshine duration, large temperature difference between day and night, small amount of precipitation and pest diseases, both are premium vintage area. Xinjiang region is very wide, so there are differences in geography and climate conditions in these three places. Fukang and Manasi areas have extremely low temperature in winter and extremely high temperature in summer. Affected by strong wind in early spring and early summer, frequent hail in late summer and sudden drop of temperature in fall, Manasi area may have different microbial diversity from Fukang area. Changji area has relatively milder climate. Besides temperature, microbes in Changji may also affected by the strong wind, sand and dust weather in spring and early summer. Because of the specific climate and soil conditions, Manasi area is one of the best grape production areas in China. However, Xinjiang is relatively un-development compare to other cities in China. Currently, we did not find reports about microbial resources and their biodiversity in the vineyard from Xinjiang region, interaction between microbial community and grape plants in Xinjiang region was also poorly understood.
In this study, Illumina MiSeq was used to study microbial community diversity of soil, grape, grapes leaves, grape juice and wine. The results will enhance our understanding of the microbiome in the vineyard from in Xinjiang region, and help to establish correlation between wine quality and regional microbiome.
Materials and methods
Sampling
Samples were collected from three different winery regions in Xinjiang province, China, named Fukang (A), Manasi (B) and Changji (C) area, both are cabernet sauvignon samples. A total of 20 samples were taken up in July and October, 2015, categorized as vineyard soils (T), grape (P), grape leaves (Y), grape juice (Z) and wine (J) (Table 1). Soil samples were collected in randomly-chosen plots around each vine root, collected at 5-10cm depth using a shovel and sieved to remove plant residues, macrofaunal, and stones. Undamaged grape samples were collected from several bunches with their pedicels attached. Grape leaves were collected from the vines to prevent cross contamination. Grape juices were sampled during grape crushing period. Wine samples from winery were collected from the final wine product. After collection in triplicates, each sample was stored immediately at -20°C for further study. The samples were collected from the private properties, permissions from the owners were received before sample collection.
Table 1. Sample list.
| Sample types | Region and name | Time |
|---|---|---|
| Soil | A(T1, T3), B(T6), C(T8) | July |
| A(T4, T5), B(T7), C(T9) | October | |
| Grape leaves | A(Y1, Y5), B(Y11), C(Y15) | July |
| A(Y9, Y10), B(Y13), C(Y17) | October | |
| Grape | A(P1), B(P2) | October |
| Grape juice | A(Z1) | October |
| Wine | A(J1) | October |
Sample preparation for grape and grape leaves
4g of grape skin or grape leaves (no flesh and fruit stalk), was taken into 50mL sterilized centrifuge tube (L1), and 6mL of TENP buffer was added and mixed vigorously in vortex for 10min. The mixture was then centrifuged at 3000×g for 5min, supernatant was transferred to another 50mL centrifuge tube (L2). This step was repeated thrice, and a total of around 18ml supernatant was transferred to L2. L2 was then centrifuged at 9000×g for 10min. The supernatant was discarded and pellet was stored at -20°C for DNA extraction [21].
Sample preparation for grape juice and wine
About 4mL of grape juice or wine were taken into 50mL sterilized centrifuge tube (L1), vigorously shake to mix, then vortex for 1min. The mixture was then centrifuged at 3000×g for 5min, and supernatant was discarded. For the pellet, 4mL of sterile water was added and vortexed for 1min. Mixture was centrifuged at 3000×g for 5min and supernatant was discarded. This wash step was repeater thrice, and the pellet was finally stored at -20°C for DNA extraction.
DNA extraction and amplification
DNA from soil samples were extracted using Mag-Bind Soil DNA Kit (OMEGA) following manufacturer’s instructions. DNA from grape, grape leaves, grape juice and wine were extracted using Fast DNA SPIN Kit for Soil (MP) based on the manufacturer’s instructions. The quantity and quality of extracted DNA were assessed by spectrophotometry (Eppendorf, Germany) and agarose gel (1%) electrophoresis, respectively.
For fungal ITS regions, PCR amplification was performed using ITS 1F (5′-CTTGGTCATTTAGAGGAGTAA-3′) and ITS 1R (5′-GCTGCGTTCTTCATCGATGC-3′) as primers and genome DNA as template. PCR was performed at a final volume of 50μL mixture containing 4μL dNTPs mixture, 5μL of 10×PCR buffer (Mg2+ plus), 5μL of template DNA, 1μL of each primer, 0.25μL Ex Taq, and added to final volume of 50μL using ddH2O. PCR conditions were 3min at 98°C for initial, followed by 35 cycles at 98°C for 45s, annealing at 53°C for 30s, and extension at 72°C for 45s, and final extension at 72°C for 8 min. PCR products were stored at -20°C.
For bacterial 16s rRNA gene region, PCR amplification was performed using 16S 515F(5′-GTGCCAGCMGCCGCGGTAA-3′) and 16S 806R(5′-GGACTACHVGGGTWTCTAAT-3′) as primers and genome DNA as template. PCR was performed at a final volume of 50μL mixture containing 4μL dNTPs mixture, 5μL 10×PCR buffer (Mg2+ plus), 5μL template DNA, 1μL of each primer, 0.25μL Ex Taq, and added to final volume using ddH2O. PCR conditions were 3min at 98°C for initial, followed by at 95°C for 45s, annealing at 50°C for 30s, and extension at 72°C for 45s, and final extension at 72°C for 5 min. PCR products were stored at -20°C. Both fungal and bacterial PCR products were analyzed with 1% gel using DL2000 marker for quality examination.
High-throughput sequencing and statistical analysis
High-throughput sequencing was performed using an Illumina Miseq platform at the International Joint Research Center of National Liquor Quality and Safety in Chinese Food Fermentation Industry Research Institute, Beijing, China. After obtaining the sequencing result and calculation of operational taxonomic units (OTUs) matrix, statistical analysis was applied using alpha indices (Shannon, Simpson, Chao 1 and ACE), heatmap of genera, principal coordinate analysis (PCoA) and UPGMA. The alpha diversity index (Shannon Index), and the species richness estimator (Chao1) were calculated by using the R package phyloseq from the OTU matrix. Principal coordinate analysis (PCoA) was performed using statistical software based on the Bray-Curtis dissimilarities calculated for the composition of the bacterial or fungal communities at the genus level.
Results and discussion
Abundance and diversity of members of the bacterial and fungal microbiota
Illumina Miseq sequencing generated 1,043,102 high quality fungal sequences with an average of 52,155 sequences per sample, and a total of 2,422,188 high quality bacterial sequences with an average of 121,109 sequences per sample. However, the fungal population in T1, T5, and J1 was not detected. The sequencing results were shown in Supporting Information(S1 and S2 Figs, S1 and S2 Tables). Though the rarefaction curve was not parallel with the x-axis, the Good’s coverage of fungal and bacterial reached 99.9% and 97.7% respectively, with majority of microbial diversity being captured. As we applied the Chao1 index and ACE index, it reflected the species richness of sample communities, while the Shannon and Simpson index reflected the species diversity. The Chao1 and ACE scores from 139.565 to 2941.088 and from 150.4 to 3354.01, respectively. The Shannon and Simpson scores ranged from 1.538 to 8.939, and 0.241 to 0.994, respectively.
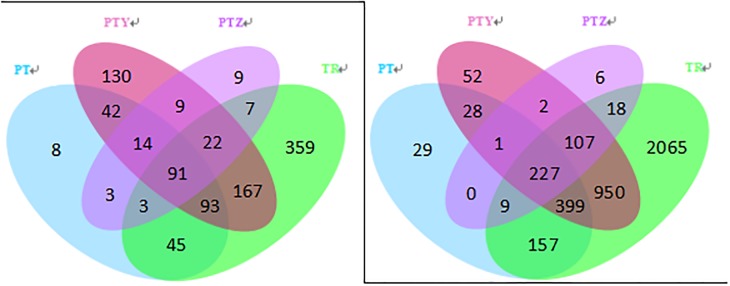
Sequences were grouped into 4597 OTUs for fungi and 23,458 OTUs for bacteria (both at the 97% similarity level). As shown in Fig 1a and 1b, after removing singletons, OTUs number was 91 for fungi and 227 for bacteria, which were similar in the T, Y, P and Z samples. Fungal OTUs were 787, 568, 299 and 158. Bacterial OTUs were 3932, 1766, 850 and 370 in T, Y, P and Z respectively.
Fig 1. Venn of fungal (a) and bacterial (b) of T, P, Y, and Z.
Earlier works suggested that soil provides essential nutrition for microbial growth, such as carbon sources (including amino acids, organic acids, and carbohydrates), nitrogen sources, and growth factors [22]. The physical and chemical properties of the soil, such as soil texture, mineral composition and organic matter, affect the growth and distribution of microbes [23–24]. Suitable growth environment favors the growth of most microbes, which in-turn alters the microbial community composition to larger extent in the soil and the plant [25]. And in contrast, external factors like pollution by heavy metals, organic pollutants, pesticide fertilizers, domestic sewage, and factory waste will directly alter the soil quality, causing major changes in the microbial communities [26–28]. It is described that microbes in vineyard soil are the source of primary inoculum to affect the structure of the microbial community on vine’s aerial parts [29], and this pattern is observed in our study. Diversity indices indicated that the diversity of fungal and bacterial community in T were significantly higher than Y, P, Z, J in October. The inoculation effect can be extended to wine brewing, as microbial community in P, Y and J were very similar with some minor differences according to Fig 1. Besides brewing microbes added in wine production process, these minor differences are possibly come from picking, transportation, crushing and other factors in grape crushing process, or fermenters and oak barrels in fermentation process [30–33].
The comparison of microbial communities in soil and vine plants suggests an inoculation effect between soil microbes and vine plants. But with the growth of vine plants and climate change, interaction between soil microbe and plants should be more complex. Comparison of soil samples collected from July and October indicate that fungal microbes in soil have higher diversity in July, while bacterial populations changed little in our study. As the weather turns gradually colder, temperature and humidity can inhibit fungal species that are not tolerant to these environmental challenges, while bacterial species showed higher adaptation in this environment. Besides, vine plants were found to limit the growth of bacteria by limiting nutrients in the early stage of growth [2, 5, 34–36]. And in response to biotic stress, stilbenes are produced in vine plants to effectively inhibit microbial activity [37–38]. To better understand such interaction and impact of soil microbes on vine plants, more samples should be taken gradually to provide systematic and detail results on microbial communities.
Comparison of fungal communities in soil, grape, grapes leaves, grape juice and wine
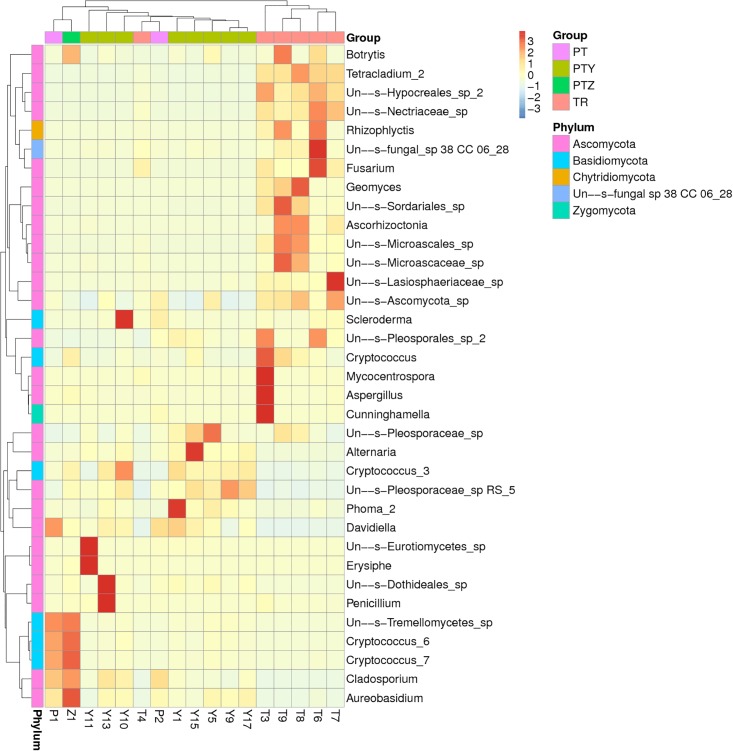
Five fungal phyla identified were Ascomycota, Basidiomycot, Chytridiomycota, Un—s-fungal sp CC 06_28 and Zygomycota. Heatmap revealed that Ascomycota and Basidiomycot were found as predominant phyla in T, P, Y, Z and J (Fig 2). This finding was consistent with previous reports [39–40]. Majority of OTUs in T and Y were Ascomycota, and similar pattern was discovered across the samples in T and Y. Basidiomycot remained less abundant in T and Y, but its abundance was found to be higher in P and Z samples. Chytridiomycota, Un—s-fungal sp CC 06_28, and Zygomycota has less abundance in T samples, but rarely detected in Y, P and Z samples. The results suggested that Ascomycota adapted to the T and Y environment, and Basidiomycot adapted to the P and Z environment.
Fig 2. Heatmap and dendrogram of abundant fungi phyla in the microbial community of samples (excepted T1, T5 and J1).
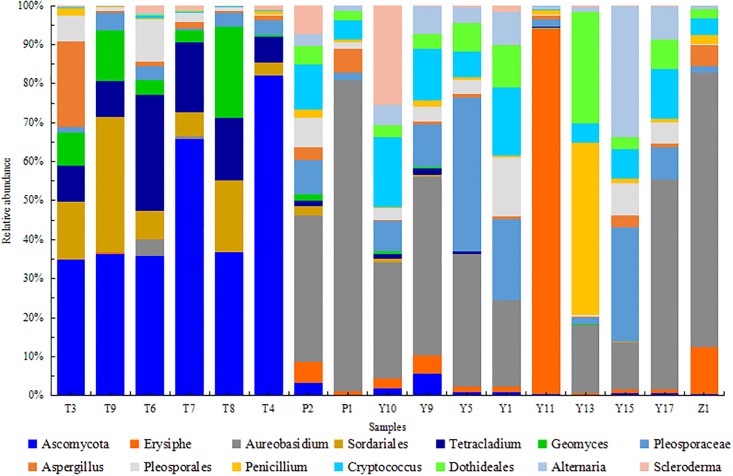
At the genus level, 271 fungal genera were detected in T, P, Y, Z and J, in which 14 fungal genera which had their relative abundance greater than 1% were selected for further analysis (Fig 3). Ascomycota, Sordariales, Tetracladium and Geomyces were the predominant genera in T samples, Aureobasidium, Pleosporaceae, Cryptococcus and Dothideales were the predominant genera in Y, P and Z samples. Other major genera were Aspergillus, Pleosporales, Penicillium, Erysisphe, Alternaria and Scleroderma. Our finding is consistent with previous reports [41]. The Ascomycota was sharply increased from July to October in A and B, however this was not obvious in C. In contrast, the Sordariales, Tetracladium and Geomyces of A, B and C were decreased in October, and Sordariales in C was found increased. Such difference might be due to interspecies competition. For Y samples, Aureobasidium, Cryptococcus and Dothideales in A, B and C have higher abundances in Octoberexcept Dothideales in A. In contrast, Pleosporaceae was found to have lower abundance in October. It is worth noticing that Erysisphe declined sharply in B, from 93% (Y11) in July to 1% (Y13) in October. This finding was consistent with an earlier report, which indicated that grapevine powdery mildew is one of the most damaging fungal diseases and it often occurs during July [42], suggesting that grapevine powdery mildew in B is quite serious and needs proper preventive measures [43–44]. Meanwhile, Erysisphe was found in Z. Interestingly, the Penicillium increased from 2% (Y11) to 44% (Y13), had become a dominant genus in B of Y samples in October. Consistent with other reports, Aspergillus, Penicillium, and Alternaria are other discovered major genera and might play an important role in T, P and J [45–46]. Studies have pointed out that ochratoxin A (OTA) produced by Aspergillus is a predominant global wine contaminant causing health hazards, the safe limit of OTA in wine is established at 2ug/L [47–49].
Fig 3. Relative abundance of fungi at genus levels of T, P, Y, and Z.
Our results indicated that fungal community changes in the grape leaves and grapes are more complex than changes in soil. This may be due to the competition between species, or natural conditions such as light intensity, light time, wind, rain, etc. or insects, human activities that causing microbial migration [29, 36]. In this metagenomics analysis, Brettanomyces bruxellensis has not been detected. It is able to convert hydroxycinnamic acids into volatile phenols, create ‘spicy’, ‘barnyard’, ‘animal’, ‘horse sweat’ and ‘medicinal’ odors in final wine product. Though a large number of culture-dependent techniques are available to assess the presence of this undesired yeast during the vinification processes, in several cases Brettanomyces is undetectable. It is been reveal that while keep alive and maintain the metabolic activities, Brettanomyces cells were tend to enter in a Viable But Not Culturable (VBNC) state. This could be the reason for not been detected in our wine samples.
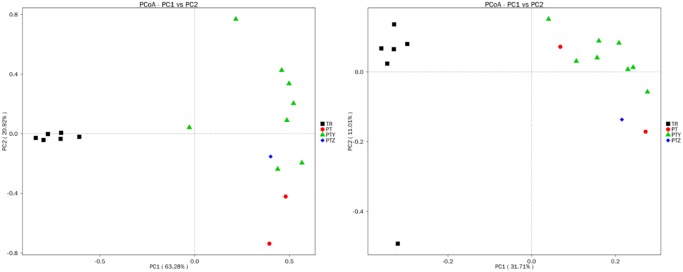
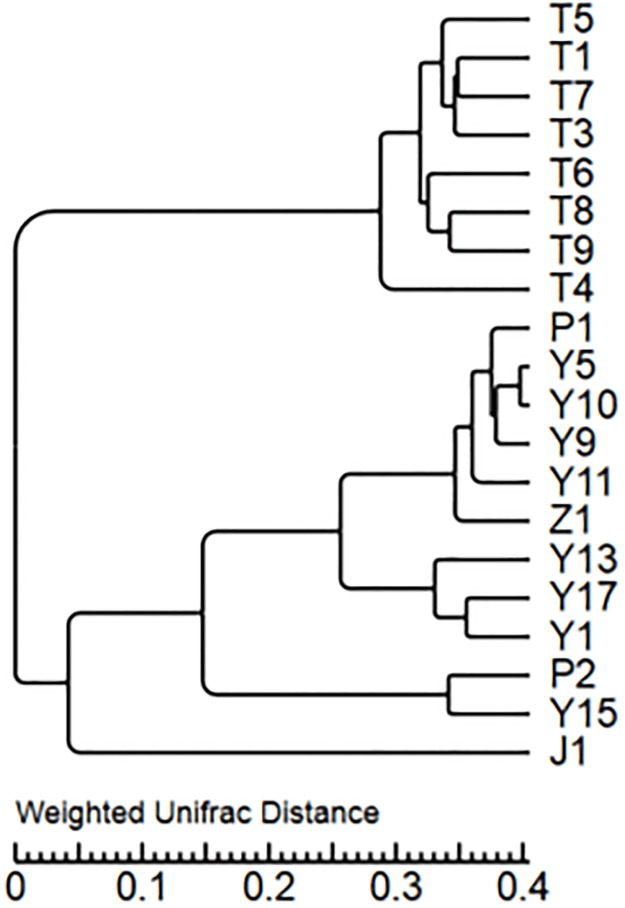
PCoA and cluster analyses were performed to evaluate similarities in fungal communities of T, Y, P, Z and J. One weighted (PC1 variance = 63.28%, PC2 variance = 20.92%) and another weighted (PC1 variance = 31.71%, PC2 variance = 11.01%) PCoA were performed (Fig 4). UPGMA clustering obtained a phylogenetic tree by using unweighted group averaging method (Fig 5). The branch of the sample intuitively reflected the similarity between the samples. Result indicates that same type of samples showed high similarity of fungal communities, as the soil samples and leave samples formed two big clusters. Soil samples from the same region each formed a small cluster, while similarity of leave samples from the same vineyard is relatively weaker. Compare to soil, grape leaves are highly exposed in environment. Microbes on leave are easier to be affected by natural conditions like insects, pathogens and human activities. For soil from vineyards, it is reported that fertilizing the soil, applying pesticides, no-tillage, continuous cropping and rotation, etc., affects the structure and physicochemical properties of the soil caused by human measures which in-turn change the composition and distribution of microbial communities [50–53]. The management of the vineyard may also be a factor in the composition of the microbial community.
Fig 4. Principal coordinate analysis of fungi microbial communities of samples.
Fig 5. Cluster analysis of the fungal microbiota.
Comparison of bacterial communities in soil, grape, grapes leaves, grape juice and wine
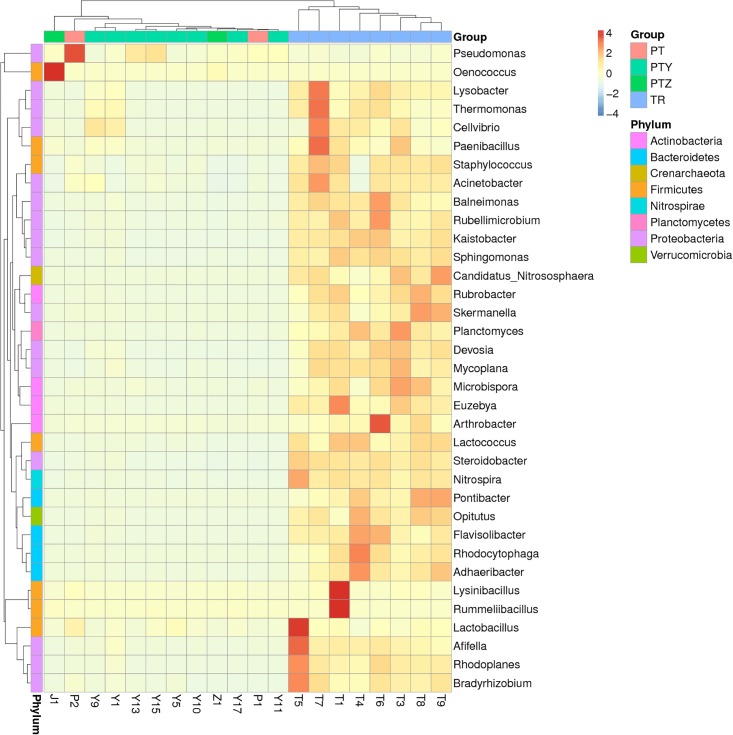
Compared to fungal population variation, the overall diversity of the bacterial microbiota in T, P, Y, Z and J samples were higher, especially in P samples. Grapes tend to mature quickly by sunlight, rain, and other conditions, as these factors can easily cause them to rot. And in juice samples, the high sugar and nutrients can increase bacterial growth [29,36]. Detected major bacteria phyla include Actinobacteria, Bacteroidetes, Crenarchaeota, Firmicutes, Nitrospirae, Planctomycetes, Proteobacteria and Verrucomicrobia (Fig 6). Heatmap revealed that Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria were the predominant phyla in T, P, Y, Z and J (Fig 6). Proteobacteria and Firmicutes were found in all the samples especially in T1, T5 and J1, and Bacteroidetes and Actinobacteria were mainly found in T samples. Crenarchaeota, Nitrospirae, Planctomycetes and Verrucomicrobia were rarely detected except in T samples. The results suggested that Proteobacteria and Firmicutes adapt better to all environment, and Bacteroidetes and Actinobacteria adapted well in soil environment.
Fig 6. Heatmap and dendrogram of abundant bacteria phyla in the microbial community of samples.
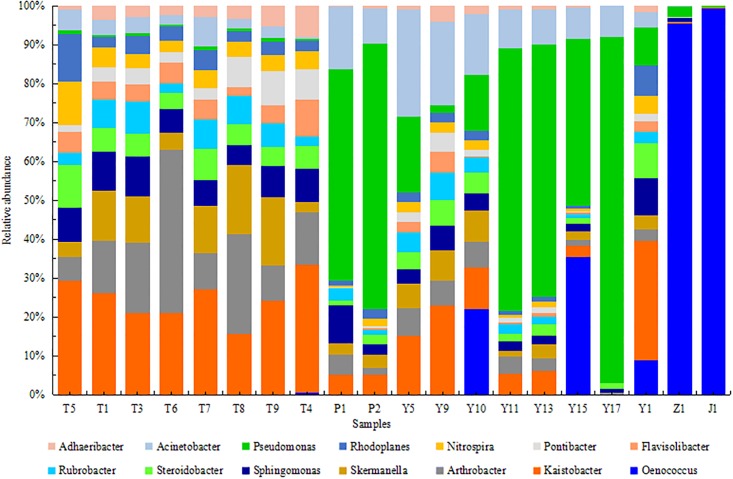
At the genus level, 317 bacterial genera were detected in T, P, Y, Z and J samples. The 14 bacterial genera with their relative abundance greater than 1% were selected (Fig 7). Kaistobacter, Arthrobacter, Skermanella and Sphingomonas were the predominant genera in T, Pseudomonas, Acinetobacter and Kaistobacter were the predominant genera in Y and P, and Oenococcus was the predominant genera in Z and J. Other major genera in samples include Steroidobacter, Rubrobacter, Flavisolibacter, Pontibacter, Nitrospira, Rhodoplanes and Adhaeribacter. These findings were consistent with previous reports [54–55]. The Kaistobacter abundance in October T samples was found increased compare with July sasmples, and Arthrobacter abundance was found declined. In contrast, no significant changes of Skermanella and Sphingomonas abundances in T samples were found. For Y and P samples collected in July and October, Kaistobacter abundance was not consistent, Pseudomonas and Acinetobacter in Y of A and B decreased in October. C sample was an exception that Pseudomonas and Acinetobacter have higher abundance in October samples, which might be due to interspecific completion that Oenococcus decreased from 35%(Y15) to 0%(Y17). Oenococcus was the dominant genus in Z and J samples, increased sharply to 95% in Z1 and 98% in J1. Oenococcus sp is a slow growing lactic acid bacterium. It is a necessary bacterium in winemaking process with the function of malolactic fermentation. It’s accumulation in wine is a natural process, and similar phenomenon was also observed in other reported studies [56]. Lactobacillus plantarum is another well studied bacterium with some strains been commercially used as malolactic fermentation (MLF) starter cultures. It is able to conduct MLF under high pH condition and in co-inoculation with yeasts. It has not been detected in this metagenomics analysis, possibly because of the special climate conditions in Xinjiang region is more suitable for Oenococcus sp.
Fig 7. Relative abundance of bacteria at genus levels of T, P, Y, Z and J.
It is discussed that the geographical location, natural climatic conditions [57], host plant phenology [58], physical and chemical properties of the soil [59–60], economic characteristics of the phyllosphere or soil [61], artificial vineyard management model, external pollution and other factors both can affected the T, Y and P microbial community composition to a certain extent, which in-turn determines the community species composition, quantity and distribution. The metagenomics analysis in this study indicates a strong correlation between grape and leave samples, differences of microbial communities in various regions were also discovered.
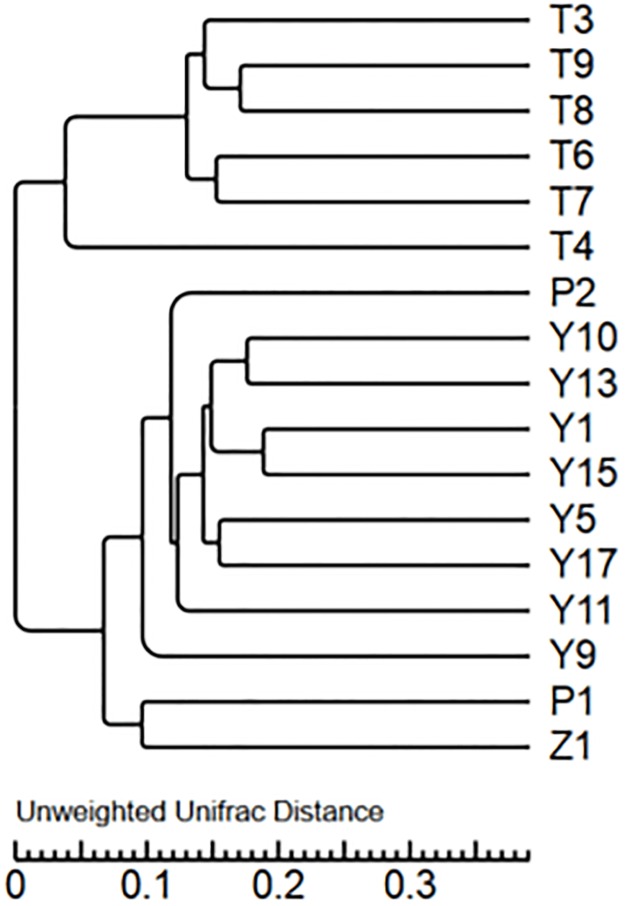
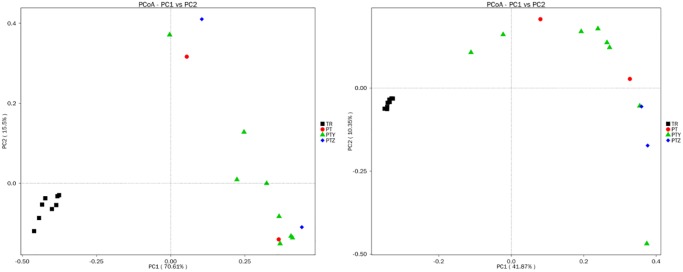
PCoA of bacterial communities in T, Y, P, Z and J samples was performed using one weighted (PC1 variance = 70.61%, PC2 variance = 15.50%) and another weighted (PC1 variance = 41.87%, PC2 variance = 10.35%) (Fig 8). UPGMA cluster map (Fig 9) reflected the bacterial microbial community structure, indicate that the similarity of bacteria in all soils were higher except T4. P, Y, Z and J had some similarity in the bacterial microbial community, though differences were existed between vineyard management in the A, B, C region.
Fig 8. Principal coordinate analysis of bacteria microbial communities of samples.
Fig 9. Cluster analysis of the bacterial microbiota.
Conclusions
The specific microbes in Xinjiang region served as autochthonous starter cultures, which play a major role in local winery production. In this study, microbial diversity of soil, grape, grape leaves, grape juice and wine were studied by high-throughput sequencing and identified the dominant genera and phyla. It is the first study to apply this technology on the winery research in Xinjiang region of China, aimed to understand the microbial diversity in Xinjiang region, and identify microorganisms that could improve the quality and flavor of wine product. The extensive revealing of microbial diversity in Xinjiang region is helpful in building wine microbial germplasm repository. In addition, this study identified various microbes which are beneficial for the wine production. Further identification and research on these microbes can help to figure out their indigenous beneficial nature, which will in-turn help the winery to improve quality and local characteristics of their wine product.
Supporting information
(TIF)
(TIF)
(XLS)
(XLS)
Acknowledgments
This work was supported by Major Technology Projects of Xinjiang (2017A01001-2) and Natural Science Foundation of China-regional fund (31360406).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Xinjiang Autonomous Region Science and Technology Plan (201431113), by the Major Technology Projects of Xinjiang (2017A01001-2 to YW), and by the Natural Science Foundation of China-regional fund (31360406 to YW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zarraonaindia I, Owens SM, Weisenhorn P, West K, Hampton-Marcell J, Lax S, et al. The soil microbiome influences grapevine-associated microbiota. mBio 2015, 6: e02527–14. doi: 10.1128/mBio.02527-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barata A, Malfeito-Ferreira M, Loureiro V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153(3): 243–259. [DOI] [PubMed] [Google Scholar]
- 3.Swiegers JH, Bartowsky EJ, Henschke PA, Pretorius IS. Yeast and bacterial modulation of wine aroma and flavor. Aust J Grape Wine Res. 2010,11(2): 139–173. [Google Scholar]
- 4.Leonetti D, Soleti R, Clere N, Vergori L, Jacques C, Duluc L, et al. Estrogen receptor α participates to the beneficial effect of red wine polyphenols in a mouse model of obesity-related disorders. Front Pharmacol. 2016,7:529 doi: 10.3389/fphar.2016.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estruch R, Ros E, Salas-Salvadó J, Covas MI, Corella D, Arós F, et al. Primary prevention of cardiovascular disease with a Mediterranean Diet. N Engl J Med. 2013,368(14):1279–1290. doi: 10.1056/NEJMoa1200303 [DOI] [PubMed] [Google Scholar]
- 6.Cueva C, Gil-Sánchez I, Ayuda-Durán B, González-Manzano S, González-Paramás AM, Santos-Buelga C, et al. An Integrated View of the Effects of Wine Polyphenols and Their Relevant Metabolites on Gut and Host Health. Molecules. 2017,22(1):99–105. doi: 10.3390/molecules22010099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dueñas M, Cueva C, Muñoz-González I, Jiménez-Girón A, Sánchez-Patán F, Santos-Buelga C, et al. Studies on Modulation of Gut Microbiota by Wine Polyphenols: From Isolated Cultures to Omic Approaches. Antioxidants (Basel). 2015,4(1):1–21. doi: 10.3390/antiox4010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barata A, Malfeito-Ferreira M, Loureiro V. The microbial ecology of wine grape berries. International Journal of Food Microbiology. 2011,153(3):243–259. doi: 10.1016/j.ijfoodmicro.2011.11.025 [DOI] [PubMed] [Google Scholar]
- 9.Ultee A, Wacker A, Kunz D, Löwenstein R, König H. Microbial Succession in Spontaneously Fermented Grape Must Before, During and After Stuck Fermentation. South African Journal of Enology & Viticulture. 2013,34(1):68–78. [Google Scholar]
- 10.Piao H, Hawley E, Kopf S, DeScenzo R, Sealock S, et al. Insights into the bacterial community and its temporal succession during the fermentation of wine grapes. Front Microbiol. 2015,6(809):809–819. doi: 10.3389/fmicb.2015.00809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Renouf V, Claisse O, Lonvaud-Funel A. Inventory and monitoring of wine microbial consortia. Applied Microbiology & Biotechnology. 2007,75(1):149–164. doi: 10.1007/s00253-006-0798-3 [DOI] [PubMed] [Google Scholar]
- 12.Kilmartin PA. 15–Understanding and controlling non-enzymatic wine oxidation. Managing Wine Quality Oenology & Wine Quality. 2010,144(5):432–458. [Google Scholar]
- 13.Moreno-Arribas MV, Smit AY, Toit MD. 17–Biogenic amines and the winemaking process. Managing Wine Quality Oenology & Wine Quality. 2010:494–522. [Google Scholar]
- 14.Ribereau-Gayon P, Dubourdieu D, Doneche B. Cytology, taxonomy and ecology of grape and wine yeasts In:Handbook of Enology. New York: John Wiley and Sons; 2005, p1–49. [Google Scholar]
- 15.Crump AM, Johnson TE, Wilkinson KL, Bastian SE. Influence of oak maturation regimen on composition, sensory properties, quality, and consumer acceptability of cabernet sauvignon wines. J Agric Food Chem. 2015, 63(5):1593–1600. doi: 10.1021/jf5044025 [DOI] [PubMed] [Google Scholar]
- 16.Vigentini I, Maghradze D, Petrozziello M, Bonello F, Mezzapelle V, Valdetara F, et al. Indigenous Georgian Wine-Associated Yeasts and Grape Cultivars to Edit the Wine Quality in a Precision Oenology Perspective. Front Microbiol. 2016,7:352 doi: 10.3389/fmicb.2016.00352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garofalo C, Tristezza M, Grieco F, Spano G, Capozzi V. From grape berries to wine: population dynamics of cultivable yeasts associated to "Nero di Troia" autochthonous grape cultivar. World J Microbiol Biotechnol. 2016,32(4):59 doi: 10.1007/s11274-016-2017-4 [DOI] [PubMed] [Google Scholar]
- 18.Cocolin L, Bisson LF, Mills DA. Direct profiling of the yeast dynamics in wine fermentations. FEMS Microbiol Lett. 2000,89(1):81–87. [DOI] [PubMed] [Google Scholar]
- 19.Prakichaiwattana CJ, Fleet GH, Heard GM. Application and evaluation of denaturing gradient gel electrophoresis to analyse the yeast ecology of wine grapes. FEMS Yeast Res. 2004,4(8):865–877. doi: 10.1016/j.femsyr.2004.05.004 [DOI] [PubMed] [Google Scholar]
- 20.Renouf V, Claisse O, Lonvaud-Funel A. Inventory and monitoring of wine microbial consortia. Appl Microbiol Biotechnol. 2007,75(1):149–164. doi: 10.1007/s00253-006-0798-3 [DOI] [PubMed] [Google Scholar]
- 21.Song PY, Ma L. TENP—PBS pretreatment application in soil DNA extraction.Journal of Anhui Agricultural (Chinese Literature). 2010, 38:12978–12980. [Google Scholar]
- 22.Wawrik B, Kerkhof L, Kukor J, Zylstra G. Effect of different carbon sources on community composition of bacterial enrichments from soil. Appl Environ Microbiol. 2005,71(11):6776–6783. doi: 10.1128/AEM.71.11.6776-6783.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sessitsch A, Weilharter A, Gerzabek MH, Kirchmann H, Kandeler E. Microbial population structures in soil particle size fractions of a long-term fertilizer field experiment. Appl Environ Microbial. 2001,67(9):4215–4224. doi: 10.1128/AEM.67.9.4215-4224.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girvan MS, Bullimore J, Pretty JN, Osborn AM, Ball AS. Soil type is primary determinant of the composition of the total and active Bacterial communities in arable soil. Appl Environ Microbial. 2003,69(3):1800–1809. doi: 10.1128/AEM.69.3.1800-1809.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshua PS, Jay MG, Joy SC. Moisture cffects on microbial activity and community structure in decomposing birch litter in the Alaskan taiga. Soil Biol Biochem. 1999,31(6):831–838. [Google Scholar]
- 26.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great Bacterial diversity and high metal toxicity in soil. Science, 2005,309:1387–1390. doi: 10.1126/science.1112665 [DOI] [PubMed] [Google Scholar]
- 27.Stephen JR, Chang YJ, Gan YD, Peacock A, Pfiffner SM, Barcelona MJ, et al. Microbial characterization of a JP-4 fuel-contaminated site using a combined lipid biomarker/polymerase chain reaction-denaturing gradient gel electrophoresis(PCR-DGGE)-based approach. Environ Microbial. 1999,1(3):231–241. [DOI] [PubMed] [Google Scholar]
- 28.Dell’Amico E, Mazzocchi M, Cavalca L, Allievi L, Andreoni V. Assessment of bacterial community structure in a long-term copper-polluted ex-vineyard soil. Microbiol Res. 2008,163(6):671–683. doi: 10.1016/j.micres.2006.09.003 [DOI] [PubMed] [Google Scholar]
- 29.Martins G, Lauga B, Miot-Sertier C, Mercier A, Lonvaud A, Soulas ML, et al. Characterization of epiphytic bacterial communities from grapes, leaves, bark and soil of grapevine plants grown, and their relations. PLOS ONE. 2013,8(8):e73013 doi: 10.1371/journal.pone.0073013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albergaria H, Arneborg N. Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: role of physiological fitness and microbial interactions. Appl Microbiol Biotechnol. 2016,100(5):2035–2046. doi: 10.1007/s00253-015-7255-0 [DOI] [PubMed] [Google Scholar]
- 31.Bartowsky EJ, Borneman AR. Genomic variations of Oenococcus oeni strains and the potential to impact on malolacticfermentation and aroma compounds in wine. Appl Microbiol Biotechnol. 2011,92(3):441–447. doi: 10.1007/s00253-011-3546-2 [DOI] [PubMed] [Google Scholar]
- 32.Sainz J, Pizarro F, Pérez-Correa JR, Agosin E. Modeling of yeast metabolism and process dynamics in batch fermentation. Biotechnol Bioeng. 2003,81(7):818–828. doi: 10.1002/bit.10535 [DOI] [PubMed] [Google Scholar]
- 33.Bastard A, Coelho C, Briandet R, Canette A, Gougeon R, Alexandre H, et al. Effect of Biofilm Formation by Oenococcus oeni on Malolactic Fermentation and the Release of Aromatic Compounds in Wine. Front Microbiol. 2016,7:613 doi: 10.3389/fmicb.2016.00613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilber-Bodmer M, Schmid M, Ahrens CH, Freimoser FM, Andrews JH. Competition assays and physiological experiments of soil and phyllosphere yeasts identify Candida subhashii as a novel antagonist of filamentous fungi. BMC Microbiol. 2017,17(1):4 doi: 10.1186/s12866-016-0908-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews JH. Biological control in the phyllosphere. Annu. Rev. Phytopathol, 1992, 30: 603–635. doi: 10.1146/annurev.py.30.090192.003131 [DOI] [PubMed] [Google Scholar]
- 36.Fortes AM, Agudelo-Romero P, Silva MS, Ali K, Sousa L, Maltese F, et al. Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding thedynamics of grape ripening. BMC Plant Biol. 2011,11:149 doi: 10.1186/1471-2229-11-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown JC, Jiang X. Activities of muscadine grape skin and polyphenolic constituents against Helicobacter pylori. J Appl Microbiol.2013,114(4):982–991. doi: 10.1111/jam.12129 [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Nan L, Liu J, Yan H, Zhang D, Han X. Isolation and identification of resveratrol-producing endophytes from wine grape Cabernet Sauvignon. Springerplus. 2016,5(1):1029 doi: 10.1186/s40064-016-2571-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinto C, Pinho D, Sousa S, Pinheiro M, Egas C, Gomes AC. Unravelling the diversity of grapevine microbiome. PLoS One. 2014, 9(1):e85622 doi: 10.1371/journal.pone.0085622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevanato P, Bertaggia M, Stellin F. Soil biological and biochemical traits linked to nutritional status in grapevine. Journal of Soil Science and Plant Nutrition. 2014,14(3):469–479. doi: 10.4067/S0718-95162014005000033 [Google Scholar]
- 41.Renouf V, Claisse O, Lonvaud A. Understangding tne microbial ecosystem on the grape herry surfur through numeration and identification of yeast and Bacteria. Australian Journal of Grape and Wine Research. 2005,11(30):316–327. [Google Scholar]
- 42.Han L, Weng K, Ma H, Xiang G, Li Z, Wang Y, et al. Identification and Characterization of Erysiphe necator-Responsive MicroRNAs in Chinese Wild Vitis pseudoreticulata by High-Throughput Sequencing. Front Plant Sci. 2016,7:621 doi: 10.3389/fpls.2016.00621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao YR, Han YT, Zhao FL, Li YJ, Cheng Y, Ding Q, et al. Identification and utilization of a new Erysiphe necator isolate NAFU1 to quickly evaluate powdery mildew resistance in wild Chinese grapevine species using detached leaves. Plant Physiol Biochem. 2016,98:12–24. doi: 10.1016/j.plaphy.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 44.Amrine KC, Blanco-Ulate B, Riaz S, Pap D, Jones L, Figueroa-Balderas R, et al. Comparative transcriptomics of Central Asian Vitis vinifera accessions reveal distinct defense strategies against powdery mildew. Hortic Res. 2015, 2:15037 doi: 10.1038/hortres.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenzini M, Cappello MS, Logrieco A, Zapparoli G. Polymorphism and phylogenetic species delimitation in filamentous fungi from predominant mycobiota in withered grapes. Int J Food Microbiol. 2016,238:56–62. doi: 10.1016/j.ijfoodmicro.2016.08.039 [DOI] [PubMed] [Google Scholar]
- 46.Prendes LP, Zachetti VG, Pereyra A, Morata de Ambrosini VI, Ramirez ML. Water activity and temperature effects on growth and mycotoxin production by Alternaria alternata strains isolated from Malbec wine grapes. J Appl Microbiol. 2017,122(2):481–492. doi: 10.1111/jam.13351 [DOI] [PubMed] [Google Scholar]
- 47.Garmendia G, Vero S. Occurrence and biodiversity of Aspergillus section Nigri on ‘Tannat’ grapes in Uruguay. Int J Food Microbiol. 2016,216:31–39. doi: 10.1016/j.ijfoodmicro.2015.08.020 [DOI] [PubMed] [Google Scholar]
- 48.Ferrari V, Dellacassa E, Coniberti A, Disegna E. Role of grapevine vegetative expression on Aspergillus spp. incidence and OTA accumulation in wines produced in a temperate humid climate. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2017,34(2):299–306. doi: 10.1080/19440049.2016.1252064 [DOI] [PubMed] [Google Scholar]
- 49.Qi TF, Renaud JB, McDowell T, Seifert KA, Yeung KK, Sumarah MW. Diversity of mycotoxin-producing Black Aspergilli in Canadian vineyards. J Agric Food Chem. 2016,64(7):1583–1589. doi: 10.1021/acs.jafc.5b05584 [DOI] [PubMed] [Google Scholar]
- 50.Cordero-Bueso G, Arroyo T, Serrano A, Tello J, Aporta I, Vélez MD, et al. Influence of the farming system and vine variety on yeast communities associated with grape berries. Int J Food Microbiol. 2011,145(1):132–139. doi: 10.1016/j.ijfoodmicro.2010.11.040 [DOI] [PubMed] [Google Scholar]
- 51.Tan Y, Cui Y, Li H, Kuang A, Li X, Wei Y, et al. Rhizospheric soil and root endogenous fungal diversity and composition in response to continuous Panax notoginseng cropping practices. Microbiol Res. 2017,194:10–19. doi: 10.1016/j.micres.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 52.Cordero-Bueso G, Arroyo T, Serrano A, Valero E. Influence of different floor management strategies of the vineyard on the natural yeast population associated with grape berries. Int J Food Microbiol. 2011,148(1):23–29. doi: 10.1016/j.ijfoodmicro.2011.04.021 [DOI] [PubMed] [Google Scholar]
- 53.Kamma M, Mburu H, Blanchart. Effects of organic and inorganic fertilization on soil Bacterial and fungal microbial diversity in the Kabete long-term trial, Kenya. Biology and fertility of soils. 2011,47(3):315–321. doi: 10.1007/s00374-011-0539-3 [Google Scholar]
- 54.Hirano SS, Upper CD. Bacteria in the Leaf Ecosystem with Emphasis on Pseudomonas syringae–a Pathogen, Ice Nucleus, and Epiphyte. Microbiol Mol Biol Rev. 2000,64(3):624–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park JY, Park JY, Han SH, Lee JH, Han YS, Lee YS, et al. Draft genome sequence of the biocontrol bacterium Pseudomonas putida B001, an oligotrophic bacterium that induces systemic resistance to plant diseases. J Bacteriol. 2011,193(23):6795–6796. doi: 10.1128/JB.06217-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cuixia Li. The separation and identification of fine Oenococcus in China.[D]. Yang Ling: Northwest Agriculture and Forestry University, 2011.
- 57.Portillo MD, Franquès J, Araque I, Reguant C, Bordons A. Bacterial diversity of Grenache and Carignan grape surface from different vineyards at Priorat wineregion (Catalonia, Spain). Int J Food Microbiol. 2015,219:56–63. doi: 10.1016/j.ijfoodmicro.2015.12.002 [DOI] [PubMed] [Google Scholar]
- 58.Martins G, Miot-Sertier C, Lauga B, Claisse O, Lonvaud-Funel A, Soulas G, et al. Grape berry bacterial microbiota: Impact of the ripening process and the farming system. Int J Food Microbiol. 2012,158(2):93–100. doi: 10.1016/j.ijfoodmicro.2012.06.013 [DOI] [PubMed] [Google Scholar]
- 59.O’Brien RD, Lindow SE. Effect of plant species and environmental conditions on ice nucleation activity of Pseudomonas syringae on leaves. Appl Environ Microbiol. 1988,54(9):2281–2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berg G, Smalla K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol. 2009,68(1):1–13. doi: 10.1111/j.1574-6941.2009.00654.x [DOI] [PubMed] [Google Scholar]
- 61.Compant S, Duffy B, Nowak J, Cle´ment C, Barka EA. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, Mechanisms of Action, and Future Prospects. Appl Environ Microbiol. 2005,71(9):4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.