Abstract
Hydrogen peroxide (H2O2) acts as a signaling messenger by triggering the reversible oxidation of redox-regulated proteins. It remains unclear how proteins can be oxidized by signaling levels of H2O2 in the presence of peroxiredoxins, which are highly efficient peroxide scavengers. Here we show that the rapid formation of disulfide bonds in cytosolic proteins is enabled, rather than competed, by cytosolic 2-Cys peroxiredoxins. Under the conditions tested, the combined deletion or depletion of cytosolic peroxiredoxins broadly frustrated H2O2-dependent protein thiol oxidation, which is the exact opposite of what would be predicted based on the assumption that H2O2 oxidizes proteins directly. We find that peroxiredoxins enable rapid and sensitive protein thiol oxidation by relaying H2O2-derived oxidizing equivalents to other proteins. Although these findings do not rule out the existence of Prx-independent H2O2 signaling mechanisms, they suggest a broader role for peroxiredoxins as sensors and transmitters of H2O2 signals than hitherto recognized.
H2O2 acts as a second messenger in signal transduction1. Basal cytosolic steady state H2O2 concentrations are estimated to lie in the low nanomolar range (≈1–10 nM)2 and rise transiently to the upper nanomolar range during oxidative signaling events (≈500–700 nM)3. The elevation of H2O2 levels is known to trigger the oxidation of thiols on redox-regulated proteins4. Reversible thiol oxidation causes transient changes in protein function that are relevant to signal transduction, such as kinase activation.
Notably, the specificity and efficiency of H2O2 as a signaling molecule has always been difficult to explain5–7. The first part of the problem is that typical redox-regulated proteins, namely those that are activated or inactivated by thiol oxidation for signaling purposes, including phosphatases, kinases and transcription factors, have been found to exhibit modest intrinsic H2O2 reactivity (k ≈101–102 M−1 s−1)8. Moreover, many of these proteins are expressed at low levels9. It is not obvious how low-reactivity thiols on low-abundance proteins can be oxidized by signaling concentrations of H2O2 within timescales appropriate for cell signaling. The second part of the problem is that the most prominent group of thiol peroxidases, the peroxiredoxins (Prxs), are expected to capture most of the H2O2 generated inside cells. This is because the intrinsic H2O2 thiol reactivity of Prxs is up to seven orders of magnitude higher compared to redox-regulated proteins (k ≈105–108 M−1 s−1)10. Additionally, Prxs are highly expressed proteins, possibly amounting to ≈1% of the total soluble cellular protein content11. Because of the combination of high abundance and high reactivity, Prxs can be expected to outcompete other thiols9. Given these considerations, there is currently no consensus on how thiols on redox-regulated proteins are actually oxidized.
There are two major schools of thought aiming to explain the phenomenon of H2O2 signaling. One school posits that H2O2 directly reacts with thiols on redox-regulated target proteins; Prxs are seen as competing H2O2 scavengers. It is therefore reasoned that Prxs need to be inactivated, temporarily and locally, to allow the local accumulation of H2O2 to reach concentrations that enable the direct oxidation of protein thiols with low intrinsic H2O2 reactivity8. This ‘direct oxidation’ model predicts that the experimental deletion or depletion of Prxs should lead to an increase of H2O2-induced protein thiol oxidation.
The other school posits that thiol peroxidases, in particular Prxs, because of their abundance and exceptional H2O2 reactivity, are almost always the primary reactants for H2O2, outcompeting other potential target thiols12. It is therefore reasoned that redox-regulated target proteins must receive oxidizing equivalents from thiol peroxidases to become oxidized. Prxs are postulated to relay oxidizing equivalents to redox-regulated proteins. Hence, they are seen as enablers of protein thiol oxidation, not as competitors13. Notably, this ‘peroxidase redox relay’ concept makes a prediction that is in direct opposition to the one made by the direct oxidation model: upon deletion of Prxs, protein thiol oxidation should be decreased, not increased.
It has been known for almost 15 years that thiol-peroxidase-based redox relays do exist in nature. For a long time, such redox relays were only known in the fungal domain. In budding yeast, the first peroxidase recognized to support a redox signaling relay was Orp1, a member of the glutathione peroxidase family. Orp1 acts as a receptor for H2O2 and forwards oxidative equivalents to the transcription factor Yap1 (ref. 14). In fission yeast, the typical 2-Cys peroxiredoxin Tpx1 was found to mediate the oxidation and activation of the protein kinase Sty1 (ref. 15) and the transcription factor Pap1 (ref. 16). Only very recently, similar evidence has emerged in human cells. In particular, Prx1 facilitates oxidation of the kinase ASK1 (ref. 17), and Prx2 facilitates oxidation of the transcription factor STAT3 (ref. 18). Hence, though it is known that Prx-based redox relays do exist in mammalian cells, very few well-documented examples have been reported so far. Thus, the key question is whether Prx-based relays are common and responsible for the majority of thiol oxidation events in mammalian cells.
In this study, we explored the abovementioned experimental predictions made by the two models, i.e., direct versus peroxidase-mediated protein thiol oxidation. If protein thiol oxidation is predominantly direct, and therefore opposed by competing Prxs, then the experimental deletion of Prxs should increase overall protein thiol oxidation. However, if thiol oxidation is predominantly mediated by Prxs, the opposite outcome can be expected, namely a decrease in protein thiol oxidation. Hence, in our study we aimed to investigate the influence of Prx expression on protein thiol oxidation. To this end, we increased endogenous H2O2 levels in cells that were either proficient or deficient in cytosolic typical 2-Cys Prxs and monitored the impact on protein thiol oxidation.
Our specific approach was based on three decisions. First, we chose to conduct our experiments under defined conditions of H2O2 delivery and cellular uptake, as characterized previously19. Second, we chose to specifically monitor protein thiol oxidation in the cytosolic–nuclear compartment of human cells. This compartment is dominated by two typical 2-Cys Prxs (Prx1 and Prx2), which together are estimated to consume most of the H2O2 that emerges inside the cytosol from either internal or external sources9. Therefore, the combined ablation of these two peroxidases is expected to have a substantial impact on cytosolic protein thiol oxidation, in one direction or the other, depending on the role of these Prxs as either competitors or mediators of thiol oxidation. The combined deletion of both Prxs also avoids potential problems of redundancy, as Prx1 and Prx2 are highly homologous and may compensate for each other when deleted individually. Third, we chose to use protein–thioredoxin (Trx) disulfide exchange interactions as a readout for protein thiol oxidation. We made use of the fact that many (if not most) redox-regulated proteins form transient disulfide bonds that can be recognized and reduced by Trx20–22. Thus, our approach specifically focused on assessing the formation of oxidative thiol modifications visible to the Trx system.
Using the above strategy, we show here that the deletion or depletion of cytosolic Prxs suppresses overall H2O2-dependent protein thiol oxidation, at least under the specific conditions tested in this study. Furthermore, we provide evidence that Prxs directly oxidize other proteins. We highlight the role of Prxs as sensitive and abundant forwarders of H2O2-derived oxidizing equivalents, and thus as highly efficient enablers of protein thiol oxidation and redox signaling, as predicted and anticipated previously9,12,23–26.
Results
H2O2-induced protein thiol oxidation depends on Prxs
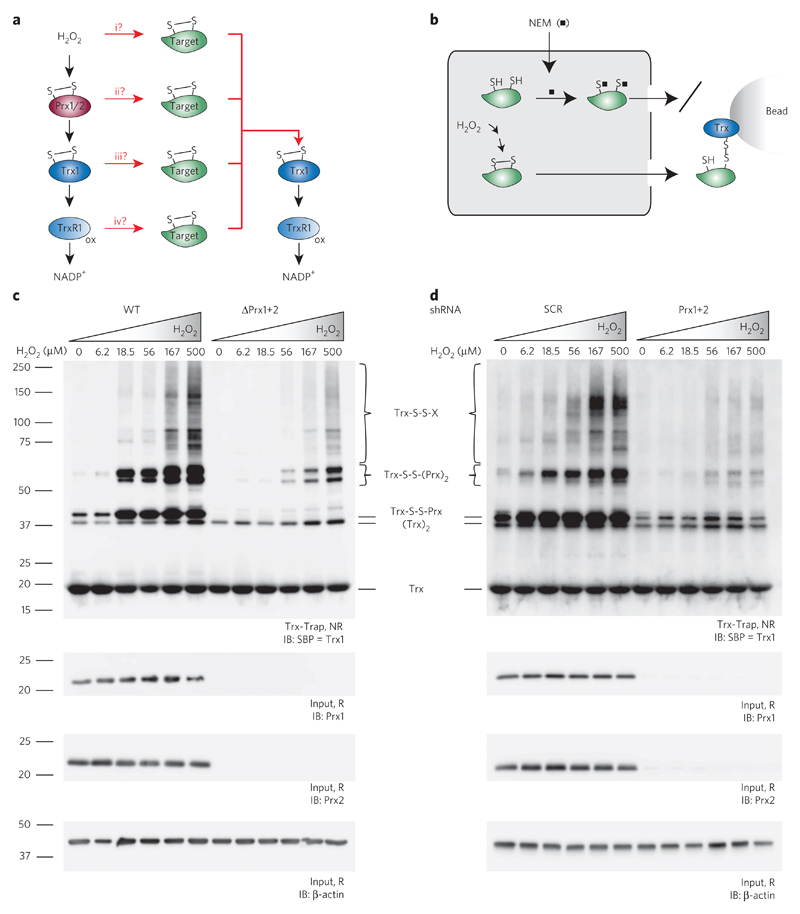
It is widely assumed that the primary function of Prxs is to scavenge H2O2 and deliver the oxidizing equivalents to the Trx system for reductive elimination27. Thus, the expected cytosolic flow of oxidizing equivalents from H2O2 to NADPH leads through Prx1 and Prx2, thioredoxin-1 (Trx1) and thioredoxin reductase-1 (TrxR1; Fig. 1a, left side). Given the H2O2–Prx–Trx–TrxR–NADPH pathway, there are four theoretical possibilities for how H2O2-derived oxidizing equivalents can find their way to a redox-regulated target protein to cause reversible thiol oxidation, typically disulfide bond formation (Fig. 1a, center). First (i), some H2O2 may escape capture by Prxs and directly oxidize target proteins. Second (ii), oxidized Prxs may transmit oxidizing equivalents to target proteins. Third (iii), oxidized Trx may transmit oxidizing equivalents to target proteins. Fourth (iv), oxidized TrxR may transmit oxidizing equivalents to target proteins. No matter how a disulfide bond is introduced into a target protein, it can be expected to be eventually reduced by the Trx system (Fig. 1a, right side).
Figure 1. H2O2-induced cytosolic protein thiol oxidation depends on cytosolic peroxiredoxins.
(a) Scheme depicting theoretical possibilities for H2O2-derived oxidizing equivalents to reach and oxidize redox-regulated proteins. Left column, canonical flow of oxidizing equivalents from H2O2 to NADPH through Prx1/2, Trx1 and TrxR1. Middle column, branch points (i–iv) potentially involved in the delivery of oxidizing equivalents to redox-regulated target proteins. Right column, reduction of oxidized target proteins by the thioredoxin system. All molecules are shown in the oxidized state. (b) Scheme depicting the mechanism-based kinetic trapping approach used to monitor cytosolic protein thiol oxidation. Prior to cell lysis, free thiols are blocked with N-ethylmaleimide (NEM; black squares). Cells are osmotically disrupted to release cytosolic proteins. Disulfide-containing proteins are selectively captured by the immobilized recombinant trapping mutant of human Trx1. (c,d) HAP1 cells proficient (wild type, WT) or deficient (ΔPrx1+2) in Prx1+2 expression (c) and HEK293T cells induced to express either scrambled (SCR) or specific (Prx1+2) shRNA (d) were exposed to the indicated concentrations of H2O2 for 15 s. Overall protein thiol oxidation, reflected by the formation of Trx-S-S-X conjugates, was assessed by the kinetic trapping approach and analyzed by immunoblotting against the SBP tag of the Trx1 trapping mutant. In c and d, different types of Trx conjugates are indicated. The Trx-S-S-Prx and Trx-S-S-(Prx)2 conjugates seen in cells lacking Prx1+2 represent trapping of other members of the Prx family (see Supplementary Fig. 3). Uncropped blots for c and d are shown in Supplementary Figure 4. IB, immunoblotting; NR, nonreducing; R, reducing conditions; SBP, streptavidin-binding peptide; Trx-Trap, eluate from Trx trapping beads. Blots are representative of ≥3 independent experiments.
To find out which of the four oxidation pathways is predominantly responsible for overall cytosolic protein disulfide bond formation in a given experimental situation, we made use of the fact that proteins with disulfide bonds are typically recognized and reduced by Trx. Cells were osmotically disrupted to release soluble cytosolic proteins into the supernatant (Fig. 1b, left side). Disulfide bond–containing proteins were then selectively captured with an immobilized recombinant human Trx1 mutant lacking its resolving cysteine (so-called ‘mechanism-based kinetic trapping’28; Fig. 1b, right side).
Using the Trx-based substrate trapping strategy, we first asked how the presence or absence of Prxs influences overall protein thiol oxidation, as triggered by the exogenous delivery of H2O2. Exposure of cells to H2O2 was restricted to 15 s, causing highly transient and fully reversible protein thiol oxidation (Supplementary Fig. 1). We compared wild-type HAP1 cells to isogenic counterparts in which the expression of both cytosolic Prxs (Prx1 and Prx2) was abolished by CRISPR–Cas9-mediated genome editing. Titration with exogenous H2O2 revealed that wild-type cells were more susceptible to protein thiol oxidation than cells lacking cytosolic Prxs (Fig. 1c and Supplementary Fig. 2a). A similar result was obtained with a different cell line (HEK293T) and a different method of disrupting Prx expression, inducible short hairpin RNA (shRNA)-mediated depletion (Fig. 1d and Supplementary Fig. 2b).
If it is generally true that proteins are directly oxidized by H2O2 (Fig. 1a, path i), their oxidation should not be decreased upon deletion or depletion of Prxs. On the contrary, deletion or depletion of Prxs would be expected to increase general protein thiol oxidation, because the lack of competition by Prxs should increase the availability of H2O2. Thus, the result suggested that the direct reaction between H2O2 and target proteins (Fig. 1a, path i) is unlikely to play a substantial role in protein thiol oxidation, at least under the given experimental conditions.
Trx1 and TrxR1 are not needed for protein thiol oxidation
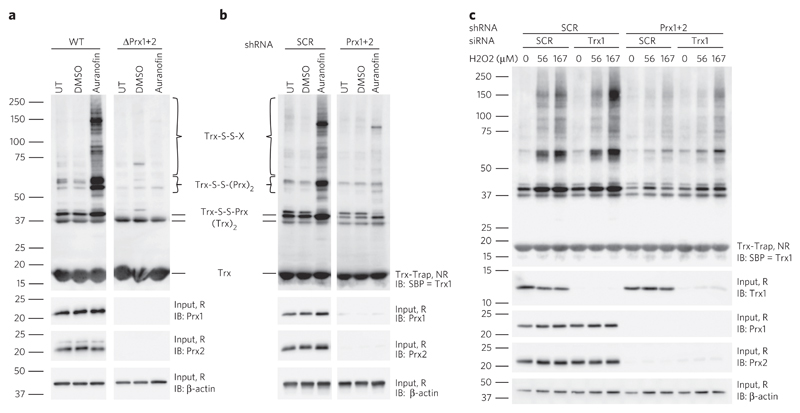
Next, we inhibited TrxR1 with the small-molecule inhibitor auranofin to block the flow of oxidizing equivalents along the whole Trx pathway and to create a pro-oxidative environment from inside the cell. As expected, treatment with auranofin led to pronounced cytosolic protein thiol oxidation (Fig. 2a,b). This finding showed that TrxR activity is not needed for protein thiol oxidation to occur, ruling out hypothetical pathway iv as a major contributor to protein thiol oxidation (Fig. 1a, path iv). However, deletion (Fig. 2a and Supplementary Fig. 5a) or depletion (Fig. 2b and Supplementary Fig. 5b) of the cytosolic Prxs almost completely abolished auranofin-induced protein thiol oxidation. Hence, overall protein thiol oxidation again depended on the presence of Prxs, which in this situation was caused by endogenous H2O2, known to accumulate when the Prx–Trx–TrxR cascade is blocked by auranofin29. Having ruled out paths i and iv, the remaining question was whether protein thiol oxidation is mediated directly by Prxs (path ii) or through Trx (path iii), which in principle can also act as a protein oxidant30,31.
Figure 2. Trx1 and Trxr1 are not required for transmission of oxidative equivalents to cytosolic proteins.
(a,b) HAP1 cells proficient (WT) or deficient (ΔPrx1+2) in Prx1+2 expression (a) and HEK293T cells induced to express scrambled (SCR) or specific (Prx1+2) shRNA (b) were treated for 1 h with 10 μM (a) or 20 μM (b) auranofin or solvent control (DMSO), or were left untreated (UT). Overall protein thiol oxidation was assessed by kinetic trapping and analyzed by immunoblotting (IB). (c) HEK293T cells induced to express scrambled or specific (Prx1+2) shRNA were transfected with scrambled or specific (Trx1) siRNA and exposed to the indicated concentrations of H2O2 for 15 s. Overall protein thiol oxidation was assessed by kinetic trapping and analyzed by immunoblotting against the SBP tag of the Trx1 trapping mutant. Uncropped blots are shown in Supplementary Figure 7. NR, nonreducing; R, reducing conditions; SBP, streptavidin binding peptide; Trx-Trap, eluate from Trx trapping beads. Blots are representative of ≥ 3 independent experiments.
To answer this question, we combined inducible shRNA-mediated Prx1 and Prx2 depletion with siRNA-mediated Trx1 depletion. If Trx1 acts as a protein oxidase, fewer proteins would be oxidized upon Trx1 depletion. However, Trx1 depletion caused a slight increase in overall protein thiol oxidation in response to H2O2, both in the presence and the absence of Prxs, but did not affect basal Prx redox state (Fig. 2c and Supplementary Fig. 6). In contrast, depletion of Prxs again led to strongly diminished overall protein disulfide formation (Fig. 2c). This result suggested that, in the given experimental context, the vast majority of cytosolic protein thiol oxidation is directly mediated by cytosolic Prxs (path ii) and is not the result of Trx1 oxidase activity (path iii).
H2O2 induces Prx disulfide conjugates with other proteins
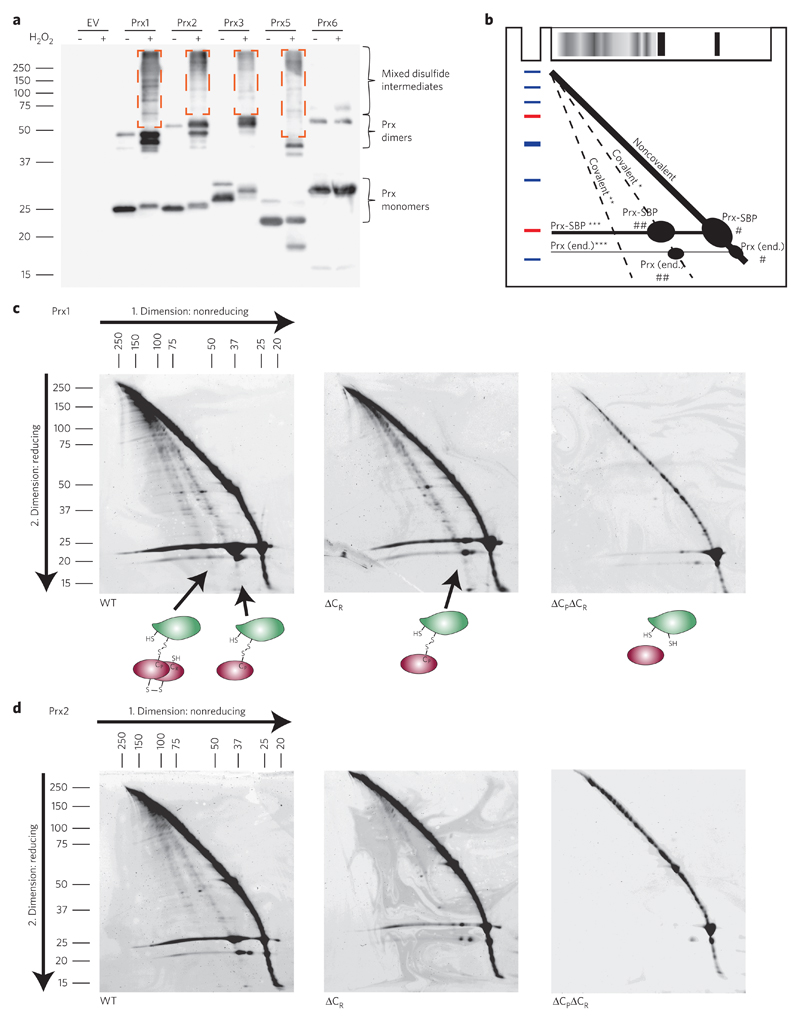
If Prxs broadly mediate H2O2-induced protein thiol oxidation, they should form transient mixed disulfides with many proteins, as previously observed in the specific case of STAT3 (ref. 18). To see whether this is indeed the case, we tagged Prxs with a high-affinity streptavidin-binding peptide (SBP; Kd ≈109), expressed them at levels similar to those of endogenous Prxs and applied a short H2O2 pulse to induce Prx oxidation; this was followed by rapid quenching of free thiols with high concentrations (100 mM) of N-ethylmaleimide (NEM). Analysis of affinity-purified Prxs by conventional nonreducing SDS–PAGE revealed that Prx1 and Prx2 form numerous highly transient disulfide exchange intermediates upon H2O2 treatment (Fig. 3a). Similar behavior was observed for the mitochondrial typical 2-Cys peroxiredoxin Prx3 and the atypical 2-Cys peroxiredoxin Prx5, but not for the 1-Cys peroxiredoxin Prx6 (Fig. 3a).
Figure 3. Upon H2O2 exposure, peroxiredoxins form transient disulfide exchange intermediates with other proteins.
(a) HEK293T cells expressing SBP-tagged Prx family members (1, 2, 3, 5 and 6) were exposed to 100 μM H2O2 for 1 min (+) or left untreated (−). Free thiols were blocked by NEM before cell lysis. Mixed disulfide intermediates (outlined by dashed-line rectangles) were visualized on nonreducing gels by immunoblotting against the streptavidin binding peptide (SBP) tag. The blot is representative of three independent experiments. EV, empty vector. (b) Scheme explaining the pattern of proteins on two-dimensional nonreducing/reducing diagonal gels. *, interaction partners (X) released from X-S-S-Prx conjugates; **, interaction partners (X) released from X-S-S-(Prx)2 conjugates; ***, Prx-SBP or Prx (end.) released from X-S-S-Prx or X-S-S-(Prx)2 conjugates; #, Prx-SBP or Prx (end.) that was originally in the monomeric form; ##, Prx-SBP or Prx (end.) that was originally in the dimeric form; end., endogenous. Red and blue marks indicate molecular weight markers. (c,d) 2 × 108 HEK293T cells expressing either wild-type (WT), resolving cysteine-deficient (ΔCR) or double (resolving and peroxidatic) cysteine-deficient (ΔCPΔCR) Prx1-SBP (c) or Prx2-SBP (d) were exposed to 100 μM H2O2 for 3 min. Following thiol blocking and affinity purification, covalent interactions were analyzed by two-dimensional nonreducing/reducing diagonal SDS–PAGE. In c, the cartoons illustrate the nature of the disulfide linked complexes that lead to the formation of the two lower diagonals. In c and d, the arrows indicate the direction of protein migration.
Focusing on cytosolic peroxiredoxins Prx1 and Prx2, we repeated the experiment on a larger scale. Analysis by two-dimensional nonreducing/reducing diagonal SDS–PAGE confirmed that wild-type Prx1 and Prx2 do engage in disulfide interactions with other proteins (Fig. 3b–d and Supplementary Fig. 8a, diagonals below main diagonal). The mixed disulfide intermediates involved either monomeric (Fig. 3c,d, left panels, first lower diagonal) or dimeric Prx (second lower diagonal). As expected, mutation of the resolving cysteine (CR) prevented formation of conjugates involving dimeric Prx (i.e., Prx-S-S-Prx-S-S-X), but still allowed formation of 1:1 conjugates (i.e., Prx-S-S-X), as reflected by the selective loss of the second lower diagonal (Fig. 3c,d, middle panels). Mutation of both peroxidatic (CP) and resolving cysteines abolished all conjugate formation with other proteins (Fig. 3c,d, right panels). The single mutation of CP also abolished conjugate formation (Supplementary Fig. 9), indicating that CP is both necessary and sufficient for forming transient covalent complexes with other proteins. We confirmed that the conjugation of Prxs to other proteins takes place under conditions of minimal cellular H2O2 exposure (10 μM for 15 s) (Supplementary Fig. 8b–d) and identified proteins located on the lower diagonals by MS. We also identified proteins captured by the Trx-trapping mutant under the same conditions of minimal H2O2 exposure (10 μM for 15 s; Supplementary Fig. 8e). We found that many proteins forming transient disulfide intermediates with Prx1 and/or Prx2 inside cells also form a disulfide bond with the Trx1 trapping mutant (Supplementary Table 1), confirming the notion that proteins oxidized by Prxs are subsequently reduced by Trx.
Oxidation of individual proteins depends on Prxs
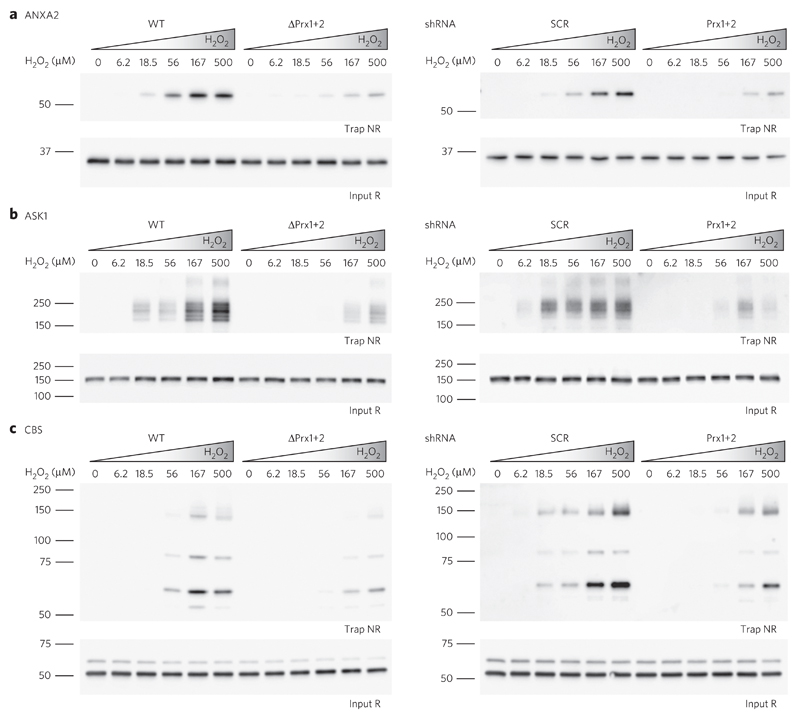
We finally asked whether the observed global effects on protein thiol oxidation can be confirmed on the level of individual proteins. To this end, we selected five example proteins: the transcription factor STAT3, apoptosis signal-regulating kinase 1 (ASK1), annexin A2 (ANXA2) and the collapsin response mediator protein 2 (CRMP2), all of which were previously reported to be redox-regulated21,22,32,33, and cystathionine β-synthase (CBS). STAT3 and ASK1 were already shown to be oxidized in a Prx-dependent manner; therefore, we included them as positive controls17,18 (Supplementary Table 1). Using specific antibodies, we reconfirmed that the selected proteins form mixed disulfide intermediates with Prxs (Supplementary Fig. 10). Furthermore, all five proteins were trapped by Trx1 upon cellular exposure to H2O2, confirming that these proteins form oxidation products that are targeted by Trx. As expected, the deletion or depletion of cytosolic Prxs diminished oxidation of all five proteins (Fig. 4 and Supplementary Fig. 11). Hence, what was observed at the level of the overall cytosolic protein pool was also reproducible at the level of individual proteins.
Figure 4. Oxidation of individual redox-regulated proteins depends on the presence of Prxs.
(a–c) HAP1 cells proficient (WT) or deficient (ΔPrx1+2) in Prx1+2 expression (left panels) and HEK293T cells induced to express scrambled (SCR) or specific (Prx1+2) shRNA (right panels) were exposed to the indicated concentrations of H2O2 for 15 s. Protein thiol oxidation of ANXA2 (a), ASK1 (b) and CBS (c) was assessed by kinetic trapping and analyzed by immunoblotting. Additional examples are shown in Supplementary Figure 11. Uncropped blots are shown in Supplementary Figure 12. NR, nonreducing; R, reducing conditions. Blots are representative of ≥2 independent experiments.
Discussion
The two major models of H2O2-dependent protein thiol oxidation (direct versus mediated oxidation) are not mutually exclusive. In principle, it is clear that both modes of thiol oxidation do occur in living cells. Evidence suggests that at least some redox-regulated proteins can be directly oxidized by H2O2 (refs. 34,35). From the viewpoint of cell signaling requirements, it seems likely that those redox-regulated proteins required to respond to H2O2 within tens of seconds (<1 min) need a peroxidase-mediated oxidation mechanism. By contrast, if a redox-regulated protein can fulfill its signaling role by becoming oxidized over many minutes, direct oxidation may be sufficient36. Yet, the feasibility of direct oxidation under signaling conditions is still under debate. For example, a recently developed mathematical model suggests that even H2O2 concentrations that are attained at the apex of gradients near endogenous supply sites (for example, mitochondria or NOX enzymes) may be too low to directly oxidize redox-regulated target proteins37.
In any case, it is likely that the spatiotemporal distribution of H2O2 and the location of the redox-regulated protein play key roles in determining by which mechanism thiol oxidation takes place. In addition, there may be features intrinsic to a protein that either promote or prevent a particular mode of thiol oxidation. For example, some proteins may exhibit an interaction site that promotes colocalization with a Prx, potentially making them preferred targets of mediated oxidation. Conversely, redox-regulated thiols located within narrow or deep cavities (like the active site cysteines of PTP1B or GAPDH) may be inaccessible for mediated oxidation mechanisms unless major structural changes are involved.
In setting up our experiments, we anticipated that Prx-mediated thiol oxidation is most likely to play a major role when redox-regulated proteins respond rapidly (within 60 s) to small increases in H2O2. Based on this premise, we exposed cells to small quantities of exogenous H2O2 for 15 s under experimental conditions we previously characterized in detail19. We observed that mixed disulfide intermediates between Prxs and other proteins were triggered by a 15 s exposure to a 10 μM H2O2 bolus (Supplementary Fig. 8), which, under the given conditions, delivered ≈280 attomoles of H2O2 to each cell. This amount is of the same order as the (conservatively) estimated number of Prx1 + Prx2 molecules per cell (see Online Methods). Thus, it seems that Prxs rapidly transfer oxidation to other proteins under very mild pro-oxidative conditions, which are unlikely to impose a substantial burden on the reductive capacity of the Prx–Trx–TrxR system.
We then tested exposure to auranofin, a drug that inhibits TrxR and elevates intracellular oxidant levels, perhaps also by additional mechanisms38. Interestingly, we observed that the influence of Prx deletion on auranofin-induced protein thiol oxidation was even more pronounced, leading to almost complete abolishment of protein thiol oxidation. Although we lack a quantitative spatiotemporal understanding of how endogenous H2O2 levels are affected by auranofin treatment in our system, this finding suggests that thiol oxidation by endogenously generated H2O2 depends even more strongly on Prxs than thiol oxidation caused by exogenously supplied H2O2.
Overall, our results strengthen the notion that Prxs have a more general and widespread role in transmitting oxidation to redox-regulated proteins than hitherto recognized39. Nevertheless, our findings should not be prematurely extrapolated to indicate that cytosolic protein thiol oxidation is always predominantly mediated by Prx. In our study we investigated two kinds of pro-oxidative situations (one based on short pulses of exogenous H2O2 and one based on auranofin) and exclusively monitored oxidative thiol modifications that are ‘visible’ to Trx1 (predominantly disulfides). These experiments cannot be expected to be representative of all pro-oxidative situations that can occur in biological systems, as they are known to vary widely in terms of intensity, duration and spatiotemporal dynamics (from signaling to ‘oxidative stress’). Moreover, not all kinds of oxidative thiol modifications can be monitored with a Trx trapping mutant. Hence, it will be an important task for the future to further define which proteins in which subcellular locations are oxidized by which mechanism under which conditions.
Several recent studies have already indicated a more general role for Prx-mediated protein thiol oxidation. For example, an experiment conducted in yeast showed that all sensing of H2O2 for the purpose of gene regulation is mediated by thiol peroxidases. Yeast cells lacking all eight thiol peroxidases were unable to alter gene expression in response to H2O2 (ref. 40). Thus, the yeast thiol peroxidases, including Prxs, are the primary sensors for H2O2, and direct oxidation of redox-regulated proteins does not seem to play a substantial role in transcriptional regulation. More recently, the extraordinary prowess of Prxs in relaying oxidation to other proteins was also indicated by the observation that Prxs mediate oxidation of redox-sensitive GFP (roGFP) with a sensitivity that allows monitoring of in vivo fluctuations of baseline H2O2 levels41.
The pro-oxidative relay function of Prxs should not be seen to contradict the long-recognized and well-established antioxidative functions of Prxs27,42. During an episode of H2O2 elevation, the majority of Prx molecules can be expected to deliver oxidizing equivalents to Trx and thus counteract H2O2 elevation and nonspecific oxidation events. Yet, at the same time, local subpopulations of Prx molecules would selectively reroute oxidative equivalents to proximal redox-regulated proteins39. Thus, the concept emerges that Prxs can protect the majority of proteins against oxidation, while actively oxidizing a subset of proteins, depending on site-specific interactions.
Finally, it is interesting to note that the deregulation of Prx expression has been associated with tumorigenesis, neurodegenerative and inflammatory diseases43,44. These associations are usually discussed in relation to the anti-oxidative function of Prxs. Our findings suggest that pro-oxidative functions of Prxs, i.e., Prx-mediated protein thiol oxidation and redox signaling, should also be considered to play a role in the disease context.
Online Methods
Cell lines, antibodies and reagents
HAP1 cells were obtained from Horizon Discovery. HAP1 ΔPrx1ΔPrx2 double-knockout cells were generated by CRISPR–Cas9-mediated gene editing. HAP1 cells were cultured in Iscove′s Modified Dulbecco′s Medium (IMDM; Life technologies), supplemented with 10% (vol/vol) bovine calf serum (Life Technologies) and 50 units/mL of penicillin and streptomycin (Life Technologies). HEK293T cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Life Technologies), supplemented with 10% (vol/vol) bovine calf serum (Life Technologies), 2 mM l-glutamine (Life Technologies) and 50 units/mL of penicillin and streptomycin (Life Technologies). All cell lines were authenticated in regular intervals by SNP-based Multiplex Human Cell Line Authentication to exclude the possibility of cross-contamination. All cell lines were furthermore confirmed to be free of mycoplasma and viral infections. Primary antibodies used in this study were rabbit anti-Prx1 (Cell Signaling; #8499), goat anti-Prx2 (R&D Systems; AF3489), mouse anti-Prx3 (Abcam; ab16753), rabbit anti-Prx4 (GeneTex, Inc; GTX15574), goat anti-Prx5 (R&D Systems; AF3774), mouse anti-SBP (Santa Cruz; sc-101595), rabbit anti-Trx1 (Cell Signaling; #2429), mouse anti-annexin A2 (BD Transduction Laboratories; BD610069), rabbit anti-ASK1 (Abcam; ab45178), rabbit anti-CBS (Abcam; ab135626), rabbit anti-CRMP2 (Abcam; ab129082), mouse anti-STAT3 (Cell Signaling; #9139) and mouse anti-β-actin (Sigma; A5441). Secondary antibodies used in this study were donkey anti-goat IgG-HRP (Santa Cruz; sc2020), peroxidase-conjugated AffiniPure goat anti-rabbit IgG (Jackson ImmunoResearch; 111-035-144) and peroxidase-conjugated AffiniPure goat anti-mouse IgG (Jackson ImmunoResearch; 115-035-146). Auranofin, N-ethylmaleimide (NEM) and dimethyl sulfoxide (DMSO) were from Sigma. Hydrogen peroxide (30%) was from Roth, and Dulbecco′s phosphate buffered saline (DPBS) was from Life Technologies.
Plasmids, shRNA constructs and siRNAs
Gateway donor vectors for human peroxiredoxins 1, 2, 3, 5 and 6 were obtained from the DKFZ Genomics and Proteomics Core Facility. The NEBuilder Assembly Tool was used for primer design. Open reading frames and the streptavidin binding peptide (SBP) tag were amplified by PCR and ligated into pcDNA3.1(−) using the Gibson Assembly Cloning Kit (New England BioLabs). Mutations were introduced by using the QuikChange Site-Directed Mutagenesis Kit (Agilent). Plasmids used in this study were pQE-60 hTrx1(CSAAA)-SBP-His6, pcDNA3.1(−), pcDNA3.1 Prx1-SBP, pcDNA3.1 Prx1(C52A)-SBP, pcDNA3.1 Prx1(C173A)-SBP, pcDNA3.1 Prx1(C52+173A)-SBP, pcDNA3.1 Prx2-SBP, pcDNA3.1 Prx2(C51A)-SBP, pcDNA3.1 Prx2(C172A)-SBP, pcDNA3.1 Prx2(C51+172A)-SBP, pcDNA3.1 Prx3-SBP, pcDNA3.1 Prx5-SBP, pcDNA3.1 Prx6-SBP, pTRIPZ Prdx1 shRNA (V2THS_152607; Dharmacon), modified pTRIPZ Prdx2 shRNA (V2THS_197737; Dharmacon; AmpR–, BleoR+), pTRIPZ nonsilencing shRNA control (RHS4743; Dharmacon), psPAX2 and pMD2.G. siRNA against Trx1 (5′-AUGACUGUCAGGAUGUUGC-3′) and control siRNA (5′-GAAUGCUCAUGUUGAAUCA -3′) were from Eurofins.
Transfection of HEK293T cells
1 × 106 HEK293T cells were seeded into 150 cm2 cell culture dishes and grown overnight. The next day, cells were transfected using the polyethylenimine (PEI) method. For each dish, 12.5 μg of DNA was mixed with 37.5 μg PEI (Polysciences) in 2 mL sterile TBS (pH 7.4). After vortexing and incubation for 30 min at room temperature, the mixture was added dropwise to the cell culture medium. The medium was replaced after 5 h. For siRNA transfection of HEK293T cells the Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific) was used, following the manufacturer’s instructions.
Generation of cell lines stably expressing doxycycline-inducible shRNAs
HEK293T were transfected with pTRIPZ vectors for control shRNA, Prx1 shRNA or Prx2 shRNA together with pMD2.G and psPAX2 plasmids (second generation lentiviral system) for viral production. Fresh HEK293T cells were infected with the viral supernatants. The procedure was repeated on the following day to enhance infection efficiency. Cells were selected with 1.5 μg/mL puromycin (Sigma) until colonies formed (7–14 d). Cells stably expressing the Prx1 shRNA construct were infected to also express the Prx2 shRNA construct. After double selection with puromycin and 150 μg/mL zeocin (Invitrogen), cells were expanded and used for experiments.
Production of recombinant thioredoxin trapping mutant
We used the human Trx1 mutant ‘CSAAA’ (i.e., C32 is unchanged while all other cysteines are mutated: C35S, C62A, C69A, C73A) in all mechanism-based kinetic trapping experiments45. The mass of the hTrx1(CSAAA)-SBP-His protein is 17123 Da. Its amino acid sequence is: MVKQIESKTAFQEALDAAGDKLVVVDFSATWCGPSKMIKPFFHSLSEKYSNVIFLEVDVDDAQDVASEAEVKAMPTFQFFKKGQKVGEFSGANKEKLEATINELVRSMDEKTTGWRGGHVVEGLAGELEQLRARLEHHPQGQREPGSHHHHHH.
The single cysteine and mutated residues are indicated in bold, the SBP tag is underlined, and the hexahistidine tag is indicated in italic.
Chemically competent Escherichia coli BL21(DE3) cells were transformed with bacterial expression plasmid pQE-60 encoding hTrx1(CSAAA)-SBP-His. LB medium (Luria/Miller, Roth) supplemented with 0.1 mg/mL ampicillin (Sigma) was inoculated with a colony of transformed E. coli and grown overnight at 37 °C. The next day, 100 mL of the preculture were diluted into 900 mL fresh LB medium supplemented with ampicillin and grown at 34 °C until reaching an optical density (OD) of 0.7–0.8. Recombinant protein expression was then induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG, AppliChem). After 3–4 h of shaking at 34 °C, cells were lysed in 20 mL bacterial protein extraction reagent (B-PER, Thermo Fisher Scientific), and the recombinant protein was affinity purified from the supernatant using 1 mL streptavidin sepharose high performance beads (SA beads; 50% slurry; GE Healthcare). After 2–3 h of incubation, SA beads were washed three times with 20 mM dithiothreitol (DTT; Sigma) in Tris-based saline (TBS, pH 7.4) and kept in the same buffer at 4 °C for at most 24 h before usage.
H2O2 bolus treatment
All experiments involving H2O2 were conducted under the same conditions (i.e., cell densities and amounts of H2O2 per cell), previously established and characterized in detail19. For example, under these conditions, a 10 μM H2O2 bolus provides a total of 14 femtomoles of H2O2 to each cell. The experimentally determined exponential decay constant of H2O2 in the supernatant of HEK293T cells (λ = 1.2–1.5 × 10−3 s−1)19 corresponds to a half-life of externally applied H2O2 between 460 and 580 s, and to a rate constant of ≈2 × 10−12 s−1 cell−1 L, which is typical and close to the average for human cell lines46. In contrast to previous experiments, we applied H2O2 boli for just 15 s, which means that only a minor fraction (2%) of the provided H2O2 is actually taken up (for example, ≈280 attomoles/cell in the case of a 10 μM bolus). This also implies that the gradient-driven flux of H2O2 into the cytosol is nearly constant over the 15 s time window (≈20 attomoles cell−1 s−1 in the case of a 10 μM bolus). Using this information, it is also possible to estimate the stoichiometric relationship between H2O2 and peroxiredoxin molecules. The volume (cytosol plus nucleus) of HEK293T cells has been reported as ≈1,800 μm3 (1.8 × 10−12 L)47. The intracellular concentration of human Prx1 has been estimated as 15–60 μM48 and that of human Prx2 as 20 μM6. The combined concentration of cytosolic peroxiredoxins may thus be estimated as 50 μM. This means that each cell holds ≈90 attomoles of cytosolic Prxs. Hence, a 15 s 10 μM H2O2 bolus in total delivers just thrice as many H2O2 molecules to one cell than there are Prx molecules in one cell.
Mechanism based kinetic trapping
7 × 106 HEK293T or HAP1 cells were seeded into 150 cm2 cell culture dishes to achieve ≈75% confluency on the next day. Following treatment (for example, H2O2), medium was aspirated and cells were incubated for 5 min with 10 mM NEM in DPBS. Cells were washed three times with DPBS and then lysed in 4 mL hypotonic lysis buffer (20 mM HEPES, 2 mM EGTA and 2 mM MgCl2, pH 7.4) supplemented with Complete protease inhibitor cocktail tablets (Roche). SA beads loaded with immobilized recombinant hTrx1(CSAAA)-SBP-His were washed three times with TBS just before use. To allow trapping of disulfide bond–containing proteins, the cytosolic efflux from one dish was incubated with 80 μL of hTrx1(CSAAA)-loaded SA beads at 4 °C on a rotary wheel. The trapping reaction was stopped after 1 h by adding 80 mM NEM. After 10 min, beads were washed with 1% Triton X-100, 0.5 mM NaCl, 1 M urea and 1 mM NEM in TBS, then with 1% Triton X-100 and 1 mM NEM in TBS and finally with 0.1% Triton X-100 in TBS. Proteins were eluted from SA beads with 135 μL of 4 mM biotin in TBS and stored at −20 °C until analysis.
Affinity purification of SBP-tagged peroxiredoxins
1 × 106 HEK293T cells were seeded in 150 cm2 cell culture dishes to be transfected on day two and used for experiments on day four. After exposure of cells to H2O2, the medium was aspirated and cells were immersed in 100 mM NEM in DPBS for 5 min. Cells were then lysed in 2 mL of hypotonic lysis buffer supplemented with Complete protease inhibitor cocktail tablets (Roche). Supernatants were incubated with 30 μL SA-beads (50% slurry) for 4 h. Beads were washed with 1% Triton X-100, 0.5 mM NaCl, 1 M urea and 1 mM NEM in TBS, then with 1% Triton X-100 and 1 mM NEM in TBS and finally with 0.1% Triton X-100 in TBS. Protein was eluted from the beads with 135 μL of 4 mM biotin in TBS and stored at −20 °C until analysis.
Immunoblot analysis
Protein samples in SDS sample buffer were equally divided into nonreduced and reduced (20 mM DTT) fractions. Samples were run on SDS–PAGE gels and proteins transferred to polyvinyl difluoride (PVDF) membranes (Immobilon-P; Millipore) using a tank transfer unit (TE22, Hoefer). Membranes were probed with appropriate antibodies and analyzed using SuperSignal West Femto chemiluminescent substrate (Thermo Scientific).
Two-dimensional nonreducing/reducing (diagonal) SDS–PAGE
Twenty 150 cm2 cell culture dishes were seeded with 2 × 106 HEK293T cells each to be transfected on day two. On day three, cells were exposed to H2O2 (10 μM or 100 μM) for 15 s or 3 min. Following NEM treatment and lysis, the combined cytosolic efflux was incubated with 200 μL of SA beads (50% slurry). Beads were eluted twice with 175 μL of 4 mM biotin in TBS. The eluate was run under nonreducing conditions on a 4%–12% Novex NuPAGE Bis–Tris gel (Life Technologies) in MOPS buffer (50 mM MOPS, 50 mM Tris Base, 0.1% SDS, 1 mM EDTA, pH 7.7). Gel lanes from the first-dimension run were cut out and incubated with 250 mM DTT in sample buffer at 65 °C for 20 min. After washing with sample buffer, the gel slice was incubated with 100 mM NEM in sample buffer for 20 min at room temperature with mild agitation. After another washing step, the gel slice was placed horizontally on a 4–12% Novex NuPAGE Bis–Tris 2D-well gel. The resulting 2D gels were stained with NOVEX Colloidal Blue Staining Kit (Invitrogen) and imaged with an Odyssey infrared imaging system (LI-COR).
Mass spectrometry
Gel pieces covering the diagonals were excised from the gel, washed with 50% acetonitrile, treated with 40 mM DTT and alkylated with 50 mM iodoacetamide following digestion with trypsin in 0.01% trifluoroacetic acid using a Digest proMS liquid handling system (Intavis Bioanalytical Instruments, Cologne, Germany). Tryptic peptides were extracted with 50% acetonitrile and 10% formic acid. Acetonitrile was removed with a vacuum concentrator, and peptides were mixed with 1% trifluoroacetic acid and analyzed with a NanoHPLC UltiMate (Dionex, Sunnyvale, CA, USA) coupled to an ESI–LTQ Orbitrap Mass Spectrometer (Thermo Finnigan, San Jose, CA, USA). Resulting mass spectra were searched against the SwissProt database using MASCOT (Matrix Science, London, UK) and the following parameters: significance level of P < 0.01, fragment ion mass tolerance of 0.5 Da and a parent ion tolerance of 100 p.p.m. Fixed modifications: +57 on C (carbamidomethyl). Variable modifications: +1 on NQ (deamidated), +16 on M (oxidation). Scaffold (Proteome Software, Portland, OR, USA) was used for analysis. Proteins were considered identified if at least two peptides were assigned with 99% probability.
Life sciences reporting summary
Further information on experimental design and reagents is available in the Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
We thank G. Kuntz for general technical assistance. We thank S. Merker and R. Hardt for technical assistance with mass spectrometry. We thank B. Jovanovic and G. Stöcklin for technical advice. We acknowledge support by the Deutsche Forschungsgemeinschaft (SFB1036 to T.P.D. and T.R.) and by the European Research Council (742039 to T.P.D.).
Footnotes
Data availability. The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
Sarah Stöcker: 0000-0002-6145-7173
Tobias P Dick: 0000-0003-1367-973X
Author contributions
T.P.D. and S.S. designed the project and wrote the paper. S.S. performed the experiments. M.M. contributed to Figure 4. T.R. analyzed mass spectrometry data.
Competing financial interests
The authors declare no competing financial interests.
Additional information
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Holmström KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–421. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 2.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone JR, Yang S. Hydrogen peroxide: a signaling messenger. Antioxid Redox Signal. 2006;8:243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 4.García-Santamarina S, Boronat S, Hidalgo E. Reversible cysteine oxidation in hydrogen peroxide sensing and signal transduction. Biochemistry. 2014;53:2560–2580. doi: 10.1021/bi401700f. [DOI] [PubMed] [Google Scholar]
- 5.Stone JR. An assessment of proposed mechanisms for sensing hydrogen peroxide in mammalian systems. Arch Biochem Biophys. 2004;422:119–124. doi: 10.1016/j.abb.2003.12.029. [DOI] [PubMed] [Google Scholar]
- 6.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 7.Brigelius-Flohé R, Flohé L. Basic principles and emerging concepts in the redox control of transcription factors. Antioxid Redox Signal. 2011;15:2335–2381. doi: 10.1089/ars.2010.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marinho HS, Real C, Cyrne L, Soares H, Antunes F. Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2014;2:535–562. doi: 10.1016/j.redox.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Winterbourn CC, Peskin AV. Kinetic approaches to measuring peroxiredoxin reactivity. Mol Cells. 2016;39:26–30. doi: 10.14348/molcells.2016.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chae HZ, Kim HJ, Kang SW, Rhee SG. Characterization of three isoforms of mammalian peroxiredoxin that reduce peroxides in the presence of thioredoxin. Diabetes Res Clin Pract. 1999;45:101–112. doi: 10.1016/s0168-8227(99)00037-6. [DOI] [PubMed] [Google Scholar]
- 12.Randall LM, Ferrer-Sueta G, Denicola A. Peroxiredoxins as preferential targets in H2O2-induced signaling. Methods Enzymol. 2013;527:41–63. doi: 10.1016/B978-0-12-405882-8.00003-9. [DOI] [PubMed] [Google Scholar]
- 13.Winterbourn CC, Hampton MB. Redox biology: signaling via a peroxiredoxin sensor. Nat Chem Biol. 2015;11:5–6. doi: 10.1038/nchembio.1722. [DOI] [PubMed] [Google Scholar]
- 14.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 15.Veal EA, et al. A 2-Cys peroxiredoxin regulates peroxide-induced oxidation and activation of a stress-activated MAP kinase. Mol Cell. 2004;15:129–139. doi: 10.1016/j.molcel.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 16.Calvo IA, et al. Dissection of a redox relay: H2O2-dependent activation of the transcription factor Pap1 through the peroxidatic Tpx1-thioredoxin cycle. Cell Rep. 2013;5:1413–1424. doi: 10.1016/j.celrep.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 17.Jarvis RM, Hughes SM, Ledgerwood EC. Peroxiredoxin 1 functions as a signal peroxidase to receive, transduce, and transmit peroxide signals in mammalian cells. Free Radic Biol Med. 2012;53:1522–1530. doi: 10.1016/j.freeradbiomed.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 18.Sobotta MC, et al. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol. 2015;11:64–70. doi: 10.1038/nchembio.1695. [DOI] [PubMed] [Google Scholar]
- 19.Sobotta MC, et al. Exposing cells to H2O2: a quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Radic Biol Med. 2013;60:325–335. doi: 10.1016/j.freeradbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Schwertassek U, et al. Reactivation of oxidized PTP1B and PTEN by thioredoxin 1. FEBS J. 2014;281:3545–3558. doi: 10.1111/febs.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madureira PA, Waisman DM. Annexin A2: the importance of being redox sensitive. Int J Mol Sci. 2013;14:3568–3594. doi: 10.3390/ijms14023568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadeau PJ, Charette SJ, Toledano MB, Landry J. Disulfide bond-mediated multimerization of Ask1 and its reduction by thioredoxin-1 regulate H2O2-induced c-Jun NH2-terminal kinase activation and apoptosis. Mol Biol Cell. 2007;18:3903–3913. doi: 10.1091/mbc.E07-05-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrer-Sueta G, et al. Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem Res Toxicol. 2011;24:434–450. doi: 10.1021/tx100413v. [DOI] [PubMed] [Google Scholar]
- 24.Flohé L. The impact of thiol peroxidases on redox regulation. Free Radic Res. 2016;50:126–142. doi: 10.3109/10715762.2015.1046858. [DOI] [PubMed] [Google Scholar]
- 25.Forman HJ, Maiorino M, Ursini F. Signaling functions of reactive oxygen species. Biochemistry. 2010;49:835–842. doi: 10.1021/bi9020378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Netto LE, Antunes F. The roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol Cells. 2016;39:65–71. doi: 10.14348/molcells.2016.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins A, Nelson KJ, Parsonage D, Poole LB, Karplus PA. Peroxiredoxins: guardians against oxidative stress and modulators of peroxide signaling. Trends Biochem Sci. 2015;40:435–445. doi: 10.1016/j.tibs.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwertassek U, Weingarten L, Dick TP. Identification of redox-active cell-surface proteins by mechanism-based kinetic trapping. Sci STKE. 2007;2007 doi: 10.1126/stke.4172007pl8. pl8. [DOI] [PubMed] [Google Scholar]
- 29.Nordberg J, Arnér ES. Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic Biol Med. 2001;31:1287–1312. doi: 10.1016/s0891-5849(01)00724-9. [DOI] [PubMed] [Google Scholar]
- 30.Debarbieux L, Beckwith J. On the functional interchangeability, oxidant versus reductant, of members of the thioredoxin superfamily. J Bacteriol. 2000;182:723–727. doi: 10.1128/jb.182.3.723-727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.García-Santamarina S, et al. Is oxidized thioredoxin a major trigger for cysteine oxidation? Clues from a redox proteomics approach. Antioxid Redox Signal. 2013;18:1549–1556. doi: 10.1089/ars.2012.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, Cheung SH, Evans EL, Shaw PE. Modulation of gene expression and tumor cell growth by redox modification of STAT3. Cancer Res. 2010;70:8222–8232. doi: 10.1158/0008-5472.CAN-10-0894. [DOI] [PubMed] [Google Scholar]
- 33.Morinaka A, et al. Thioredoxin mediates oxidation-dependent phosphorylation of CRMP2 and growth cone collapse. Sci Signal. 2011;4:ra26. doi: 10.1126/scisignal.2001127. [DOI] [PubMed] [Google Scholar]
- 34.Peralta D, et al. A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat Chem Biol. 2015;11:156–163. doi: 10.1038/nchembio.1720. [DOI] [PubMed] [Google Scholar]
- 35.Truong TH, et al. Molecular basis for redox activation of epidermal growth factor receptor kinase. Cell Chem Biol. 2016;23:837–848. doi: 10.1016/j.chembiol.2016.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antunes F, Brito PM. Quantitative biology of hydrogen peroxide signaling. Redox Biol. 2017;13:1–7. doi: 10.1016/j.redox.2017.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travasso RDM, Sampaio Dos Aidos F, Bayani A, Abranches P, Salvador A. Localized redox relays as a privileged mode of cytoplasmic hydrogen peroxide signaling. Redox Biol. 2017;12:233–245. doi: 10.1016/j.redox.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anestål K, Prast-Nielsen S, Cenas N, Arnér ES. Cell death by SecTRAPs: thioredoxin reductase as a prooxidant killer of cells. PLoS One. 2008;3:e1846. doi: 10.1371/journal.pone.0001846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stöcker S, Van Laer K, Mijuskovic A, Dick TP. The conundrum of hydrogen peroxide signaling and the emerging role of peroxiredoxins as redox relay hubs. Antioxid Redox Signal. 2017 doi: 10.1089/ars.2017.7162. [DOI] [PubMed] [Google Scholar]
- 40.Fomenko DE, et al. Thiol peroxidases mediate specific genome-wide regulation of gene expression in response to hydrogen peroxide. Proc Natl Acad Sci USA. 2011;108:2729–2734. doi: 10.1073/pnas.1010721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan B, et al. Real-time monitoring of basal H2O2 levels with peroxiredoxin-based probes. Nat Chem Biol. 2016;12:437–443. doi: 10.1038/nchembio.2067. [DOI] [PubMed] [Google Scholar]
- 42.Rhee SG, Kil IS. Multiple functions and regulation of mammalian peroxiredoxins. Annu Rev Biochem. 2017;86:749–775. doi: 10.1146/annurev-biochem-060815-014431. [DOI] [PubMed] [Google Scholar]
- 43.Park MH, Jo M, Kim YR, Lee CK, Hong JT. Roles of peroxiredoxins in cancer, neurodegenerative diseases and inflammatory diseases. Pharmacol Ther. 2016;163:1–23. doi: 10.1016/j.pharmthera.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins A, Poole LB, Karplus PA. Tuning of peroxiredoxin catalysis for various physiological roles. Biochemistry. 2014;53:7693–7705. doi: 10.1021/bi5013222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwertassek U, et al. Selective redox regulation of cytokine receptor signaling by extracellular thioredoxin-1. EMBO J. 2007;26:3086–3097. doi: 10.1038/sj.emboj.7601746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner BA, Witmer JR, van ’t Erve TJ, Buettner GR. An assay for the rate of removal of extracellular hydrogen peroxide by cells. Redox Biol. 2013;1:210–217. doi: 10.1016/j.redox.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kempe H, Schwabe A, Crémazy F, Verschure PJ, Bruggeman FJ. The volumes and transcript counts of single cells reveal concentration homeostasis and capture biological noise. Mol Biol Cell. 2015;26:797–804. doi: 10.1091/mbc.E14-08-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park JW, Piszczek G, Rhee SG, Chock PB. Glutathionylation of peroxiredoxin I induces decamer to dimers dissociation with concomitant loss of chaperone activity. Biochemistry. 2011;50:3204–3210. doi: 10.1021/bi101373h. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.