Abstract
Activated macrophages undergo metabolic reprogramming which drives their pro-inflammatory phenotype, but the mechanistic basis for this remains obscure. Here we demonstrate that upon lipopolysaccharide (LPS) stimulation macrophages shift from producing ATP by oxidative phosphorylation to glycolysis, while also increasing succinate levels. We show that increased mitochondrial oxidation of succinate via succinate dehydrogenase (SDH) and an elevation of mitochondrial membrane potential combine to drive mitochondrial ROS production. RNA sequencing reveals that this combination induces a pro-inflammatory gene expression profile, while an inhibitor of succinate oxidation, dimethyl malonate (DMM), promotes an anti-inflammatory outcome. Blocking ROS production with rotenone, by uncoupling mitochondria, or by expressing the alternative oxidase (AOX) inhibits this inflammatory phenotype, with AOX protecting mice from LPS lethality. The metabolic alterations that occur upon activation of macrophages therefore repurpose mitochondria from ATP synthesis to ROS production in order to promote a pro-inflammatory state.
Introduction
Macrophages have two key roles: to respond rapidly to infection and injury and then to help repair the tissue damage that occurs as a result of this response. This requires macrophages to initially adopt a pro-inflammatory phenotype and then later, when the immediate danger has passed, to acquire an anti-inflammatory phenotype to promote resolution and repair. The factors that drive and sustain these changes are not fully understood. However, multiple lines of evidence implicate alterations in mitochondrial function, reactive oxygen species (ROS) production and related metabolic pathways in this phenotypic switch (O'Neill and Pearce, 2016). For example, pro-inflammatory macrophages are more glycolytic, produce more ROS and accumulate succinate to a greater extent than resting macrophages (O'Neill and Pearce, 2016). Even so, the mechanisms and significance of these changes are obscure. The electron transport chain (ETC) is a major component of mitochondrial metabolism and resting macrophages utilize this efficient form of oxidative metabolism to generate ATP. However, once macrophages are activated (e.g. with the Toll-like receptor 4 (TLR4) agonist lipopolysaccharide (LPS)) oxidative phosphorylation is suppressed and cells favor glycolysis as an alternative, but less energetically efficient, mode of ATP generation (Tannahill et al., 2013). In dendritic cells (DCs) the switch to glycolysis and away from oxidative phosphorylation supports fatty acid synthesis required for the expansion of organelles necessary for the production and secretion of key proteins essential for DC activation (Everts et al., 2014) and to prevent apoptosis in the face of a lowered level of ATP production by oxidative phosphorylation (Everts et al., 2012). TLR4 stimulation will also lead to mitochondrial ROS generation from complex I through an unknown mechanism (West et al., 2011). On the other hand, activated T cells maintain oxidative phosphorylation in conjunction with glycolysis but whether oxidative metabolism is required for the production of biosynthetic precursors and ATP following stimulation or to generate a ROS signal is unclear (Sena et al., 2013). Here we set out to determine the mechanistic rationale underlying the mitochondrial, ROS and metabolic changes in macrophages that determine the inflammatory status of the cell, noting that all are linked by electron transfer in the ETC. We have found that following stimulation with LPS macrophages repurpose their mitochondria from ATP production to succinate-dependent ROS generation, with glycolysis taking on the role of ATP generation, enabling mitochondria to sustain a high membrane potential. Thus, the enhanced production of succinate is a critical regulator of the pro-inflammatory response to LPS, both through the generation of ROS following oxidation by the ETC and also via HIF-1α stabilization. Concurrently, anti-inflammatory gene expression is decreased. These events demonstrate a role for succinate oxidation by the ETC in the pro-inflammatory response to LPS and provide insights into the previously elusive mechanism by which macrophages generate mitochondrial ROS. We have identified a process whereby mitochondrial metabolism is intimately linked to the profound gene expression changes that occur in macrophages in order for them to fulfill their dual role in infection and inflammation.
Results
Succinate drives IL-1β production and limits the production of IL-1RA and IL-10
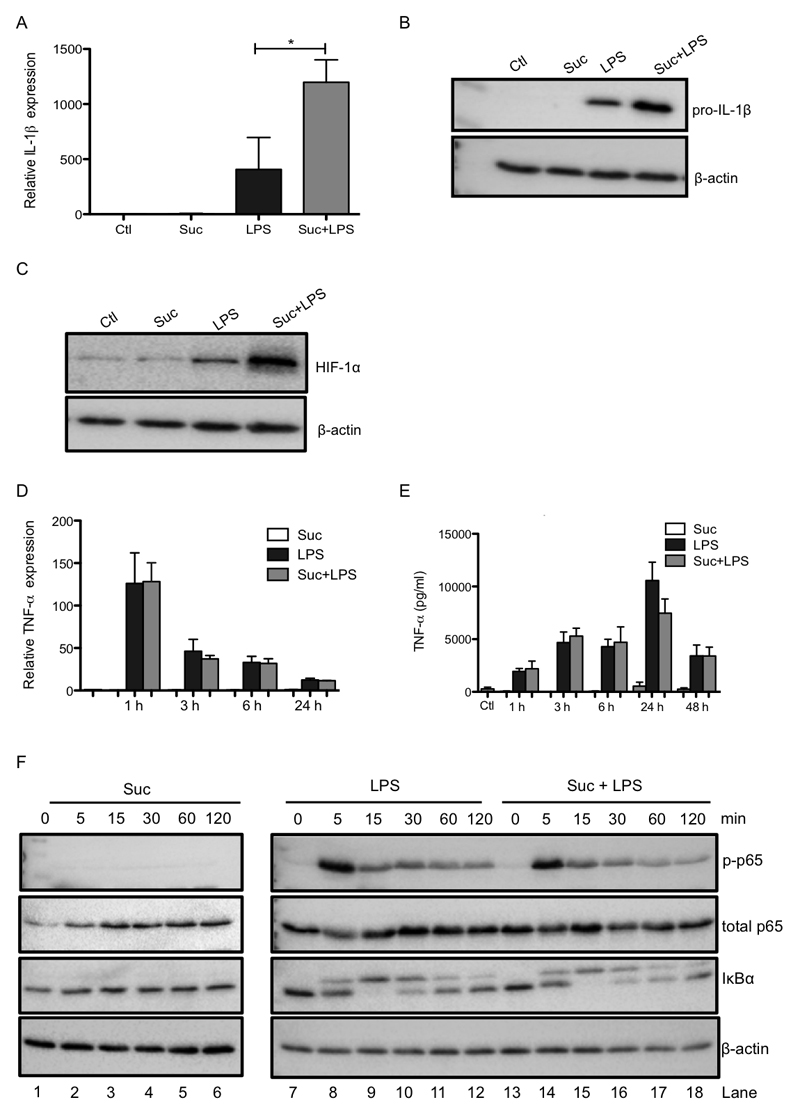
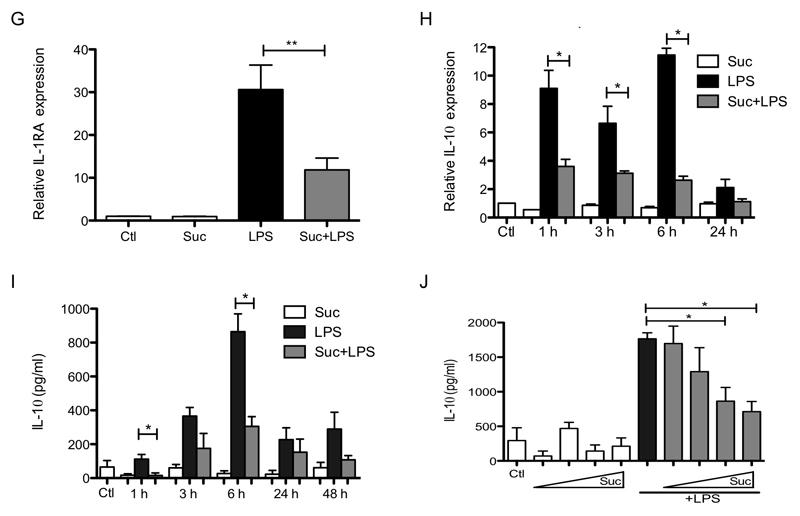
Succinate is a well established pro-inflammatory metabolite which is known to accumulate during macrophage activation, the levels of which affect HIF-1α activity, a key transcription factor in the expression of pro-inflammatory genes (Tannahill et al., 2013). We therefore examined the effects of pretreatment of LPS-activated bone marrow derived macrophages (BMDMs) with cell-permeable diethyl succinate (hereafter referred to as succinate), which greatly increases succinate in the cytosol and mitochondrial matrix. Succinate enhanced LPS-induced IL-1β mRNA and pro-IL-1β protein with an accompanying boost in HIF-1α protein levels (Figure 1A-C), as previously shown (Tannahill et al., 2013). Succinate also boosted LPS-induced glycolysis (Supplementary Figure 2A). Importantly, this synergistic effect of exogenous succinate and LPS did not occur for another pro-inflammatory cytokine, tumor necrosis factor-α (TNF-α) (Figure 1D, E) demonstrating the specificity of this response. Succinate alone had no effect on cytokine production, nor did it affect LPS-induced NFκB activation either on its own (Figure 1F, left-hand side) or in combination with LPS (right-hand side), as measured by p65 phosphorylation (top panel) or IκB degradation (third panel). Conversely, succinate inhibited the induction of the anti-inflammatory cytokines IL-1RA (Figure 1G) and IL-10 (Figure 1H) in response to LPS. The effect of succinate on IL-10 was rapid, being evident from 1 h, and sustained for both mRNA (Figure 1H) and protein (Figure 1I) induction, and was concentration-dependent (Figure 1J). While two other Krebs cycle metabolites, α-ketoglutarate (α-KG) and fumarate, increased LPS-induced IL-1β message their effects on LPS-induced pro-IL-1β were far less robust than for succinate (Supplementary Figure 1A, B), and only succinate decreased LPS-induced IL-10 production (Supplementary Figure 1C, left-hand side). Furthermore, fumarate also boosted LPS-induced TNF-α suggesting its pro-inflammatory effects are mechanistically distinct from succinate (Supplementary Figure 1C, right-hand side). Moreover, diethyl butyl malonate (DEBM), an inhibitor of the mitochondrial succinate transporter, which causes endogenous succinate to accumulate, boosted LPS-induced IL-1β and limited IL-10 with no effect on TNF-α (Supplementary Figure 1D-F). These findings suggest that succinate acts within the cell to enhance and sustain endogenous pro-inflammatory gene expression, while at the same time inhibiting anti-inflammatory gene expression.
Figure 1. Succinate drives IL-1β production and limits the production of IL-1RA and IL-10.
BMDMs were pretreated for 3 h with succinate (Suc; 5 mM; A-I, or 0.2 – 5 mM, J) before being stimulated with LPS (100 ng/ml) for 48 h (A - C, G, J), or the indicated times (D – F, H, I). mRNA was extracted from total cell lysates and analyzed by qPCR for IL-1β (A), TNF-α (D) IL-1RA (G) and IL-10 (H) expression. Whole cell lysates were analyzed by western blotting for pro-IL-1β, HIF-1α, phospho-p65, total p65, IκBα and β-actin (B, C, F). Supernatants were analyzed by ELISA for TNF-α (E) and IL-10 production (I, J). The data in (A, D, E, G - J) represent mean ± S.E.M., n=3, *p<0.05, **p < 0.01. The blots in (B, C F) are representative of 3 independent experiments.
Mitochondrial succinate oxidation alters the expression of pro- and anti-inflammatory genes
To examine how succinate within macrophages might be having its opposing effects on pro- and anti-inflammatory pathways, we investigated two of the ways in which succinate could act within the cell: either by enhancing HIF-1α activation by inhibiting prolyl hydroxylase (PHD) function or as a result of oxidation by mitochondrial succinate dehydrogenase (SDH). Succinate was found to robustly boost LPS-induced HIF-1α protein levels, an effect that was not observed by any other metabolite tested (Supplementary Figure 2D). Moreover, the HIF-1α activator dimethyloxalylglycine (DMOG) mimicked the effect of succinate on IL-1β (Supplementary Figure 2F, G). Succinate did not alter the expression of cMyc, another transcription factor with a known role in the regulation of lymphocyte function, nor the expression of its downstream target CD71 (Supplementary Figure 2H, I). We therefore conclude that succinate’s pro-inflammatory effects require HIF-1α stabilization.
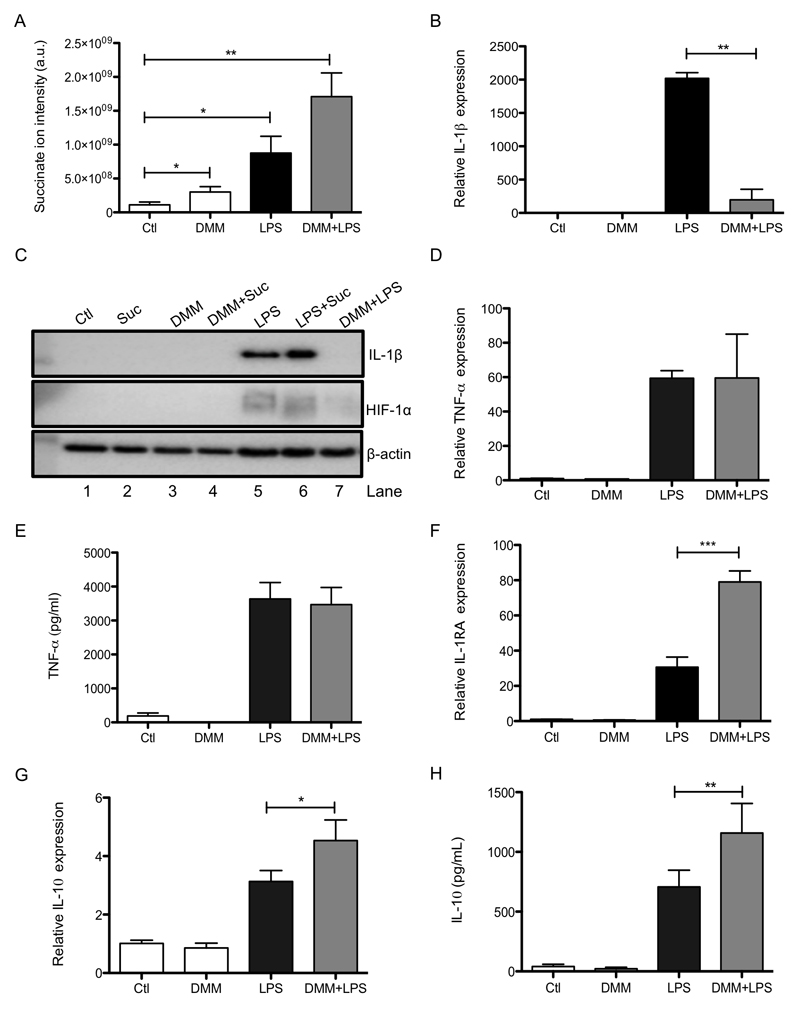
We also tested the cell permeable molecule dimethyl malonate (DMM), which is rapidly hydrolysed within the cell to generate malonate, a potent competitive inhibitor of succinate oxidation by SDH (Dervartanian and Veeger, 1964). DMM led to an increase in succinate in the cytosol, presumably by impairing the oxidation of succinate to fumarate, and also boosted LPS-induced succinate accumulation (Figure 2A). Importantly, succinate and malonate were the only two metabolites significantly altered by the addition of DMM, demonstrating specificity (Supplementary Figure 3). DMM should inhibit effects of succinate that act through SDH in the mitochondria, but enhance succinate action through PHD in the cytosol (as a result of the succinate boost evident by this agent). Intriguingly, in direct opposition to succinate, DMM abrogated LPS-induced IL-1β mRNA (Figure 2B), pro-IL-1β protein and HIF-1α (Figure 2C, compare lane 5 to lane 7), while leaving TNF-α unaffected (Figure 2D, E). At earlier time points it appeared that DMM limited LPS-induced cMyc (Supplementary Figure 2H) however this was not significant and at later time points, when DMM displays effects on IL-1β, cMyc did not appear to have a role (Supplementary Figure 2I). Again in opposition to succinate, DMM boosted LPS-induced IL-1RA (Figure 2F) and IL-10 expression and production (Figure 2G, H). These data indicate that, in addition to direct effects on PHDs in the cytosol, the oxidation of succinate by mitochondria is central to the pro-inflammatory response of LPS-activated macrophages.
Figure 2. Inhibition of succinate dehydrogenase impairs LPS-induced IL-1β production and boosts IL-1RA and IL-10.
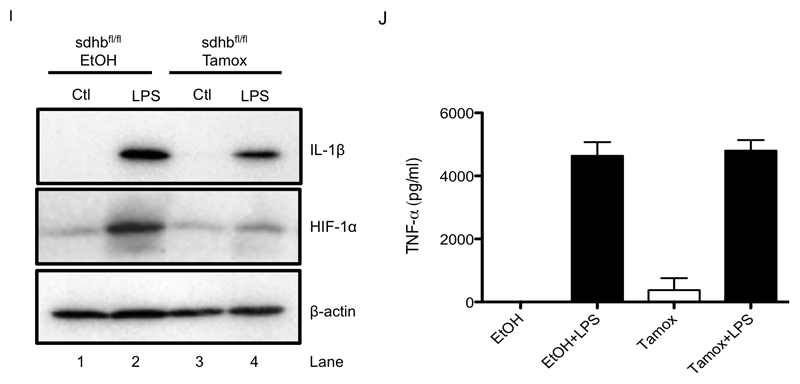
BMDMs were pretreated for 3 h with dimethyl malonate (DMM; 10 mM) prior to stimulation with LPS (100ng/ml) for 24 h (A), 48 h (B, C, E, G, H), or 4 h (D, F). SDHB-proficient (SDHBfl/fl EtOH) and SDHB-deficient (SDHBfl/fl Tamox) BMDMs were untreated (Ctl) or treated with LPS (100 ng/μl) for 24 h (I, J). Lysed cells were analyzed by liquid chromatography-mass spectrometry (LC-MS) to determine succinate levels (A). mRNA from total cell lysates was analyzed by qPCR for IL-1β (B), TNF-α (D), IL-1RA (F) and IL-10 (G) expression. Whole cell lysates were analyzed by Western blotting for pro-IL-1β, HIF-1α and β-actin (C, I). Supernatants were analyzed by ELISA for TNF-α (E, J) and IL-10 production (H). The data in (A, B, D - H) represent mean ± S.E.M., n=3. The data in (J) represent mean ± S.E.M., n=6, *p < 0.05, **p < 0.01, ***p < 0.001. The blots are representative of 3 (C) or 5 (J) independent experiments.
As IL-10 and IL-1β were reciprocally regulated by DMM we examined whether IL-10 itself may be responsible for the regulation of IL-1β by this compound. This was not the case as DMM still inhibited LPS-induced pro-IL-1β in the presence of an IL-10 receptor blocking antibody (Supplementary Figure 2J).
To further examine a role for SDH activity in the pro-inflammatory effects of succinate we next employed macrophages from mice lacking the B subunit of SDH. The B subunit is required for succinate to reduce ubiquinone (Guzy et al., 2008) and its absence will therefore block succinate oxidation. LPS-induced pro-IL-1β and HIF-1α were decreased in SDHB-deficient BMDMs (Figure 2I, upper and middle panels respectively, compare lane 2 to lane 4) while TNF-α was unchanged (Figure 2J).
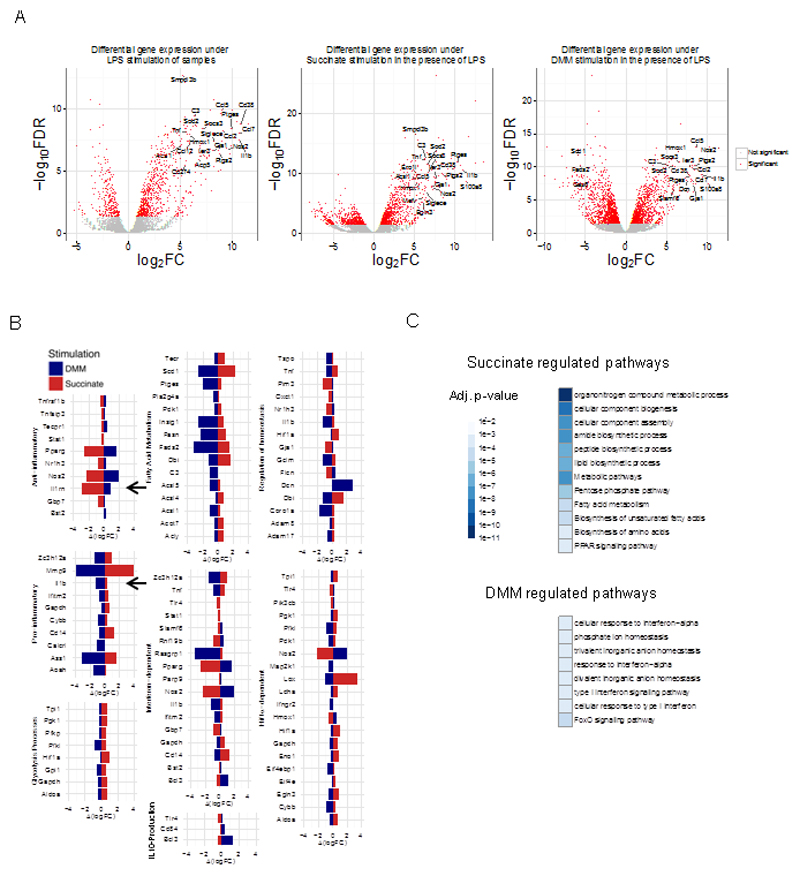
To gain further insight into how the manipulation of mitochondrial succinate oxidation might alter the phenotype of macrophages we next determined the effects of succinate and DMM on the macrophage genome by performing RNA sequencing analysis. This revealed whole sets of LPS-induced genes that were altered by DMM or succinate pretreatment (Figure 3, Supplementary Figure 3K-M and Supplementary Tables 1-6). The gene expression values in different samples were modeled with a generalized linear model (details in Methods). Of particular relevance is that many genes that were down-regulated by DMM were reciprocally up-regulated by succinate and vice versa (Supplementary Figure 3M), further suggesting that the metabolism of succinate by SDH is crucial for its pro-inflammatory action. These gene sets were related to immune response pathways including interferon (IFN) signaling, the cellular response to hypoxia, fatty acid metabolism, glycolytic processes and positive and negative regulation of homeostasis (illustrated in Figure 3B). A direct comparison of LPS stimulation under pretreatment with DMM or succinate confirmed this, as several biological pathways related to homeostasis, metabolism and inflammation were affected (Figure 3C) even under this stricter comparison. Most importantly, DMM suppressed expression of genes associated with inflammation (including those encoding IL-1β, HIF-1α-dependent genes and genes involved in fatty acid synthesis) whilst boosting anti-inflammatory gene expression (such as the genes encoding IL-1RA and several type I IFN-inducible genes) (Figure 3B, blue histobars). Succinate had the opposite effect, boosting pro-inflammatory gene expression whilst decreasing anti-inflammatory gene expression (Figure 3B, red histobars). The reciprocal regulation of IL-1β and IL-1RA (both indicated with an arrow) is especially noteworthy. Type I IFNs are also of interest as they inhibit LPS-induced pro-IL-1β production and boost IL-10 in BMDMs (Guarda et al., 2011) providing further mechanistic evidence for the regulation of IL-10, and indeed of IL-1β, by these agents. As expected, RNA sequencing revealed that succinate induced, while DMM limited, HIF-1α activity. These findings suggest that HIF-1α is an important part of the mechanism by which succinate and DMM regulate IL-1β, and confirm that the extent of succinate oxidation by mitochondrial SDH controls LPS-induced gene expression. When active, SDH boosts inflammatory gene expression and when inhibited an anti-inflammatory phenotype ensues.
Figure 3. Limiting succinate oxidation induces an anti-inflammatory response.
BMDMs were pretreated with dimethyl malonate (DMM; 10 mM) or succinate (5 mM) for 3 h before being stimulated with LPS (100 ng/μl) for 4 or 48 h. RNA was isolated and RNA sequencing was performed. The gene expression in the different stimulations and time-points was modeled with a generalized linear model, and fold-changes and FDR-adjusted p-values were calculated. Fig 3(A) shows the distribution of the fold changes (log2FC) and the FDR – adjusted p-values (log FDR) for the comparison between LPS-treated samples and control – without and with pretreatment with succinate and DMM. A large number of genes were found to be oppositely regulated when pretreated with succinate as compared to when they were pretreated with DMM. Significant changes are coloured red, while the insignificant changes are coloured grey. Fig 3(B) shows the difference in fold changes when the BMDMs were pretreated with either DMM or succinate, as compared to when they were not. The genes are annotated for the immune pathways they belong to. Fig 3(C) shows the results of functional enrichment of our gene expression analysis in the Kegg and Reactome pathway databases. The heatmap represents the statistical significance (FDR-adjusted p-value) of the different pathways found to be enriched in our analysis, with the darker colours denoting pathways enriched with higher confidence.
Taken together these results indicate that SDH is a critical regulator of the macrophage phenotype. Inhibiting SDH causes succinate to accumulate but importantly prevents the induction of a range of pro-inflammatory factors typified by IL-1β, whilst enhancing a range of anti-inflammatory factors, typified by IL-1RA and IL-10. Increased oxidation of succinate by SDH in mitochondria is therefore required for the induction of pro-inflammatory genes, whilst simultaneously limiting the induction of anti-inflammatory ones.
Inhibition of succinate dehydrogenase in vivo is anti-inflammatory
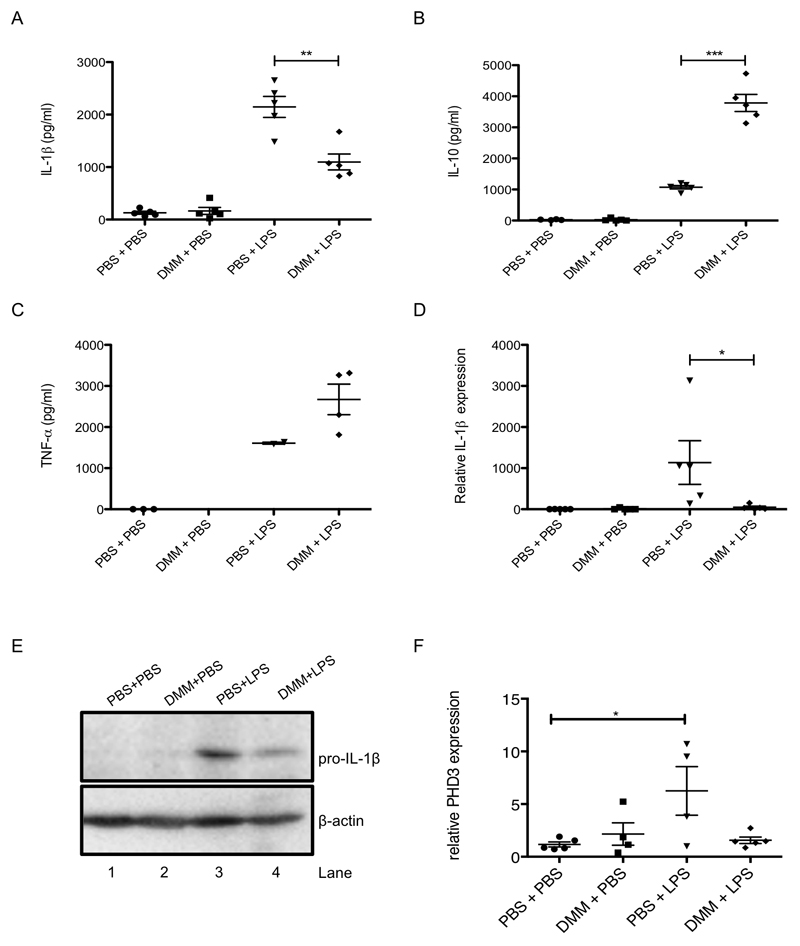
To examine if SDH activity was also involved in the pro-inflammatory response in vivo we next investigated the effect of DMM on LPS action in mice. DMM was effective in an LPS-induced sepsis model, where it decreased serum levels of IL-1β (Figure 4A) and boosted IL-10 (Figure 4B), but had no significant effect on TNF-α (Figure 4C). DMM reduced IL-1β (Figure 4D, E) and PHD3 expression (Figure 4F) in the spleen. SDH activity is therefore critical for determining the inflammatory phenotype, both in vitro and in vivo.
Figure 4. Inhibition of succinate dehydrogenase in vivo is anti-inflammatory.
Mice were injected intraperitoneally (i.p.) with DMM (160 mg/kg) or PBS for 3 h, followed by PBS or LPS (15 mg/kg) for 2 h. Serum was isolated from whole blood and IL-1β (A), IL-10 (B) and TNF-α (C) production were measured by ELISA. Spleens were isolated and IL-1β (D) and PHD3 (F) expression were analyzed by qPCR and pro-IL-1β and β-actin were measured by Western blotting (E). The data in (A-D, F) represent mean ± S.E.M., n=5 per group, *p < 0.05, **p < 0.01, ***p < 0.001. Blots are representative of 1 sample from each treatment group.
Glycolytic ATP production facilitates an increase in mitochondrial membrane potential that is required for the pro-inflammatory effects of LPS
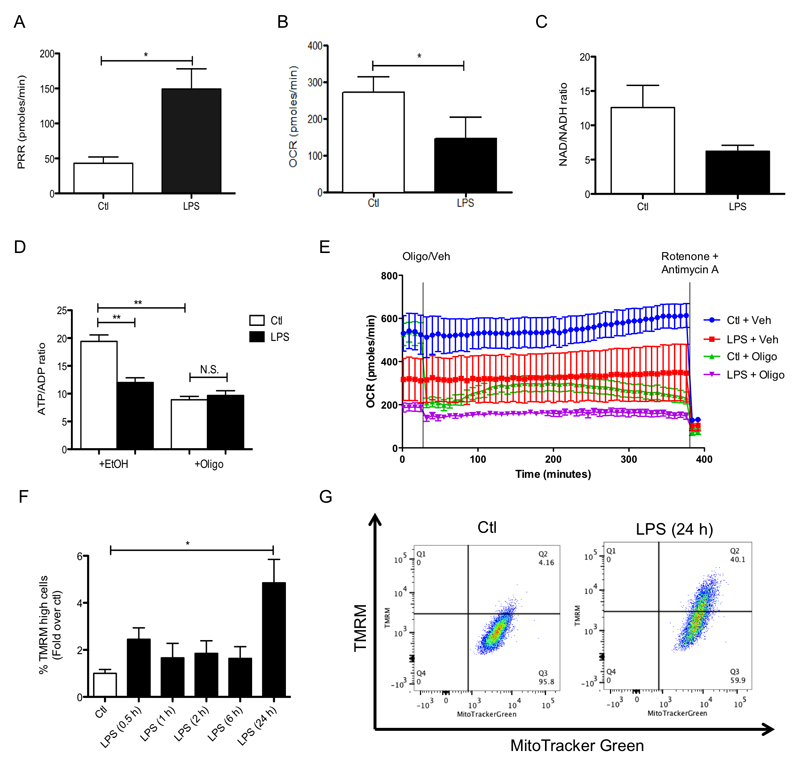
To further explore the mechanism by which SDH might affect the macrophage phenotype, we next considered how SDH activity interacts with other metabolic changes occurring in LPS-activated macrophages. A hallmark of the pro-inflammatory phenotype of macrophages is their switch away from ATP production by oxidative phosphorylation to glycolytic metabolism (O'Neill and Pearce, 2016; Tannahill et al., 2013). Previous work showed that inhibiting glycolysis with 2-deoxyglucose (2DG) prevented activation of HIF-1α and induction of IL-1β in LPS-treated macrophages yet had no effect on TNF-α production (Tannahill et al., 2013) (Supplementary Figure 4A-D). These data suggested that the enhanced glycolytic ATP production upon activation may act in combination with the stimulation of SDH to sustain the pro-inflammatory phenotype. We hypothesized that the increased ATP generated by glycolysis in the cytosol would significantly decrease the requirement for mitochondrial oxidative phosphorylation to supply ATP to the cell. Such a shift would decrease resting oxygen consumption by the mitochondrial respiratory chain, as has been shown previously (Tannahill et al., 2013). Here we demonstrate that glycolysis is boosted by LPS (Figure 5A) and oxygen consumption is decreased (Figure 5B), yet not abrogated, by LPS (Figure 5E). Addition of oligomycin to control cells dramatically decreased respiration by inhibition of mitochondrial ATP synthesis, while the addition of oligomycin had little further effect on respiration to cells treated with LPS (Figure 5E). LPS decreased the ATP/ADP ratio consistent with a shift from ATP synthesis by oxidative phosphorylation to ATP synthesis by glycolysis (Figure 5D). Importantly, addition of oligomycin, which will abolish any contribution to ATP synthesis by the ETC, did not affect the ATP/ADP ratio in LPS-treated cells (Figure 5D). Taken together these data indicate that following LPS treatment cells were not making ATP by mitochondrial oxidative phosphorylation. It is interesting to note that inhibition of the ATP synthase with oligomycin decreased LPS-induced IL-1β suggesting that some level of ATP synthesis within the mitochondrial matrix may be necessary for the generation of a pro-inflammatory response (Supplementary Figure 5). The NAD+/NADH ratio also increased with LPS treatment (Figure 5C) consistent with a decrease in mitochondrial NADH oxidation and complex I forward activity.
Figure 5. Glycolytic ATP production facilitates an increase in mitochondrial membrane potential that is required for the pro-inflammatory effects of LPS.
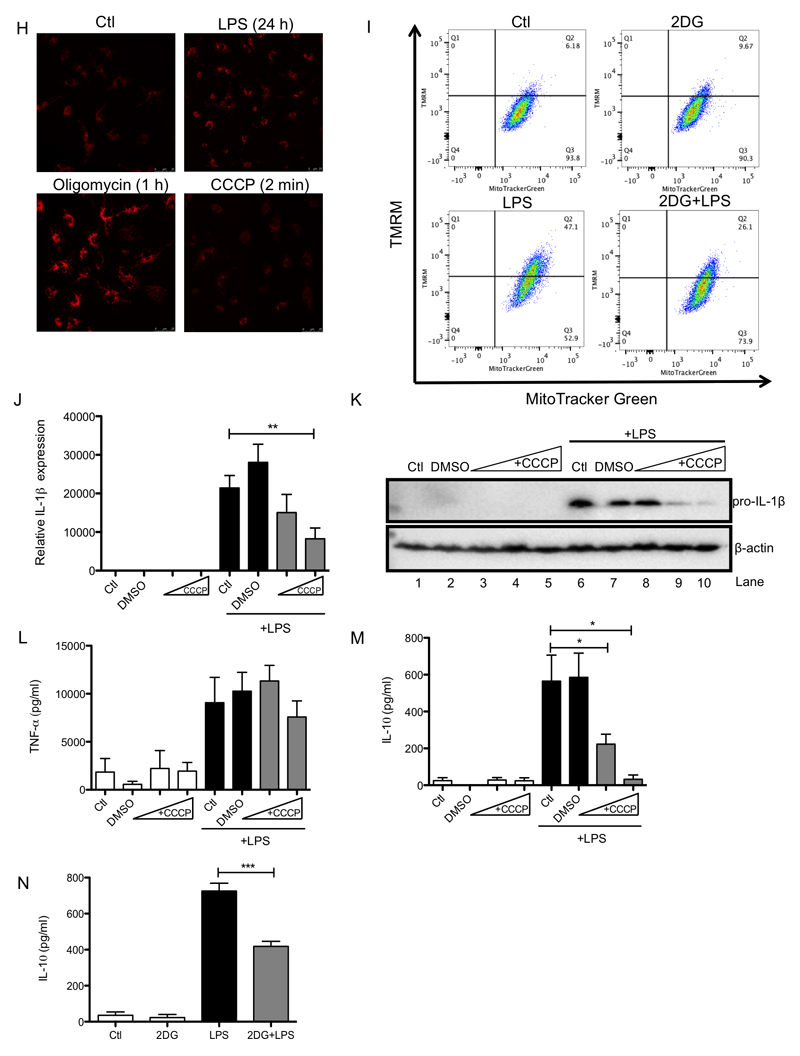
(A – E) BMDMs were stimulated with LPS (100 ng/ml) for 48 h (A, B) or 24 h (C, D). Oxygen consumption rate (OCR) and proton production rate (PPR) were analyzed as read-outs for for oxidative phosphorylation and glycolysis, respectively, using the Seahorse XF-24. The NAD+/NADH ratio in cell lysates was determined using an NAD+/NADH assay kit (C). The ATP/ADP ratio in cell lysates was determined using an ATP/ADP assay kit (D). BMDMs were untreated (Ctl) or stimulated with LPS (100 ng/ml) for 24 h before OCR analysis using the Seahorse XF-24 (E). During the Seahorse run, BMDMs were first injected with oligomycin (Oligo; 10 μM) or vehicle (EtOH), and OCR was measured for the following 6 h. At this point, rotenone (Rot; 100 nM) and antimycin A (4 μM) were injected to all wells to abolish OCR. (F – I) BMDMs were untreated (Ctl), treated with LPS (100 ng/ml) for the indicated times (F, G) or pretreated with 2-deoxyglucose (2DG; 1 mM) for 3 h prior to LPS for 24 h (I). Cells were costained with TMRM (20 nM) and MitoTracker Green (50 nM) for 30 min and then analyzed by FACS to quantify the membrane potential. The intensity of TMRM staining reflects the membrane potential. To analyse the membrane potential by confocal microscopy BMDMs were untreated (Ctl), treated with LPS (100 ng/ml) for 24 h, oligomycin (Oligo: 5 μM) for 1 h or with carbonylcyanide m-chlorophenylhydrazone (CCCP; 10 μM) for 2 min (H). Cells were stained with TMRM (20 nM) for 30 min prior to imaging. The intensity of TMRM staining reflects the membrane potential. (J - M) BMDMs were also pretreated with CCCP (0.5-10 μM; J - M), or 2DG (1 mM; N) for 3 h before being stimulated with LPS (100 ng/ml) for 4 h (J - M) or 48 h (N). mRNA was extracted from total cell lysates and analyzed by qPCR for IL-1β expression (J) and whole cell lysates were analyzed by Western blotting for pro-IL-1β and β-actin (K). Supernatants were analyzed by ELISA for TNF-α (L) and IL-10 (M, N) production. The data in (A-D, J - M) represent mean ± S.E.M., n=3, *p=0.05, **p < 0.01, ***p < 0.001. The data in (F) shows quantification of TMRM high cells and represents mean ± S.E.M., *p<0.05. The cytometric dot plots in (G, I) are representative from 3 (G) or 4 (I) separate experiments. Images in (H) are representative from 5 separate experiments. The blots in (K) are representative of three independent experiments. The Seahorse OCR data in (E) is representative of 4 independent experiments.
Cells that are making ATP by glycolysis and not by mitochondrial oxidative phosphorylation, as in the case of LPS activation, are expected to have a higher mitochondrial membrane potential (ΔΨm). This is because the ΔΨm, which is generated by proton pumping through complexes I, III and IV across the mitochondrial inner membrane, is no longer being used by the ATP synthase to make ATP. We found that LPS increased ΔΨm as measured by potential-sensitive TMRM fluorescence, by flow cytometry and by confocal microscopy (Figure 5F-H). The percentage of cells with an elevated ΔΨm increased from 6.18% to 47.1% in response to LPS and this was decreased to 26.1% when glycolysis was inhibited with 2DG (Figure 5I). This suggests that the increased ATP supply by glycolysis and the consequential decrease in requirement for mitochondrial ATP production leads to an increase in the ΔΨm that is important for LPS signaling. To determine if there was indeed a requirement for increased membrane potential in LPS signaling we next explored the effect of the uncoupler carbonylcyanide m-chlorophenylhydrazone (CCCP) on LPS-activated macrophages. As expected, addition of CCCP led to a decrease in ΔΨm (Figure 5H) and a boost in respiration, which reached its maximum effect on these variables at 5-10 μM CCCP indicating complete uncoupling (Supplementary Figure 4F). Concentrations of CCCP sufficient to uncouple mitochondria also decreased LPS-induced IL-1β expression (Figure 5J), and pro-IL-1β levels (Figure 5K, compare lane 7 to lanes 8, 9, 10), but only affected TNF-α at higher concentrations where cellular viability was impaired (Supplementary Figure 4H). Dissipation of the mitochondrial membrane potential with CCCP or 2DG also impaired LPS-induced IL-10 production (Figure 5M and N, respectively) suggesting that coupled mitochondria are required for the production of this anti-inflammatory cytokine. Importantly, the lack of effect by both CCCP and 2DG on TNF-α demonstrates that this cytokine is regulated differently than IL-1β or IL-10, most likely by a mitochondria-independent mechanism. Together these data suggest that the shift from oxidative phosphorylation to glycolysis and the consequent elevation in ΔΨm, in conjunction with the increased succinate, together provide a pro-inflammatory signal.
Increased membrane potential and succinate oxidation induce the generation of mitochondrial ROS that drives IL-1β
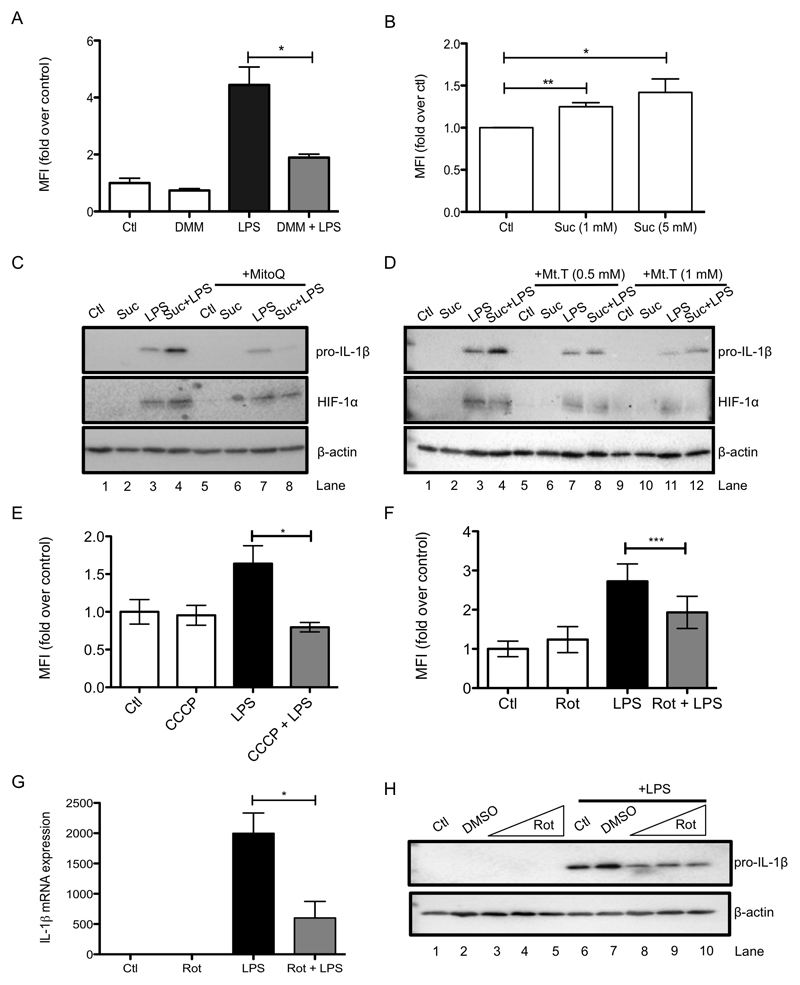
As mitochondrial ROS production is greatly enhanced by both an increase in ΔΨm and an increase in SDH activity (Chouchani et al., 2016) we next examined whether ROS could act as a redox signal emanating from mitochondria to drive IL-1β production. Addition of LPS led to an increase in ROS as measured by both MitoSOX and CellROX (Supplementary Figure 6A and Figure 6A). DMM limited LPS-induced ROS (Figure 6A and Supplementary Figure 6D) while succinate boosted ROS production (Figure 6B and Supplementary Figure 6B). This ROS signal led to increased IL-1β expression as evidenced by the use of a variety of ROS scavengers. The mitochondria-targeted antioxidants MitoQ (Figure 6C, compare lanes 3 and 4 to lanes 7 and 8) and MitoTEMPO (Figure 6D, compare lanes 3 and 4 to lanes 7 and 8, and 11 and 12) and the thiol reductant N-acetylcysteine (Supplementary Figure 6C), all inhibited the effect of succinate on LPS-induced pro-IL-1β and HIF-1α suggesting that this ROS signal was critical for IL-1β production in response to succinate. In addition, LPS-induced ROS production was prevented by dissipating the mitochondrial membrane potential with CCCP (Figure 6E and Supplementary Figure 6E). Together these data suggest that increased succinate oxidation and an elevation of the mitochondrial membrane potential are both required to generate a pro-inflammatory mitochondrial ROS signal.
Figure 6. Inhibition of ROS production by impairing complex I or II activity or by dissipating the membrane potential limits IL-1β production in LPS-activated macrophages.
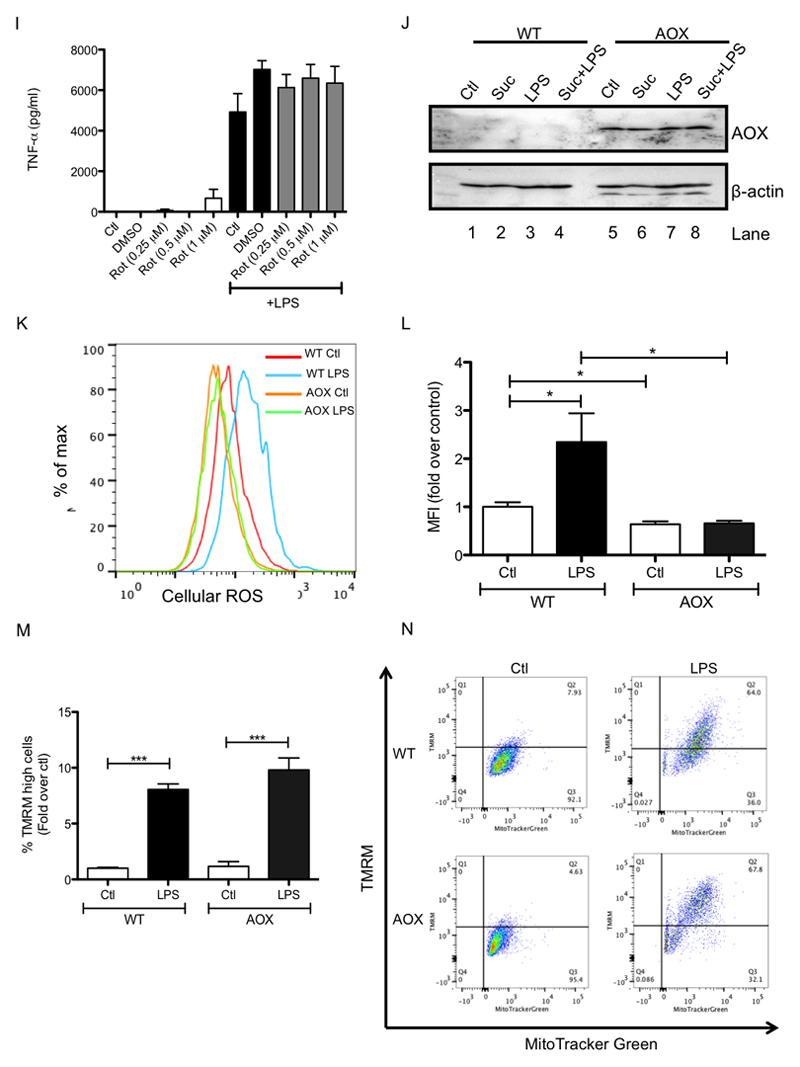
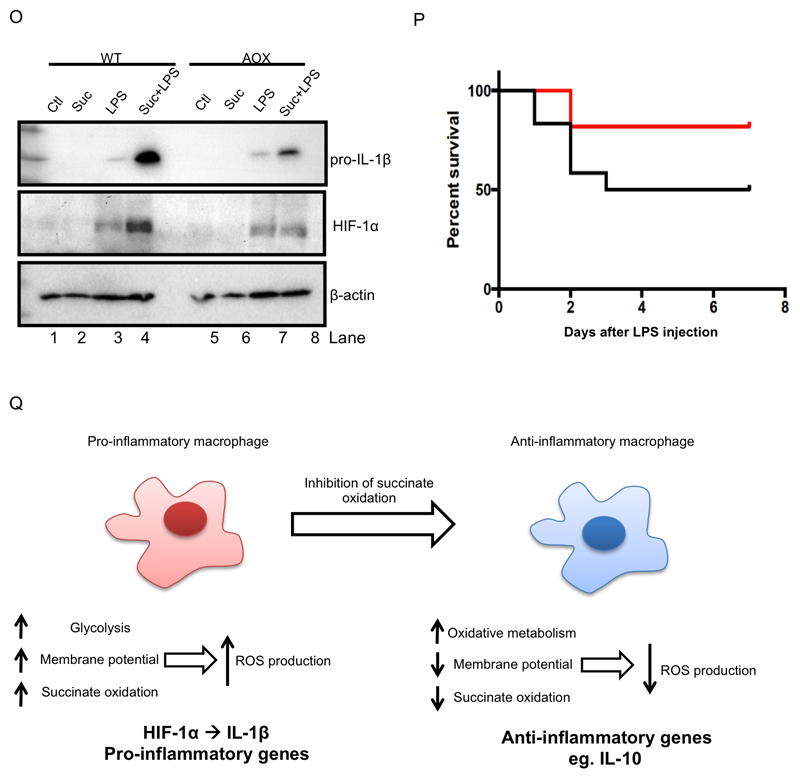
(A, B, E, F) BMDMs were pretreated for 3 h with dimethyl malonate (DMM; 10 mM; A), carbonylcyanide m-chlorophenylhydrazone (CCCP; 7.5 μM; E), or rotenone (Rot; 0.5 μM; F) before being stimulated with LPS (1 μg/ml; A, E, F) for 24 h (A, F) or 4 h (E), or were treated with succinate (Suc; 1 - 5 mM) for 24 h (B). Live cells were analyzed by FACS and mean fluorescence intensity (MFI) was quantified as a measure of cellular reactive oxygen species production. (C, D, G, H, I) BMDMs were pretreated with MitoQ (500 nM; C) or MitoTEMPO (Mt.T; 0.5 - 1 mM; D) for 1 h prior to the addition of succinate (Suc; 5 mM) for 3 h before stimulation with LPS (100 ng/ml) for 48 h. BMDMS were also pretreated for 3 h with rotenone (Rot; 0.1 – 1 μM; G - I) prior to stimulation with LPS (100 ng/ml) for 24 h. (J – N) Wild-type and AOX-expressing BMDMs were untreated (Ctl) or pretreated for 3 h with succinate (Suc; 5 mM; J, M) before being stimulated with LPS (100 ng/μl; J, M-O or 1 μg/ml; K, L) for 48 h. Whole cell lysates were analyzed by Western blotting for pro-IL-1β, HIF-1α, β-actin and AOX (C, D, H, J, O). mRNA was extracted from total cell lysates and analyzed by qPCR for IL-1β expression (G). Supernatants were analyzed by ELISA for TNF-α production (I). Live cells were analyzed by FACS and mean fluorescence intensity (MFI) was quantified as a measure of cellular reactive oxygen species production (K, L) or cells were costained with TMRM (20 nM) and MitoTracker Green (50 nM) for 30 min and then analyzed by FACS to quantify the membrane potential (M, N). The intensity of TMRM staining reflects the membrane potential. The cytometric dot plots in (N) are representative from 5 independent experiments. (P) Wild-type (WT) and alternative oxidase (AOX)-expressing mice were injected i.p. with LPS (10 mg/kg); survival rate was monitored. AOX group n=11, WT group n=12. The data in (A, E – G, I) represents mean ± S.E.M., n=3, or n=5 for (L, M) *p<0.05, **p < 0.01, ***p < 0.001. The blots in (C, D, H, J, K, O) are representative of 3 independent experiments. (Q) shows a schematic diagram illustrating the proposed mechanism by which metabolic alterations govern the inflammatory phenotype of macrophages.
The complex I inhibitor rotenone significantly decreased LPS-induced ROS (Figure 6F, right-hand side, and Supplementary Figure 6F) while also decreasing LPS-induced IL-1β mRNA expression (Figure 6G) and pro-IL-1β (Figure 6H, compare lane 7 to lanes 8, 9 and 10) yet had no effect on TNF-α (Figure 6I), supporting a role for complex I activity in ROS production in this system. To further investigate the role of mitochondrial ROS in LPS-activated macrophages we used BMDMs from mice expressing an alternative oxidase (AOX) from Ciona intestinalis (El-Khoury et al., 2013). AOX provides a pathway to oxidise excess electrons that build up in the ubiquinone (CoQ) pool, for example as a result of succinate accumulation, that can contribute to mitochondrial ROS production. AOX was confirmed to be present in BMDMs from these mice (Figure 6J). AOX-expression in BMDMs impaired the boost in ROS production following LPS treatment (Figure 6K, L) and strongly impaired the boost in LPS-induced IL-1β and HIF-1α with succinate (Figure 6O, compare lanes 3 and 4 to lanes 7 and 8). There was no difference in the observed LPS-induced boost in membrane potential between wild-type and AOX-expressing BMDMs (Figure 4M, N). Crucially, AOX expression increased the survival of mice injected with LPS. There was 50% mortality in wild-type mice while in AOX-expressing mice survival remained above 80% (Figure 6P). These data suggest that limiting ROS production either pharmacologically or using genetic approaches limits IL-1β levels and is protective against LPS lethality, supporting a role for mitochondrial ROS production in inflammation.
Discussion
Alterations in mitochondrial metabolism following LPS treatment of macrophages are now understood to be vital for an appropriate immune response (Mills and O'Neill, 2016). Studies dating back to the 1970s demonstrated that LPS attenuates macrophage respiration by inhibiting complexes II and III (Kato, 1972) and slowing state III respiration (McGivney and Bradley, 1979). LPS also alters the Krebs cycle, effectively breaking it at two points: after citrate and after succinate (Jha et al., 2015). Furthermore, previous studies have shown that LPS increases mitochondrial ROS production (West et al., 2011) as an important response for bacterial killing in macrophages, but the mechanism was obscure. Our study shows that mitochondrial ROS generation following the oxidation of succinate is central to determining the inflammatory phenotype of macrophages.
This study provides a model that explains the many disparate aspects of the metabolic changes that occur upon activation of macrophages with LPS (Figure 6O). Together these findings suggest that an important consequence of the shift to glycolytic ATP production upon activation of macrophages is the release of mitochondria from their requirement to produce ATP by oxidative phosphorylation, thereby enabling the mitochondrial membrane potential to increase. This is coupled with the remodeling of metabolism to funnel metabolites to succinate, the oxidation of which we have demonstrated is critical in the regulation of the inflammatory state of macrophages. We demonstrate that succinate oxidation and a high mitochondrial membrane potential generate a redox signal that can alter HIF-1α activity (illustrated in Figure 6O). This supports a recent study performed in keratinocytes demonstrating a critical role for ROS derived from the ETC in HIF-1α stabilization (Hamanaka et al., 2016). Crucially we report that these events alter the expression levels of a range of pro and anti-inflammatory genes. The inflammatory phenotype of the macrophage, as indicated by increased inflammatory gene expression and reciprocally decreased anti-inflammatory gene expression, is governed by succinate oxidation by SDH. We demonstrate that ROS drive expression of cytokines such as IL-1β via HIF-1α. Precisely how succinate inhibits anti-inflammatory gene expression is under investigation. This may involve succinate-induced regulation of other members of the α-KG-dependent dioxygenase family such as those involved in epigenetic changes occurring in macrophages (Yang and Pollard, 2013). We are currently investigating this possibility. From our data it is clear, nevertheless, that SDH is a key arbiter for the inflammatory response in macrophages. Furthermore, our observation that TNF-α is not controlled by SDH points to specificity in the role of the mitochondria in the regulation of cytokine production. In a recent study by Sancho and colleagues, live bacteria were shown to alter the assembly of the ETC via the sensing of bacterial RNA, further emphasizing the importance of mitochondrial alterations in innate immunity (Garaude et al., 2016).
There are a number of possible sources for the ROS signal emanating from mitochondria in response to succinate oxidation and an elevated membrane potential, including from complex I and complex III (Chandel et al., 2000). We favour the hypothesis that the ROS signal is generated by reverse electron transport (RET) at complex I of the ETC. Complex I can produce large amounts of ROS by the precisely regulated process of RET when the CoQ pool is highly reduced and the mitochondrial membrane potential is high, together providing the thermodynamic driving force to push electrons in reverse to the ROS-producing site within complex I (Chance and Hollunger, 1961). Importantly, succinate has long been known to drive RET and recently has been shown to generate ROS production by RET during ischemia-reperfusion (I/R) injury (Chouchani et al., 2014). The data we present in this study indicate that the ROS signal is sensitive to uncoupling with CCCP, prevention of succinate oxidation by DMM, oxidation of the CoQ pool by expression of AOX (Perales-Clemente et al., 2008) and inhibition of complex I with rotenone, all of which are consistent with RET at complex I as the source of the pro-inflammatory ROS signal in LPS-activated macrophages. The decrease in LPS-induced ROS with the complex I inhibitor rotenone is particularly intriguing as this inhibitor only lowers ROS by complex I if RET is occurring (Votyakova and Reynolds, 2001), and otherwise induces ROS production (Barrientos and Moraes, 1999). The role of RET in metabolic signaling has been implicated in a number of recent studies, such as I/R injury (Chouchani et al., 2014), mitochondrial ROS production during ageing in Drosophila, (Scialo et al., 2016) and in hypoxia sensing in the carotid body (Fernandez-Aguera et al., 2015). Furthermore, previous reports demonstrated that metformin, which inhibits complex I, decreases IL-1β in response to LPS (Kelly et al., 2015). Also of interest is the recent identification of TLR4 as a driver of ROS and subsequent systemic inflammation in mice deficient in the complex I subunit Ndufs4 (Jin et al., 2014), further emphasizing modulation of complex I by TLR4. While further work is required to assess the source of mitochondrial ROS during macrophage activation, we feel that RET at complex I is a strong candidate.
This central role for mitochondria in immune signaling and the innate immune response fits with the function of this organelle in many other aspects of cell death and signaling. Indeed, the endosymbiotic origins of mitochondria may not only reflect the requirement for eukaryotes to evolve to respire oxidatively but these organelles might also have evolved to serve as signaling hubs, which can influence the phenotype of immune cells, and possibly other cell types, to aid in the response to infection.
Finally, the finding that succinate oxidation is an important regulator of inflammatory signaling opens up a number of new therapeutic opportunities. For example, DMM may be useful in the treatment of inflammatory diseases in which IL-1β has been linked to pathogenesis (Dinarello, 2011). Promisingly, inhibition of SDH with DMM has shown efficacy here in a mouse model of acute LPS-induced sepsis and previously in mouse models of Escherichia coli infection (Garaude et al., 2016) and I/R injury (Chouchani et al., 2014). Treatment with DMM or derivatives may therefore improve disease outcome by restoring the balance between IL-1β and IL-1RA, or indeed by increasing IL-10 production, which can both decrease IL-1β and enhance IL-1RA levels (Cassatella et al., 1994) and suppress inflammation more generally (Couper et al., 2008). The shift in macrophages away from inflammatory gene expression towards anti-inflammatory gene expression within an inflammatory environment (such as in our LPS model) is especially noteworthy since such an environment will prevail in inflammatory diseases where inhibitors of SDH might prove especially useful.
Experimental Procedures
Reagents
LPS used in in vitro studies was from Escherichia coli, serotype EH100 (Alexis). LPS for in vivo trials was from Escherichia coli, serotype O55:B5 (Sigma-Aldrich). Alpha-ketoglutarate (αKG), carbonyl cyanide m-chlorophenyl hydrazine (CCCP), 2-deoxyglucose (2DG), diethyl succinate, diethyl butyl malonate (DEBM), dimethyl malonate (DMM), MitoTEMPO, dimethyloxalylglycine (DMOG), methyl pyruvate (pyr), triethyl citrate, oligomycin, rotenone, 4-hydroxytamoxifen, glucose, antimycin A and the ADP/ATP ratio assay kit were from Sigma-Aldrich. Dimethyl fumarate was from Santa Cruz. MitoQ was a kind gift from Prof. Rob Smith (University of Otago, New Zealand). The AOX antibody was a kind gift from Dr Mike Murphy (University of Cambridge, United Kingdom). Antibodies used were goat anti-mouse IL-1β (R&D Systems), mouse anti-β-actin (Sigma-Aldrich), rabbit anti-HIF-1α (Novus), rabbit anti-phospho-p65, rabbit anti-total p65, and rabbit anti-IκBα (all Cell Signalling). Secondary horseradish peroxidase-conjugated anti-mouse IgG, anti-rabbit IgG and anti-goat IgG were from Jackson ImmunoResearch Inc. MitoSOX, CellROX, MitoTracker Green (MTG) and Aqua Live/Dead stains were all from Molecular Probes. Tetramethylrhodamine, methyl ester (TMRM) was from Biosciences. FACS tubes were purchased from BD. XF assay buffer, Seahorse cell culture plates, utility plates and calibrant solution were obtained from Seahorse Biosciences. NAD+/NADH Quantification Colorimetric Kit was purchased from BioVision and the required 10 kDa molecular weight cut-off filters were from Merck Millipore. LDH assay kits were purchased from Promega. IL-10 receptor blocking antibody and isotype control were purchased from BD Biosciences.
Mouse strains
Wild type C57Bl/6 mice were from Harlan U.K. and Harlan Netherlands. Animals were maintained under specific pathogen-free conditions in line with Irish and European Union regulations. Experiments were approved by local ethical review and were carried out under the authority of Ireland’s project license. C57Bl/6 mice carrying a single copy of Ciona intestinalis AOX gene in the Rosa26 locus were generated by T. Braun, H. T. Jacobs and M. Szibor (full details to be published elsewhere). Legs from AOX-expressing mice were prepared by Prof. Mike Murphy (University of Cambridge, UK). Legs from SDHB-deficient mice were a gift from Prof. Eyal Gottlieb (University of Glasgow, UK).
Generation of bone marrow-derived macrophages (BMDMs)
Mice were euthanized in a CO2 chamber and death was confirmed by cervical dislocation. Bone marrow cells were extracted from the leg bones and differentiated in DMEM (containing 10% foetal calf serum, 1% penicillin streptomycin and 20% L929 supernatant) for 6 days, at which time they were counted and replated for experiments. Unless stated, 5 X 106 BMDMs per millilitre were used in in vitro experiments.
Real-time PCR
Total RNA was isolated using the RNeasy Plus Mini kit (Qiagen) and quantified using a Nanodrop 2000 UV-visible spectrophotometer. cDNA was prepared using 20–100 ng/μl total RNA by a reverse transcription-polymerase chain reaction (RT-PCR) using a high capacity cDNA reverse transcription kit (Applied Biosystems), according to the manufacturer’s instructions. Real-time quantitative PCR (qPCR) was performed on cDNA using SYBR Green probes specific for IL-1β, IL-10, TNF-α, IL-1RA and Rps18, or Taqman probes specific for HIF-1α, PHD3 and Rps18. qPCR was performed on a 7900 HT Fast Real-Time PCR System (Applied Biosystems) using Kapa fast master mix high ROX (Kapa Biosystems, for SYBR probes) or 2X PCR fast master mix (Applied Biosystems, for Taqman probes). The SYBR primer pair sequences were as follows: Il1b, FW 5’-TGGCAACTGTTCCTG-3’, RV 5’-GGAAGCAGCCCTTCATCTTT-3’; Il10, FW 5’-AGGCGCTGTCATCG-ATTT-3’, RV 5’-CACCTTGGTCTTGGAGCTTAT-3’; Tnfa, FW 5’-GCCTCTTCT-CATTCCTGCTT-3’, RV 5’-TGGGAACTTCTCATCCCTTTG-3’; Rps18, FW 5’-GGATGTGAAGGATGGGAAGT-3’, RV 5’-CCCTCTATGGGCTCGAATTT-3’; Il-1ra, FW 5’-TTGTGCCAAGTCTGGAGATG-3’, RV 5’-CTCAGAGCGGATGAAGGTAAAG-3’; Phd3, FW 5’-TGCTGAAGAAAGGGCAGAAG-3’; RV 5’-GCACACCACAGTCAGTCTTTA -3’; cMyc, FW 5'-CCA CCA GCA GCG ACT CTG-3', RV 5'-GAG ATG AGC CCG ACT CCG-3'; CD71, FW 5'-AAGTGACGTAGATCCAGAGGG-3', RV 5'-GACAATGGTTCCCCACCAAA-3'. Fold changes in expression were calculated by the Delta Delta Ct method using mouse Rps18 as an endogenous control for mRNA expression. All fold changes are expressed normalized to the untreated control.
Western blotting
Protein samples from cultured cells were prepared by direct lysis of cells in 5X Laemmli sample buffer, followed by heating at 95°C for 5 min. For spleen samples, 30 mg of spleen was homogenized in RIPA buffer using the Qiagen TissueLyserII system. The resulting homogenate was centrifuged at 14000 rpm for 10 min at 4°C, and supernatants were used for SDS-PAGE. Protein samples were resolved on 8% or 12% SDS-PAGE gels and were then transferred onto polyvinylidene difluoride (PVDF) membrane using either a wet or semi-dry transfer system. Membranes were blocked in 5% (w/v) dried milk in Tris-buffered saline-Tween (TBST) for at least one hour at room temperature. Membranes were incubated with primary antibody, followed by the appropriate horseradish peroxidase-conjugated secondary antibody. They were developed using LumiGLO enhanced chemiluminescent (ECL) substrate (Cell Signalling). Bands were visualized using the GelDoc system (Biorad).
Enzyme-linked immunosorbent assay (ELISA)
Cytokine concentrations in cell supernatants were measured using enzyme-linked immunosorbent assay (ELISA) Duoset kits for mouse IL-10 and TNF-α, according to the manufacturer’s instructions. Cytokine concentrations in serum samples isolated from whole blood were measured using Quantikine ELISA kits for mouse IL-1β, IL-10 and TNF-α. Duoset and Quantikine kits were from R&D Systems. Optical density values were measured at a wavelength of 450 nm, using a FLUOstar Optima plate reader (BMG Labtech). Concentrations were calculated using a 4-parameter fit curve.
FACS Analysis of reactive oxygen species (ROS)
BMDMs were seeded at 0.5x106 cells/ml. One well was seeded for each of the following controls: unstained cells, single-stained cells and dead cells. For cellular ROS measurements cells were treated and stimulated as normal. 2 hours prior to staining, 100% EtOH was added to the dead cell control well. 30 minutes prior to the end of the stimulation, CellROX (5 μM) was added directly into the cell culture medium. Supernatants of cells that were to be stained with Aqua Live/Dead were removed, and an Aqua Live/Dead dilution (1 ml; 1 in 1000 in PBS) was added to each well. Cells were incubated in tinfoil at 37°C for 30 min. For mitoROS measurements MitoSOX (5 μM) and Aqua Live/Dead were each added for 30 min prior to stimulation. The media was then removed and replaced with stimulus-containing media. The supernatant was removed and cells were scraped in PBS (1 ml), before being transferred to polypropylene FACS tubes. Cells were centrifuged at 2000 rpm for 3 min. Cells were washed in PBS and centrifuged two further times, and were finally resuspended in PBS (500 μl). BMDMs were analyzed using a Dako CyAn flow cytometer, and data was analyzed using FlowJo software.
NAD+/NADH measurement
BMDMs were plated at 0.5x106 cells/ml in 10 cm non-cell culture-treated dishes (10 ml/dish) and treated as required. NAD+/NADH was assayed using an NAD+/NADH quantification colorimetric kit (BioVision) according to the manufacturer’s instructions.
ATP/ADP measurement
BMDMs were plated at 0.5x106 cells/ml in white 96-well plates (100 μl/well) and treated as required. ATP/ADP was assayed using an ATP/ADP quantification bioluminescent kit (Sigma) according to the manufacturer’s instructions.
LDH assay
Cells were plated at 0.5x106 cells/ml in white 24-well plates (500 μl/well) and treated as required. Cytotoxicity, as determined by LDH release, was assayed using CytoTox96 Non-radioactive Cytotoxicity Assay kit (Promega) according to the manufacturer’s instructions.
Metabolite measurements
Cell culture medium – cell culture supernatant (50 μl) was centrifuged at maximum speed for 5 min at 4°C. The supernatant was transferred to a fresh microcentrifuge tube and resuspended in extraction buffer (750 μl; EB; 50% methanol, 30% acetonitrile, 20% dH2O, 100 ng/ml HEPES). Samples were agitated for 15 min at 4°C and then centrifuged at maximum speed for 10 min at 4°C. The resultant supernatant was transferred into autosampler vials and stored at -80°C prior to analysis by liquid chromatography-mass spectrometry (LC-MS).
Cells – Cells were washed three times with ice-cold PBS, with all of the PBS being removed after the last wash. EB was added (1 ml per 1x106 cells). Samples were agitated for 15 min at 4°C. The resultant suspension was transferred to ice-cold microcentrifuge tubes and centrifuged at maximum speed for 10 min at 4°C. The supernatant was transferred into autosampler vials and stored at -80°C prior to analysis by LC-MS.
LC-MS analysis of sample extracts was performed on a QExactive Orbitrap mass spectrometer coupled to a Dionex UltiMate 3000 Rapid Separation LC system (Thermo). The liquid chromatography system was fitted with a SeQuant Zic-HILIC (150 mm × 4.6 mm, 5 µm) with guard column (20 mm × 2.1 mm, 5 µm) from Merck (Darmstadt, Germany). The mobile phase was composed of 0.1% formic acid in water (solvent A), and 0.1% formic acid in acetonitrile (solvent B), and the flow rate set at 300 μL x min-1. The mass spectrometer was operated in full MS and polarity switching mode. Samples were randomized in order to avoid machine drift, and blinded to the operator. The acquired spectra were analyzed using XCalibur Qual Browser and XCalibur Quan Browser software (Thermo Scientific) by referencing to an internal library of compounds.
Seahorse analysis of oxygen consumption and lactate production
Cells were plated at 0.2x106 cells/well of a 24-well Seahorse plate with one well per row of the culture plate containing only supplemented media without cells, as a negative control. Cells were treated and stimulated as normal. A utility plate containing calibrant solution (1 ml/well) together with the plates containing the injector ports and probes was placed in a CO2-free incubator at 37°C overnight. The following day media was removed from cells and replaced with glucose-supplemented XF assay buffer (500 μl/well) and the cell culture plate was placed in a CO2-free incubator for at least 0.5 h. Inhibitors (Oligomycin, carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP), 2DG, Rotenone; 70 μl) were added to the appropriate port of the injector plate. This plate together with the utility plate was run on the Seahorse for calibration. Once complete, the utility plate was replaced with the cell culture plate and run on the Seahorse XF-24.
Endotoxin-induced model of sepsis
Mice were treated i.p. ± dimethyl malonate (160 mg/kg) or PBS for 3 h prior to stimulation with LPS (15 mg/kg) i.p. for 2 h. Mice were euthanized in a CO2 chamber and peritoneal cells, spleens and whole blood samples were harvested. AOX expressing mice were treated i.p. with LPS (10 mg/kg) on day 0 and then followed up for seven days.
Mitochondrial membrane potential measurement-TMRM confocal assay
Cells were plated at 0.3x106 cells/ml in DMEM containing 10% FCS and 1% P/S in CELLview cell culture dishes containing four compartments per dish (500 μl/compartment). TMRM cell-permeable fluorescent dye was either added before or after stimulation depending on the length and nature of treatment. In the case of LPS-treated cells following stimulation media was removed and replaced with TMRM-containing media (20 nM) and incubated at 37° in the dark for 30 min. Oligomycin or CCCP was added after the addition of TMRM for 1 h or 2 min respectively. Cells were imaged on a Leica SP8 confocal microscope with an excitation laser of 550 nm and detection set for 560-650 nM using a 40x oil-objective lens. A number of images were taken for each treatment.
Mitochondrial membrane potential measurement-TMRM FACS assay
Cells were plated at 0.5x106 cells/ml in 12-well plates. The following control cell samples were accounted for: unstained, single-stained TMRM, MitoTracker Green (MTG) and Aqua Live/Dead stain and 100% dead cells (treated with 100% ethanol for 1 h). Cells were prepared and treated as normal. TMRM stock was made up in DMSO (10 mM). MTG and Aqua Live/Dead stain were prepared by the addition of DMSO (75 μl or 50 μl/vial respectively). Supernatants were removed from all cells that were to be stained with Aqua Live/Dead (i.e. all samples except single-stained and unstained controls) and this was replaced with PBS (1 ml) containing Aqua Live/Dead stain (1 μl) and incubated at 37°C in the dark for 30 min. A mix of PBS containing MTG (50 nM) and TMRM (20 nM) was made up. Supernatant was removed from cells to be stained with MTG and TMRM (i.e. all samples except single-stained and unstained controls) and this was replaced with PBS containing MTG and TMRM (1 ml) and incubated at 37°C in the dark for 30 min. Supernatants were removed and cells were washed with PBS (1 ml). PBS (500 μL) was added per well and cells were removed from the plate surface using a cell scraper and transferred to polypropylene FACS tubes. Cells were then analyzed using a LSRFortessa flow cytometer, and data were analyzed using FlowJo software.
In vitro deletion of SDHB
BMDMs from mice carrying a SDHB fl/fl allele were utilized. Ablation of SDHB was achieved by adding 4-hydroxytamoxifen (600 nM) on day 4 of macrophage differentiation and again for a further 24 h when cells were replated on day 6. Ethanol was used as a vehicle control for the 4-hydroxytamoxifen treatment.
Transcriptomics analysis
Total RNA was isolated using the RNeasy Plus Mini kit (Qiagen) and quantified using a Nanodrop 2000 UV-visible spectrophotometer. cDNA libraries were generated using the Smart-seq2 protocol modified for 50 ng of total RNA input and sequenced on a MiSeq (Illumina, San Diego, CA) per manufacturer’s instructions. Sequences obtained from the RNASeq pipeline were aligned against the Mus musculus genome using TopHat + Bowtie. HTSeq-count was used to count the transcripts associated with each gene, and a counts matrix containing the number of counts for each gene across different samples and stimulations was obtained. The counts were normalized using the TMM method (DOI: 10.1186/gb-2010-11-3-r25). To analyze differential expression across different stimulations, the counts matrix was fitted with a generalized linear model, using the edgeR package. A nested interaction model involving LPS stimulation and stimulation with DMM or succinate as the two factors was used to analyze the gene expression. Three comparisons were made using this model to explore in detail the effects of DMM and succinate on LPS-activated macrophages -
1. Differential gene expression under stimulation with LPS alone in the base case was compared to LPS stimulation when the samples were pretreated with DMM or succinate, and the strength and direction of regulation of expression by these two compounds was compared to the base case (Figure 3A). A list of genes which showed statistically significant regulation in opposite directions in both the base case and with pretreatment (e.g. genes found to have higher expression when stimulated with succinate + LPS compared with LPS alone, and lower expression with DMM + LPS compared with LPS alone) was obtained - and analyzed for biological pathway enrichment in the GO and Kegg databases (Figure 3B).
2. A direct comparison was made between LPS-activated macrophages pretreated with succinate and LPS-activated macrophages pretreated with DMM to find very high confidence pathways that were oppositely regulated under both comparisons (Figure 3C).
3. Further, samples which were treated with LPS at different time points (4 h or 48 h) were analyzed separately to identify genes which were being expressed differently at different stages of the LPS stimulation (Supplemental Figure 1).
For all the comparisons, an FDR-adjusted p-value of 0.05 was considered to be the threshold for statistical significance, where the Benjamini-Hochberg test was used for multiple testing correction.
Statistical analysis
One- or two-tailed t-tests measured significance of expression of genes with different treatments.
Supplementary Material
Acknowledgements
We thank Henry Däbritz (Cancer Metabolism Research Unit, Cancer Research UK, Beatson Institute, Garscube Estate, Switchback Road, Bearsden, Glasgow, G61 1BD) for bones from SDHB floxed mice. We thank Ariel Lefkovith and Bihua Li for their contribution to the library preparation for the RNA sequencing analysis. We thank Praveen K. Dhandapani and Kira Holmström for assisting with the provision of the AOX mouse tissues.
This work was supported by funds and grants from the UK MRC MC_U105663142 to MPM and from Science Foundation Ireland, European Research Council and Irish Research Council to LAON. Funding was provided by Academy of Finland (CoE grant 272376 to HTJ), the European Research Council (advanced grant 232738 to HTJ), Tampere University Hospital Medical Research Fund, and the Sigrid Juselius Foundation (to HTJ). CF and ASHC were supported by an MRC Core Fund to the MRC Cancer Unit.
Footnotes
Author Contributions
E. L. Mills and B. Kelly designed and performed experiments and analyzed the data. E. L. Mills wrote the manuscript. A. Logan performed and analyzed experiments measuring the membrane potential. A. S. H. Costa and C. Frezza performed and analyzed experiments measuring succinate levels. S. C. Corr assisted with the in vivo LPS trial. D. Ryan assisted in experiments comparing the effects of Krebs cycle metabolites on LPS-induced cytokines. G. McManus aided in confocal microscopy experiments. J. H. M. Däbritz and E. Gottlieb provided the SDHB-deficient bones. M. Varma4 performed the RNA sequencing analysis. I. Latorre coordinated RNA sequencing work. R. J. Xavier provided guidance and advice. T. Braun, H. T. Jacobs and M. Szibor generated AOX-expressing mice and provided data on the LPS in vivo trial. M. P. Murphy provided advice, reagents and oversaw a portion of the work. L. A. O’Neill conceived ideas and oversaw the research programme.
The authors declare competing financial interests. MPM and CF have a patent application on the therapeutic application of SDH inhibitors.
References
- Barrientos A, Moraes CT. Titrating the effects of mitochondrial complex I impairment in the cell physiology. The Journal of biological chemistry. 1999;274:16188–16197. doi: 10.1074/jbc.274.23.16188. [DOI] [PubMed] [Google Scholar]
- Cassatella MA, Meda L, Gasperini S, Calzetti F, Bonora S. Interleukin 10 (IL-10) upregulates IL-1 receptor antagonist production from lipopolysaccharide-stimulated human polymorphonuclear leukocytes by delaying mRNA degradation. The Journal of experimental medicine. 1994;179:1695–1699. doi: 10.1084/jem.179.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B, Hollunger G. The interaction of energy and electron transfer reactions in mitochondria. I. General properties and nature of the products of succinate-linked reduction of pyridine nucleotide. The Journal of biological chemistry. 1961;236:1534–1543. [PubMed] [Google Scholar]
- Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, Gaude E, Aksentijevic D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord EN, Smith AC, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouchani ET, Pell VR, James AM, Work LM, Saeb-Parsy K, Frezza C, Krieg T, Murphy MP. A Unifying Mechanism for Mitochondrial Superoxide Production during Ischemia-Reperfusion Injury. Cell metabolism. 2016;23:254–263. doi: 10.1016/j.cmet.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Couper KN, Blount DG, Riley EM. IL-10: the master regulator of immunity to infection. J Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- Dervartanian DV, Veeger C. Studies on Succinate Dehydrogenase. I. Spectral Properties of the Purified Enzyme and Formation of Enzyme-Competitive Inhibitor Complexes. Biochim Biophys Acta. 1964;92:233–247. [PubMed] [Google Scholar]
- Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Khoury R, Dufour E, Rak M, Ramanantsoa N, Grandchamp N, Csaba Z, Duvillie B, Benit P, Gallego J, Gressens P, et al. Alternative oxidase expression in the mouse enables bypassing cytochrome c oxidase blockade and limits mitochondrial ROS overproduction. PLoS genetics. 2013;9:e1003182. doi: 10.1371/journal.pgen.1003182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKvarepsilon supports the anabolic demands of dendritic cell activation. Nat Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, van der Windt GJ, Freitas TC, Chott R, Yarasheski KE, Pearce EL, Pearce EJ. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Aguera MC, Gao L, Gonzalez-Rodriguez P, Pintado CO, Arias-Mayenco I, Garcia-Flores P, Garcia-Perganeda A, Pascual A, Ortega-Saenz P, Lopez-Barneo J. Oxygen Sensing by Arterial Chemoreceptors Depends on Mitochondrial Complex I Signaling. Cell metabolism. 2015;22:825–837. doi: 10.1016/j.cmet.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Garaude J, Acin-Perez R, Martinez-Cano S, Enamorado M, Ugolini M, Nistal-Villan E, Hervas-Stubbs S, Pelegrin P, Sander LE, Enriquez JA, et al. Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nature immunology. 2016 doi: 10.1038/ni.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarda G, Braun M, Staehli F, Tardivel A, Mattmann C, Forster I, Farlik M, Decker T, Du Pasquier RA, Romero P, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Guzy RD, Sharma B, Bell E, Chandel NS, Schumacker PT. Loss of the SdhB, but Not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol Cell Biol. 2008;28:718–731. doi: 10.1128/MCB.01338-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Weinberg SE, Reczek CR, Chandel NS. The Mitochondrial Respiratory Chain Is Required for Organismal Adaptation to Hypoxia. Cell Rep. 2016 doi: 10.1016/j.celrep.2016.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Jin Z, Wei W, Yang M, Du Y, Wan Y. Mitochondrial complex I activity suppresses inflammation and enhances bone resorption by shifting macrophage-osteoclast polarization. Cell metabolism. 2014;20:483–498. doi: 10.1016/j.cmet.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M. Site of action of lipid A on mitochondria. Journal of bacteriology. 1972;112:268–275. doi: 10.1128/jb.112.1.268-275.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B, Tannahill GM, Murphy MP, O'Neill LA. Metformin Inhibits the Production of Reactive Oxygen Species from NADH:ubiquinone Oxidoreductase to Limit Induction of IL-1beta, Boosts IL-10 in LPS-activated Macrophages. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.M115.662114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivney A, Bradley SG. Action of bacterial endotoxin and lipid A on mitochondrial enzyme activities of cells in culture and subcellular fractions. Infection and immunity. 1979;25:664–671. doi: 10.1128/iai.25.2.664-671.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills EL, O'Neill LA. Reprogramming mitochondrial metabolism in macrophages as an anti-inflammatory signal. European journal of immunology. 2016;46:13–21. doi: 10.1002/eji.201445427. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales-Clemente E, Bayona-Bafaluy MP, Perez-Martos A, Barrientos A, Fernandez-Silva P, Enriquez JA. Restoration of electron transport without proton pumping in mammalian mitochondria. Proc Natl Acad Sci U S A. 2008;105:18735–18739. doi: 10.1073/pnas.0810518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scialo F, Sriram A, Fernandez-Ayala D, Gubina N, Lohmus M, Nelson G, Logan A, Cooper HM, Navas P, Enriquez JA, et al. Mitochondrial ROS Produced via Reverse Electron Transport Extend Animal Lifespan. Cell Metab. 2016;23:725–734. doi: 10.1016/j.cmet.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena LA, Li S, Jairaman A, Prakriya M, Ezponda T, Hildeman DA, Wang CR, Schumacker PT, Licht JD, Perlman H, et al. Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity. 2013;38:225–236. doi: 10.1016/j.immuni.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votyakova TV, Reynolds IJ. DeltaPsi(m)-Dependent and -independent production of reactive oxygen species by rat brain mitochondria. Journal of neurochemistry. 2001;79:266–277. doi: 10.1046/j.1471-4159.2001.00548.x. [DOI] [PubMed] [Google Scholar]
- West AP, Brodsky IE, Rahner C, Woo DK, Erdjument-Bromage H, Tempst P, Walsh MC, Choi Y, Shadel GS, Ghosh S. TLR signalling augments macrophage bactericidal activity through mitochondrial ROS. Nature. 2011;472:476–480. doi: 10.1038/nature09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Pollard PJ. Succinate: a new epigenetic hacker. Cancer Cell. 2013;23:709–711. doi: 10.1016/j.ccr.2013.05.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.