Abstract
Missense mutations in the TP53 gene lead to accumulation of dysfunctional TP53 proteins in tumors, showing oncogenic gain-of-function (GOF) activities. Stabilization of mutant TP53 (mutp53) is required for the GOF; however, the mechanisms by which mutp53 promotes cancer progression and how mutp53 stability is regulated are not completely understood. Recent work from our laboratory has identified statins, inhibitors of the mevalonate pathway, as degraders of conformational mutp53. Specific reduction of mevalonate-5-phosphate (MVP), a metabolic intermediate in the mevalonate pathway, by statins or mevalonate kinase (MVK) knockdown triggers CHIP ubiquitin ligase-mediated degradation of conformational mutp53 by inhibiting interaction between mutp53 and DNAJA1, a Hsp40 family member. Thus, the mevalonate pathway contributes to mutp53 stabilization. Given that mutp53 is shown to promote cancer progression by upregulating mRNA expression of mevalonate pathway enzymes by binding to the sterol regulatory element-binding protein 2 (SREBP2) and subsequently increasing activities of mevalonate pathway-associated oncogenic proteins (e.g., Ras, Rho, YAP/TAZ), there is a positive-feedback loop between mutp53 and the mevalonate pathway. Here, we summarize recent evidence linking the mevalonate pathway-mutp53 axis with cancer progression and further discuss the clinical relevance of this axis.
Subject terms: Cancer metabolism, Drug development
Facts
Stabilization of mutp53 greatly contributes to its GOF activities.
Mutp53 enhances expression of mevalonate pathway enzymes by directly interacting with SREBP2.
Enhanced mevalonate pathway activity potentiates the malignant properties of cancer cells through activation of oncogenic proteins Ras, Rho, and YAP/TAZ.
Inhibition of the mevalonate pathway by statins or MVK knockdown induces degradation of misfolded mutp53.
Reduced cellular mevalonate-5-phosphate (MVP), an intermediate metabolite in the mevalonate pathway, triggers degradation of misfolded mutp53 by disrupting the mutp53-DNAJA1 interaction.
Inhibition of the mevalonate pathway reduces malignant properties of cancer cells via multiple mechanisms, including mutp53 degradation, inhibition of mutp53 GOF, reduction in protein prenylation of Ras and Rho, and decreased activity of YAP/TAZ.
Open questions
Which cancer types are regulated by or addicted to the mevalonate pathway-mutp53 axis.
Whether all conformational or misfolded mutp53 can be degraded by statins and how the cellular contexts could impact statins’ effects on misfolded mutp53 degradation and/or cancer cell progression.
Detailed mechanisms by which reduction in MVP inhibits the mutp53-DNAJA1 interaction to induce mutp53 degradation.
Whether statins or other inhibitors of the mevalonate pathway could reduce spontaneous tumor growth in mutp53 knock-in mouse models.
Whether combination of mevalonate pathway inhibitors with standard-of-care chemotherapy drugs could have cooperative effects on inhibiting malignant progression of mutp53-expressing cancer cells.
Introduction
TP53 functions as a tumor suppressor mainly by inducing transcription of numerous downstream target genes which regulate DNA repair, cell cycle arrest, senescence, apoptosis, and metabolism [1, 2]. TP53 also induces apoptosis by directly interacting with the multi-domain members of the Bcl-2 family at the mitochondrial membrane [3]. Hence, loss of TP53 activity in normal cells causes uncontrolled cell proliferation and death, as well as metabolic reprogramming, leading to immortalization and ultimately cancer [4–6]. This is well supported by the fact that TP53 is the most frequently mutated gene in human cancers [7]. The majority of TP53 mutations are missense mutations in the DNA-binding domain, resulting in loss of the transcriptional and tumor suppressive activities of TP53 [7]. When cells express both the wild-type TP53 (wtp53) and mutant TP53 (mutp53), mutp53 can inactivate wtp53 by its dominant-negative activity through oligomerization or aggregation with wtp53 [4, 8–10]. Additionally, mutp53 shows the gain-of-function (GOF) activities and promotes tumor progression, metastasis, and drug resistance by binding to or co-aggregating with tumor suppressive (e.g., p63, p73, MRN complex) or oncogenic (e.g., Ets2, NF-Y) proteins even in cells lacking wtp53 and only expressing mutp53 [11–17]. Indeed, many TP53 mutants lose the wild-type conformation, and/or acquire aggregation properties, thus showing the GOF activity [14]. Consequently, the presence of mutp53 is not simply the same as the absence of the wtp53 allele. Many TP53 mutants are inherently unstable, and therefore stabilization of mutp53 in tumors is crucial for showing the oncogenic GOF activities [16, 18, 19]. Importantly, knockdown of mutp53 has been shown to reduce malignant properties of cancer cells [20–22]. Yet, molecular mechanisms underlying mutp53 GOF, as well as mutp53 stabilization or degradation, are not completely understood.
To understand the mechanism of mutp53 stabilization/degradation and identify workable strategies that induce mutp53 degradation, we recently performed high-throughput screening of chemical libraries. This screening identified “statins”, a class of drugs that inhibit hydroxyl-methylglutaryl coenzyme A reductase (HMGCoAR) and hence reduce cholesterol production through the mevalonate pathway, as degradation inducers of conformational or misfolded mutp53 at a concentration of as low as 4 µM; statins have a minimal impact on wtp53 and DNA-contact mutp53 with native structure [23]. Specifically, reduction of cellular mevalonate-5-phosphate (MVP), but not other metabolic intermediates in the mevalonate pathway, triggers misfolded mutp53 degradation in a protein prenylation-independent manner [23]. Mechanistically, decreased MVP, by statins or mevalonate kinase (MVK) knockdown, inhibits mutp53’s binding to a molecular chaperone of the Hsp40 family, DNAJA1, leading to CHIP ubiquitin ligase-mediated mutp53 degradation [23]. Increasing evidence indicates that inhibition of mevalonate pathway by statins, zoledronic acid (also known as bisphosphonate), and protein prenylation inhibitors results in the suppression of multiple types of cancer [24–28]. Thus, our recent findings, in addition to a report by Freed-Pastor et al. [29] in which mutp53 upregulates expression of mevalonate pathway enzymes, significantly advance our understanding of the involvement of the mevalonate pathway in cancer progression, further encouraging strategies that target the mevalonate pathway for cancer therapy. Here, we mainly summarize recent findings that link mutp53 and the mevalonate pathway with regard to cancer progression.
The mevalonate pathway and cancer
The mevalonate pathway is an essential lipogenic pathway that uses acetyl-CoA to produce isoprenoids and cholesterol [30]. Isoprenoids are required for protein prenylation/lipidation (farnesylation and geranylgeranylation), which enables target proteins, including Ras and Rho small guanosine triphosphatases (GTPases), to anchor to the cell membrane [31]. Cholesterol is used as an important hydrophobic precursor to bile acids, hormones, and lipoproteins [32].
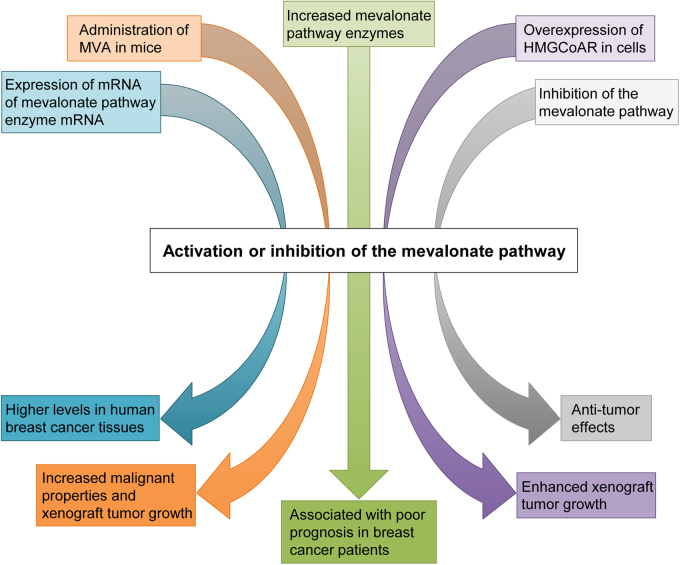
Accumulating evidence has suggested involvement of the mevalonate pathway in cancer progression. For instance, human breast cancer tissues express mRNA of several mevalonate pathway enzymes at higher levels, when compared with normal breast tissues (Fig. 1) [33, 34]. These enzymes include HMGCoAR, the rate-limiting step enzyme, and farnesyl diphosphate synthase (FDPS), a key branch point enzyme. Also, administration of mevalonic acid (MVA), a metabolite produced by HMGCoAR, enhances tumor growth in a breast cancer xenograft mouse model [34]. Moreover, increased expression of mevalonate pathway-associated proteins is correlated with poor prognosis in breast cancer patients [35]. Overexpression of HMGCoAR in a human hepatocellular carcinoma cell line HepG2 and a non-tumorigenic breast epithelial cell line MCF10A increases their anchorage-independent cell growth, as well as tumor growth of HepG2 cells in a xenograft model [35]. Meanwhile, numerous clinical studies support antitumor effects of statins and other inhibitors of the mevalonate pathway (Fig. 1) [24, 36]. However, the underlying mechanisms by which inhibition of the mevalonate pathway suppresses tumor progression remain unclear. Increasing evidence indicates functional association between the mevalonate pathway and oncogenic proteins including mutp53, Ras, Rho, and YAP/TAZ.
Fig. 1.
Association between the mevalonate pathway activity and cancer progression. The mevalonate pathway is associated with increased tumor malignancy through several in vitro and in vivo observations. The use of statins to reduce the mevalonate pathway activity is correlated with decreased tumor malignancy.
The mevalonate pathway-mutp53 axis
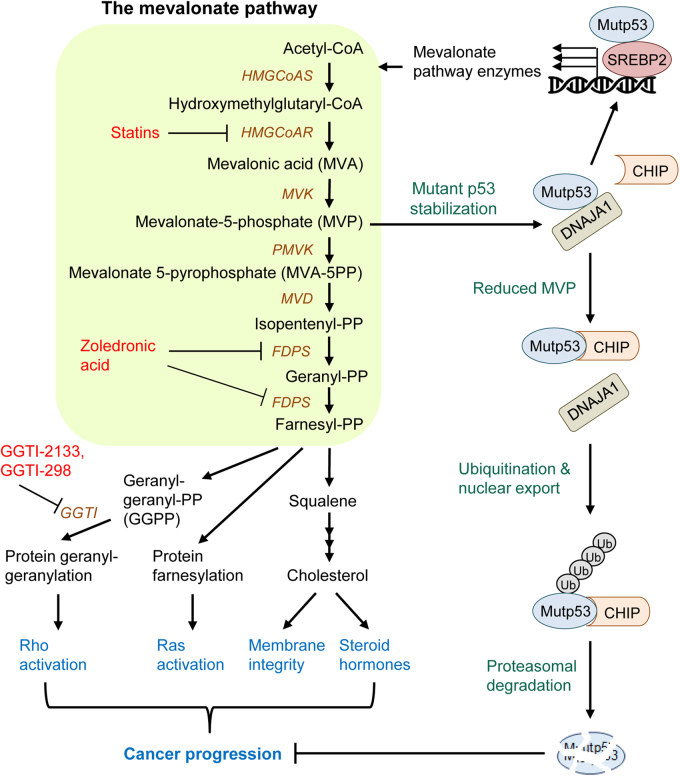
Expression of lipogenic enzymes, including mevalonate pathway enzymes, is mainly regulated by sterol regulatory element-binding proteins (SREBPs), basic helix-loop-helix leucine zipper transcription factors. SREBP2 is shown to bind with oncogenic mutp53, leading to increase in the expression of various mevalonate pathway enzymes, such as HMGCoAR, MVK, and FDPS (Fig. 2) [29]. Indeed, blockade of this pathway by statins, a geranylgeranyl transferase I (GGTI) inhibitor, or other compounds rescues disrupted mammary architecture of breast cancer cells carrying mutp53 in three-dimensional (3-D) culture. Knockdown of mutp53 also nullifies disrupted mammary architecture of breast cancer cells in 3-D culture [29]. Importantly, the presence of p53 mutations in human breast cancer tissues is correlated with high expression of mevalonate pathway enzymes and poor prognosis in patients with breast cancer [29]. These results strongly suggest that oncogenic GOF mutp53 contributes to breast cancer progression through the mevalonate pathway.
Fig. 2.
The interplay between the mevalonate pathway and mutp53. The mevalonate pathway can enhance cancer progression through several molecular mechanisms, including protein prenylation/lipidation (Ras farnesylation, Rho geranylgeranylation), cholesterol synthesis, and stabilization of mutp53. Mevalonate-5-phosphate (MVP), a metabolic intermediate in the mevalonate pathway, promotes interaction between conformational mutp53 and DNAJA1, which inhibits CHIP ubiquitin ligase-mediated ubiquitination and proteasomal degradation of mutp53, leading to mutp53 stabilization. Mutp53 is shown to bind to SREBP2, increasing the expression of mevalonate pathway enzymes and subsequently MVP levels, which in turn stabilizes mutp53, thus forming a positive-feedback loop. Therefore, inhibition of the mevalonate pathway with statins, zoledronic acid, or GGTI inhibitors (GGTI-2133, GGTI-298) may represent a novel therapeutic strategy of targeting cancers with mutp53 by inhibiting mutp53 GOF activities, prenylation of oncogenes, and cholesterol synthesis, as well as inducing mutp53 degradation. HMGCoAS hydroxylmethylglutaryl coenzyme A syntase, HMGCoAR hydroxylmethylglutaryl coenzyme A reductase, MVK mevalonate kinase, PMVK phosphomevalonate kinase, MVD mevalonate pyrophosphate decarboxylase, FDPS farnesyl diphosphate synthase, GGTI geranylgeranyl transferase I, PP pyrophosphate
Recently, through high-throughput screening of chemical compound libraries, we have identified statins as drugs that deplete mutp53 [23]. Intriguingly, statins mainly deplete conformational or misfolded mutp53 with minimal impact on wtp53 and DNA-contact mutp53 that maintain native protein structure. Statin’s effect is specific to the mevalonate pathway, because mutp53 depletion by statins is rescued by supplementation with MVA or MVP, but not mevalonate-5-pyrophosphate (MVA-5PP). Also, zoledronic acid (25 µM) and GGTI-2133 (4 µM) that inhibit downstream enzymes in the mevalonate pathway have minimal effects on mutp53 levels [23]. Knockdown of MVK, which converts MVA to MVP, also reduces levels of conformational mutp53 (Fig. 2) [23]. The observed reduction in mutp53 levels is regulated via ubiquitination by CHIP (C-terminus of Hsc70-interacting protein), since statins or MVK knockdown triggers ubiquitination, nuclear export, and degradation of mutp53, which is rescued by CHIP knockdown [23]. Thus, our study indicates that MVP, a metabolic intermediate in the mevalonate pathway, plays a key role in the regulation of conformational mutp53 stabilization, and statin’s effect on mutp53 degradation is independent of prenylation or cholesterol synthesis (Fig. 2).
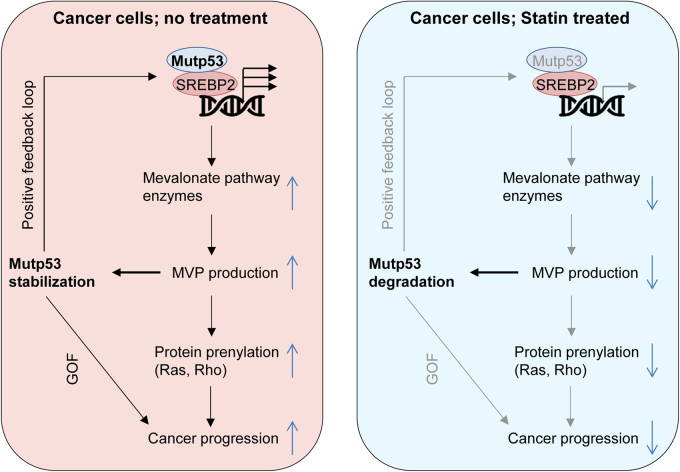
The key question is how MVP regulates stability of conformational or misfolded mutp53. We initially suspected involvement of Hsc70/Hsp70, since altered structure of conformational mutp53 leads to exposure of a binding site for Hsc70 (high affinity heat shock cognate 71 kDa protein) in the DNA-binding domain [37] and also CHIP binds to Hsp70/Hsc70 [38]; however, knockdown of Hsc70 fails to induce degradation of p53R156P and p53R175H distinct from statins, suggesting that Hsc70 is not involved in the statin-mediated mutp53 degradation [23]. Other proteins involved in protein refolding and degradation of structurally distorted proteins are Hsp90 and Hsp40. Hsp90 is known to promote tumorigenesis and stabilize both conformational and DNA-contact mutp53 [20, 39–41]. However, statins affect levels of neither Hsp90 nor its client proteins, such as Raf1, EGFR (epidermal growth factor receptor), and Her2/Neu, suggesting that the mevalonate pathway does not likely affect Hsp90 activity [23]. Since Her2/Neu is ubiquitinated and degraded by CHIP [42], this result also suggests that mutp53 degradation by statins is not simply due to non-specific activation of CHIP. Hence, we next examined involvement of Hsp40 which is known to bind to mutp53 [43] and competitively inhibits CHIP-mediated refolding [44]. Similar to statins, downregulation of DNAJA1, but not another Hsp40 member, DNAJB1, induces CHIP-mediated ubiquitination, nuclear export, and proteasomal degradation of several conformational p53 mutants with minimal effects on wtp53 and DNA-contact mutp53, while overexpression of DNAJA1 inhibits statin’s effects on conformational mutp53 [23]. Intriguingly, reduced MVP by statins or MVK knockdown leads to inhibited binding between mutp53 and DNAJA1, while binding of mutp53 with CHIP increases following statin treatment (Fig. 2) [23]. These observations strongly suggest that DNAJA1 protects misfolded mutp53 from CHIP-mediated degradation, and when the mutp53-DNAJA1 interaction is inhibited by reduced cellular MVP, it allows CHIP to ubiquitinate, nuclear export, and induce degradation of mutp53 (Fig. 2). Thus, the mevalonate pathway-DNAJA1 axis controls the fate (degradation or stabilization) of misfolded mutp53. Given that mutp53 upregulates expression of mevalonate pathway enzymes [29] and that the mevalonate pathway contributes to stabilization of mutp53 [23], there is a positive-feedback loop between mutp53 and the mevalonate pathway in cancer cells carrying misfolded mutp53 (Figs. 2 and 3).
Fig. 3.
The mevalonate pathway-mutp53 positive-feedback loop and statin’s inhibitory effects on this loop and cancer progression. Cancer cells expressing mutp53 have increased expression of the mevalonate pathway enzymes due to the mutp53-SREBP2 interaction, leading to enhanced activity of the mevalonate pathway. Activated mevalonate pathway increases the levels of MVP, leading to mutp53 stabilization (positive-feedback loop) and also promotes prenylation of Ras and Rho proteins, accelerating cancer progression. Thus, the mevalonate pathway promotes cancer progression by increasing activities of oncogenic proteins mutp53, Ras, and Rho (left). Inhibition of the mevalonate pathway activity by statins reduces MVP levels and protein prenylation of Ras and Rho, which in turn induces mutp53 degradation and decreases activities of Ras and Rho, thus inhibiting cancer progression (right).
Biologically, statins inhibit viable cell proliferation and/or tumor growth of KHOS/NP (p53R156P) osteosarcoma cell line, CAL33 (p53R175H) head and neck cancer cell line, and mouse embryonic fibroblasts (MEFs) expressing p53R172H (equivalent with human p53R175H) more efficiently than cells expressing wtp53 or DNA-contact mutp53 and those null for p53 [23]. Statin treatment also reduces mutp53 levels in tumors in xenograft mouse models. These results strongly suggest that the mevalonate pathway promotes tumor progression by increasing activities of oncogenes by protein prenylation (Ras and Rho) and stabilizing mutp53 (Fig. 3). Since cancer cells are frequently addicted to oncogenes (mutp53, Ras, Rho), tumor suppressive effects of statins are likely due to reduced activities of these oncogenes by degradation of misfolded mutp53 and reduced prenylation of Ras and Rho (Fig. 3). Intriguingly, statins are also shown to inhibit histone deacetylases (HDACs 1, 2, and 3) activities, increase acetylation of histone H3, enhance p21WAF/CIP expression, and reduce cell proliferation or tumor formation in multiple cancer cell lines regardless of p53 status, since these observations are reported using cells having wtp53, mutp53, and null for p53 [45–47]. However, it is unclear whether these observations are caused by inhibition of HMGCoAR or direct binding of statins to HDACs.
Regulation of Ras activity by the mevalonate pathway and mutp53
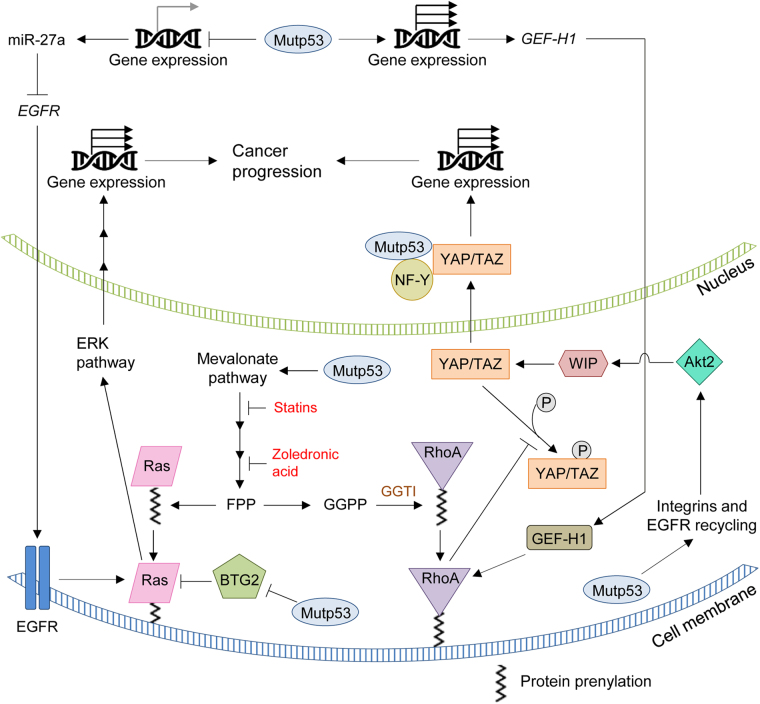
Protein prenylation plays crucial roles in multiple biological process, such as cell proliferation, migration, and survival [32]. Specifically, farnesylation of Ras GTPase, which is regulated by the mevalonate pathway, plays crucial roles in Ras activation and tumor progression [48, 49]. Co-overexpression of HMGCoAR and Ras is shown to enhance foci formation of primary mouse embryonic fibroblast (MEFs) [35], suggesting that increase in the mevalonate pathway activity cooperates with Ras to transform normal cells. Furthermore, inhibition of HMGCoAR and FDPS, both of which contribute to production of farnesyl pyrophosphate (FPP) [50], suppresses viable cell proliferation with decreased Ras farnesylation in pancreatic and breast cancer cells; the observed inhibition of cell proliferation is partially rescued by supplementation with FPP [33, 51]. These data provide evidence that the mevalonate pathway-mediated Ras farnesylation plays a crucial role in cancer progression (Fig. 4). In this line, the recent finding that mutp53 upregulates expression of mevalonate pathway enzymes and, in consequence, increases the activity of this pathway [29] suggests that mutp53 could indirectly increase Ras activity as one of its oncogenic GOF. Additionally, conformational p53R175H and p53H179R are shown to increase oncogenic H-RasV12 activity by binding with B-cell translocation gene 2 (BTG2), a negative regulator of H-RasV12 (Fig. 4) [52].
Fig. 4.
Functional association between mutp53 and mevalonate pathway-associated oncogenic proteins (Ras, RhoA, YAP/TAZ). Activated mevalonate pathway promotes prenylation and activities of Ras and RhoA, leading to their translocation into the plasma membrane and activation of the ERK and YAP/TAZ signaling pathways, respectively. Nuclear translocated ERK and YAP/TAZ promote transcription of downstream target genes involved in cancer cell proliferation and migration. Additionally, mutp53 increases activities of Ras, RhoA, and YAP/TAZ signaling through protein-protein interactions (BTG2, NY-F) and enhancing recycling of integrins/EGFR.
Oncogenic Ras activates the extracellular signal-regulated protein kinase 1/2 (ERK1/2) pathway (Raf-MEK1/2/-ERK1/2) [53]. Activation of membrane tyrosine kinase receptors (RTKs) results in Ras-mediated ERK1/2 nuclear translocation and activation of different transcription factors involved in proliferation, migration and cancer progression (Fig. 4) [54]. Additionally, mutp53 (p53R273H) is shown to suppress the expression of miR-27a, leading to the increase in the expression of EGFR and activation of the Ras-ERK1/2 pathway in breast (MDA-MB-231) and lung (H1299) cancer cells (Fig. 4) [55]. Also, downregulation of mutp53 in MDA-MB-231 cells results in reduction in the EGFR expression, ERK1/2 activity, cell proliferation, and tumor growth [55]. Together, these studies suggest that mutp53 could increase Ras activity through mevalonate pathway-dependent and -independent mechanisms.
Regulation of Rho activity by the mevalonate pathway and mutp53
The Ras homolog family (Rho) of GTPases is a group of cell signaling transducers that regulate cytoskeleton reorganization, vesicle trafficking, cell polarity, and cell cycle [56]. Like Ras, prenylation-dependent docking of Rho GTPases to the cell membrane is required for their normal functions [56]. Production of geranylgeranyl pyrophosphate (GGPP) through the mevalonate pathway is required for Rho genarylgenarylation [48]. Hence, activation of the mevalonate pathway leads to increase in Rho prenylation, which in turn promotes cancer progression (Fig. 4). Ginestier et al. [57] show that breast cancer cells growing in spheroid conditions (serum-free, non-adherent) have an increase in the expression of mevalonate pathway enzymes, and statins inhibit spheroid formation accompanied with reduction in the stem cell population. The observed reduction in stemness by statins is independent of cholesterol production or Ras farnesylation, but is dependent on Rho geranylgeranylation, since supplementation with GGPP rescues the stemness reduced by statins [57]. Also, pharmacologic inhibition of GGTI, an enzyme required for Rho geranylgeranylation, reduces the stem cell population of breast cancer cells and inhibits tumor establishment of a breast cancer patient-derived xenograft in mice [57]. Additionally, blocking the mevalonate pathway with statins or zoledronic acid in pancreatic cancer cells inhibits viable cell proliferation and induces apoptosis, which is nullified by supplementation with GGPP [33]. Thus, geranylgeranylation of Rho is crucial for cancer progression (Fig. 4). Given that mutp53 upregulates mevalonate pathway enzymes, mutp53 may play an important role in prenylation and activation of Rho.
Additionally, Mizuarai et al. [58] show that inducible expression of several p53 mutants (p53V157F, p53R175H, and p53R248Q) in U2OS cells significantly upregulates GEF-H1, a guanine exchange factor-H1 for RhoA, and subsequently activates RhoA (Fig. 4). Moreover, knockdown of mutp53 inhibits viable cell proliferation of several cancer cells with reduced GEF-H1 expression [58]. Also, cells expressing mutp53 tend to have higher mRNA expression of GEF-H1 when compared with those expressing wtp53 or null for p53 [58]. Thus, Rho activity is indirectly regulated by mutp53 through the mevalonate pathway and GEF-H1 (Fig. 4). This mutp53-RhoA axis could be a therapeutic target for mutp53-expressing cancers.
Regulation of YAP/TAZ activity by the mevalonate pathway and mutp53
Yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ) are transcriptional coactivators and downstream effectors of the Hippo signaling pathway which regulates organ growth, tissue regeneration, and stemness [50]. Also, the roles of YAP and TAZ in cancer progression are well appreciated [59]. A recent study using fluorescence microscopy-based high-throughput drug screening reveals that statins inhibit nuclear localization of YAP/TAZ in MDA-MB-231 cells [60]. Also, blockade of the mevalonate pathway by statins, zoledronic acid, or GGTI-298 (a GGTI inhibitor) inhibits YAP/TAZ nuclear localization as well as transcription of YAP/TAZ target genes in multiple cancer cell lines [60]. Furthermore, statins and zoledronic acid suppress proliferation and tumor growth of breast cancer cells, which is associated with reduction in the YAP/TAZ activity [60]. Consistent with this finding, Wang et al. [61] also report that YAP activity is inhibited by stains and knockdown of Rho, which decreases mRNA expression of RHAMM (receptor for hyaluronan-mediated motility), leading to inhibited phosphorylation of ERK1/2 and migration/invasion in MDA-MB-231 breast cancer cells. Since Rho inhibits phosphorylation of YAP through LATS1/2 [62], these observations suggest that nuclear localization and activity of YAP/TAZ are regulated through the mevalonate pathway and Rho activity (Fig. 4). Thus, the mevalonate pathway-Rho-YAP/TAZ axis plays a role in breast cancer progression and metastasis.
Besides the indirect effect on YAP/TAZ activation mediated through the mevalonate pathway-Rho axis, mutp53 can physically interact with YAP in breast cancer cells, leading to enhanced transcription of pro-proliferative genes [63]. Mechanistically, mutp53 forms a complex with YAP and NF-Y to bind to the NF-Y responsive elements in the downstream target genes, such as cyclin A, cyclin B, and cyclin-dependent kinase 1 (CDK1) genes, leading to an increase in their transcription (Fig. 4) [63]. Knockdown of mutp53 or YAP in SK-Br-3 and CAL27 breast cancer cells reduces expression of the NF-Y target genes and inhibits viable cell proliferation [63]. In human breast tumors, increased levels of mutp53 correlate with high YAP activity signature and high levels of YAP target genes, cyclin A, cyclin B, and CDK1 [63].
Additionally, mutp53 is shown to increase stabilization and nuclear translocation of YAP/TAZ in glioblastoma and breast cancer cells; this is mediated by enhanced recycling efficacy of EGRF and integrins by mutp53 and subsequent increase in the activity of Akt2-WIP (WASP interacting protein) signaling, which in turn promotes YAP/TAZ nuclear translocation (Fig. 4) [64]. Biologically, overexpression of mutp53 increases stem cell-like properties in astrocytes with increased YAP/TAZ activity, while knockdown of mutp53 or WIP reduces stem-cell markers expression, proliferation, and tumor growth in mutp53-expressing glioblastoma and breast cancer cells [64]. These results indicate a novel mutp53 GOF which contributes to stem-like malignant properties of cancer cells via activation of the Akt2-WIP-YAP/TAZ axis by increasing integrin/EGFR recycling (Fig. 4).
Clinical relevance of statins in cancer progression
Clinical studies demonstrating the efficacy of statins on human cancer suppression remain controversial [24, 65]. Statins have been suggested to improve overall survival in patients with multiple types of cancer, including high-grade endometrial cancer [66], colorectal cancer [67], kidney cancer [68], and multiple myeloma [69]. Statin use is also associated with decreased risk of all-cause mortality in cancer patients [70] and risk of insulin-related cancer development in patients with diabetes [71]. Moreover, post-diagnostic use of statins is correlated with favorable recurrence-free survival when compare with pre-diagnostic statin use [70]. However, there are controversial observations, including (1) no correlation between statin use and risk of breast cancer [72] and (2) an increased risk of developing prostate cancer by statin use [73]. Thus, although many clinical studies support the positive roles of statins in human cancer suppression or patient’s prognosis, statin’s efficacy might be different among type or dose of statins, cancer type, and type of genetic alterations in tumors. Indeed, our in vivo mouse model studies suggest that tumors carrying conformational/misfolded p53 mutants respond better to statins as compared with those with wtp53, p53-null, and DNA-contact p53 mutants [23]. Our studies also suggest that a higher dose of statins (near maximum dose of statins clinically allowed) is required for suppressing tumor growth in mice [23]. Hence, status of p53 and dose of statins may affect clinical outcome of statin’s effects on tumor suppression. To the best of our knowledge, there is no clinical study analyzing statin’s effects on tumor suppression, based on the p53 status in tumors or dose of statins used for patients.
Other inhibitors for the mevalonate pathway have been tested in clinical trials with promising results. Zoledronic acid improves clinical outcome of patients with multiple myeloma [25]. Other clinical studies with zoledronic acid include metastatic breast cancer, prostate cancer, and pre-existing bone metastases [26, 74]. Also farnesyl transferase inhibitors (FTIs) and GGTI inhibitors have shown promising results in cancer preclinical models; however, in clinical trials their efficacy appears to be not as good as expected [28, 75, 27]. It should be noted that none of these studies consider p53 status in the tumors, which could be an important variable to predict the response to mevalonate pathway inhibitors.
Conclusion and future perspectives
Increasing evidence indicates tumor progressive roles of the mevalonate pathway as well as tumor suppressive roles of statins and other mevalonate pathway inhibitors [23]. Our recent findings also highlight the importance of identifying p53 status (conformational/misfolded mutp53 or not) in tumors to determine efficacy of statins to suppress cancer progression. However, there are several remaining questions to be answered in the future. These include (1) how MVP enhances the interaction between conformational/misfolded mutp53 and DNAJA1 to stabilize mutp53, (2) if any other molecular chaperones could stabilize conformational/misfolded mutp53 like DNAJA1, (3) which other p53 mutants can be degraded by statins, (4) whether statins reduce growth of spontaneous tumors in mutp53 knock-in mouse models, (5) whether combination of statins with standard-of-care chemotherapy drugs has cooperative effects on inhibiting malignant progression of mutp53-expressing cancer cells, (6) if statins or Hsp90 inhibitors that deplete mutp53 show equivalent or better effects on cancer suppression as compared with agents that restore wtp53 activity from mutp53, and (7) whether cancer patients taking high-dose statins have more benefits to their prognosis than those taking low-dose statins or without statin treatment. Studies to address these concerns should be performed in order to fully understand the mechanisms underlying mutp53 stabilization or degradation, determine efficient use of statins for cancer therapy, and discover novel therapeutic strategies for cancer carrying mutp53.
Acknowledgements
We thank Atul Ranjan and Satomi Yamamoto for editing the manuscript and helpful discussion. This manuscript is supported by NIH R01 CA174735 (T.I.) grant.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Edited by G. Melino
References
- 1.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harbor Perspect Biol. 2010;2:a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levav-Cohen Y, Goldberg Z, Tan KH, Alsheich-Bartok O, Zuckerman V, Haupt S, et al. The p53-Mdm2 loop: a critical juncture of stress response. Subcell Biochem. 2014;85:161–86. doi: 10.1007/978-94-017-9211-0_9. [DOI] [PubMed] [Google Scholar]
- 3.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochim Biophys Acta. 2009;1787:414–20. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parrales A, Iwakuma T. Targeting oncogenic mutant p53 for cancer therapy. Front Oncol. 2015;5:288. doi: 10.3389/fonc.2015.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 6.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab. 2013;18:617–33. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rivlin N, Brosh R, Oren M, Rotter V. Mutations in the p53 tumor suppressor gene: important milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–74. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol. 2011;223:116–26. doi: 10.1002/path.2784. [DOI] [PubMed] [Google Scholar]
- 9.Costa DC, de Oliveira GA, Cino EA, Soares IN, Rangel LP, Silva JL. Aggregation and prion-like properties of misfolded tumor suppressors: is cancer a prion disease? Cold Spring Harbor Perspect Biol. 2016;8:10. doi: 10.1101/cshperspect.a023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silva JL, De Moura Gallo CV, Costa DC, Rangel LP. Prion-like aggregation of mutant p53 in cancer. Trends Biochem Sci. 2014;39:260–67. doi: 10.1016/j.tibs.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Di Agostino S, Strano S, Emiliozzi V, Zerbini V, Mottolese M, Sacchi A, et al. Gain of function of mutantp53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Ferraiuolo M, Di Agostino S, Blandino G, Strano S. Oncogenic Intra-p53 Family Member Interactions in Human Cancers. Front Oncol. 2016;6:77. doi: 10.3389/fonc.2016.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Do PM, Varanasi L, Fan S, Li C, Kubacka I, Newman V, et al. Mutant p53 cooperates with ETS2 to promote etoposide resistance. Genes Dev. 2012;26:830–45. doi: 10.1101/gad.181685.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, et al. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell. 2014;54:960–74. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat Chem Biol. 2011;7:285–95. doi: 10.1038/nchembio.546. [DOI] [PubMed] [Google Scholar]
- 16.Adhikari AS, Iwakuma T. Mutant p53 gain of oncogenic function: in vivo evidence, mechanism of action and its clinical implications. Fukuoka Igaku Zasshi. 2009;100:217–28. [PubMed] [Google Scholar]
- 17.Freed-Pastor WA, Prives C. Mutantp53: one name, many proteins. Genes Dev. 2012;26:1268–86. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–44. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwakuma T, Lozano G, Flores ER. Li-Fraumeni syndrome: a p53 family affair. Cell Cycle (Georgetown, Tex) 2005;4:865–7. doi: 10.4161/cc.4.7.1800. [DOI] [PubMed] [Google Scholar]
- 20.Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA, et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature. 2015;523:352–6. doi: 10.1038/nature14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer SV, Parrales A, Begani P, Narkar A, Adhikari AS, Martinez LA, et al. Allele-specific silencing of mutant p53 attenuates dominant-negative and gain-of-function activities. Oncotarget. 2016;7:5401–15. doi: 10.18632/oncotarget.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–9. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- 23.Parrales Alejandro, Ranjan Atul, Iyer Swathi V., Padhye Subhash, Weir Scott J., Roy Anuradha, Iwakuma Tomoo. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nature Cell Biology. 2016;18(11):1233–1243. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies JT, Delfino SF, Feinberg CE, Johnson MF, Nappi VL, Olinger JT, et al. Current and emerging uses of statins in clinical therapeutics: a review. Lipid Insights. 2016;9:13–29. doi: 10.4137/LPI.S37450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aviles A, Neri N, Huerta-Guzman J, Nambo MJ. Randomized clinical trial of zoledronic acid in multiple myeloma patients undergoing high-dose chemotherapy and stem-cell transplantation. Curr Oncol. 2013;20:e13–20. doi: 10.3747/co.20.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman RE, Seaman JJ. The role of zoledronic acid in cancer: clinical studies in the treatment and prevention of bone metastases. Semin Oncol. 2001;28(2Suppl 6):11–16. doi: 10.1016/S0093-7754(01)90260-X. [DOI] [PubMed] [Google Scholar]
- 27.Appels NM, Beijnen JH, Schellens JH. Development of farnesyl transferase inhibitors: a review. Oncologist. 2005;10:565–78. doi: 10.1634/theoncologist.10-8-565. [DOI] [PubMed] [Google Scholar]
- 28.Lobell RB, Liu D, Buser CA, Davide JP, DePuy E, Hamilton K, et al. Preclinical and clinical pharmacodynamic assessment of L-778,123, a dual inhibitor of farnesyl:protein transferase and geranylgeranyl:protein transferase type-I. Mol Cancer Ther. 2002;1:747–58. [PubMed] [Google Scholar]
- 29.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–58. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurnher M, Gruenbacher G, Nussbaumer O. Regulation of mevalonate metabolism in cancer and immune cells. Biochim Biophys Acta. 2013;1831:1009–1015. doi: 10.1016/j.bbalip.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Thurnher M, Nussbaumer O, Gruenbacher G. Novel aspects of mevalonate pathway inhibitors as antitumor agents. Clinical Cancer Res. 2012;18:3524–31. doi: 10.1158/1078-0432.CCR-12-0489. [DOI] [PubMed] [Google Scholar]
- 32.Buhaescu I, Izzedine H. Mevalonate pathway: a review of clinical and therapeutical implications. Clin Biochem. 2007;40:575–84. doi: 10.1016/j.clinbiochem.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 33.Gobel A, Thiele S, Browne AJ, Rauner M, Zinna VM, Hofbauer LC, et al. Combined inhibition of the mevalonate pathway with statins and zoledronic acid potentiates their anti-tumor effects in human breast cancer cells. Cancer Lett. 2016;375:162–71. doi: 10.1016/j.canlet.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Duncan RE, El-Sohemy A, Archer MC. Mevalonate promotes the growth of tumors derived from human cancer cells in vivo and stimulates proliferation in vitro with enhanced cyclin-dependent kinase-2 activity. J Biol Chem. 2004;279:33079–84. doi: 10.1074/jbc.M400732200. [DOI] [PubMed] [Google Scholar]
- 35.Clendening JW, Pandyra A, Boutros PC, El Ghamrasni S, Khosravi F, Trentin GA, et al. Dysregulation of the mevalonate pathway promotes transformation. Proc Natl Acad Sci USA. 2010;107:15051–56. doi: 10.1073/pnas.0910258107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallianou NG, Kostantinou A, Kougias M, Kazazis C. Statins and cancer. Anticancer Agents Med Chem. 2014;14:706–12. doi: 10.2174/1871520613666131129105035. [DOI] [PubMed] [Google Scholar]
- 37.Fourie AM, Hupp TR, Lane DP, Sang BC, Barbosa MS, Sambrook JF, et al. HSP70 binding sites in the tumor suppressor protein p53. J Biol Chem. 1997;272:19471–79. doi: 10.1074/jbc.272.31.19471. [DOI] [PubMed] [Google Scholar]
- 38.Edkins AL. CHIP: a co-chaperone for degradation by the proteasome. Subcell Biochem. 2015;78:219–42. doi: 10.1007/978-3-319-11731-7_11. [DOI] [PubMed] [Google Scholar]
- 39.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18:1904–13. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Chen J. Phosphorylation and hsp90 binding mediate heat shock stabilization of p53. J Biol Chem. 2003;278:2066–71. doi: 10.1074/jbc.M206697200. [DOI] [PubMed] [Google Scholar]
- 41.Peng Y, Chen L, Li C, Lu W, Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem. 2001;276:40583–90. doi: 10.1074/jbc.M102817200. [DOI] [PubMed] [Google Scholar]
- 42.Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci U S A. 2002;99:12847–52. doi: 10.1073/pnas.202365899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King FW, Wawrzynow A, Hohfeld J, Zylicz M. Co-chaperones Bag-1, Hop and Hsp40 regulate Hsc70 and Hsp90 interactions with wild-type or mutant p53. EMBO J. 2001;20:6297–6305. doi: 10.1093/emboj/20.22.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kampinga HH, Kanon B, Salomons FA, Kabakov AE, Patterson C. Overexpression of the cochaperone CHIP enhances Hsp70-dependent folding activity in mammalian cells. Mol Cell Biol. 2003;23:4948–58. doi: 10.1128/MCB.23.14.4948-4958.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin YC, Lin JH, Chou CW, Chang YF, Yeh SH, Chen CC. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 2008;68:2375–83. doi: 10.1158/0008-5472.CAN-07-5807. [DOI] [PubMed] [Google Scholar]
- 46.Karlic H, Thaler R, Gerner C, Grunt T, Proestling K, Haider F, et al. Inhibition of the mevalonate pathway affects epigenetic regulation in cancer cells. Cancer Genet. 2015;208:241–252. doi: 10.1016/j.cancergen.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warita K, Warita T, Beckwitt CH, Schurdak ME, Vazquez A, Wells A, et al. Statin-induced mevalonate pathway inhibition attenuates the growth of mesenchymal-like cancer cells that lack functional E-cadherin mediated cell cohesion. Sci Rep. 2014;4:7593. doi: 10.1038/srep07593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mullen PJ, Yu R, Longo J, Archer MC, Penn LZ. The interplay between cell signalling and the mevalonate pathway in cancer. Nat Rev Cancer. 2016;16:718–31. doi: 10.1038/nrc.2016.76. [DOI] [PubMed] [Google Scholar]
- 49.Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 50.Likus W, Siemianowicz K, Bienk K, Pakula M, Pathak H, Dutta C, et al. Could drugs inhibiting the mevalonate pathway also target cancer stem cells? Drug Resistance Updates. 2016;25:13–25. doi: 10.1016/j.drup.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 51.Elsayed M, Kobayashi D, Kubota T, Matsunaga N, Murata R, Yoshizawa Y, et al. Synergistic antiproliferative effects of zoledronic acid and fluvastatin on human pancreatic cancer cell lines: an in vitro study. Biol Pharm Bull. 2016;39:1238–46. doi: 10.1248/bpb.b15-00746. [DOI] [PubMed] [Google Scholar]
- 52.Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet. 2012;44:1104–10. doi: 10.1038/ng.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buganim Y, Solomon H, Rais Y, Kistner D, Nachmany I, Brait M, et al. p53 Regulates the Ras circuit to inhibit the expression of a cancer-related gene signature by various molecular pathways. Cancer Res. 2010;70:2274–84. doi: 10.1158/0008-5472.CAN-09-2661. [DOI] [PubMed] [Google Scholar]
- 54.Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002;12:9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- 55.Wang W, Cheng B, Miao L, Mei Y, Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574. doi: 10.1038/cddis.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–70. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ginestier C, Monville F, Wicinski J, Cabaud O, Cervera N, Josselin E, et al. Mevalonate metabolism regulates Basal breast cancer stem cells and is a potential therapeutic target. Stem Cells. 2012;30:1327–37. doi: 10.1002/stem.1122. [DOI] [PubMed] [Google Scholar]
- 58.Mizuarai S, Yamanaka K, Kotani H. Mutant p53 induces the GEF-H1 oncogene, a guanine nucleotide exchange factor-H1 for RhoA, resulting in accelerated cell proliferation in tumor cells. Cancer Res. 2006;66:6319–26. doi: 10.1158/0008-5472.CAN-05-4629. [DOI] [PubMed] [Google Scholar]
- 59.Moroishi T, Hansen CG, Guan KL. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer. 2015;15:73–9. doi: 10.1038/nrc3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sorrentino G, Ruggeri N, Specchia V, Cordenonsi M, Mano M, Dupont S, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–66. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Wu Y, Wang H, Zhang Y, Mei L, Fang X, et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci USA. 2014;111:E89–98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–43. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Di Agostino S, Sorrentino G, Ingallina E, Valenti F, Ferraiuolo M, Bicciato S, et al. YAP enhances the pro-proliferative transcriptional activity of mutant p53 proteins. EMBO Rep. 2016;17:188–201. doi: 10.15252/embr.201540488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Escoll M, Gargini R, Cuadrado A, Anton IM, Wandosell F. Mutant p53 oncogenic functions in cancer stem cells are regulated by WIP through YAP/TAZ. Oncogene. 2017;36:3515–27. doi: 10.1038/onc.2016.518. [DOI] [PubMed] [Google Scholar]
- 65.Alfaqih MA, Allott EH, Hamilton RJ, Freeman MR, Freedland SJ. The current evidence on statin use and prostate cancer prevention: are we there yet? Nat Rev Urol. 2017;14:107–119. doi: 10.1038/nrurol.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feng CH, Miller CM, Tenney ME, Lee NK, Yamada SD, Hasan Y. Statin use significantly improves overall survival in high-grade endometrial cancer. Int J Gynecol Cancer. 2016;26:1642–49. doi: 10.1097/IGC.0000000000000819. [DOI] [PubMed] [Google Scholar]
- 67.Gray RT, Coleman HG, Hughes C, Murray LJ, Cardwell CR. Statin use and survival in colorectal cancer: results from a population-based cohort study and an updated systematic review and meta-analysis. Cancer Epidemiol. 2016;45:71–81. doi: 10.1016/j.canep.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 68.Nayan M, Punjani N, Juurlink DN, Finelli A, Austin PC, Kulkarni GS, et al. Statin use and kidney cancer survival outcomes: A systematic review and meta-analysis. Cancer Treat Rev. 2017;52:105–116. doi: 10.1016/j.ctrv.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 69.Sanfilippo Kristen Marie, Keller Jesse, Gage Brian F., Luo Suhong, Wang Tzu-Fei, Moskowitz Gerald, Gumbel Jason, Blue Brandon, O’Brian Katiuscia, Carson Kenneth R. Statins Are Associated With Reduced Mortality in Multiple Myeloma. Journal of Clinical Oncology. 2016;34(33):4008–4014. doi: 10.1200/JCO.2016.68.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mei Z, Liang M, Li L, Zhang Y, Wang Q, Yang W. Effects of statins on cancer mortality and progression: A systematic review and meta-analysis of 95 cohorts including 1,111,407 individuals. Int J Cancer. 2017;140:1068–81. doi: 10.1002/ijc.30526. [DOI] [PubMed] [Google Scholar]
- 71.Kautzky-Willer A, Thurner S, Klimek P. Use of statins offsets insulin-related cancer risk. J Intern Med. 2017;281:206–16. doi: 10.1111/joim.12567. [DOI] [PubMed] [Google Scholar]
- 72.Undela K, Srikanth V, Bansal D. Statin use and risk of breast cancer: a meta-analysis of observational studies. Breast Cancer Res Treat. 2012;135:261–9. doi: 10.1007/s10549-012-2154-x. [DOI] [PubMed] [Google Scholar]
- 73.Nordstrom T, Clements M, Karlsson R, Adolfsson J, Gronberg H. The risk of prostate cancer for men on aspirin, statin or antidiabetic medications. Eur J Cancer. 2015;51:725–33. doi: 10.1016/j.ejca.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 74.Alliance for Clinical Trials in Oncology. URL: https://clinicaltrials.gov/ct2/results?cond=&term=zoledronic+acid&cntry1=&state1=&recrs=
- 75.Dudakovic A, Tong H, Hohl RJ. Geranylgeranyl diphosphate depletion inhibits breast cancer cell migration. Invest New Drugs. 2011;29:912–20. doi: 10.1007/s10637-010-9446-y. [DOI] [PubMed] [Google Scholar]